Abstract
A direct method has been developed for the in vitro synthesis of stable DNA – protein cross-links (DPC’s) between guanine and amino acids (lysine, arginine). This method employs the combination of guanine neutral radicals, G(-H)•, and side-chain C-centered amino acid radicals. The latter were generated indirectly after first causing the selective photoionization of 2-aminopurine (2AP) embedded in the oligonucleotide, 5’-d(CC[2AP]TCGCTACC) by intense nanosecond 308 nm excimer laser pulses. The 2AP radical cation deprotonates rapidly to form the 2AP(-H)• neutral radical which, in turn, oxidizes the nearby guanine to form the neutral guanine G(-H)• radical, as described previously (Shafirovich et al. J. Phys. Chem. B, 2001, 105, 8431). In parallel, the hydrated electrons generated by the photoionization of 2AP, are scavenged by nitrous oxide to generate hydroxyl radicals. In the presence of a large excess of the amino acids, the hydroxyl radicals oxidize the latter to produce C-centered amino acid radicals that combine with the G(-H)• radicals to form the guanine-amino acid cross-linked oligonucleotide product. Analogous products were generated by photoionizing the free nucleoside, 2’,3’,5’-tri-O-acetylguanoise, (tri-O-Ac-Guo) using intense nanosecond 266 nm Nd: Yag laser pulse irradiation. The guanine – amino acid cross-links thus produced either site-specifically positioned in oligonucleotides, or in the free nucleoside tri-O-Ac-Guo were isolated by HPLC methods and identified by high resolution LC-TOF/MS and LC-MS/MS methods. The possibility is discussed that analogous guanine-amino acid cross-linked products could be formed in vivo under single hit radical generation mechanisms during oxidative stress.
Introduction
Oxidatively generated damage to cellular DNA is genotoxic and has been correlated with the development of many human cancers.1 There is growing evidence that complex DNA lesions, so-called tandem and clustered DNA lesions are potentially more harmful than single oxidized nucleobases.2 The complex modifications, which can be induced by single radical hits of hydroxyl radicals or one-electron oxidants, include intra- and interstrand cross-links and DNA-protein cross-links (DPCs).3 Formation of DPCs has been clearly demonstrated in cellular DNA4 exposed to ionizing radiation,5, 6 photosensitization,7–10 strong oxidizing agents prepared chemically,11 or generated in situ by photochemical methods.12, 13 However, structural information on DPCs is scarce, although several examples are in existence. Dizdaroglu et al. have characterized thymine – amino acid cross-links produced by ionizing radiation in solutions of DNA – histone protein non-covalent complexes.14, 15 Other examples are guanine – lysine cross-links discovered by Perrier et al.16 The C. J. Burrows group reported cross-links between 8-oxo-7,8-dihydroguanine (8-oxoGua), the major lesion formed by the oxidation of guanine in double-stranded DNA, and lysine residues of proteins produced by the one-electron oxidant Ir(IV).17, 18
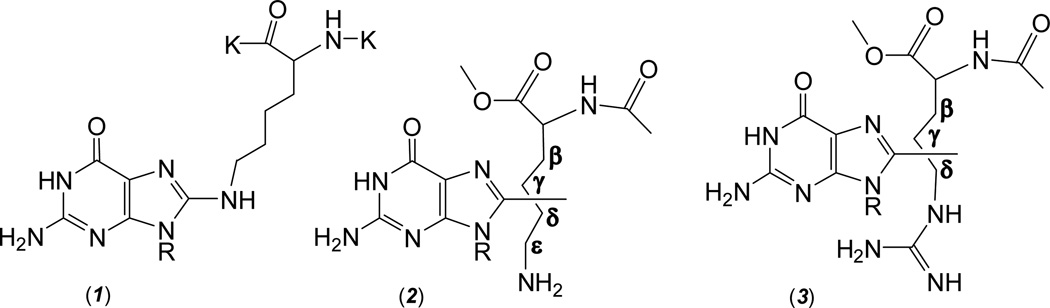
The key step in the oxidatively induced cross-linking reaction is the formation of the covalent bond between the nucleobase and the amino acid that, at the mechanistic level, requires abstraction of two electrons and two-protons. Perrier et al.16 showed that the one-electron oxidation of guanine, the most easily oxidizable DNA base,19 and hence the primary target of oxidatively generated damage to DNA,20 is followed by the nucleophilic addition of the ε-amino group of the lysine residue to the C8 atom of the guanine radical cation, G•+; the radical adduct thus formed is readily oxidized by weak oxidants such as oxygen to the Nε-(guanin-8-yl)-lysine cross-link (Figure 1).
Figure 1.
Guanine – lysine cross-links derived from the nucleophilic addition of the ε-amino group of lysine residue to the C8 atom of G•+ radicals (1) and combination of G(-H)• and C-centered radicals produced by H-atom abstraction from methyl esters of N-acetyl-lysine (2) and N-acetyl-arginine (3). The cross-links (2 and 3) are the mixtures of the isomers in which C8-atom of guanine is linked to one of the side-chain C-atoms (β, γ, δ, ε).
In this work we developed a new approach for the in vitro generation of stable DPC’s between guanine and amino acids (lysine, arginine) based on the combination of guanine neutral radicals, G(-H)• and side-chain C-centered amino acid radicals. This pair of radicals was generated by a laser flash photolysis method developed earlier by our group for the synthesis of oxidized and nitrated derivatives of guanine.21, 22 This method employs the selective two-photon ionization of nucleobases by intense nanosecond laser pulses.23 The guanine radical cations are generated directly by one-electron oxidation,24 while the hydrated electrons thus produced are scavenged by nitrous oxide to generate hydroxyl radicals.25 The latter rapidly abstract hydrogen atoms from amino acid added present in large excess relative to the nucleic acids, to produce side-chain C-centered amino acid radical.26 Here, we successfully synthesized guanine – amino acid cross-links (Figure 1) with guanine residues either embedded site-specifically in oligonucleotides, or with the free nucleoside, 2’,3’,5’-tri-O-acetylguanoise, (tri-O-Ac-Guo).
Experimental
Materials
All chemicals (analytical grade) were used as received. Methyl esters of Nα-acetyl-l-lysine (Ac-Lys-Me) and Nα-acetyl-l-argenine (Ac-Arg-Me), both in the form of hydrochlorides were obtained from Bachem (Torrance, CA). The 2’-deoxyribooligonucleotide, 5'-d(CC[2AP]TCGTACC) with a single 2-aminopurine residue (2AP), was synthesized by standard automated phosphoramidite chemistry techniques. Phosphoramidites and other chemicals required for the oligonucleotide synthesis were obtained from Glen Research (Sterling, VA). The tritylated oligonucleotides were deprotected overnight at 55 °C employing concentrated aqueous ammonia solutions. The crude oligonucleotides were purified by reversed-phase HPLC, detritylated in 80% acetic acid according to standard protocols, and desalted using reversed-phase HPLC. The integrity and composition of the oligonucleotides were confirmed by MALDI-TOF mass spectrometry.
