Abstract
A critical requirement for mitosis is the distribution of genetic material to the two daughter cells. The central player in this process is the macromolecular kinetochore structure, which binds to both chromosomal DNA and spindle microtubule polymers to direct chromosome alignment and segregation. This review will discuss the key kinetochore activities required for mitotic chromosome segregation, including the recognition of a specific site on each chromosome, kinetochore assembly and the formation of kinetochore–microtubule connections, the generation of force to drive chromosome segregation, and the regulation of kinetochore function to ensure that chromosome segregation occurs with high fidelity.
During mitosis, the chromosomes are tethered to spindle microtubule polymers via a kinetochore. Careful assembly and regulation of these macromolecular structures are key to ensuring that chromosome segregation occurs with high fidelity.
A key objective for cell division is to physically distribute the genomic material to the two new daughter cells. Achieving proper chromosome segregation requires three primary things (Fig. 1): (1) the ability to specifically recognize and detect each unit of DNA; (2) a physical connection between the DNA and other cellular structures to mediate their distribution; and (3) a force-generating mechanism to drive the spatial movement of the DNA to the daughter cells. Although this article focuses on how these processes are achieved during mitosis in eukaryotic cells, these key principles are required for DNA segregation in all organisms, including bacteria. Perhaps the simplest DNA distribution machine is the partitioning system that segregates the small, circular bacterial R1 plasmid (Fig. 1). The R1 partitioning system uses just a single component for each of the three key activities listed above (reviewed in Salje et al. 2010). First, a 160-bp sequence-specific DNA element termed parC allows the partitioning system to recognize a specific region of the plasmid. Second, the DNA-binding protein ParR associates with the parC DNA sequence. ParR can then mediate connections between the plasmid DNA and third factor—the filament forming protein ParM. ParM polymerization is capable of generating force to drive the separation of two replicated copies of the R1 plasmid. The R1 plasmid partitioning system is both simple and elegant, and it demonstrates that it is possible to achieve DNA segregation with only two proteins and a short DNA sequence.
Figure 1.
Core requirements for DNA segregation. Cartoon diagram showing the core activities required for DNA segregation of the bacterial R1 plasmid or eukaryotic chromosomes highlighting the recognition of DNA, physical connections, and force.
In striking contrast to the R1 plasmid partitioning system, chromosome segregation in eukaryotes (Fig. 1) requires hundreds of different proteins. Given the ability of the simple R1 partitioning system to efficiently mediate DNA segregation in bacteria, it raises the question of why this added complexity is present in eukaryotes. Importantly, there are significant limitations to the bacterial system that would prevent such a system from working in eukaryotes. For example, bacteria are ∼1–2-µm long, whereas vertebrate cells can be ∼10–50 µm in diameter creating a larger spatial requirement to move the DNA (Fig. 1). In addition, although only a single R1 plasmid is present in each bacterium, human cells have 46 different units of DNA (23 from each parent), which are packaged into chromosomes. Each chromosome must be distributed properly during every cell division. Independently recognizing each of these units to ensure their accurate distribution represents a complex challenge. Indeed, adding even one additional R1 plasmid causes the system to break down, with ParM polymers acting indefinitely, pushing the two most closely positioned units of DNA apart to opposite ends of a cell (Campbell and Mullins 2007). Finally, eukaryotic cells require that chromosome segregation occur with high fidelity to ensure that the two replicated units of DNA are distributed accurately to the two new daughter cells. Even a single chromosome mis-segregation event in a multicellular organism has the potential to lead to lethality, lead to developmental disorders, or contribute to cancer progression (Holland and Cleveland 2009; Gordon et al. 2012), placing a high premium on the accuracy of this process.
Despite the differences in complexity between bacterial plasmid partitioning systems and the eukaryotic chromosome segregation machinery, the fundamental requirements for distributing DNA to two new cells are remarkably similar (Fig. 1). First, it is necessary to have a region of each chromosome that is “recognized” by the chromosome segregation machinery. In eukaryotes, this region of DNA is termed the centromere. Second, a group of proteins must assemble on this DNA element to facilitate its “connections” to other structures in the cell. In eukaryotes, this physical connection is provided by a macromolecular structure termed the kinetochore. The kinetochore is an impressive molecular machine that requires the coordinated functions of more than 100 different protein components (Cheeseman and Desai 2008). Third, the kinetochore must interact with additional structures that provide the “force” to move the chromosomes. Chromosome segregation in eukaryotes requires microtubule polymers that generate force primarily through their depolymerization.
In this review, I will discuss the molecular mechanisms that underlie kinetochore function, including the recognition of a specific site on each chromosome, the formation of the physical kinetochore–microtubule connections, and the forces that drive chromosome segregation during mitosis in eukaryotes, as well as the mechanisms that regulate kinetochore function.
RECOGNITION: CENTROMERES AND KINETOCHORE SPECIFICATION
For chromosome segregation to occur, a unit of DNA must first be recognized by the chromosome segregation machinery. In most eukaryotes, the centromere is restricted to a single region of each chromosome (termed monocentric). In the absence of a functional centromere, a kinetochore will not assemble on the DNA and that chromosome will fail to segregate during mitosis. In contrast, if more than one centromere forms at distal sites on a single chromosome, that chromosome can form multiple independent attachments to spindle microtubules and can be fragmented by spindle forces during mitosis. Thus, specifying the position of the centromere is a key challenge for the cell. Although all eukaryotes require this recognition process, the nature and size of this centromere DNA varies dramatically between organisms. For example, the centromere sequences in the budding yeast Saccharomyces cerevisiae are only 125 bp in length and contain a sequence-specific DNA element. This sequence is bound by the CBF3 complex (Biggins 2013), a protein complex found exclusively in budding yeast species. Even small base pair changes within the budding yeast centromere prevent CBF3 binding, eliminate centromere function, and prevent a chromosome from segregating properly. In contrast, the size of most eukaryotic centromeres is much larger (Cleveland et al. 2003), encompassing 40–100 kb in the fission yeast Schizosaccharomyces pombe up to megabases of repetitive DNA in some animal and plant species. Finally, in some cases, such as the nematode Caenorhabditis elegans, chromosomes are holocentric with the kinetochore assembling along the entire length of each chromosome (Maddox et al. 2004). Although there are often specific DNA sequences that are associated with the centromere, such as a 171-bp α-satellite repeat in humans (Masumoto et al. 1989), most organisms do not have a specific centromere DNA sequence requirement. The most striking evidence for the sequence-independent nature of the vertebrate centromere comes from individuals in which centromere has relocated to a region of the chromosome lacking α-satellite repeats (termed a “neocentromere”) (Amor et al. 2004). In some cases, the α-satellite sequences are still present on a chromosome, but no longer behave as functional centromeres, indicating that these DNA repeat sequences are neither necessary nor sufficient for centromere specification.
