Abstract
It has been demonstrated that loss of heterozygosity (LOH) was frequently observed on chromosomes 8p22-p23 in hepatocellular carcinoma (HCC) and was associated with metastasis and prognosis of HCC. However, putative genes functioning on this chromosomal region remain unknown. In this study, we evaluated LOH status of four genes on 8p22-p23 (MCPH1, TUSC3, KIAA1456, and ZDHHC2). LOH on ZDHHC2 was associated with early metastatic recurrence of HCC following liver transplantation and was correlated with tumor size and portal vein tumor thrombi. Furthermore, our results indicate that ZDHHC2 expression was frequently decreased in HCC. Overexpression of ZDHHC2 could inhibit proliferation, migration, and invasion of HCC cell line Bel-7402 in vitro. These results suggest an important role for ZDHHC2 as a tumor suppressor in metastasis and recurrence of HCC.
1. Introduction
Hepatocellular carcinoma (HCC) is one of the most common human malignancies worldwide, with a high rate of incidence and mortality [1, 2]. Over the past decades, many efforts have been made to improve the overall survival rate of HCC, of which both liver transplantation (LT) and surgical resection are considered potentially curative treatments for early-stage HCC. However, the long-term survival of HCC patients after surgery still remains poor due to the frequent metastasis and recurrence. Therefore, it is essential to explore the molecular pathogenesis of HCC metastasis and recurrence and detect novel molecular targets in HCC.
Loss of heterozygosity (LOH) refers to the loss of one of the two alleles at one or more loci in a heterozygote [3]. LOH in a chromosomal fragment implies the presence of putative tumor suppressor genes (TSGs) and can be used as an indirect way to identify TSGs [4]. Inactivation of TSGs was thought to be correlated with the metastasis and recurrence of primary cancer [5, 6]. Previous reports have revealed that frequent allelic losses on chromosomes 1p, 4q, 6q, 8p, 9p, 10q, 11p, 13q, 14q, 16q, and 17p are commonly observed in HCC patients [3, 4, 7–10]. Notably, loss of 8p is one of the most frequent chromosomal alterations in a variety of cancers including HCC [11–16]. Several studies have demonstrated that deletions of allele on 8p22-p23 are associated with metastasis and prognosis of HCC [7, 11, 17]. This suggests that 8p22-p23 might harbor one or more TSGs that are important in the progression, especially in the metastasis and recurrence of HCC. Until now, only few critical TSGs have been found on 8p22-p23, and it is necessary to investigate the LOH status of candidate genes in order to identify TSGs within this region. Additionally, studies about the roles of LOH in progression and prognosis of HCC in patients with LT are limited in number.
In this study, we selected the four most noteworthy genes on 8p22-p23 (MCPH1, TUSC3, KIAA1456, and ZDHHC2) to reveal the potential relationships between LOH of these genes and the clinical characteristics in HCC after LT. We found that the LOH on ZDHHC2 was associated with early metastatic recurrence of HCC following LT and was correlated with tumor size and portal vein tumor thrombi (PVTT). Further study provides the evidence that ZDHHC2 has important role as a tumor suppressor in metastasis and recurrence of HCC.
2. Materials and Methods
2.1. Clinical Specimens
This study involved three independent cohorts of HCC patients. All 3 cohorts were from the First Affiliated Hospital, Zhejiang University School of Medicine, Zhejiang, China. Cohort 1 was a total of 40 randomly selected HCC patients with LT performed during 2006 and 2009; cohort 2 and cohort 3 were, respectively, a total of 55 and 23 HCC patients following partial hepatectomy performed during 2010 and 2011. This study was approved by the Ethical Review Committee of the First Affiliated Hospital, Zhejiang University School of Medicine, and informed consent was obtained according to the Declaration of Helsinki.
2.2. Sample Collection
Primary HCC and adjacent normal liver tissues were obtained during the operation. All tissue samples were immediately cut into small pieces, snap-frozen in liquid nitrogen, and stored at −80°C. Diagnosis for all samples was confirmed by histopathological examination. Genomic DNA was extracted using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA) in accordance with the manufacturer's instructions.
2.3. SNP-Based LOH Analysis
Five SNPs (rs2023627, rs2517064, rs966473, rs3750249, and rs6997380) that resided in ZDHHC2, six SNPs (rs352808, rs1362865, rs1429087, rs352750, rs10081561, and rs1579451) in TUSC3, six SNPs (rs1055749, rs2305023, rs2442502, rs2442468, rs2959804, and rs921291) in MCPH1, and four SNPs (rs9644018, rs2203836, rs2977097, and rs17179360) in KIAA1456 were selected to determine LOH. Sequences of these SNPs were obtained from the Human SNP Database. The methods for multiplex polymerase chain reaction (PCR) and LOH analysis were described previously [3], and SNPs were amplified under the same PCR conditions as microsatellite markers. Information for each SNP on these candidates is shown in Figure 1. LOH was defined as the value of LOH index within a range between 0.7 and 1.5 [18]. LOH on a single gene was determined to be positive when one or more SNPs on each gene detected LOH.
