The RAD18 E3 ligase requires SIVA1 as an accessory protein for binding to its substrate PCNA during translesion DNA synthesis.
Abstract
Translesion DNA synthesis (TLS) is a universal DNA damage tolerance mechanism conserved from yeast to mammals. A key event in the regulation of TLS is the monoubiquitination of proliferating cell nuclear antigen (PCNA). Extensive evidence indicates that the RAD6–RAD18 ubiquitin-conjugating/ligase complex specifically monoubiquitinates PCNA and regulates TLS repair. However, the mechanism by which the RAD6–RAD18 complex is targeted to PCNA has remained elusive. In this study, we used an affinity purification approach to isolate the PCNA-containing complex and have identified SIVA1 as a critical regulator of PCNA monoubiquitination. We show that SIVA1 constitutively interacts with PCNA via a highly conserved PCNA-interacting peptide motif. Knockdown of SIVA1 compromised RAD18-dependent PCNA monoubiquitination and Polη focus formation, leading to elevated ultraviolet sensitivity and mutation. Furthermore, we demonstrate that SIVA1 interacts with RAD18 and serves as a molecular bridge between RAD18 and PCNA, thus targeting the E3 ligase activity of RAD18 onto PCNA. Collectively, our results provide evidence that the RAD18 E3 ligase requires an accessory protein for binding to its substrate PCNA.
Introduction
The genomes of all living cells are under continuous attack from a variety of endogenous and exogenous DNA-damaging agents, which lead to many types of DNA lesions. These DNA lesions can block genome replication and transcription and, if not repaired, yield mutations or wilder scale genome aberrations that threaten the survival of the individual cells and the whole organism. To counteract the deleterious effects of damaged DNA, organisms have evolved a variety of surveillance and repair mechanisms (Jackson and Bartek, 2009; Ciccia and Elledge, 2010). Translesion DNA synthesis (TLS) is a universal DNA damage tolerance mechanism conserved from yeast to mammals and performed by a class of specialized DNA polymerases (Friedberg, 2005; Lehmann et al., 2007; Moldovan et al., 2007). These TLS polymerases possess a spacious active site and are capable of accommodating a variety of DNA lesions that would block the high-fidelity replicative DNA polymerases (Prakash et al., 2005). Most of the TLS polymerases belong to the Y family, which includes Polη (polymerase η), Polκ, Polι, and Rev1 (Ohmori et al., 2001; Sale et al., 2012). Studies have shown that TLS is accomplished by the concerted action of multiple TLS polymerases. Remarkably, human DNA Polη is able to replicate past a cis-syn thymine–thymine (TT) dimer, a major photoproduct induced by UV irradiation, as efficiently as past undamaged DNA (McCulloch et al., 2004). Inactivation of Polη in humans causes the variant form of the skin cancer-prone syndrome xeroderma pigmentosum (XP-V; Johnson et al., 1999; Masutani et al., 1999a,b; Bienko et al., 2010). Cells from XP-V individuals are deficient in the replication of UV-damaged DNA and show hypermutability after UV exposure (Lehmann et al., 1975; Maher et al., 1976; Masutani et al., 1999b; Bienko et al., 2010).
A key event in the regulation of TLS is the monoubiquitination of proliferating cell nuclear antigen (PCNA), a homotrimeric protein that acts as an auxiliary factor for DNA polymerases (Hoege et al., 2002; Stelter and Ulrich, 2003; Moldovan et al., 2007). In response to DNA damage and/or replication stress, PCNA is monoubiquitinated at the lysine 164 residue by the E2 ubiquitin-conjugating enzyme RAD6 and the E3 ubiquitin ligase RAD18 (Hoege et al., 2002; Watanabe et al., 2004; Lehmann, 2011). PCNA can also be monoubiquitinated by the CRL4(Cdt2) E3 ubiquitin ligase in the absence of external DNA damage (Terai et al., 2010). In mammals, monoubiquitinated PCNA has been reported to have a much higher affinity than unmodified PCNA for Polη (Haracska et al., 2001a; Kannouche et al., 2004; Watanabe et al., 2004; Terai et al., 2010). This is in line with the identification of ubiquitin-binding domains in all Y family polymerases that might contribute to the increased interaction of PCNA and TLS polymerases after UV irradiation (Haracska et al., 2001b, 2002; Bienko et al., 2005; Plosky et al., 2006; Schmutz et al., 2010). Because untimely DNA synthesis by low-fidelity TLS polymerases could result in a higher mutagenesis rate, monoubiquitination of PCNA is kept in check by the USP1–UAF1 deubiquitinase complex (Huang et al., 2006; Cohn et al., 2007; Kim et al., 2009). Depletion of USP1 or UAF1 in human cells results in increased levels of monoubiquitinated PCNA both in the presence and absence of DNA damage (Huang et al., 2006; Cohn et al., 2007; Kim et al., 2009). There is growing evidence that the level of monoubiquitinated PCNA is closely linked with the DNA damage bypass to protect cells from a high level of mutagenesis. However, it still remains unclear how the level of monoubiquitinated PCNA is regulated.