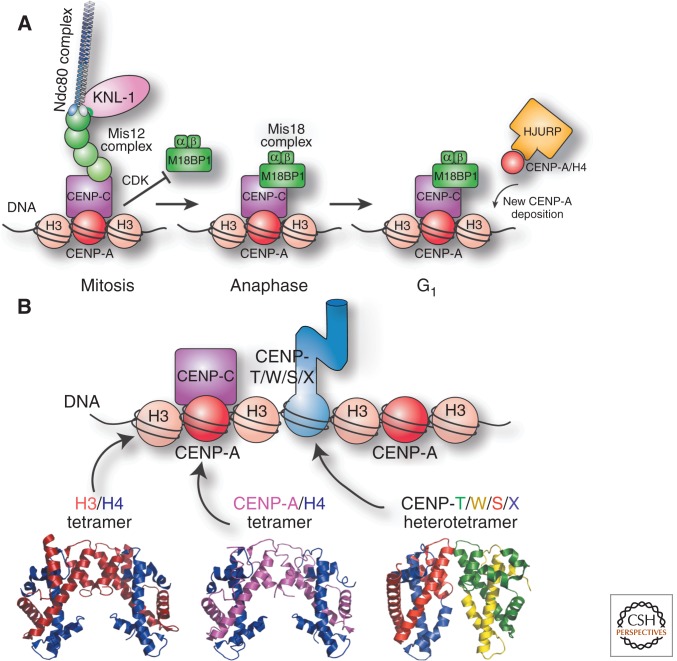
Because of the DNA sequence–independent nature of the vertebrate centromere, it is instead thought that the centromere is defined epigenetically. The key player in this process is the histone H3 variant CENP-A (Fig. 2), which forms specialized nucleosomes found exclusively at the centromeres (Palmer et al. 1987, 1991). Although there has been an ongoing debate about the precise composition of this centromeric nucleosome (Black and Cleveland 2011; Dunleavy et al. 2013), recent work has agreed on several fundamental defining features that make CENP-A ideally suited to specify centromere identity. CENP-A is required for the localization of all known kinetochore proteins in vertebrate cells (Regnier et al. 2005; Liu et al. 2006), as well as most other eukaryotes, indicating that it is the core player for defining the site of a functional kinetochore. In addition, CENP-A is stably associated with centromeres (Jansen et al. 2007). Indeed, the existing population of CENP-A remains associated with chromosomes even during DNA replication, when it is passed conservatively to the two newly replicated chromosomes. Finally, using a specialized set of deposition factors described below, new CENP-A is only deposited at sites where preexisting CENP-A is present. The combination of these three properties allows CENP-A to act as an epigenetic mark for centromere specification.
Figure 2.
Recognition. (A) Diagram showing the key players and processes required for the deposition of the specialized CENP-A-containing nucleosomes at centromeres. (B) (Top) Diagram of the key components of centromeric chromatin. (Bottom) Crystal structures of the H3/H4 tetramer based on data from Davey et al. (2002), CENP-A/H4 tetramer based on data from Tachiwana et al. (2011), and CENP-T/W/S/X heterotetramer based on data from Nishino et al. (2012).
Although the critical role for CENP-A in defining centromere identity is well established, understanding the mechanisms that underlie the stability and propagation of CENP-A at centromeres is still a work in progress. Recent work has identified both intrinsic features of the CENP-A nucleosome and extrinsic associated factors that play a key role in this process. For CENP-A to act as a mark for the centromere, it must be structurally and functionally distinct from the other histones that associate with chromosomes (Fig. 2). Indeed, a region within the sequence of CENP-A, termed the CENP-A targeting domain (CATD), provides distinct structural properties (Black et al. 2004, 2007) that allow this nucleosome to be recognized by specialized deposition factors (Foltz et al. 2009; Hu et al. 2011). To ensure that CENP-A is restricted to centromeres, several mechanisms act together to ensure the proper deposition of CENP-A nucleosomes (Fig. 2A). In vertebrate cells, CENP-A deposition occurs during G1 (Jansen et al. 2007), not during S phase when canonical histone H3-containing nucleosomes are deposited in a replication-coupled manner. This cell cycle restriction occurs at least in part through the negative regulation of CENP-A deposition by cyclin-dependent kinase (CDK) activity (Silva et al. 2012), which is high during mitosis, but declines at mitotic exit. During G1, a series of factors act to incorporate CENP-A at centromeres (Fig. 2A). This includes a specialized histone chaperone, HJURP, that associates with soluble CENP-A/histone H4 dimers to assemble them into complete nucleosomes (Dunleavy et al. 2009; Foltz et al. 2009) and the Mis18 complex (Mis18α, Mis18β, and M18BP1/KNL2) (Hayashi et al. 2004; Fujita et al. 2007; Maddox et al. 2007), which acts to recruit HJURP to centromeres (Barnhart et al. 2011). These factors are targeted to existing centromeres to ensure that CENP-A deposition occurs exclusively at active centromeres. To achieve this, CENP-A nucleosomes interact with the inner kinetochore CENP-C (Moree et al. 2011; Dambacher et al. 2012), which in turn acts as the centromere receptor for the Mis18 complex incorporation machinery (Fig. 2A), ensuring that CENP-A-containing chromatin is able to propagate itself. Although CENP-A incorporation occurs with high fidelity, in cases where CENP-A is inappropriately incorporated into noncentromeric chromatin, these nucleosomes are removed from chromatin and targeted for degradation (Collins et al. 2005; Hewawasam et al. 2010; Ranjitkar et al. 2010). Together, these factors act to ensure that there is a single site on each chromosome that is marked by CENP-A to direct assembly of the kinetochore structure.
Beyond CENP-A, additional DNA-binding proteins also localize to centromeres. An intriguing player in this process is the recently identified CENP-T-W-S-X complex (Hori et al. 2008; Gascoigne et al. 2011; Nishino et al. 2012). Although the CENP-T-W-S-X complex lacks sequence homology with canonical nucleosomes, these proteins are structurally similar to the histones within a nucleosome (Fig. 2B) (Nishino et al. 2012). Current data suggest that these proteins form a specialized nucleosome-like structure that wraps DNA around its surface (Nishino et al. 2012). Although CENP-T-W-S-X localization occurs downstream from CENP-A (Hori et al. 2008), CENP-A is not sufficient to direct CENP-T localization (Gascoigne et al. 2011) suggesting that additional factors may also control the localization of these nucleosome-like particles. Together, these DNA-binding proteins ensure that the kinetochore forms stable interactions with a single region of each chromosome such that the segregation machinery can “recognize” each chromosome.
PHYSICAL CONNECTIONS: BUILDING THE CHROMOSOME–MICROTUBULE ATTACHMENT
Once the site of kinetochore assembly is specified on each chromosome, the next critical task is to construct a multiprotein structure that is capable of associating with dynamic microtubule polymers. The kinetochore is composed of more than 100 different proteins in vertebrate cells, each of which is present in multiple copies per kinetochore. Based on the relative spatial localization of these proteins and their different functions, they can be grouped into three main categories: (1) inner kinetochore proteins that are required to form the connection with chromosomal DNA and provide a platform to assemble the kinetochore, (2) outer kinetochore proteins that form connections with microtubules, and (3) regulatory proteins that monitor or control the activities of the kinetochore.
Assembling the Kinetochore Structure
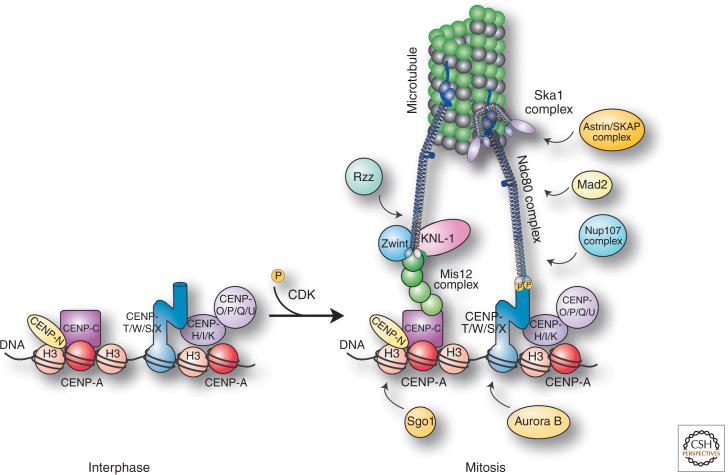
A key challenge for a structure composed of more than 100 different proteins is to assemble its component parts in a controlled and organized manner (Gascoigne and Cheeseman 2011). In the case of the kinetochore, this building process is also carefully regulated over the course of the cell cycle (Fig. 3). Sixteen kinetochore proteins reside at centromeric DNA throughout the cell cycle (termed the constitutive centromere-associated network—CCAN) (Cheeseman and Desai 2008). As a cell enters mitosis, outer kinetochore proteins are rapidly assembled on this platform of inner kinetochore proteins within a time frame of <20 min (Gascoigne and Cheeseman 2013). Finally, as the cell exits mitosis, these outer kinetochore proteins rapidly disassemble. Thus, the kinetochore is a highly dynamic, cell cycle–regulated assembly that also displays an impressive structural stability with the ability to resist forces from the spindle microtubules during mitosis (Rago and Cheeseman 2013). This makes the kinetochore quite different from other large molecular machines, such as the ribosome or the proteasome, that are stably maintained once they are assembled.