Figure 1.
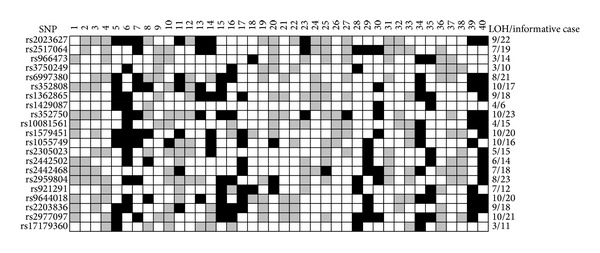
Status and frequency of LOH on each SNP in 40 HCC cases. Black box: LOH; gray box: retention; white box: noninformative case.
2.4. qRT-PCR
Total RNA from HCC samples was isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions; then cDNA was synthesized. qPCR reactions were performed by the ABI7500 system (Applied Biosystems, CA) and SYBR green dye (TaKaRa Biotechnology, Dalian, China). GAPDH was used as an internal control. The primers' sequences used in this study were listed in Supplementary Table 1 available online at http://dx.doi.org/10.1155/2014/832712. Every sample was tested in duplicate.
2.5. Immunohistochemistry
Immunohistochemistry (IHC) of paraffin-embedded tissue sections was performed using the antibody ZDHHC2 (AP5592a, Abgent, CA, USA) as we have previously described [19]. Evaluation and semiquantitative estimation of IHC results were performed independently by two pathologists without knowledge of the clinicopathological outcomes. Intensity was graded as 0 (no staining), 1 (weak staining), 2 (moderate staining), or 3 (strong staining). The abundance of positive cells was graded from 0 to 4 (0, <5% positive cells; 1, 5–25%; 2, 26–50%; 3, 51–75%; 4, >75%). The composite score obtained by multiplying the two values was analyzed.
2.6. Cell Culture
Normal liver cell line L-02; liver cancer cell lines Hep3B, HuH-7, and Bel-7402; and the metastasis-capable human HCC cell lines MHCC97L and HCCLM3 were purchased from American Type Culture Collection (Manassas, VA), Shanghai Institute of Cell Biology (Shanghai, China), and Liver Cancer Institute of Fudan University (Shanghai, China). All of the cell lines were maintained in the recommended culture condition and incubated at 37°C humidified environment containing 5% CO2.
2.7. Overexpression of ZDHHC2
The plasmid of the coding sequence (CDS) length ZDHHC2 with EGFP-tag and the negative control plasmid only containing EGFP-tag were completed by the Genechem Company (Shanghai, China). A PCR-amplified CDS-length ZDHHC2 fragment was inserted into the XhoI/KpnI sites of GV230 vector (Genechem, Shanghai, China), using the primers forward, 5′-TCCGCTCGAGATGGCGCCCTCGGGCC-3′ and reverse, 5′-ATGGGGTACCGTAGTCTCATTTTCCATGGTTAATG-3′. Transfection of the vectors was performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions.
2.8. Western Blotting Analysis
Protein extraction and western blotting analysis were performed as previously described [20]. The following antibodies were used: anti-ZDHHC2, SantaCruz (sc-292338); anti-b-actin, Sigma-Aldrich (A1978); anti-GFP-Tag, Abmart (M20004).
2.9. Cell Proliferation Assay
Cell growth was determined by Cell Counting Kit-8 (CCK-8) cell proliferation assay (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer's instructions as previously described [21].
2.10. Transwell Migration and Invasion Assay
For invasion assay, 48 hours after transfection, 5 × 10E4 Bel-7402 cells in serum-free RPMI1640 were seeded into the upper chambers of each well (24-well insert, 8-mm pore size, Millipore, Billerica, MA, USA) coated with Matrigel (BD Bioscience, San Jose, CA, USA). For migration assay, the upper chambers were not coated with Matrigel, and cells were seeded after 24-hour transfection. RPMI1640 containing 10% FBS was placed in the lower chambers as a chemoattractant. After 24 hours of incubation, cells on the upper membrane surface were wiped off, and the cells that invaded across the Matrigel membrane were fixed with 100% methanol and stained with 0.2% crystal violet. The number of invasive cells was then counted (five randomly chosen high-power fields for each membrane) under a microscope.