Generation of Guanine – Amino Acid Cross-Links
The 0.250 mL N2O-saturated samples containing 0.025 mM 5'-d(CC[2AP]TCGTACC, 10 mM methyl ester of Nα-acetyl-l-lysine (or Nα-acetyl-l-argenine) and 100 mM NaCl in 5 mM phosphate buffer solution (pH 7.5), were photolyzed by a train of 308 nm XeCl excimer laser pulses (~ 12 ns pulse width, 10 Hz repetition rate, 60 mJ/pulse/cm2) for a fixed period of time (10 s) that provides energy dosage of ~6 J/cm2. The 0.250 mL N2O-saturated samples containing 0.1 mM tri-O-Ac-Guo, 10 mM methyl ester of Nα-acetyl-l-lysine (or Nα-acetyl-l-argenine) and 100 mM NaCl in 5 mM phosphate buffer solution (pH 7.5) were photolyzed by a train of 266 nm Nd: YAG laser pulses (~ 6 ns pulse width, 20 Hz repetition rate, ~200 mJ/pulse/cm) for a fixed period of time (15 s) that provides energy dosage of ~60 J/cm2. After irradiation, the samples were immediately subjected to HPLC analysis.
HPLC Isolation of Cross-Linked Products
The separation of the oligonucleotide reaction products was performed on an anion-exchange analytical column (250 mm × 4.6 mm i.d., DNAPac PA-100 Dionex, Sunnyvale, CA, USA). The elution buffer solution was a 25–60% linear gradient of solvent B (10% acetonitrile and 90% 1.5M ammonium acetate) in solvent A (10% acetonitrile and 90% water) for 30 min, followed by an isocratic phase (100% B) for 15 min at a flow rate of 1 mL/min (detection of products at 260 nm).
The nucleoside reaction products were separated on a reversed-phase analytical (250 mm × 4.6 mm i.d.) Microsorb-MV C18 column (Varian, Walnut Creek, CA) employing a 2 – 30% linear gradient of methanol in 25 mM ammonium formate in water (pH 7) for 60 min at a flow rate of 1 mL/min (detection of products at 260 nm). The HPLC fractions were evaporated under vacuum to remove methanol and purified by a second HPLC cycle. The guanine – amino acid cross-linked adducts were desalted by reversed-phase HPLC using the following mobile phases: 5 mM ammonium acetate for 5 min, deionized water for 15 min and water/acetonitrile = 1: 1 for 15 min. The desalted samples were concentrated under vacuum and subjected to LC-MS analysis.
High Resolution LC-TOF/MS and LC-MS/MS
An Agilent 1100 Series binary LC system coupled to an Agilent LC/MSD TOF time of flight mass spectrometer equipped with a dual electrospray ion source was used to perform exact mass measurements (nebulizer gas 40 psig, drying gas 8 L/min, gas temperature 350 °C, fragmentor 175 V, scan range m/z 100 to 3500, transient length 104992 points at 1 GHz, 10,000 transients/scan, reference flow 5 µL/min, internal reference ESI positive ion 121.050873 and 922.009798 or ESI negative ion 119.036320 and 966.000725). The MS/MS experiments were performed with an Agilent 1100 Series capillary LC/MSD Ion Trap XCT mass spectrometer equipped with an electrospray ion source (nebulizer gas 40 psig, drying gas 8 L/min, gas temperature 350 °C, acquisition range m/z 100–2200, target mass 600, compound stability 10%, trap drive 100%, manual MS/MS, fragmentation amplitude 1.0 V, smart frag 30–200%, fragmentation time 10 ms, fragmentation width 4 amu.). All experiments were performed using 2 uL loop injection flowing water/acetonitrile = 1: 1 with 0.1 % formic acid in the positive ion mode or water/acetonitrile = 1: 1 containing 15 mM imidazole in the negative ion mode.
Results and discussion
Generation of Guanine and C-Centered Amino Acid Radicals by a Two-Photon Ionization Mechanism
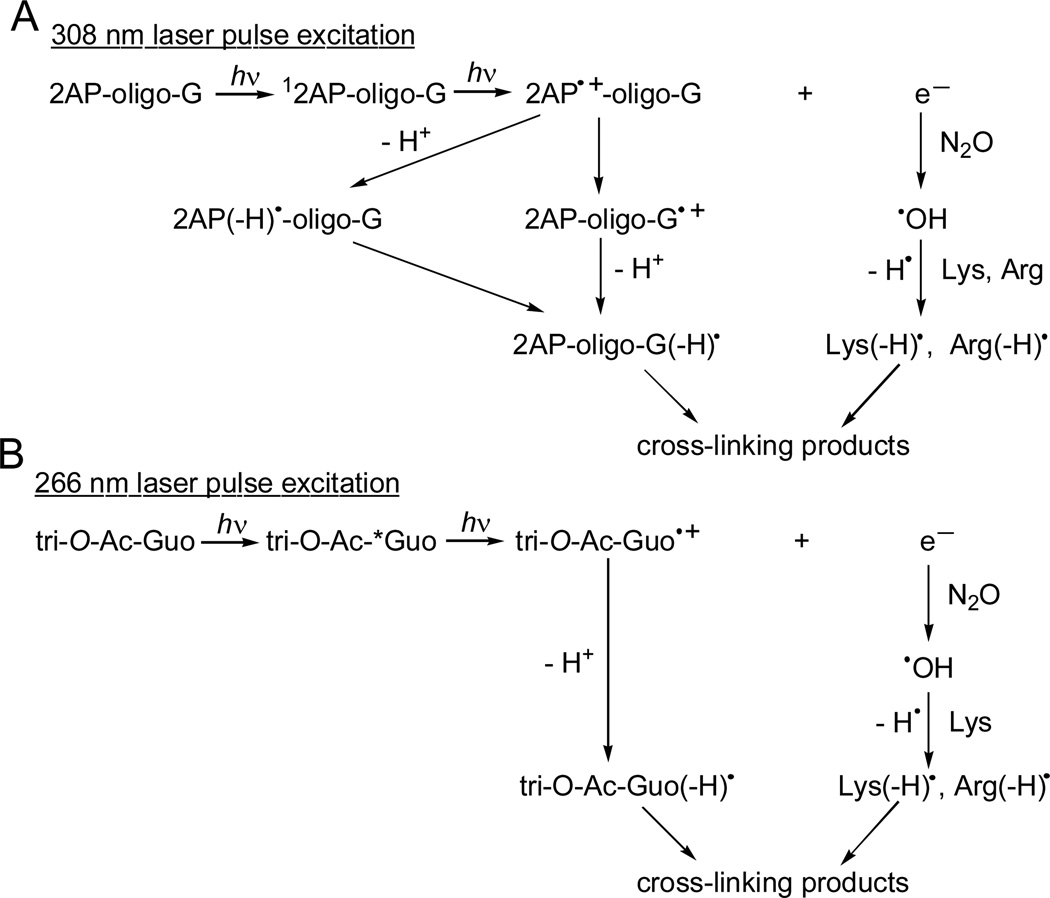
In the case of DNA, this method involves the site-specific incorporation of a single 2-aminopurine (2AP) residue that is characterized by an absorption band with a maximum at 305 nm.27 This nucleic acid base analog either in single- or double-stranded oligonucleotides is selectively photoionized with intense nanosecond 308 nm excimer laser pulses by a two-photon absorption mechanism.23 Absorption of the first photon results in the formation of the 2AP singlet excited state (12AP), and the absorption of the second photon causes its photoionization (Figure 2A).