Figure 3.
Attachment. Diagram showing kinetochore structure and organization during interphase and mitosis. At mitotic entry, CDK/Cyclin B phosphorylation promotes outer kinetochore assembly on a platform of constitutive kinetochore proteins. For information on the additional kinetochore proteins shown in the figure, see Cheeseman and Desai (2008).
The precise molecular connectivity within the kinetochore is a focus of ongoing research. However, there appear to be two main branches within the kinetochore that connect the DNA to the microtubules in vertebrate cells (Fig. 3). The first of these branches involves CENP-C, which binds directly to CENP-A nucleosomes (Carroll et al. 2010; Kato et al. 2013) and also interacts with the four-subunit Mis12 complex (Gascoigne et al. 2011; Przewloka et al. 2011; Screpanti et al. 2011). The Mis12 complex, in turn, interacts with KNL1 and the four-subunit Ndc80 complex (Cheeseman et al. 2004, 2006; Obuse et al. 2004; Petrovic et al. 2010), a key microtubule-binding protein at kinetochores (see below). The second branch involves the DNA-binding CENP-T-W-S-X complex. In addition to associating with DNA, CENP-T interacts directly with the Ndc80 complex (Gascoigne et al. 2011; Nishino et al. 2013). Each of these branches is essential to generate proper connections with microtubules in vertebrate cells. In addition, consistent with a key role for these pathways in directing downstream kinetochore assembly, artificial targeting of CENP-T or CENP-C to an ectopic site on a chromosome arm results in the assembly of functional kinetochore-like structures (Gascoigne et al. 2011; Schleiffer et al. 2012; Hori et al. 2013). In vertebrate cells, the assembly of both the CENP-C and CENP-T pathways is controlled in a cell cycle–regulated manner using a combination of phosphorylation downstream from CDK (Fig. 3) and exclusion of the Ndc80 complex from the nucleus during interphase (Gascoigne and Cheeseman 2013). At mitotic entry, the nuclear envelope breaks down, allowing the Ndc80 complex access to the chromosomes for assembly at kinetochores. Phosphorylation of CENP-T and other targets by CDK promotes protein–protein interactions to drive kinetochore assembly (Fig. 3). However, defining the complete molecular connectivity within the kinetochore required to link DNA and microtubules and construct a higher-order kinetochore structure remains a key ongoing challenge.
Binding to Spindle Microtubules
During mitosis, the kinetochore must form a direct physical connection with the microtubule polymers from the mitotic spindle. Indeed, there are a large number of outer kinetochore proteins that have been shown to bind directly to microtubules. Of these kinetochore proteins, the key, conserved player in forming robust interactions with microtubules is the four-subunit Ndc80 complex (Figs. 3 and 4) (DeLuca and Musacchio 2012). The Ndc80 complex forms an extended rod-shaped structure (Ciferri et al. 2005; Wei et al. 2005) that binds directly to microtubule polymers (Cheeseman et al. 2006) through a Calponin homology domain and a positively charged, unstructured amino-terminal tail (Wei et al. 2007; Ciferri et al. 2008). When Ndc80 complex function is disrupted in cells, this results in severe defects in the ability of microtubules to attach to chromosomes leading to extensive chromosome mis-segregation (DeLuca et al. 2002; Desai et al. 2003), or even the complete failure of chromosome segregation (Wigge et al. 1998; Wigge and Kilmartin 2001).
Figure 4.
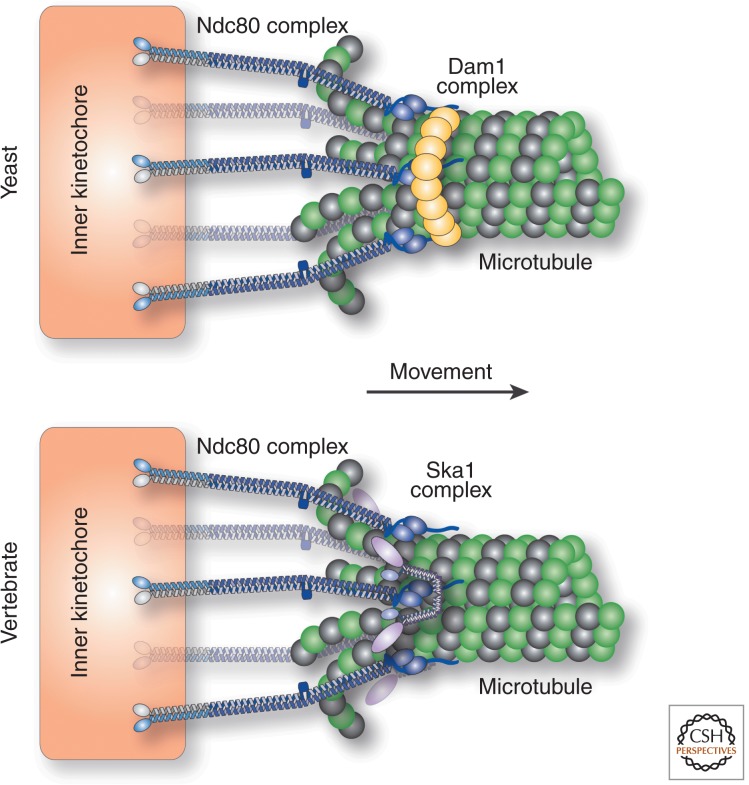
Force. Diagrams showing the kinetochore associating with depolymerizing microtubules and harnessing the force induced by microtubule depolymerization to direct chromosome movement. The Ndc80 complex is the core player in forming kinetochore–microtubule interactions, but it requires additional interactions with the Dam1 complex (fungi, top) or the Ska1 complex (vertebrates, bottom).