2.11. Statistical Analysis
Comparisons between LOH and clinicopathological data were analyzed using chi-square test or Fisher exact test. Cumulative recurrence-free survival was assessed by Kaplan-Meier method and univariate analysis by log-rank test. Variables were brought into multivariate analysis when there was statistical significance in univariate analysis. Multivariate analysis was assessed using Cox proportional hazard model to identify variables that were independent predictors of clinical outcome. Independent Student's t-test was used to analyze the differences between two groups. Data are presented as the mean±SD, unless otherwise indicated. P < 0.05 was considered statistically significant. All statistical analyses were performed with the use of SPSS version 16.0 software (SPSS, Chicago, IL).
3. Results
3.1. LOH of ZDHHC2 Associates with Recurrence of Hepatocellular Carcinoma following Liver Transplantation
DNA from forty individuals with HCC who had received a liver transplants was analyzed for twenty-one SNPs of these four specific genes. Their mean heterozygosity was 87.5%. The heterozygosity on ZDHHC2, MCPH1, TUSC3, and KIAA1456 was correspondingly 95% (38 of 40), 92.5% (37 of 40), 85% (34 of 40), and 77.5% (31 of 40). Distribution of LOH on each SNP is shown in Figure 1. The LOH frequencies at all selected genes were so high, with a mean value of 50.3%. The frequencies of LOH on ZDHHC2, MCPH1, TUSC3, and KIAA1456 were 45% (17 of 38), 54% (20 of 37), 50% (17 of 34), and 52% (16 of 31), respectively.
To study the association of LOH on each gene with clinicopathological characteristics and to explore its potential biological role in HCC initiation, development, and progression, we compared frequencies of LOH based on clinicopathological findings including age, preoperative serum AFP level, tumor number, tumor size, PVTT and histopathologic grading. All of the results between clinicopathological features and LOH in HCC were shown in Table 1.
Table 1.
Clinicopathological correlation with LOH in HCC cases (cohort 1).
| Parameters | Genes | LOH/informative case | P value | |
|---|---|---|---|---|
| Age | ≤50 year | >50 year | ||
| ZDHHC2 | 12/17 | 5/21 | 0.004 | |
| TUSC3 | 10/16 | 7/18 | 0.169 | |
| MCPH1 | 11/17 | 9/20 | 0.231 | |
| KIAA1456 | 10/16 | 6/15 | 0.210 | |
|
| ||||
| Preoperative AFP level | ≤400 ng/mL | >400 ng/mL | ||
| ZDHHC2 | 4/15 | 13/23 | 0.070 | |
| TUSC3 | 5/12 | 12/22 | 0.473 | |
| MCPH1 | 6/14 | 14/23 | 0.286 | |
| KIAA1456 | 5/11 | 11/20 | 0.611 | |
|
| ||||
| Tumor number | Single | Multiple | ||
| ZDHHC2 | 4/15 | 13/23 | 0.070 | |
| TUSC3 | 5/12 | 12/22 | 0.473 | |
| MCPH1 | 4/14 | 16/23 | 0.015 | |
| KIAA1456 | 4/11 | 12/20 | 0.208 | |
|
| ||||
| Tumor size | ≤5 cm | >5 cm | ||
| ZDHHC2 | 6/23 | 11/15 | 0.004 | |
| TUSC3 | 9/22 | 8/12 | 0.151 | |
| MCPH1 | 9/22 | 11/15 | 0.052 | |
| KIAA1456 | 8/18 | 8/13 | 0.347 | |
|
| ||||
| PVTT | Absent | Present | ||
| ZDHHC2 | 11/31 | 6/7 | 0.031 | |
| TUSC3 | 14/29 | 3/5 | 1.000 | |
| MCPH1 | 14/30 | 6/7 | 0.097 | |
| KIAA1456 | 11/24 | 5/7 | 0.394 | |
|
| ||||
| Histopathologic grading | Well + moderately | Poorly | ||
| ZDHHC2 | 5/16 | 12/22 | 0.154 | |
| TUSC3 | 3/11 | 14/23 | 0.067 | |
| MCPH1 | 9/16 | 11/21 | 0.815 | |
| KIAA1456 | 5/14 | 11/17 | 0.108 | |
AFP: alpha-fetoprotein; PVTT: portal vein tumor thrombi.