Figure 2.
Generation of G(-H)• and Lys(-H)• and Arg(-H)• radicals in oligonucleotides (A) and free nucleosides (B) by two-photon ionization mechanism. *Guo is a first excited state (singlet and/or triplet).
Previous laser flash photolysis studies have shown that the primary products of the laser pulse excitation are the 2AP•+ radical cations and hydrated electrons (Figure 2A).23 The yields of hydrated electrons are proportional to the square of the laser fluence rate (photon s−1 cm−2) as expected for the two-photon processes. The 2AP•+ radicals rapidly deprotonate to yield the neutral 2AP(-H)• radicals. Both the radical cation and the neutral radical are strong one-electron oxidants that are capable of oxidizing guanines by proton-coupled electron transfer mechanism to form G(-H)• radicals when the 2AP and G residues are separated by 1, 2, or even 3 intervening bases.21–24, 28
The normal DNA nucleobases have a negligible absorbance at 308 nm in comparison with 2AP and are not ionized by the 308 nm excimer laser excitation.23 In the case of free guanine nucleosides, the two-photon-induced ionization of G-residues was initiated by shorter-wavelength intense nanosecond 266 nm Nd: YAG laser pulses (Figure 2B).29, 30 The primary products of the laser pulse excitation are G•+ radical cations and hydrated electrons detected by transient absorption spectroscopy.29 In neutral solutions, the guanine radical cations, Guo•+ (pKa =3.9), rapidly deprotonate to form neutral Guo(-H)• radicals31, 32
In N2O-saturated solutions, the hydrated electrons derived from the photoionization of nucleobases are rapidly and quantitatively scavenged by N2O to form •OH radicals.25 The latter abstract H-atoms from the amino acids with the formation of C-centered amino acid radicals.25 This reaction competes with the direct oxidation of guanine (or DNA) by •OH radicals.33 Both reactions of •OH radicals are extremely rapid and occur with rate constants in the (1–5)×109 M−1s−1 range that are close to the diffusion controlled limit.26, 33–35 Here, we used ~100- or 400-fold excess of amino acids relative to tri-O-Ac-Guo or 5'-d(CC[2AP]TCGTACC) concentrations, to ensure that the competitive direct reactions of •OH radicals with these molecules are minimized. Thus, two-photon photoexcitation of DNA strands with site-specifically positioned 2AP and G bases in N2O-saturated aqueous solutions, provides a very convenient approach for the generation of similar fluxes of guanine and C-centered amino acid radicals, which can then combine to generate the cross-linked products (Figure 2).
Identification of Oligonucleotide - Amino Acid Cross-links
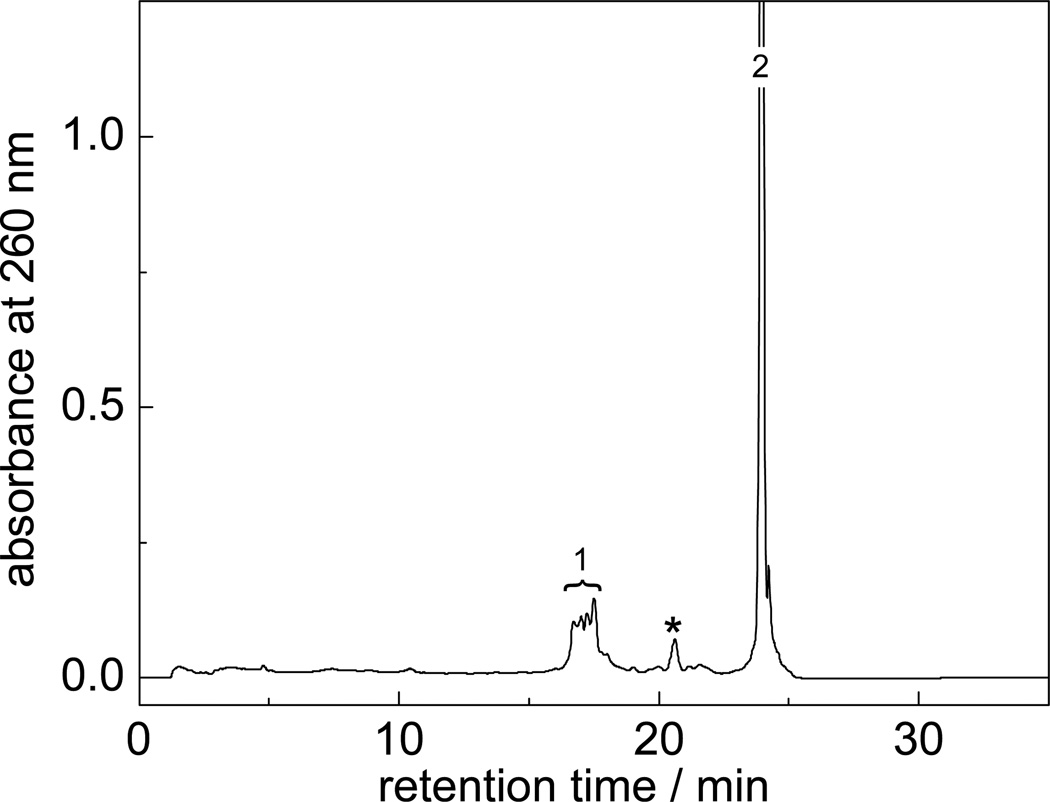
The stable products generated by the two-photon excitation of the 5'-d(CC[2AP]TCGTACC) sequence by intense nanosecond 308 nm laser pulses in N2O-saurated buffer solutions (pH 7.5) containing Nα-acetyl-l-lysine, were separated by anion-exchange HPLC methods (Figure 3).
Figure 3.
Anion-exchange HPLC elution profile of the end products generated by two-photon ionization of the 0.025 mM 5'-d(CC[2AP]TCGTACC) sequence with a 10 s train of 308 nm laser pulses in N2O-saturated buffer solution (pH 7.5) containing 10 mM Nα-acetyl-l-lysine. The cross-linked products elute at 16 – 18 min (fraction 1), and the unmodified sequence 5'-d(CC[2AP]TCGTACC) at 24.0 min (peak 2). The 20 min peak indicated by an asterisk is an impurity that was present in the starting sequence.
The partially resolved reaction products are observed at 16 – 18 min (fraction 1), whereas the unmodified oligonucleotide elutes at 24.0 min. Each of these fractions was then desalted, concentrated under vacuum, and subjected to LC/MS analysis.
The negative ion trap mass spectra of these four fractions exhibit triply and doubly charged ions at m/z 1144.2 and 1716.8. In contrast, the unmodified standard sequence is characterized by triply and doubly negatively charged ions at m/z 1077.5 and 1616.8. The deconvolution of these spectra showed that the masses of the modified oligonucleotides were 200 Da greater than the mass of the unmodified parent oligonucleotide (M). Thus, the masses of the products in each of the four fractions were equal to M+200 Da, which corresponds to the sum of the masses of the oligonucleotide and the lysine methyl ester minus 2 Da.