Although the Ndc80 complex provides the core microtubule attachment activity at kinetochores, additional proteins act to modulate and enhance microtubule interactions. Interacting with microtubules is an especially difficult task because microtubules are not static polymers, but instead undergo constant changes in their length through polymerization and depolymerization (Desai and Mitchison 1997), such that the kinetochore must remain stably bound to growing or shrinking microtubule polymers. The Ndc80 complex has a relatively modest intrinsic affinity for microtubule polymers (∼2–3 µm) (Cheeseman et al. 2006). On its own in solution, the Ndc80 complex does not remain attached to shrinking microtubules (Schmidt et al. 2012). However, the presence of multiple Ndc80 complexes at kinetochores bound to each spindle microtubule (Joglekar et al. 2006) could allow the kinetochore to remain processively associated with a depolymerizing microtubule as long as at least a single Ndc80 complex was bound to the microtubule at a given time. In fact, artificial clustering of the Ndc80 complex in vitro will allow it to remain associated with a depolymerizing microtubule (Powers et al. 2009). In addition, other factors may act together with the Ndc80 complex to mediate such processive interactions. In fungi, the 10-subunit Dam1 complex appears to act as a key processivity factor at kinetochores (Fig. 4). This is particularly important in budding yeast (S. cerevisiae), where there is only a single microtubule bound to each kinetochore (Winey et al. 1995), in contrast to the 15 to 20 microtubules per vertebrate kinetochore (McEwen et al. 2001), placing a high premium on maintaining the interaction with this microtubule. The Dam1 complex is capable of forming a ring-like structure around a microtubule polymer (Miranda et al. 2005; Westermann et al. 2005) such that the depolymerization-induced peeling away of the microtubule protofilaments would cause the Dam1 complex to slide down the microtubule (Grishchuk et al. 2008), but remain attached. In vitro, the Dam1 complex has the ability to remain associated with the end of a depolymerizing microtubule (Asbury et al. 2006; Westermann et al. 2006). Importantly, the Dam1 complex can impart this activity to the Ndc80 complex (Lampert et al. 2010; Tien et al. 2010), thereby generating an integrated, processive microtubule interface. Although the Dam1 complex is not conserved outside of fungi, recent work has suggested that the Ska1 complex is a functional counterpart to the Dam1 complex (Fig. 4) (Hanisch et al. 2006; Daum et al. 2009; Gaitanos et al. 2009; Raaijmakers et al. 2009; Theis et al. 2009; Welburn et al. 2009; Schmidt et al. 2012). The Ska1 complex is present in metazoa including vertebrates, plants, and nematodes, but not fungi. Although the structure (Jeyaprakash et al. 2012; Schmidt et al. 2012) and mechanistic basis (Schmidt et al. 2012) by which the Ska1 complex acts is likely to be quite different from the Dam1 complex, the Ska1 complex also displays the ability to remain associated with the end of a depolymerizing microtubule (Welburn et al. 2009; Schmidt et al. 2012), a property that it can confer to the Ndc80 complex (Schmidt et al. 2012). As it is critical for kinetochores to maintain proper connections with spindle microtubules, kinetochores likely utilize and coordinate multiple microtubule binding activities to achieve this important goal.
FORCE: DRIVING CHROMOSOME MOVEMENT
The kinetochore–microtubule interaction is not a simple, static physical attachment. For chromosome movement to occur, kinetochores must use their interactions with microtubule polymers to generate force. During prometaphase, kinetochores are captured by microtubule polymers and moved to align them in the middle of the cell at the metaphase plate. During metaphase, paired sister chromatids are attached to microtubules from opposite spindle poles (termed bi-oriented). The opposing forces acting on these bi-oriented sister kinetochores drive oscillatory chromosome movements and act to signal proper attachments (see below). Finally, during anaphase A, kinetochores are pulled toward the spindle poles to segregate the chromosomes. Defining the mechanisms by which kinetochores generate the force required to align and segregate chromosomes has been a key challenge.
Generating Force to Drive Chromosome Segregation
There are two different ways in which a kinetochore can attach to a microtubule polymer. First, it can form lateral interactions with the microtubule polymer such that it binds to the side of a microtubule. Based on parallels to vesicle transport along axonal microtubules in neurons, it was initially assumed that microtubule-based molecular motors would act to move a kinetochore “cargo” along the tracks provided by the microtubule polymers through such lateral interactions. In fact, multiple microtubule-based motors including the plus-end directed kinesin CENP-E (Yen et al. 1992) and the microtubule minus-end directed motor cytoplasmic dynein (Pfarr et al. 1990; Steuer et al. 1990) localize to kinetochores. In the case of the CENP-E, at least one role for this motor is to ferry chromosomes located near the spindle poles to the middle of the spindle using lateral interactions with adjacent microtubule polymers (Kapoor et al. 2006). In the absence of CENP-E, the majority of the chromosomes align at the metaphase plate, but at least a subset of chromosomes remain clustered around the spindle pole (Putkey et al. 2002). Dynein may also contribute to chromosome movement along microtubule fibers, as well as playing roles in microtubule capture, kinetochore signaling, and spindle organization (Howell et al. 2001; Bader and Vaughan 2010). In addition to kinetochore-localized microtubule motors, DNA bound chromokinesins also provide force through lateral interactions to push the chromosomes away from the pole, contributing to chromosome congression to the metaphase plate (Mazumdar and Misteli 2005).
Although lateral kinetochore–microtubule interactions allow a kinetochore to initially capture a microtubule and be moved along that polymer using the motors described above, robust kinetochore–microtuble interactions require end-on attachments such that the microtubule plus end is embedded in the kinetochore. Although microtubule-based motors make important contributions to chromosome segregation and the regulation of kinetochore–microtubule interactions, they are not strictly required for the physical movement and distribution of the chromosomes. Instead, when kinetochores form end-on attachments to microtubules, it is thought that the microtubule polymer itself functions as the motor to power chromosome movement (McIntosh et al. 2010). Indeed, microtubule depolymerization can generate substantial force. Within the lattice of the microtubule polymer, tubulin dimers are trapped in a straight conformation. However, as a microtubule begins to disassemble, the tubulin can adopt a bent conformation (Nogales and Wang 2006), causing it to peel backward. An individual bending protofilament can generate a power stroke of up to 5 pN during microtubule depolymerization (Grishchuk et al. 2005). Based on these measurements, a typical microtubule (composed of 13 protofilaments) could generate up to 65 pN of force, a significant amount of force on a subcellular scale. When paired sister kinetochores make end-on attachments to microtubules from opposite spindle poles, chromosome movement can be driven almost entirely by microtubule polymerization and depolymerization. However, this creates a strong requirement for the microtubule-binding proteins at kinetochores to remain attached to these dynamic microtubule polymers and use the force that is generated by their depolymerization to direct chromosome movements.
Modulating Microtubule Dynamics at Kinetochores
Microtubule dynamics play a key role in directing chromosome movement. At anaphase, when paired sister chromatids separate from each other, the cell can reel in these chromosomes toward each spindle pole through microtubule depolymerization. During metaphase, sister chromatid oscillations can also be driven by microtubule dynamics, but require the coordination of the two, paired chromatids. For paired sister kinetochores to remain attached to opposite spindle poles, as one side shortens the microtubule, the microtubules attached to the paired kinetochore must compensate by increasing in length. This coordination may involve the mechanical coupling of the two sister kinetochores using a force-based mechanism (Dumont et al. 2012).
As the polymerization state of the microtubule is primarily responsible for directing chromosome movement, it is critical for the kinetochore to control microtubule dynamics. At kinetochores, there are multiple proteins that directly influence the microtubule polymerization status. The kinesin-13 family of proteins (Kif2c/MCAK in vertebrates) (Kline-Smith et al. 2004) acts to promote microtubule catastrophe (the switch to depolymerization), and the kinesin-8 family of proteins (Kif18a in vertebrates) (Mayr et al. 2007; Stumpff et al. 2008) also modulates microtubule dynamics, possibly by acting as a depolymerase. Opposing these depolymerases is a series of polymerization-promoting factors including the CLASPs (Clasp1 and Clasp2) (Maiato et al. 2003; Al-Bassam et al. 2010) and TOG-domain containing proteins (ch-TOG in human cells) (Al-Bassam and Chang 2011), which helps to deliver tubulin dimers to the microtubule plus end. Together, these factors ensure that the kinetochore both harnesses and controls the force generated by the microtubules to precisely direct chromosome alignment and segregation.