To determine the association between gene LOH status and prognostic data, we performed univariate and multivariate survival analysis. As a result, we found that the 1-year cumulative recurrence-free survival in HCC patients with LOH on ZDHHC2 was significantly lower than that with heterozygosity retention (P = 0.022, Figure 2). However, there were no significant differences between recurrence-free survival and LOHon MCPH1 (P = 0.320), TUSC3 (P = 0.546), and KIAA1456 (P = 0.564). Furthermore, based on the univariate analysis, we investigate the relationship between 1-year cumulative recurrence-free survival and clinicopathological factors. Taken together, significant associations were found between HCC recurrence and the following variables: LOH on ZDHHC2, preoperative AFP level >400 ng/mL, tumor size >5 cm, and presence of PVTT (Table 2). Factors which were found to have prognostic value including the clinicopathologic characteristics and gene LOH status in the univariate analysis were added into the corresponding multivariate Cox model of survival, and only PVTT was identified as independent prognostic factor for recurrence-free survival in HCC patients after LT (Table 3).
Figure 2.
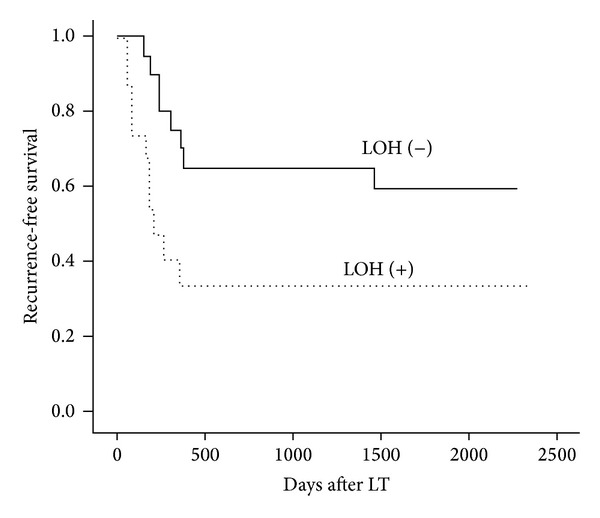
Kaplan-Meier analysis and log-rank test of the recurrence-free survival according to the LOH status of ZDHHC2 in 38 informative cases.
Table 2.
Univariate analysis of HCC recurrence.
| Variables | n | 1-year cumulative recurrence rate (%) |
P value |
|---|---|---|---|
| LOH on ZDHHC2 | |||
| Positive | 17 | 66.7 ± 12.2 | 0.022 |
| Negative | 11 | 30.0 ± 10.2 | |
| Preoperative AFP level | |||
| ≤400 ng/mL | 15 | 15.4 ± 10.0 | 0.005 |
| >400 ng/mL | 25 | 62.5 ± 9.9 | |
| Tumor size | |||
| ≤5 cm | 25 | 27.3 ± 9.5 | 0.001 |
| >5 cm | 15 | 73.3 ± 11.4 | |
| PVTT | |||
| Absent | 33 | 35.5 ± 8.6 | <0.001 |
| Present | 7 | 100 |
AFP: alpha-fetoprotein; PVTT: portal vein tumor thrombi.
Table 3.
Multivariate analysis of HCC recurrence.
| Variables | Hazard ratio |
95% confidence interval |
P value |
|---|---|---|---|
| LOH on ZDHHC2 | 1.263 | 0.405–3.935 | 0.687 |
| AFP >400 ng/mL | 4.156 | 0.864–19.994 | 0.076 |
| Tumor size >5 cm | 1.615 | 0.430–6.063 | 0.478 |
| PVTT (+) | 3.468 | 1.004–11.982 | 0.049 |
AFP: alpha-fetoprotein; PVTT: portal vein tumor thrombi.
3.2. Decreased Expression of ZDHHC2 Is Observed in HCC Tissue Samples and HCC Cell Lines
To investigate the expression of ZDHHC2 in tumor tissues and peritumor tissues from HCC samples, we conducted qRT-PCR in cohort 2 of 55 HCC samples and IHC in cohort 3 of 23 HCC samples. We found that mRNA level of ZDHHC2 expression is significantly lower in tumor tissues than peritumor tissues from the same patients (P = 0.003) (Figure 3(a)). We analyzed the correlation between ZDHHC2 mRNA expression and clinical parameters including age, preoperative serum AFP level, tumor number, tumor size, PVTT, and histopathologic grading. In accord with the correlation of LOH of ZDHHC2 and clinical parameters, lower expression of ZDHHC2 was associated with the tumor size and the presence of PVTT (Supplementary Table 2).
Figure 3.

Frequently decreased expression of ZDHHC2 in human HCC tissues. (a) In cohort 2 (55 paired HCC tissue samples), the expression levels of ZDHHC2 mRNA are significantly lower in tumor tissues than peritumor tissues from the same patients (P = 0.003). (b) Relative IHC staining of ZDHHC2 expression in paired HCC tissue samples (cohort 3, n = 23). The ZDHHC2 expression level was significantly downregulated in tumors compared with the corresponding adjacent nontumor liver tissues (P = 0.015). (c) and (d) are preventative pictures, respectively (200x).