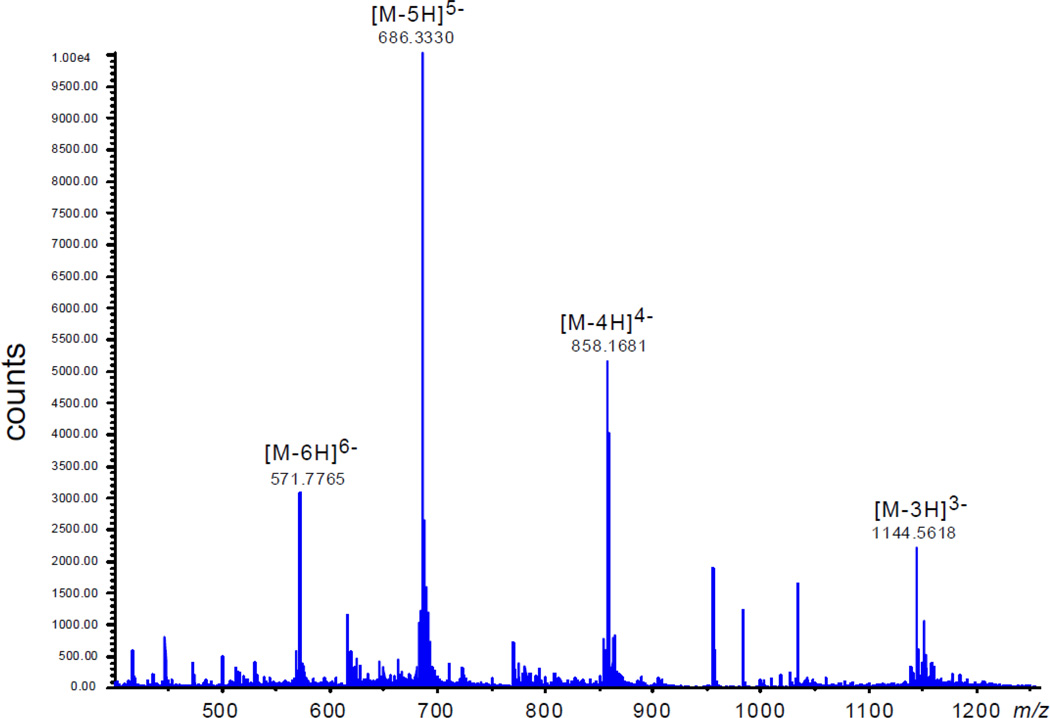
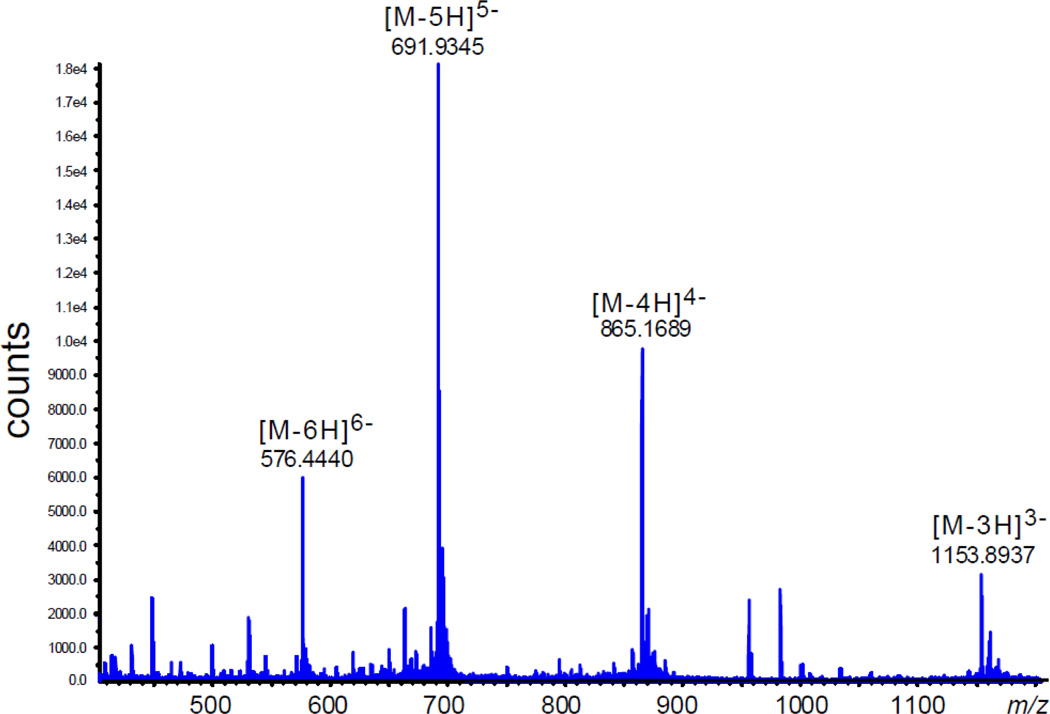
The exact mass spectra of these lysine oligonucleotide adducts exhibit a series of multiply negatively charged ions, [M-nH]n− with n = 3, 4, 5 and 6 (Figure 4).
Figure 4.
The exact mass spectrum of the 5'-d(CC[2AP]TCGTACC) - Ac-Lys-Me cross-link in the negative ion mode.
The deconvolution of this spectrum gives the monoisotopic mass of 3435.70 that corresponds to the expected formula C113H151N39O67P10 (molecular weight, MW = 3437.37, accuracy 3.8 ppm) for this oligonucleotide – lysine adduct. The negative exact mass spectrum of the unmodified sequence also shows a series of [M-nH]n− ions with n = 3, 4, 5 and 6 (data not shown). The deconvolution of this spectrum yields the monoisotopic mass of 3235.58 that corresponds to the expected formula C104H135N37O64P10 (MW = 3237.13, accuracy 2.5 ppm).
A further experiment showed that the two-photon ionization approach can be successfully used for the synthesis of the analogous arginine oligonucleotide adducts. These adducts were generated by intense nanosecond 308 nm laser pulse irradiation of N2O-saurated buffer solutions (pH 7.5) containing Nα-acetyl-l-arginine in addition to the oligonucleotide. Analysis of the photoproducts by anion-exchange HPLC methods allowed for the isolation of a series of arginine-oligonucleotide adducts with identical masses of M+232 that correspond to the addition of the arginine methyl ester residue to the oligonucleotide. The exact negative mass spectrum of the argenine oligonucleotide adduct is shown in Figure 5.
Figure 5.
The exact negative mass spectrum of the 5'-d(CC[2AP]TCGTACC) - Ac-Arg-Me cross-linked adduct.
The deconvolution of this spectrum yields the monoisotopic mass 3463.70 that corresponds to the expected formula C113H151N41O67P10 (MW = 3465.38, accuracy 4.2 ppm).
These results indicate that the two-photon ionization method generates guanine cross-links with amino acids. To further confirm this conclusion based on the exact mass spectra of these cross-linked oligonucleotide – amino acid adducts, and to obtain better insights into their structural features, we synthesized the free nucleoside 2’,3’,5’-tri-O-acetylguanosine (tri-O-Ac-Guo)-Ac-Lys-Me cross-link using the same approach.
In contrast, we did not observe any cross-link formation using continuous illumination. In this experiment the samples were irradiated with light from a 100 W high-pressure Xe lamp that was reflected from a dichroic mirror with reflectance in the 300–340 nm (oligonucleotide) or 240–290 nm (free nucleoside) spectral ranges. The energy incident on the sample was 50–100 mW/cm2 and the irradiation time was varied to achieve different energy dosages from 6 or 60 J/cm2 used in the laser excitation mode. These results indicate that single-photon absorption mechanisms responsible for DNA damage by UV-A light (290–400 nm)36 do not provide for the efficient generation of the reactive species required for the formation of guanine cross-links with amino acids under our experimental conditions.