REGULATION: ENSURING THE FIDELITY OF CHROMOSOME SEGREGATION
Even minor defects during mitosis can result in catastrophic consequences for a cell. Therefore, the kinetochore must not only move the chromosomes, but also must monitor and correct this process to ensure that it occurs with high fidelity. In the proper bi-oriented configuration, the replicated and paired sister kinetochores bind to microtubule polymers from opposite spindle poles. However, a number of potential mistakes can occur in the formation of kinetochore–microtubule attachments including the failure to attach to one or both kinetochores (unattached), both sister kinetochores attaching to the same spindle pole (syntelic), or a single kinetochore simultaneously attaching to both spindle poles (merotelic). In the presence of such errors, it is critical to have mechanisms in place to sense and correct these problems. As such, kinetochore function is tightly regulated, both to ensure the proper formation of bi-oriented kinetochore–microtubule attachments, and to delay the progression into anaphase when errors persist.
Regulating Kinetochore–Microtubule Attachments
Kinetochore function is tightly regulated to correct any errors, and coordinate its activity with cell cycle progression. Chief among the regulatory players that control kinetochore function are a series of protein kinases that act at kinetochores to control proper kinetochore–microtubule attachments. These kinases include Aurora B (Lampson and Cheeseman 2011), Polo-like Kinase 1 (Plk1) (Liu et al. 2012), Mps1 (Liu and Winey 2012), Bub1 (Elowe 2011), and CDK (Chen et al. 2008; Gascoigne and Cheeseman 2013). Each of these kinases localizes to kinetochores, phosphorylates kinetochore-bound substrates, and makes distinct contributions to regulating kinetochore function. In some cases, these kinases act globally to control kinetochore function. For example, as discussed above, CDK-dependent phosphorylation ensures that kinetochore function and assembly state change are simultaneously controlled at each kinetochore to alter its function at cell cycle transitions (Fig. 3). In other cases, kinetochore-localized kinases regulate kinetochore function depending on attachment status.
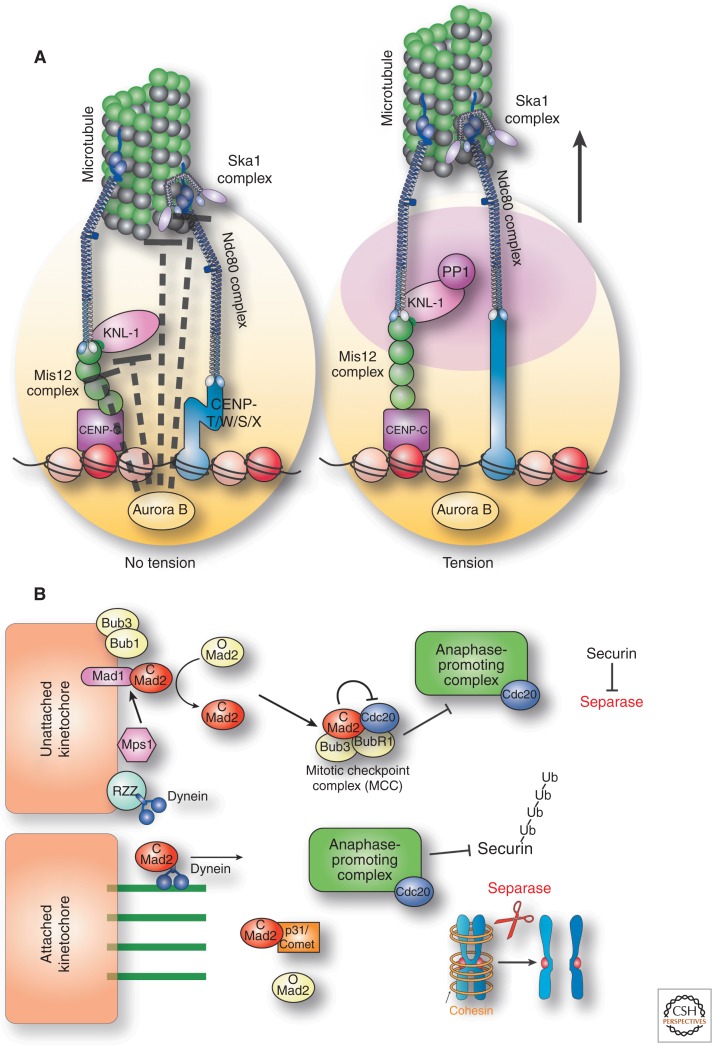
A key challenge for the regulation of kinetochore function is to eliminate inappropriate microtubule attachments. Aurora B kinase is a key player in this correction mechanism (Fig. 5A) (Biggins et al. 1999; Tanaka et al. 2002). Aurora B phosphorylation directly inhibits the activities of multiple components of the kinetochore–microtubule interface (Fig. 5A), including the Dam1 complex (Cheeseman et al. 2002), Ndc80 complex (Cheeseman et al. 2006; DeLuca et al. 2006), and Ska1 complex (Chan et al. 2012; Schmidt et al. 2012). The combined effect of this phosphorylation eliminates incorrect kinetochore–microtubule attachments, resetting the kinetochore to an unattached ground state from which new, correct attachments can be formed. Importantly, it is critical that this correction mechanism target improper attachments without affecting bi-oriented kinetochores. Only bi-oriented kinetochores are under tension, with the paired sister kinetochores pulled by the attached microtubules toward opposite spindle poles. Aurora B kinase displays tension-sensitive phosphorylation of its outer kinetochore substrates such that phosphorylation is strongly reduced on bi-oriented kinetochores (Liu et al. 2009; Welburn et al. 2010). This tension-sensitive phosphorylation appears to be achieved at least in part through the spatial separation of Aurora B from its targets (Liu et al. 2009). Aurora B and its associated proteins that comprise the chromosomal passenger complex (CPC) are primarily localized to the inner centromere region (Cooke et al. 1987; Vader et al. 2006) between the sister kinetochores, whereas many of its key functional substrates are localized to the outer kinetochore interface with microtubule (Fig. 5A). In the presence of tension, the substrates for Aurora B can be located >100 nm away at the outer kinetochore (Wan et al. 2009). Tension distorts the kinetochore structure, increasing the distance between Aurora B and its substrates and reducing the likelihood that they will be phosphorylated.
Figure 5.
Regulation. (A) Diagram showing the tension-dependent deformation of the kinetochore structure. Aurora B kinase located at the inner centromere (at the base of the kinetochore) is spatially separated from its substrates at the outer kinetochore. The presence of tension on bi-oriented kinetochores strongly reduces the ability of Aurora B to phosphorylate outer kinetochore proteins and inactivate microtubule attachments. (B) Model showing the spindle assembly checkpoint proteins preventing cell cycle progression in the absence of unattached kinetochores.
Phosphorylation plays a key role in promoting kinetochore assembly and preventing inappropriate microtubule interactions. However, it is also critical to remove this dynamic phosphorylation once its function is complete. Several counteracting phosphatases act to reverse the phosphorylation of kinetochore targets. The phosphatases PP1 (Fig. 5A) and PP2A both localize to kinetochores (Trinkle-Mulcahy et al. 2003; Foley et al. 2011), and phosphatase-targeting factors play key roles in allowing these phosphatases to dephosphorylate their kinetochore substrates at the correct time (Kim et al. 2010; Liu et al. 2010; Suijkerbuijk et al. 2012; Kruse et al. 2013). Together, these cell cycle–dependent and attachment-sensitive phosphorylation events help coordinate and control the multiple, complex activities at kinetochores to achieve high fidelity chromosome segregation.