Furthermore, by multiplying the values of staining intensity and relative abundance of positive cells, we found that the mean staining score of ZDHHC2 in tumor tissues is significantly lower than those in peritumor tissues (P = 0.015) (Figure 3(b)). The representative pictures of tumors and peritumors are shown in Figures 3(c) and 3(d).
Moreover, ZDHHC2 expression level in HCC cell lines was detected by western blotting. Compared to normal liver cell line L-02, ZDHHC2 expression levels in HCC cell lines (Hep3B, HuH-7, Bel-7402, MHCC97L, and HCCLM3) were significantly lower (Figure 4).
Figure 4.

ZDHHC2 expression levels in HCC cell lines were detected by western blot. Compared to normal liver cell line L-02, ZDHHC2 expression level in HCC cell lines was significantly lower.
3.3. Overexpression of ZDHHC2 Inhibits Cell Proliferation and Invasion in BEL-7402 In Vitro
To determine whether reduction of ZDHHC2 affects biological function in HCC and overexpression of ZDHHC2 could be used as potential consideration to treat HCC, further experimental studies were then performed. Overexpression vector and negative control vector are transiently transfected into Bel-7402 HCC cell line, then downstream analysis was performed. Transient transfection was successfully performed as shown in western blotting analysis (Figure 5). ZDHHC2 overexpression lasts stably at 24-, 48-, and 72-hour time points.
Figure 5.

Plasmids containing ZDHHC2-EGFP or EGFP were transfected into Bel-7402. The transfection was monitored by western blotting analysis at the 24-, 48-, and 72-hour time points. Bel-7402 without any treatment (BLANK), transiently transfected with negative control vector (NC), and transiently transfected with overexpression vector (ZDHHC2).
CCK-8 cell proliferation assay and transwell migration and invasion assay were conducted in Bel-7402 without any treatment (BLANK), transiently transfected with negative control vector (NC), and transiently transfected with overexpression vector (ZDHHC2). The results showed that overexpression of ZDHHC2 significantly impaired the proliferation (Figure 6(a)) and migratory and invasive capacity of Bel-7402 cell (Figures 6(b) and 6(c)) compared to both BLANK group and NC group.
Figure 6.

CCK-8 cell proliferation assay and transwell migration and invasion assay were conducted in Bel-7402 without any treatment (BLANK), transiently transfected with negative control vector (NC), and transiently transfected with overexpression vector (ZDHHC2). (a) Overexpression of ZDHHC2 significantly impaired Bel-7402 proliferation (n = 3, *P < 0.05 compared to BLANK group, **P < 0.01 compared to BLANK group, and ## P < 0.01 compared to NC group). (b) Overexpression of ZDHHC2 inhibits the migration and invasion of BEL-7402 cell (n = 5, **P < 0.01, and ***P < 0.001) compared to both BLANK group and NC group. Representative pictures are shown in (c).
4. Discussion
HCC is a frequent and highly aggressive malignancy worldwide with very poor prognosis. Recently, LT has been recognized as a potential curative treatment for early-stage HCC patients. However, the clinical outcome remains challenging because of a high incidence of tumor recurrence after LT [22]. Recurrence is principally attributable to the presence of microscopic extrahepatic metastatic foci before LT [23], which is also known as metastastic recurrence. A better understanding of the underlying molecular mechanisms governing cancer metastastic recurrence will be a great help for predicting prognosis.
Numerous studies have revealed a high-LOH frequency on 8p22-p23 in HCC [7, 9, 17, 24]. Our previous report has also supported [3]. This implies that one or several TSGs may lie within this region. Therefore, the genes on 8p22-p23 are widely investigated and several new genes have been identified as TSGs in recent years, such as CUB and Sushi multiple domains 1 (CSMD1) [9], deleted in liver cancer 1 (DLC1) [25] and mitochondrial tumour suppressor 1 (MTUS1) [26].
In the current study, we confirmed that LOH was a prevalent event on 8p22-p23 in HCC, with frequencies of LOHs on four specific genes (MCPH1, KIAA1456, TUSC3, and ZDHHC2) ranging from 45% to 54%. Interestingly, LOH on ZDHHC2 was clearly associated with early metastastic recurrence of HCC following LT, although it had the lowest LOH frequency of these four genes. In all 38 informative cases, patients with LOH had an increasing risk of early metastastic recurrence post-LT, which directly linked to patients' long-term survival. Multivariate analyses revealed that LOH on ZDHHC2 could predict the risk of HCC early recurrence together with AFP level, tumor size, and PVTT but was not an independent prognostic factor. In addition, our data showed that LOH status of ZDHHC2 was correlated with some clinicopathological parameters, including tumor size and PVTT. These data suggested that LOH on ZDHHC2 may serve as a molecular event in advanced HCC, as tumor size >5 cm and PVTT were representatives of advancement of HCC.