The alternative methods for generation DNA – protein cross-links based on single-photon absorption mechanisms employ photosensitization and photo-induced charge separation. For instance, visible light irradiation of proflavin covalently linked to oligo-α- or oligo-β-deoxynucleotides generates the photo-cross-linking species detected by gel electrophoresis.8, 9, 37 Singlet oxygen generated by photosensitization of typical type II photosensitizers (methylene blue, porphyrines and others) induces formation of the cross-links in DNA-histone complexes.10 Photo-induced one-electron oxidation of guanine by electronically-excited state of Ru-complex covalently-linked to DNA initiates formation of the cross-links with lysine-tyrosine-lysine tripeptide.12, 13
Tri-O-Ac-Guo – Ac-Lys-Me Cross-Links
The cross-linked products generated by the two-photon ionization of 2’,3’,5’-tri-O-acetylguanosine by intense nanosecond 266 nm laser pulses in N2O-saturated buffer solutions (pH 7.5) containing the methyl ester of Nα-acetyl-l-lysine, were separated by reversed-phase HPLC methods (data not shown). The positive ion trap mass spectra of these products exhibited a molecular ion, [M+H]+ at m/z 610.2. In contrast, the unmodified standard, tri-O-Ac-Guo exhibited the molecular ion at m/z 410.2. This mass difference of 200 Da clearly indicates the formation of the tri-O-Ac-Guo – Ac-Lys-Me cross-link. The exact positive mass spectra of tri-O-Ac-Guo – Ac-Lys-Me cross-links yields the monoisotope mass of 609.24 that corresponds to the expected formula C25H35N7O11 (MW = 609.59, accuracy 1.5 ppm); the unmodified standard, tri-O-Ac-Guo, yields the monoisotope mass of 409.12 that corresponds to the expected formula C16H19N5O8 (MW = 409.35, accuracy 0.3 ppm).
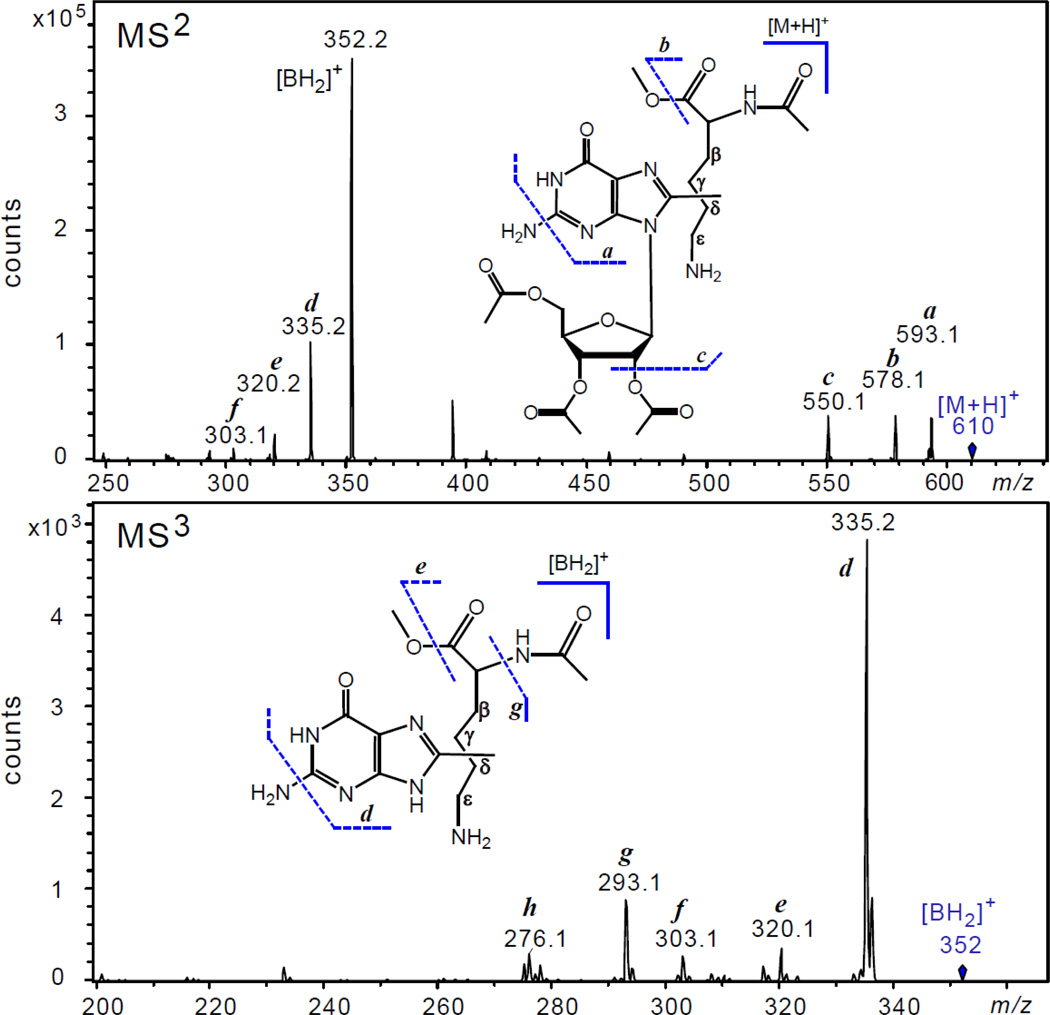
To obtain insights into the structure of the tri-O-Ac-Guo – Ac-Lys-Me cross-link, we analyzed the fragmentation patterns of the parent molecular ion, [M+H]+ at m/z 610 (MS1 data not shown). The major pathway of the [M+H]+ fragmentation (Panel MS2 in Figure 6) involves the cleavage of the N-glycosidic bond with the concomitant transfer of a hydrogen atom from the sugar residue to the base.38
Figure 6.
The positive ion spectra (MS/MS) of the tri-O-Ac-Guo – Ac-Lys cross-link shown as a mixture of the isomers in which the C8-atoms of guanine are linked to one of the different side-chain C-atoms (β, γ, δ, ε). Panel MS2: product ion spectrum obtained by fragmentation of the molecular ion, [M+H]+ at m/z 610. The product ions detected: a at m/z 593.1 [M+H-NH3]+, b at m/z 578.1 [M+H-CH3OH]+, and c at m/z 550.1 [M+H- CH3COOH]+. Panel MS3; product ion spectrum obtained by fragmentation of the aglycone ion, [BH2]+ at m/z 352. The product ions detected: d at m/z 335.2 [BH2-NH3]+, e at m/z 320.1 [BH2-CH3OH]+, f at m/z 303.1 [BH2-NH3-CH3OH]+, g at m/z 293.1 [BH2-CH3CONH2]+, h at m/z 276.1 [BH2-NH3-CH3CONH2]+.