SIVA1 is a small protein originally identified as an intracellular ligand for CD27 (Prasad et al., 1997; Xue et al., 2002). The structure of SIVA1 protein contains a death domain homology region in the central part and two zinc finger–like cysteine-rich domains in the C terminus (Prasad et al., 1997; Xue et al., 2002). SIVA1 plays a role both in extrinsic and intrinsic apoptotic pathways (Prasad et al., 1997; Xue et al., 2002; Resch et al., 2009). Recently, SIVA1 has also been shown to act as a suppressor of p53 (Du et al., 2009; Wang et al., 2013). Here, we identified SIVA1 as a critical regulator of PCNA monoubiquitination in response to UV-induced DNA damage. SIVA1 specifically interacts with RAD18 and PCNA in vivo and in vitro and displays properties of a substrate-specific adaptor. Depletion of SIVA1 compromised RAD18-dependent PCNA monoubiquitination and Polη focus formation, leading to increased UV sensitivity and mutation frequency. Our results provide new insights into the molecular mechanism that regulates PCNA monoubiquitination and TLS in response to UV damage.
Results
Identification of SIVA1 as a PCNA-binding protein
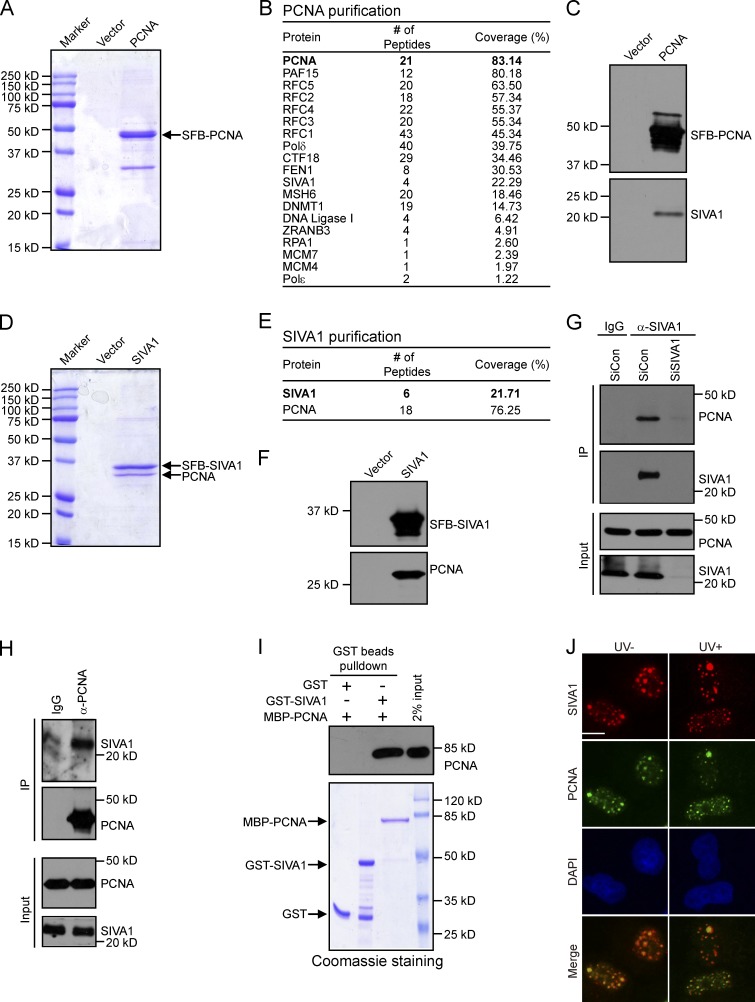
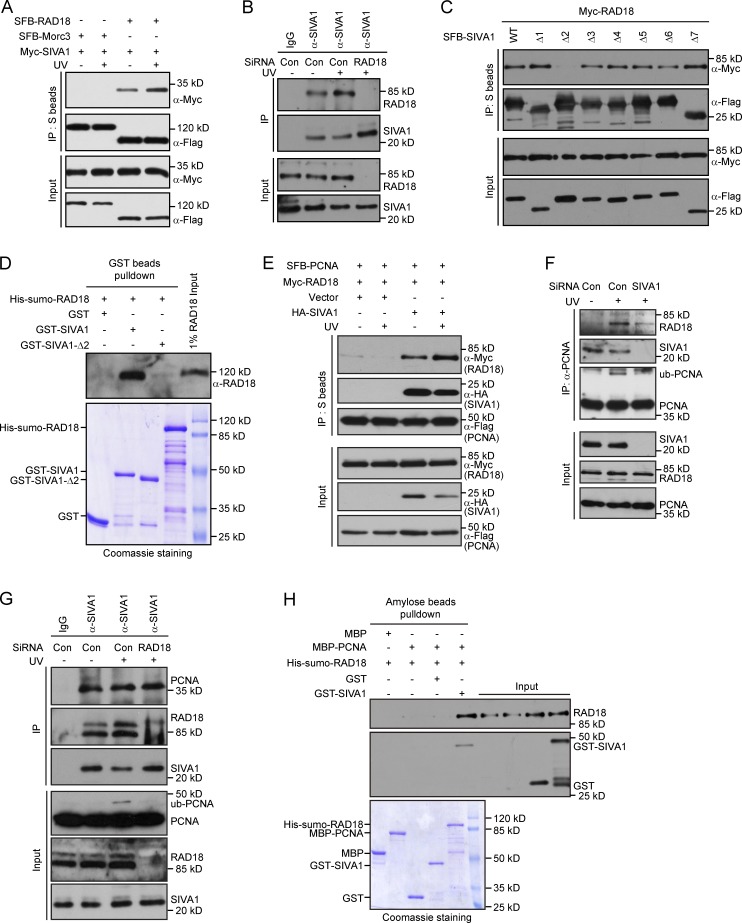
To identify previously undetected interacting partners of PCNA, we performed tandem affinity purification using lysates prepared from HEK293T cells ectopically expressing SFB (streptavidin-Flag–S protein)-tagged wild-type PCNA (Fig. 1 A). Mass spectrometry analysis identified many known PCNA-binding proteins, including PAF15, FEN1, and replication factor C (RFC) complex (Fig. 1 B; Emanuele et al., 2011; Povlsen et al., 2012). Interestingly, we also repeatedly identified SIVA1 as a putative PCNA-binding protein (Fig. 1 B). The presence of SIVA1 in PCNA–protein complexes was confirmed by Western blotting (Fig. 1 C).
Figure 1.