ZDHHC2 was originally described as REAM (reduced expression in metastasis), because its silence was associated with increased metastatic potential of cancer cells [27]. It was reported that reduced ZDHHC2 expression is observed in gastric adenocarcinoma patients and associated with lymph node metastasis and independently predicts an unfavorable prognosis [28]. However, the expression level of ZDHHC2 in HCC remains to be explored. Our study provides the evidence that ZDHHC2 expression was frequently decreased in HCC. Overexpression of ZDHHC2 could inhibit proliferation, migration, and invasion of HCC cell line Bel-7402 in vitro. These results suggest an important role for ZDHHC2 as a tumor suppressor in metastasis and recurrence of HCC.
DHHC2, encoded by ZDHHC2, is one member of a family of more than 20 palmitoyl acyltransferases (PATs) characterized by an Asp-His-His-Cys (DHHC) motif [29]. Protein palmitoylation, mediated by PATs, can affect proteins in many ways, including regulating membrane attachment, subcellular trafficking, and membrane microlocalisation [29]. Many DHHC2-substrate interactions have been identified so far, including PSD95, SNAP25, SNAP23, CKAP4, CD9, and CD151 [29]. The decrease or absence of DHHC2 expression could influence the palmitoylation of these substrates, and whatever role palmitoylation had in signaling downstream from that event would be disrupted. Of these substrates, CKAP4, CD9, and CD151 have been associated with initiation and progression of cancer. Decreased expression of DHHC2 leads to reduced palmitoylation of CKAP4, making it no longer traffick efficiently to the cell surface where it act as a receptor for antiproliferative factor (APF) [30, 31]. Therefore, the ability of APF to halt cell proliferation and suppress the expression of genes involved in tumorigenesis is inhibited. Moreover, DHHC2 could stimulate palmitoylation of tetraspanins CD9 and CD151 [32]. CD9 is a potential tumor suppressor [33], while CD151 is supposed to promote tumor metastasis [34]. Our data together with these previous results support the notion that DHHC2 and its substrates may play an important role in the process of development, metastasis, and recurrence in various cancers including HCC. Despite several substrates have been identified, it is possible that more functional substrates remain unknown. Accordingly, our further study should focus on searching novel substrates and investigating the biological function of DHHC2 and its substrates in HCC, which may well provide novel and important targets for pharmacological intervention in the progression of HCC.
5. Conclusions
LOH was frequent on ZDHHC2, MCPH1, TUSC3, and KIAA1456 in human HCC. Of these, LOH on ZDHHC2 might contribute to early metastatic recurrence of HCC after LT. Reduced expression of ZDHHC2 was detected in HCC and associates with biological function of HCC such as proliferation, migration, and invasion. These results suggest an important role for ZDHHC2 as a tumor suppressor in metastasis and recurrence of HCC.
Supplementary Material
Supplementary Table 1: RT-PCR primers.
Supplementary Table 2: Clinicopathological Correlation With ZDHHC2 mRNA expression in HCC Cases (Cohort 2).