The aglycone ion, [BH2]+ formed is detected at m/z 352.2 and has a mass that is equal to the sum of the Guo and Ac-Lys-Me masses minus 2 Da. The product ions, a, b and c formed with lower yields can be tentatively assigned to the ions derived from fragmentation of the [M+H]+ ion and are observed at m/z 593.1 [M+H-NH3]+, m/z 578.1 [M+H-CH3OH]+, and m/z 550.1 [M+H-CH3COOH]+, respectively.
The fragmentation patterns of the aglycone ion, [BH2]+ detected at m/z 352.2 are shown in Panel MS2 (Figure 6). The major product ion, d at m/z 335.2 arises from the expulsion of ammonia from [BH2]+ ion and is typically observed in many guanine derivatives.39–41 The other product ions, e, f, g, and h, formed with lower yields, were tentatively assigned to the ions derived from the fragmentation of the lysine residue and are observed at m/z 320.2 [BH2-CH3OH]+, m/z 303.1 [BH2-NH3-CH3OH]+, m/z 293.1 [BH2-CH3CONH2]+, and m/z 276.1 [BH2-NH3-CH3CONH2]+.
Structural Variants of Guanine – Lysine Cross-Links
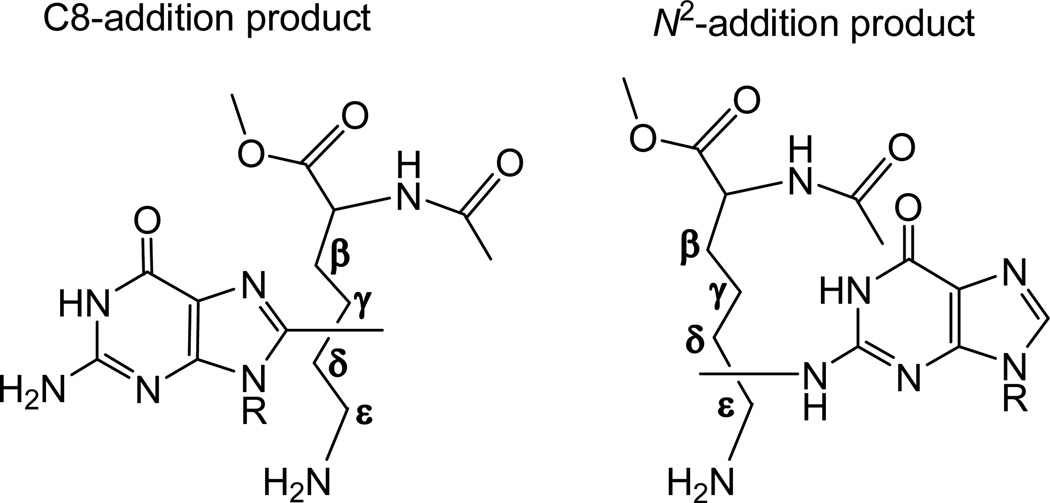
Hydrogen atom abstraction by hydroxyl radicals is an unselective process26. ESR studies of spin-trapped radicals in neutral aqueous solutions of lysine showed that •OH radicals abstract H-atoms mostly from β, γ and δ side-chain carbon atoms of lysine.42 Our previous experiments have shown that alkyl radicals (methyl, n-propyl and n-pentyl) add mostly to C8 and N2 sites of G(-H)• radicals to form C8-alkyl- and N2-alkylguanines.40, 41 Based on these considerations we can expect that six structural isomers for guanine – lysine cross-linked adducts are formed via the addition of the β, γ and δ side-chain C-centered radicals, either to C8 or N2 sites of G(-H)• radicals. Two possible structures are shown in Figure 7.
Figure 7.
The isomeric guanine – lysine cross-links derived via the addition of the C-centered (β, γ, δ, ε) side-chain lysine radicals to C8 or N2 sites of G(-H)• radicals.
This hypothesis is confirmed by the detection of a series of cross-linked products, which are partially resolved by HPLC methods (Figure 3) and that have exactly the same mass (within experimental error of 3 – 5 ppm). Selective 15N-labeling of guanine experiments have shown that the expulsion of NH3 involves opening of the pyrimidine ring,39 and hence is clearly observed in both C8- and N2-alkylguanine adducts.40 Fragmentation of the lysine residues with the formation of the product ions, e and g indicates that the end-methyl groups remain intact and are not involved in the cross-linking reaction (Figure 6).
Biological Implications
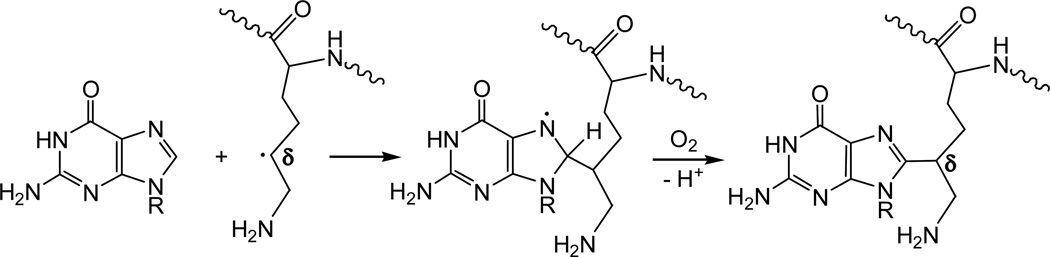
The in vitro synthetic approach described here offers a simple method for synthesizing oligonucleotide – amino acid cross-linked adducts by the combination of G(-H)• radicals with alkyl radicals produced by H-atom abstraction from side-chains of amino acids (lysine and argentine). It is well documented that •OH radicals can produce both G(-H)•35 and side-chain amino acid26 radicals. In vivo generation of hydroxyl radicals occurs via two major pathways: (1) Fenton reaction involving the reduction of H2O2 by ferrous or copper ions43, and (2) radiolysis of water molecules through the indirect effects of ionizing radiation.44 However, the probability of simultaneous formation of G(-H)• and alkyl amino acid radicals in close proximity to one another is expected to be extremely low.45 Dizdaroglu and co-workers have proposed that alkyl radicals of amino acids can directly attack nucleobases (e.g., thymine) to form reducing radical adducts, which are then readily oxidized by weak oxidants such as dioxygen.14, 15 Our experiments showed that methyl radicals directly alkylate guanine to form mostly C8-methylguanine.40 In the case of guanine – amino acid cross-links, the side-chain amino acid radicals can attack the C8-site of guanine to form radical adducts, which are readily oxidized by oxygen or other weak oxidants to form cross-linked products similar to those described in this work (Figure 8).
Figure 8.
Possible mechanism of the addition of the δ-lysine radical to C8-site of G(-H)• radical in vivo.