Talking to the Cell Cycle: Spindle Assembly Checkpoint Signaling
During mitosis, replicated sister chromatids are held together by the cohesin complex. At anaphase onset, cohesin is cleaved by the protease Separase (Nasmyth and Haering 2009), allowing the paired sister chromatids to separate and segregate to the two new daughter cells. This occurs through the cleavage of covalent peptide bonds within the cohesion subunit Scc1. Such cleavage is irreversible, so it is critical to only initiate anaphase once each pair of sister chromatids has formed proper bi-oriented attachments to spindle microtubule polymers. Therefore, it is important to not only correct errors, but also to delay anaphase onset if errors persist. The molecular players tasked with this important role are the proteins of the spindle assembly checkpoint (SAC): Mad1, Mad2, Bub1, BubR1/Mad3, Bub3, and Mps1 (Fig. 5B) (Musacchio and Salmon 2007). Checkpoint function in metazoa additionally requires the Rod/ZW10/Zwilch (RZZ) complex (Karess 2005), which acts in part to recruit dynein to kinetochores (Fig. 5B). These proteins act to detect errors in kinetochore–microtubule attachments and to translate this to a signal that arrests the cell cycle in metaphase.
The checkpoint must be able to detect and distinguish a kinetochore that lacks proper attachments from a kinetochore that is correctly attached to microtubules. In the presence of incorrect or missing attachments, the checkpoint generates a signal at kinetochores that can diffuse to the rest of the cell and inhibit the cell cycle machinery to prevent cell cycle progression (Fig. 5B). At present, it remains unclear precisely how the checkpoint proteins sense kinetochore attachment status. Several kinetochore-localized microtubule-binding proteins including KNL1/Blinkin, Ndc80/Hec1, and CENP-E have been proposed to act with checkpoint proteins to signal attachment status (Martin-Lluesma et al. 2002; Mao et al. 2003; Kiyomitsu et al. 2007). These proteins have the potential to interact with the checkpoint proteins differentially depending on the presence of microtubule attachments. However, a direct connection between microtubule binding and checkpoint signaling activity has not been demonstrated at a molecular level.
Although the mechanisms that sense attachment status are unknown, the nature of the downstream checkpoint signal is becoming increasingly clear. Elegant structural work demonstrated that a key part of this signal is the conformation state of the checkpoint protein Mad2 (Sironi et al. 2002; De Antoni et al. 2005). Mad2 can be present in either a “closed” or “open” configuration based on its three-dimensional structure (Fig. 5B). In the open conformation (termed O-Mad2), Mad2 is essentially inactive. However, Mad2 is held in the closed conformation (termed C-Mad2) at kinetochores, and C-Mad2 can cause additional Mad2 to adopt this closed conformation. Because Mad2 only localizes to kinetochores with attachment defects (Chen et al. 1996; Waters et al. 1998), C-Mad2 is propagated in the presence of incorrect attachments. C-Mad2 can then diffuse away from the kinetochore, and together with the checkpoint proteins BubR1 and Mad3, C-Mad2 can form the mitotic checkpoint complex (MCC) to bind to Cdc20 (Sudakin et al. 2001), a critical targeting subunit for the anaphase-promoting complex (APC). The APC is an E3 Ubiquitin ligase that targets diverse proteins for degradation to initiate anaphase (Peters 2006), including Securin, an inhibitory factor for the Separase cohesin protease. By holding Cdc20 in an inactive state, the MCC prevents APC activation thereby delaying anaphase onset. Once proper bi-oriented microtubule attachments are formed, additional proteins then act to reverse and turn off the checkpoint signal (Fig. 5B), including dynein-dependent removal of the checkpoint proteins from kinetochores (Howell et al. 2001) and the action of the Mad2-binding protein p31/Comet (Mapelli et al. 2006; Hagan et al. 2011). At this point, the checkpoint is termed “satisfied,” the APC is activated, Securin is degraded, Separase is activated, cohesin is cleaved, and anaphase sister chromatid segregation can occur (Fig. 5B). Thus, the kinetochore not only acts as the key molecular machine to drive forward the segregation process, but also serves as a hub for the sensory molecules that monitor the correct execution of this process and coordinate chromosome segregation with the cell cycle.
CONCLUDING REMARKS
The kinetochore is a fascinating molecular machine that plays a central role in the fundamental processes that are required for the recognition, connections, and force generation that underlie mitosis. The kinetochore was first identified cytologically more than 130 years ago as the site on each chromosome that attached to the mitotic spindle (Flemming 1880). However, it was not until the mid-1980s that the first protein components of the human kinetochore were identified (Earnshaw and Rothfield 1985). Over the past three decades, and the past 10 years in particular, there has been an explosion in the molecular identification of kinetochore components combined with the analysis of their functions. However, significant questions still remain for understanding the structure, organization, and activities of these players and how they are coordinated and integrated to achieve proper chromosome segregation. Defining these mechanisms is an important goal for understanding the faithful segregation of the genome during mitosis.
ACKNOWLEDGMENTS
I thank the members of my laboratory for critical comments and suggestions, Florencia Rago, Tom Dicesare, and Kara McKinley for help with the figures, and the many amazing people that I have been fortunate to work with as mentors, in my laboratory, as collaborators, and as colleagues. The work in our lab is supported by an award from the Leukemia & Lymphoma Society (Scholar Award), a grant from the National Institutes of Health/National Institute of General Medical Sciences (GM088313), and a Research Scholar Grant (121776) from the American Cancer Society.
Footnotes
Editors: Mitsuhiro Yanagida, Anthony A. Hyman, and Jonathon Pines
Additional Perspectives on Mitosis available at www.cshperspectives.org
REFERENCES
- Al-Bassam J, Kim H, Brouhard G, van Oijen A, Harrison SC, Chang F 2010. CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev Cell 19: 245–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J, Chang F 2011. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol 21: 604–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor DJ, Bentley K, Ryan J, Perry J, Wong L, Slater H, Choo KH 2004. Human centromere repositioning “in progress”. Proc Natl Acad Sci 101: 6542–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN 2006. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci 103: 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader JR, Vaughan KT 2010. Dynein at the kinetochore: Timing, interactions and functions. Semin Cell Dev Biol 21: 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR 2011. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol 194: 229–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S 2013. The composition, functions, and regulation of the budding yeast kinetochore. Genetics 194: 817–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev 13: 532–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Cleveland DW 2011. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell 144: 471–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Cleveland DW 2004. Structural determinants for generating centromeric chromatin. Nature 430: 578–582 [DOI] [PubMed] [Google Scholar]
- Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW 2007. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell 25: 309–322 [DOI] [PubMed] [Google Scholar]
- Campbell CS, Mullins RD 2007. In vivo visualization of type II plasmid segregation: Bacterial actin filaments pushing plasmids. J Cell Biol 179: 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Milks KJ, Straight AF 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol 189: 1143–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YW, Jeyaprakash AA, Nigg EA, Santamaria A 2012. Aurora B controls kinetochore-microtubule attachments by inhibiting Ska complex-KMN network interaction. J Cell Biol 196: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A 2008. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol 9: 33–46 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR III, Chan CS, Drubin DG, Barnes G 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111: 163–172 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR III, Oegema K, Desai A 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev 18: 2255–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A 2006. The Conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127: 983–997 [DOI] [PubMed] [Google Scholar]
- Chen RH, Waters JC, Salmon ED, Murray AW 1996. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science 274: 242–246 [DOI] [PubMed] [Google Scholar]
- Chen Q, Zhang X, Jiang Q, Clarke PR, Zhang C 2008. Cyclin B1 is localized to unattached kinetochores and contributes to efficient microtubule attachment and proper chromosome alignment during mitosis. Cell Res 18: 268–280 [DOI] [PubMed] [Google Scholar]
- Ciferri C, De Luca J, Monzani S, Ferrari KJ, Ristic D, Wyman C, Stark H, Kilmartin J, Salmon ED, Musacchio A 2005. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem 280: 29088–29095 [DOI] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. 2008. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421 [DOI] [PubMed] [Google Scholar]