Acknowledgments
This study was supported by National S&T Major Project (no. 2012ZX10002-017), NSFC for Innovative Research Group (no. 81121002), National High Technology Research and Development Program 863 of China (no. 2012AA021002), and Special Fund for Health Research in the Public Welfare (201302009). The authors thank Penghong Song, Kezhen Shen, Xiaowen Feng, Wenfeng Song, Guoqiang Cao, Leiming Chen, and Rong Su for their valuable technical assistance.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Authors' Contribution
Chuanhui Peng and Zhijun Zhang contributed equally to this work.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer Journal for Clinicians. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Zhou L, Zhou W, Wu L, Yu X, Xing C, Zheng S. The association of frequent allelic loss on 17p13.1 with early metastastic recurrence of hepatocellular carcinoma after liver transplantation. Journal of Surgical Oncology. 2010;102(7):802–808. doi: 10.1002/jso.21743. [DOI] [PubMed] [Google Scholar]
- 4.Li S, Wang H, Li J, et al. Genome-wide analyses on loss of heterozygosity in hepatocellular carcinoma in Southern China. Journal of Hepatology. 2001;34(6):840–849. doi: 10.1016/s0168-8278(01)00047-2. [DOI] [PubMed] [Google Scholar]
- 5.Schwendel A, Langreck H, Reichel M, et al. Primary small-cell lung carcinomas and their metastases are characterized by a recurrent pattern of genetic alterations. International Journal of Cancer. 1997;74(1):86–93. doi: 10.1002/(sici)1097-0215(19970220)74:1<86::aid-ijc15>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 6.Bockmühl U, Petersen S, Schmidt S, et al. Patterns of chromosomal alterations in metastasizing and nonmetastasizing primary head and neck carcinomas. Cancer Research. 1997;57(23):5213–5216. [PubMed] [Google Scholar]
- 7.Lu T, Hano H. Identification of minimal regions of deletion at 8p23.1-22 associated with metastasis of hepatocellular carcinoma. Liver International. 2007;27(6):782–790. doi: 10.1111/j.1478-3231.2007.01504.x. [DOI] [PubMed] [Google Scholar]
- 8.Okuno T, Ueda M, Tsuruyama T, et al. Loss of heterozygosity on 10q23 is involved in metastatic recurrence of hepatocellular carcinoma. Cancer Science. 2009;100(3):520–528. doi: 10.1111/j.1349-7006.2008.01056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Midorikawa Y, Yamamoto S, Tsuji S, et al. Allelic imbalances and homozygous deletion on 8p23.2 for stepwise progression of hepatocarcinogenesis. Hepatology. 2009;49(2):513–522. doi: 10.1002/hep.22698. [DOI] [PubMed] [Google Scholar]
- 10.Chou YW, Chung K, Jeng L, Chen T, Liaw Y. Frequent allelic loss on chromosomes 4q and 16q associated with human hepatocellular carcinoma in Taiwan. Cancer Letters. 1998;123(1):1–6. doi: 10.1016/s0304-3835(97)00276-0. [DOI] [PubMed] [Google Scholar]
- 11.Emi M, Fujiwara Y, Ohata H, et al. Allelic loss at chromosome band 8p21.3-p22 is associated with progression of hepatocellular carcinoma. Genes Chromosomes and Cancer. 1993;7(3):152–157. doi: 10.1002/gcc.2870070307. [DOI] [PubMed] [Google Scholar]
- 12.Fujiwara Y, Ohata H, Emi M, et al. A 3-Mb physical map of the chromosome region 8p21.3-p22, including a 600-kb region commonly deleted in human hepatocellular carcinoma, colorectal cancer, and non-small cell lung cancer. Genes Chromosomes and Cancer. 1994;10(1):7–14. doi: 10.1002/gcc.2870100103. [DOI] [PubMed] [Google Scholar]
- 13.Fujiwara Y, Emi M, Ohata H, et al. Evidence for the presence of two tumor suppressor genes on chromosome 8p for colorectal carcinoma. Cancer Research. 1993;53(5):1172–1174. [PubMed] [Google Scholar]
- 14.Anbazhagan R, Fujii H, Gabrielson E. Allelic loss of chromosomal arm 8p in breast cancer progression. The American Journal of Pathology. 1998;152(3):815–819. [PMC free article] [PubMed] [Google Scholar]
- 15.Bova GS, Carter BS, Bussemakers MJG, et al. Homozygous deletion and frequent allelic loss of chromosome 8p22 loci in human prostate cancer. Cancer Research. 1993;53(17):3869–3873. [PubMed] [Google Scholar]
- 16.Knowles MA, Shaw ME, Proctor AJ. Deletion mapping of chromosome 8 in cancers of the urinary bladder using restriction fragment length polymorphisms and microsatellite polymorphisms. Oncogene. 1993;8(5):1357–1364. [PubMed] [Google Scholar]
- 17.Pang J, Qin L, Ren N, et al. Loss of heterozygosity at D8S298 is a predictor for long-term survival of patients with tumor-node-metastasis stage I of hepatocellular carcinoma. Clinical Cancer Research. 2007;13(24):7363–7369. doi: 10.1158/1078-0432.CCR-07-0593. [DOI] [PubMed] [Google Scholar]