Other examples of purine lesions produced by C8-alkylation are the 5',8-cyclo-2'-deoxyguanosine (cdG) and 5',8-cyclo-2’-deoxyadenosine (cdA) lesions.46, 47 These lesions form via the addition of the C5'-radicals produced by H-atom abstraction from 2’-deoxyribose residues, to the C8-positions of G or A bases, followed by the oxidation of the radical adducts. The cdA and cdG lesions have been found in γ-irradiated human cultured cells,48, 49 and in humans.50 The cyclopurine adducts are highly mutagenic51, 52 and their accumulation in cellular DNA may contribute to the etiology of cancer and other diseases.49, 53–55 The guanine – thymine tandem lesions, G[8-5m]T are formed by the addition of 5-(uracilyl) methyl radicals to the C8 position of guanine to form a radical adduct that is rapidly oxidized by traces of oxygen or other weak oxidants56–60. These lesions have been found in cultured human HeLa-S3 cells exposed to ionizing radiation.61 The G[8-5m]T lesions are highly mutagenic.62–64 Thus, a single initiating oxidation event, or H-atom abstraction by •OH radicals, can potentially trigger alkylation of nucleobases in cellular DNA to form complex DNA lesions.45
Conclusion
Two-photon ionization of nucleobases in N2O-saturated aqueous solutions by intense UV laser pulses is a direct method for generation of similar fluxes of G(-H)• radicals and C-centered side chain radicals of amino acids (lysine, arginine). In the case of free guanine nucleosides, the two-photon ionization of G-residues can be directly initiated by intense nanosecond 266 nm Nd: YAG laser pulses. In 2AP-modified oligonucleotides one-electron oxidation of guanine residues occurs indirectly after first causing the selective two-photon ionization of 2AP-residues by intense nanosecond 308 nm XeCl excimer laser pulses. The hydrated electrons formed in parallel with the nucleobase radicals are scavenged by nitrous oxide to generate hydroxyl radicals. In the presence of a large excess of the amino acids, •OH radicals rapidly produce C-centered side chain amino acid radicals. The latter readily combine with the G(-H)• radicals to form site-specifically positioned guanine – amino acid cross-linked oligonucleotide products. The analogous cross-linked products could be formed in vivo under single hit radical generation mechanisms during oxidative stress. Thus, further research is warranted on the detection of these DPC’s in vivo.
Acknowledgements
We thank Dr. Kasia Janota for help in the high resolution LC-TOF/MS and LC-MS/MS measurements. This work was supported by the National Institute of Environmental Health and Sciences Grant RO1 ES 011589. Components of this work were conducted in the Shared Instrumentation Facility at NYU that was constructed with support from a Research Facilities Improvement Grant (C06 RR-16572) from the National Center for Research Resources, National Institutes of Health. The acquisition of the MALDI-TOF mass spectrometer was supported by the National Science Foundation (CHE-0958457).
References
- 1.Kryston TB, Georgiev AB, Pissis P, Georgakilas AG. Mutat. Res. 2011;711:193–201. doi: 10.1016/j.mrfmmm.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 2.Georgakilas AG, O'Neill P, Stewart RD. Radiat. Res. 2013;180:100–109. doi: 10.1667/RR3041.1. [DOI] [PubMed] [Google Scholar]
- 3.Cadet J, Bellon S, Berger M, Bourdat AG, Douki T, Duarte V, Frelon S, Gasparutto D, Muller E, Ravanat JL, Sauvaigo S. Biol. Chem. 2002;383:933–943. doi: 10.1515/BC.2002.100. [DOI] [PubMed] [Google Scholar]
- 4.Barker S, Weinfeld M, Murray D. Mutat. Res. 2005;589:111–135. doi: 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Mee LK, Adelstein SJ. Proc. Natl. Acad. Sci. U. S. A. 1981;78:2194–2198. doi: 10.1073/pnas.78.4.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olinski R, Briggs RC, Hnilica LS, Stein J, Stein G. Radiat. Res. 1981;86:102–114. [PubMed] [Google Scholar]
- 7.Helene C, Maurizot JC. CRC Crit. Rev. Biochem. 1981;10:213–258. doi: 10.3109/10409238109113600. [DOI] [PubMed] [Google Scholar]
- 8.Le Doan T, Perrouault L, Praseuth D, Habhoub N, Decout JL, Thuong NT, Lhomme J, Helene C. Nucleic Acid. Res. 1987;15:7749–7760. doi: 10.1093/nar/15.19.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Praseuth D, Le Doan T, Chassignol M, Decout JL, Habhoub N, Lhomme J, Thuong NT, Helene C. Biochemistry. 1988;27:3031–3038. doi: 10.1021/bi00408a055. [DOI] [PubMed] [Google Scholar]
- 10.Villanueva A, Canete M, Trigueros C, Rodriguez-Borlado L, Juarranz A. Biopolym. 1993;33:239–244. doi: 10.1002/bip.360330206. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton JW, Wetterhahn KE. Carcinogen. 1986;7:2085–2088. doi: 10.1093/carcin/7.12.2085. [DOI] [PubMed] [Google Scholar]
- 12.Wagenknecht HA, Stemp ED, Barton JK. Biochemistry. 2000;39:5483–5491. doi: 10.1021/bi992897m. [DOI] [PubMed] [Google Scholar]
- 13.Kurbanyan K, Nguyen KL, To P, Rivas EV, Lueras AM, Kosinski C, Steryo M, Gonzalez A, Mah DA, Stemp ED. Biochem. 2003;42:10269–10281. doi: 10.1021/bi020713p. [DOI] [PubMed] [Google Scholar]
- 14.Gajewski E, Fuciarelli AF, Dizdaroglu M. Int. J. Radiat. Biol. 1988;54:445–459. doi: 10.1080/09553008814551821. [DOI] [PubMed] [Google Scholar]
- 15.Dizdaroglu M, Gajewski E. Cancer Res. 1989;49:3463–3467. [PubMed] [Google Scholar]
- 16.Perrier S, Hau J, Gasparutto D, Cadet J, Favier A, Ravanat JL. J. Am. Chem. Soc. 2006;128:5703–5710. doi: 10.1021/ja057656i. [DOI] [PubMed] [Google Scholar]
- 17.Hickerson RP, Chepanoske CL, Williams SD, David SS, Burrows CJ. J. Am. Chem. Soc. 1999;121:9901–9902. [Google Scholar]
- 18.Xu X, Muller JG, Ye Y, Burrows CJ. J. Am. Chem. Soc. 2008;130:703–709. doi: 10.1021/ja077102a. [DOI] [PubMed] [Google Scholar]
- 19.Steenken S, Jovanovic SV. J. Am. Chem. Soc. 1997;119:617–618. [Google Scholar]
- 20.Cadet J, Douki T, Ravanat JL. Nat. Chem. Biol. 2006;2:348–349. doi: 10.1038/nchembio0706-348. [DOI] [PubMed] [Google Scholar]
- 21.Misiaszek R, Crean C, Joffe A, Geacintov NE, Shafirovich V. J. Biol. Chem. 2004;279:32106–32115. doi: 10.1074/jbc.M313904200. [DOI] [PubMed] [Google Scholar]