Identification of SIVA1 as PCNA-binding partner. (A and D) HEK293T cells stably expressing either the vector control or SFB-tagged (S tag, Flag epitope tag, and streptavidin-binding peptide tag) PCNA/SIVA1 were used for tandem affinity purification of protein complexes. The final eluates were resolved on SDS-PAGE gels and Coomassie stained. (B and E) Tables are summaries of proteins identified by mass spectrometry analysis. Letters in bold indicate the bait proteins. (C and F) Confirmation of PCNA- or SIVA1-interacting proteins by Western blotting. PCNA/SIVA1 purification products were analyzed by Western blots using specific antibodies against Flag, SIVA1, or PCNA. (G and H) Association of endogenous SIVA1 with PCNA in HeLa cells was performed by coimmunoprecipitation using the anti-SIVA1 or anti-PCNA antibody. SiRNA-treated HeLa cells were lysed in the presence of Benzonase, cell lysates were then incubated with protein A agarose beads conjugated with the indicated antibodies, and Western blot analysis was performed according to standard procedures. siCon, control siRNA. (I) Direct in vitro binding between recombinant MBP-PCNA and GST-SIVA1 purified from E. coli. GST served as negative control for PCNA binding. (top) PCNA was detected by immunoblotting. (bottom) Purified proteins visualized by Coomassie staining. (J) SIVA1 colocalizes with PCNA at nucleus foci. HeLa cells were transfected with HA-Flag–tagged SIVA1 plasmids. 24 h after transfection, cells were treated with 50 J/m2 UV or left untreated. After 1 h recovery, foci assembled by this fusion protein and by PCNA were detected by immunofluorescence using anti-Flag and anti-PCNA antibodies, respectively. HA-Flag–SIVA1 foci were detected in red, whereas PCNA foci were detected in green. A merged image shows colocalization. IP, immunoprecipitation. Bar, 10 µm.
To ensure that SIVA1 indeed forms a complex with PCNA, we performed reverse tandem affinity purification using HEK293T cells stably expressing SFB-tagged SIVA1 and identified PCNA as a major SIVA1-associated protein (Fig. 1, D and E). The presence of PCNA in SIVA1 purification was also confirmed by Western blotting (Fig. 1 F). Collectively, these results indicate that SIVA1 is a bone fide PCNA-interacting protein.
SIVA1 interacts with PCNA in vivo and in vitro
To verify our tandem affinity purification results, we performed coimmunoprecipitation assays using SFB-tagged SIVA1 and Myc-tagged PCNA. We found that SIVA1, like p21, interacts strongly with PCNA (Fig. S1 A). In contrast, the unrelated control protein Morc3 (Takahashi et al., 2007) does not interact with PCNA (Fig. S1 A). We further performed coimmunoprecipitation experiments and confirmed the interaction between endogenous SIVA1 and PCNA (Fig. 1, G and H; and Fig. S1, B and C). The SIVA1–PCNA complex formation was DNA damage independent, and this preexisting complex could be detected in HeLa cells (Fig. S1 D) as well as in other cell lines including HEK293T cells (Fig. S1 E).