- 18.Funel N, Morelli M, Giovannetti E, et al. Loss of heterozygosity status of d9s105 marker is associated with downregulation of Krüppel-like factor 4 expression in pancreatic ductal adenocarcinoma and pancreatic intraepithelial lesions. Pancreatology. 2011;11(1):30–42. doi: 10.1159/000322990. [DOI] [PubMed] [Google Scholar]
- 19.Zhang F, Wu L, Zhou L, et al. Predictive value of expression and promoter hypermethylation of XAF1 in hepatitis B virus-associated hepatocellular carcinoma treated with transplantation. Annals of Surgical Oncology. 2008;15(12):3494–3502. doi: 10.1245/s10434-008-0146-1. [DOI] [PubMed] [Google Scholar]
- 20.Cheng J, Zhou L, Xie Q, et al. The impact of miR-34a on protein output in hepatocellular carcinoma HepG2 cells. Proteomics. 2010;10(9):1557–1572. doi: 10.1002/pmic.200900646. [DOI] [PubMed] [Google Scholar]
- 21.Wu L, Yang Z, Zhou L, et al. Identification of histone deacetylase 3 as a biomarker for tumor recurrence following liver transplantation in HBV-associated hepatocellular carcinoma. PLoS ONE. 2010;5(12) doi: 10.1371/journal.pone.0014460.e14460 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Zheng S, Xu X, Wu J, et al. Liver transplantation for hepatocellular carcinoma: Hangzhou experiences. Transplantation. 2008;85(12):1726–1732. doi: 10.1097/TP.0b013e31816b67e4. [DOI] [PubMed] [Google Scholar]
- 23.Zimmerman MA, Ghobrial RM, Tong MJ, et al. Recurrence of hepatocellular carcinoma following liver transplantation: a review of preoperative and postoperative prognostic indicators. Archives of Surgery. 2008;143(2):182–188. doi: 10.1001/archsurg.2007.39. [DOI] [PubMed] [Google Scholar]
- 24.Lu T, Hano H, Meng C, Nagatsuma K, Chiba S, Ikegami M. Frequent loss of heterozygosity in two distinct regions, 8p23.1 and 8p22, in hepatocellular carcinoma. World Journal of Gastroenterology. 2007;13(7):1090–1097. doi: 10.3748/wjg.v13.i7.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seng TJ, Low JSW, Li H, et al. The major 8p22 tumor suppressor DLC1 is frequently silenced by methylation in both endemic and sporadic nasopharyngeal, esophageal, and cervical carcinomas, and inhibits tumor cell colony formation. Oncogene. 2007;26(6):934–944. doi: 10.1038/sj.onc.1209839. [DOI] [PubMed] [Google Scholar]
- 26.Rodrigues-Ferreira S, Di Tommasso A, Dimitrov A, et al. 8p22 MTUS1 gene product ATIP3 is a novel anti-mitotic protein underexpressed in invasive breast carcinoma of poor prognosis. PLoS ONE. 2009;4(10) doi: 10.1371/journal.pone.0007239.e7239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oyama T, Miyoshi Y, Koyama K, et al. Isolation of a novel gene on 8p21. 3-22 whose expression is reduced significantly in human colorectal cancers with liver metastasis. Genes, Chromosomes and Cancer. 2000;29(1):9–15. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1001>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 28.Yan S, Tang J, Huang C, et al. Reduced expression of ZDHHC2 is associated with lymph node metastasis and poor prognosis in gastric adenocarcinoma. PLoS ONE. 2013;8(2) doi: 10.1371/journal.pone.0056366.e56366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greaves J, Chamberlain LH. DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends in Biochemical Sciences. 2011;36(5):245–253. doi: 10.1016/j.tibs.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Planey SL, Ceballos C, Stevens SM, Jr., Keay SK, Zacharias DA. Identification of CKAP4/p63 as a major substrate of the palmitoyl acyltransferase DHHC2, a putative tumor suppressor, using a novel proteomics method. Molecular and Cellular Proteomics. 2008;7(7):1378–1388. doi: 10.1074/mcp.M800069-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Planey SL, Keay SK, Zhang C, Zacharias DA. Palmitoylation of cytoskeleton associated protein 4 by DHHC2 regulates antiproliferative factor-mediated signaling. Molecular Biology of the Cell. 2009;20(5):1454–1463. doi: 10.1091/mbc.E08-08-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma C, Yang XH, Hemler ME. DHHC2 affects palmitoylation, stability, and functions of tetraspanins CD9 and CD151. Molecular Biology of the Cell. 2008;19(8):3415–3425. doi: 10.1091/mbc.E07-11-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyake M, Inufusa H, Adachi M, et al. Suppression of pulmonary metastasis using adenovirally motility related protein-1 (MRP-1/CD9) gene delivery. Oncogene. 2000;19(46):5221–5226. doi: 10.1038/sj.onc.1203919. [DOI] [PubMed] [Google Scholar]
- 34.Zöller M. Gastrointestinal tumors: metastasis and tetraspanins. Zeitschrift fur Gastroenterologie. 2006;44(7):573–586. doi: 10.1055/s-2006-926795. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: RT-PCR primers.
Supplementary Table 2: Clinicopathological Correlation With ZDHHC2 mRNA expression in HCC Cases (Cohort 2).