- 22.Misiaszek R, Crean C, Geacintov NE, Shafirovich V. J. Am. Chem. Soc. 2005;127:2191–2200. doi: 10.1021/ja044390r. [DOI] [PubMed] [Google Scholar]
- 23.Shafirovich V, Dourandin A, Huang W, Luneva NP, Geacintov NE. J. Phys. Chem. B. 1999;103:10924–10933. [Google Scholar]
- 24.Shafirovich V, Dourandin A, Geacintov NE. J. Phys. Chem. B. 2001;105:8431–8435. [Google Scholar]
- 25.Buxton GV, Greenstock CL, Helman WP, Ross AB. J. Phys. Chem. Ref. Data. 1988;17:513–886. [Google Scholar]
- 26.Garrison WM. Chem. Rev. 1987;87:381–398. [Google Scholar]
- 27.Drobnik J, Augenstein L. Photochem. Photobiol. 1966;5:83–97. doi: 10.1111/j.1751-1097.1966.tb05763.x. [DOI] [PubMed] [Google Scholar]
- 28.Shafirovich V, Dourandin A, Huang W, Luneva NP, Geacintov NE. Phys. Chem. Chem. Phys. 2000;2:4399–4408. [Google Scholar]
- 29.Shafirovich V, Cadet J, Gasparutto D, Dourandin A, Geacintov NE. Chem. Res. Toxicol. 2001;14:233–241. doi: 10.1021/tx000204t. [DOI] [PubMed] [Google Scholar]
- 30.Angelov D, Spassky A, Berger M, Cadet J. J. Am. Chem. Soc. 1997;119:11373–11380. [Google Scholar]
- 31.Candeias LP, Steenken S. J. Am. Chem. Soc. 1989;111:1094–1099. [Google Scholar]
- 32.Kobayashi K, Tagawa S. J. Am. Chem. Soc. 2003;125:10213–10218. doi: 10.1021/ja036211w. [DOI] [PubMed] [Google Scholar]
- 33.Steenken S. Chem. Rev. 1989;89:503–520. [Google Scholar]
- 34.Candeias LP, Steenken S. Chem. Eur. J. 2000;6:475–484. doi: 10.1002/(sici)1521-3765(20000204)6:3<475::aid-chem475>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 35.Chatgilialoglu C, D'Angelantonio M, Kciuk G, Bobrowski K. Chem. Res. Toxicol. 2011;24:2200–2206. doi: 10.1021/tx2003245. [DOI] [PubMed] [Google Scholar]
- 36.Banyasz A, Douki T, Improta R, Gustavsson T, Onidas D, Vaya I, Perron M, Markovitsi D. J. Am. Chem. Soc. 2012;134:14834–14845. doi: 10.1021/ja304069f. [DOI] [PubMed] [Google Scholar]
- 37.Praseuth D, Perrouault L, Le Doan T, Chassignol M, Thuong N, Helene C. Proc. Natl. Acad. Sci. U. S. A. 1988;85:1349–1353. doi: 10.1073/pnas.85.5.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ni J, Mathews MAA, McCloskey JA. Rapid Commun. Mass Spectrom. 1997;11:535–540. doi: 10.1002/(SICI)1097-0231(199704)11:6<535::AID-RCM898>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 39.Gregson JM, McCloskey JA. Int. J. Mass Spectrom. 1997;165/166:475–485. [Google Scholar]
- 40.Crean C, Geacintov NE, Shafirovich V. J. Phys. Chem. B. 2009;113:12773–12781. doi: 10.1021/jp903554n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crean C, Shao J, Geacintov NE, Shafirovich V. Chem. Eur. J. 2009;15:10634–10640. doi: 10.1002/chem.200900500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rustgi S, Joshi A, Moss H, Riesz P. Int. J. Radiat. Biol. 1977;31:415–440. doi: 10.1080/09553007714550521. [DOI] [PubMed] [Google Scholar]
- 43.Wardman P, Candeias LP. Radiat. Res. 1996;145 [PubMed] [Google Scholar]
- 44.Cadet J, Loft S, Olinski R, Evans MD, Bialkowski K, Wagner JR, Dedon PC, Moeller P, Greenberg MM, Cooke MS. Free Radic. Res. 2012;48:367–381. doi: 10.3109/10715762.2012.659248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cadet J, Ravanat JL, TavernaPorro M, Menoni H, Angelov D. Cancer Lett. 2012;327:5–15. doi: 10.1016/j.canlet.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Jaruga P, Dizdaroglu M. DNA Repair (Amst) 2008;7:1413–1425. doi: 10.1016/j.dnarep.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 47.Chatgilialoglu C, Ferreri C, Terzidis MA. Chem. Soc. Rev. 2011;40:1368–1382. doi: 10.1039/c0cs00061b. [DOI] [PubMed] [Google Scholar]
- 48.Dizdaroglu M, Dirksen ML, Jiang HX, Robbins JH. Biochem. J. 1987;241:929–932. doi: 10.1042/bj2410929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez H, Jaruga P, Leber D, Nyaga SG, Evans MK, Dizdaroglu M. Biochem. 2007;46:2488–2496. doi: 10.1021/bi062022p. [DOI] [PubMed] [Google Scholar]
- 50.Jaruga P, Dizdaroglu M. Biochem. Biophys. Res. Commun. 2010;397:48–52. doi: 10.1016/j.bbrc.2010.05.050. [DOI] [PubMed] [Google Scholar]
- 51.Yuan B, Wang J, Cao H, Sun R, Wang Y. Nucleic Acid. Res. 2011;39:5945–5954. doi: 10.1093/nar/gkr159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.You C, Dai X, Yuan B, Wang J, Wang J, Brooks PJ, Niedernhofer LJ, Wang Y. Nat. Chem. Biol. 2012 doi: 10.1038/nchembio.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D'Errico M, Parlanti E, Teson M, de Jesus BM, Degan P, Calcagnile A, Jaruga P, Bjoras M, Crescenzi M, Pedrini AM, Egly JM, Zambruno G, Stefanini M, Dizdaroglu M, Dogliotti E. EMBO J. 2006;25:4305–4315. doi: 10.1038/sj.emboj.7601277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brooks PJ. DNA Repair (Amst) 2008;7:1168–1179. doi: 10.1016/j.dnarep.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirkali G, Tunca M, Genc S, Jaruga P, Dizdaroglu M. Free Radic. Biol. Med. 2008;44:386–393. doi: 10.1016/j.freeradbiomed.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 56.Box HC, Budzinski EE, Dawidzik JD, Wallace JC, Evans MS, Gobey JS. Radiat. Res. 1996;145:641–643. [PubMed] [Google Scholar]
- 57.Box HC, Dawidzik JB, Budzinski EE. Free Radic. Biol. Med. 2001;31:856–868. doi: 10.1016/s0891-5849(01)00653-0. [DOI] [PubMed] [Google Scholar]
- 58.Romieu A, Bellon S, Gasparutto D, Cadet J. Org. Lett. 2000;2:1085–1088. doi: 10.1021/ol005643y. [DOI] [PubMed] [Google Scholar]
- 59.Bellon S, Ravanat JL, Gasparutto D, Cadet J. Chem. Res. Toxicol. 2002;15:598–606. doi: 10.1021/tx015594d. [DOI] [PubMed] [Google Scholar]
- 60.Hong H, Cao H, Wang Y, Wang Y. Chem. Res. Toxicol. 2006;19:614–621. doi: 10.1021/tx060025x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang Y, Hong H, Cao H, Wang Y. Biochem. 2007;46:12757–12763. doi: 10.1021/bi7012195. [DOI] [PubMed] [Google Scholar]
- 62.Colis LC, Raychaudhury P, Basu AK. Biochem. 2008;47:8070–8079. doi: 10.1021/bi800529f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raychaudhury P, Basu AK. J. Nucleic Acid. 2010;2010:101495. doi: 10.4061/2010/101495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raychaudhury P, Basu AK. Biochem. 2011;50:2330–2338. doi: 10.1021/bi102064z. [DOI] [PMC free article] [PubMed] [Google Scholar]