- Collins KA, Castillo AR, Tatsutani SY, Biggins S 2005. De novo kinetochore assembly requires the centromeric Histone H3 variant. Mol Biol Cell 16: 5649–5660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke CA, Heck MM, Earnshaw WC 1987. The inner centromere protein (INCENP) antigens: Movement from inner centromere to midbody during mitosis. J Cell Biol 105: 2053–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambacher S, Deng W, Hahn M, Sadic D, Frohlich J, Nuber A, Hoischen C, Diekmann S, Leonhardt H, Schotta G 2012. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus 3: 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum JR, Wren JD, Daniel JJ, Sivakumar S, McAvoy JN, Potapova TA, Gorbsky GJ 2009. Ska3 is required for spindle checkpoint silencing and the maintenance of chromosome cohesion in mitosis. Curr Biol 19: 1467–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ 2002. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol 319: 1097–1113 [DOI] [PubMed] [Google Scholar]
- De Antoni A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, Mapelli M, Sironi L, Faretta M, Salmon ED, et al. 2005. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol 15: 214–225 [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Musacchio A 2012. Structural organization of the kinetochore-microtubule interface. Curr Opin Cell Biol 24: 48–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED 2002. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol 159: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127: 969–982 [DOI] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ 1997. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 13: 83–117 [DOI] [PubMed] [Google Scholar]
- Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Shevchenko A, Hyman A, Oegema K 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev 17: 2421–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S, Salmon ED, Mitchison TJ 2012. Deformations within moving kinetochores reveal different sites of active and passive force generation. Science 337: 355–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137: 485–497 [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Zhang W, Karpen GH 2013. Solo or doppio: How many CENP-As make a centromeric nucleosome? Nat Struct Mol Biol 20: 648–650 [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N 1985. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91: 313–321 [DOI] [PubMed] [Google Scholar]
- Elowe S 2011. Bub1 and BubR1: At the interface between chromosome attachment and the spindle checkpoint. Mol Cell Biol 31: 3085–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming W 1880. Beiträge zur Kenntnis der Zelle und ihrer Lebenserscheinungen. Arch Mikrosk Anat 18: 151–259 [Google Scholar]
- Foley EA, Maldonado M, Kapoor TM 2011. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat Cell Biol 13: 1265–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR 3rd, Bassett EA, Wood S, Black BE, Cleveland DW 2009. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137: 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M 2007. Priming of centromere for CENP-A recruitment by human hMis18α, hMis18β, and M18BP1. Dev Cell 12: 17–30 [DOI] [PubMed] [Google Scholar]
- Gaitanos TN, Santamaria A, Jeyaprakash AA, Wang B, Conti E, Nigg EA 2009. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J 28: 1442–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Cheeseman IM 2011. Kinetochore assembly: If you build it, they will come. Curr Opin Cell Biol 23: 102–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Cheeseman IM 2013. CDK-dependent phosphorylation and nuclear exclusion coordinately control kinetochore assembly state. J Cell Biol 201: 23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM 2011. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 145: 410–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DJ, Resio B, Pellman D 2012. Causes and consequences of aneuploidy in cancer. Nat Rev Genet 13: 189–203 [DOI] [PubMed] [Google Scholar]
- Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR 2005. Force production by disassembling microtubules. Nature 438: 384–388 [DOI] [PubMed] [Google Scholar]
- Grishchuk EL, Efremov AK, Volkov VA, Spiridonov IS, Gudimchuk N, Westermann S, Drubin D, Barnes G, McIntosh JR, Ataullakhanov FI 2008. The Dam1 ring binds microtubules strongly enough to be a processive as well as energy-efficient coupler for chromosome motion. Proc Natl Acad Sci 105: 15423–15428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan RS, Manak MS, Buch HK, Meier MG, Meraldi P, Shah JV, Sorger PK 2011. p31(comet) acts to ensure timely spindle checkpoint silencing subsequent to kinetochore attachment. Mol Biol Cell 22: 4236–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch A, Sillje HH, Nigg EA 2006. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J 25: 5504–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118: 715–729 [DOI] [PubMed] [Google Scholar]
- Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL 2010. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell 40: 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW 2009. Boveri revisited: Chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol 10: 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang W-H, Suzuki E, Okawa K, et al. 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. 135: 1039–1052 [DOI] [PubMed] [Google Scholar]
- Hori T, Shang WH, Takeuchi K, Fukagawa T 2013. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J Cell Biol 200: 45–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol 155: 1159–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, et al. 2011. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev 25: 901–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LET, Black BE, Foltz DR, Cleveland DW 2007. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 176: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaprakash AA, Santamaria A, Jayachandran U, Chan YW, Benda C, Nigg EA, Conti E 2012. Structural and functional organization of the Ska complex, a key component of the kinetochore-microtubule interface. Mol Cell 46: 274–286 [DOI] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED 2006. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol 8: 581–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A 2006. Chromosomes can congress to the metaphase plate before biorientation. Science 311: 388–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess R 2005. Rod-Zw10-Zwilch: A key player in the spindle checkpoint. Trends Cell Biol 15: 386–392 [DOI] [PubMed] [Google Scholar]
- Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y 2013. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 340: 1110–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Holland AJ, Lan W, Cleveland DW 2010. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell 142: 444–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Obuse C, Yanagida M 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev Cell 13: 663–676 [DOI] [PubMed] [Google Scholar]
- Kline-Smith SL, Khodjakov A, Hergert P, Walczak CE 2004. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol Biol Cell 15: 1146–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T, Zhang G, Larsen MS, Lischetti T, Streicher W, Kragh Nielsen T, Bjorn SP, Nilsson J 2013. Direct binding between BubR1 and B56-PP2A phosphatase complexes regulate mitotic progression. J Cell Sci 126: 1086–1092 [DOI] [PubMed] [Google Scholar]
- Lampert F, Hornung P, Westermann S 2010. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J Cell Biol 189: 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson MA, Cheeseman IM 2011. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol 21: 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Winey M 2012. The MPS1 family of protein kinases. Annu Rev Biochem 81: 561–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S-T, Rattner JB, Jablonski SA, Yen TJ 2006. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol 175: 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science 323: 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Vleugel M, Backer CB, Hori T, Fukagawa T, Cheeseman IM, Lampson MA 2010. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol 188: 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Davydenko O, Lampson MA 2012. Polo-like kinase-1 regulates kinetochore-microtubule dynamics and spindle checkpoint silencing. J Cell Biol 198: 491–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Oegema K, Desai A, Cheeseman IM 2004. “Holo”er than thou: Chromosome segregation and kinetochore function in C. elegans. Chromosome Res 12: 641–653 [DOI] [PubMed] [Google Scholar]
- Maddox PS, Hyndman F, Monen J, Oegema K, Desai A 2007. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol 176: 757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Fairley EA, Rieder CL, Swedlow JR, Sunkel CE, Earnshaw WC 2003. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell 113: 891–904 [DOI] [PubMed] [Google Scholar]
- Mao Y, Abrieu A, Cleveland DW 2003. Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell 114: 87–98 [DOI] [PubMed] [Google Scholar]
- Mapelli M, Filipp FV, Rancati G, Massimiliano L, Nezi L, Stier G, Hagan RS, Confalonieri S, Piatti S, Sattler M, et al. 2006. Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J 25: 1273–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Lluesma S, Stucke VM, Nigg EA 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297: 2267–2270 [DOI] [PubMed] [Google Scholar]
- Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T 1989. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol 109: 1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr MI, Hummer S, Bormann J, Gruner T, Adio S, Woehlke G, Mayer TU 2007. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol 17: 488–498 [DOI] [PubMed] [Google Scholar]
- Mazumdar M, Misteli T 2005. Chromokinesins: Multitalented players in mitosis. Trends Cell Biol 15: 349–355 [DOI] [PubMed] [Google Scholar]
- McEwen BF, Chan GK, Zubrowski B, Savoian MS, Sauer MT, Yen TJ 2001. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol Biol Cell 12: 2776–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Volkov V, Ataullakhanov FI, Grishchuk EL 2010. Tubulin depolymerization may be an ancient biological motor. J Cell Sci 123: 3425–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda JJ, De Wulf P, Sorger PK, Harrison SC 2005. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol 12: 138–143 [DOI] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF 2011. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol 194: 855–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED 2007. The spindle-assembly checkpoint in space and time. Nat Rev Cancer 8: 379–393 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH 2009. Cohesin: Its roles and mechanisms. Annu Rev Genet 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Nishino T, Takeuchi K, Gascoigne KE, Suzuki A, Hori T, Oyama T, Morikawa K, Cheeseman IM, Fukagawa T 2012. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell 148: 487–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, Rago F, Hori T, Tomii K, Cheeseman IM, Fukagawa T 2013. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J 32: 424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Wang HW 2006. Structural intermediates in microtubule assembly and disassembly: How and why? Curr Opin Cell Biol 18: 179–184 [DOI] [PubMed] [Google Scholar]
- Obuse C, Iwasaki O, Kiyomitsu T, Goshima G, Toyoda Y, Yanagida M 2004. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat Cell Biol 6: 1135–1141 [DOI] [PubMed] [Google Scholar]
- Palmer DK, O’Day K, Wener MH, Andrews BS, Margolis RL 1987. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol 104: 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DK, O’Day K, Trong HL, Charbonneau H, Margolis RL 1991. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci 88: 3734–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM 2006. The anaphase promoting complex/cyclosome: A machine designed to destroy. Nat Rev Mol Cell Biol 7: 644–656 [DOI] [PubMed] [Google Scholar]
- Petrovic A, Pasqualato S, Dube P, Krenn V, Santaguida S, Cittaro D, Monzani S, Massimiliano L, Keller J, Tarricone A, et al. 2010. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J Cell Biol 190: 835–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr CM, Coue M, Grissom PM, Hays TS, Porter ME, McIntosh JR 1990. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature 345: 263–265 [DOI] [PubMed] [Google Scholar]
- Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, Asbury CL 2009. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell 136: 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, Glover DM 2011. CENP-C is a structural platform for kinetochore assembly. Curr Biol 21: 399–405 [DOI] [PubMed] [Google Scholar]
- Putkey FR, Cramer T, Morphew MK, Silk AD, Johnson RS, McIntosh JR, Cleveland DW 2002. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev Cell 3: 351–365 [DOI] [PubMed] [Google Scholar]
- Raaijmakers JA, Tanenbaum ME, Maia AF, Medema RH 2009. RAMA1 is a novel kinetochore protein involved in kinetochore-microtubule attachment. J Cell Sci 122: 2436–2445 [DOI] [PubMed] [Google Scholar]
- Rago F, Cheeseman IM 2013. Review series: The functions and consequences of force at kinetochores. J Cell Biol 200: 557–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S 2010. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell 40: 455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier V, Vagnarelli P, Fukagawa T, Zerjal T, Burns E, Trouche D, Earnshaw W, Brown W 2005. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol Cell Biol 25: 3967–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salje J, Gayathri P, Lowe J 2010. The ParMRC system: Molecular mechanisms of plasmid segregation by actin-like filaments. Nat Rev Microbiol 8: 683–692 [DOI] [PubMed] [Google Scholar]
- Schleiffer A, Maier M, Litos G, Lampert F, Hornung P, Mechtler K, Westermann S 2012. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat Cell Biol 14: 604–613 [DOI] [PubMed] [Google Scholar]
- Schmidt JC, Arthanari H, Boeszoermenyi A, Dashkevich NM, Wilson-Kubalek EM, Monnier N, Markus M, Oberer M, Milligan RA, Bathe M, et al. 2012. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev Cell 23: 968–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screpanti E, De Antoni A, Alushin GM, Petrovic A, Melis T, Nogales E, Musacchio A 2011. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol 21: 391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE 2012. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell 22: 52–63 [DOI] [PubMed] [Google Scholar]
- Sironi L, Mapelli M, Knapp S, De Antoni A, Jeang KT, Musacchio A 2002. Crystal structure of the tetrameric Mad1-Mad2 core complex: Implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J 21: 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuer ER, Wordeman L, Schroer TA, Sheetz MP 1990. Localization of cytoplasmic dynein to mitotic spindles and kinetochores. Nature 345: 266–268 [DOI] [PubMed] [Google Scholar]
- Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L 2008. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell 14: 252–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol 154: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk SJ, Vleugel M, Teixeira A, Kops GJ 2012. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev Cell 23: 745–755 [DOI] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, et al. 2011. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 476: 232–235 [DOI] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJR, Nasmyth K 2002. Evidence that the Ipl1-Sli15 (Aurora Kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108: 317–329 [DOI] [PubMed] [Google Scholar]
- Theis M, Slabicki M, Junqueira M, Paszkowski-Rogacz M, Sontheimer J, Kittler R, Heninger AK, Glatter T, Kruusmaa K, Poser I, et al. 2009. Comparative profiling identifies C13orf3 as a component of the Ska complex required for mammalian cell division. EMBO J 28: 1453–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien JF, Umbreit NT, Gestaut DR, Franck AD, Cooper J, Wordeman L, Gonen T, Asbury CL, Davis TN 2010. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol 189: 713–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Andrews PD, Wickramasinghe S, Sleeman J, Prescott A, Lam YW, Lyon C, Swedlow JR, Lamond AI 2003. Time-lapse imaging reveals dynamic relocalization of PP1γ throughout the mammalian cell cycle. Mol Biol Cell 14: 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader G, Medema RH, Lens SM 2006. The chromosomal passenger complex: Guiding Aurora-B through mitosis. J Cell Biol 173: 833–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, O’Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF, et al. 2009. Protein architecture of the human kinetochore microtubule attachment site. Cell 137: 672–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Salmon ED 1998. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol 141: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RR, Sorger PK, Harrison SC 2005. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci 102: 5363–5367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC 2007. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol 14: 54–59 [DOI] [PubMed] [Google Scholar]
- Welburn JP, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR III, Cheeseman IM 2009. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell 16: 374–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn JP, Vleugel M, Liu D, Yates JR III, Lampson MA, Fukagawa T, Cheeseman IM 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell 38: 383–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, Nogales E, Barnes G 2005. Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol Cell 17: 277–290 [DOI] [PubMed] [Google Scholar]
- Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G 2006. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature 440: 565–569 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Jensen ON, Holmes S, Souès S, Mann M, Kilmartin JV 1998. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol 141: 967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kilmartin JV 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J Cell Biol 152: 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH Jr, McDonald KL, McIntosh JR 1995. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol 129: 1601–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Li G, Schaar BT, Szilak I, Cleveland DW 1992. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature 359: 536–539 [DOI] [PubMed] [Google Scholar]