We next tested the possibility that there could be a direct protein–protein interaction between PCNA and SIVA1. Pull-down assays using recombinant maltose-binding protein (MBP)–tagged PCNA and GST-tagged SIVA1 purified from Escherichia coli demonstrated that SIVA1 binds directly to PCNA in vitro (Fig. 1 I).
PCNA and several other proteins involved in DNA metabolism could form discrete foci in the nucleus. Given that SIVA1 forms a complex with PCNA, we speculate that SIVA1 may colocalize with PCNA at nucleus foci. As expected, SIVA1 could form discrete nuclear foci and colocalized with PCNA (Fig. 1 J). Collectively, these results established that SIVA1 binds PCNA and suggested an involvement for SIVA1 protein in PCNA-mediated cellular processes.
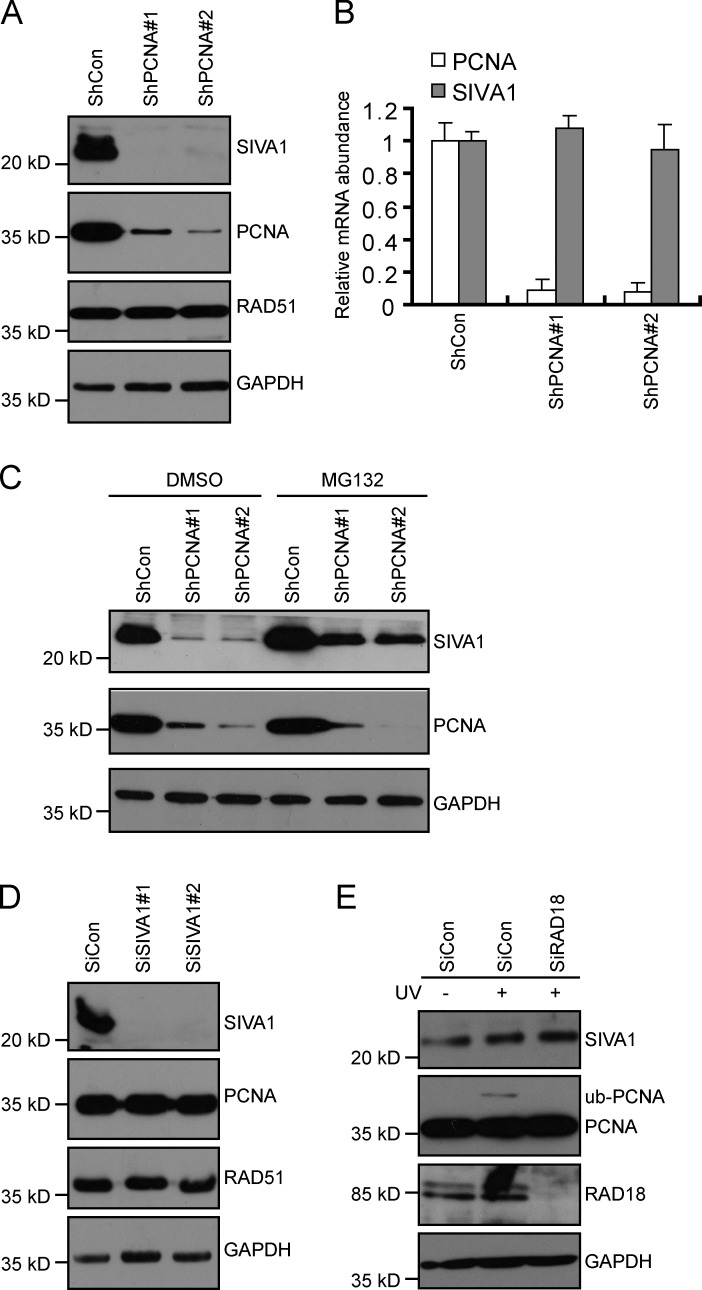
PCNA regulates SIVA1 protein stability
The stability of one component in a multiprotein complex often depends on the presence of other components so that when one protein is depleted, the others become unstable and show reduced expression levels. Indeed, reduction of PCNA expression by shRNAs in cells led to a dramatic decrease in SIVA1 protein level but not in mRNA level (Fig. 2, A and B). Moreover, this decrease was partially rescued by treatment with the proteasome inhibitor MG132 (Fig. 2 C). In contrast, depletion of SIVA1 did not affect the stability of PCNA (Fig. 2 D). Together, these results suggest that SIVA1 is normally stabilized in vivo by PCNA but not vice versa.
Figure 2.
PCNA regulates the stability of SIVA1. (A) PCNA depletion decreases the stability of SIVA1. HeLa cells were infected with lentiviruses carrying nontarget control or PCNA-specific shRNAs. 48 h later, cell lysates were subjected to immunoblotting using the indicated antibodies. (B) PCNA depletion has no effect on SIVA1 mRNA levels. HeLa cells were infected with lentiviruses carrying nontarget control or PCNA-specific shRNAs. 48 h later, the levels of specific mRNAs were determined by quantitative RT-PCR. Bars represent the mean of three experiments, and error bars are SDs. (C) MG132 treatment partially rescues the levels of SIVA1 protein in PCNA-depleted cells. HeLa cells were infected with lentiviruses carrying nontarget control or PCNA-specific shRNAs. 48 h later, cells were treated with proteasome inhibitor MG132 (10 µM) or DMSO for 6 h. Cell lysates were then subjected to immunoblotting using the indicated antibodies. (D) SIVA1 depletion has no effect on PCNA protein stability. HeLa cells were transfected twice with control siRNA or siRNAs specific for SIVA1. Cell lysates prepared 48 h after the second transfection were analyzed by immunoblotting with the indicated antibodies. (E) RAD18 depletion has no effect on SIVA1 protein stability. HeLa cells transfected with control siRNA or siRNAs specific for RAD18 were treated with 50 J/m2 UV for 1 h or left untreated. Cell lysates were then analyzed by immunoblotting with indicated antibodies. ShCon, control shRNA; SiCon, control siRNA; ub, ubiquitin.
In response to UV damage, PCNA can be monoubiquitinated at K164 by E3 ubiquitin ligase RAD18 (Hoege et al., 2002; Watanabe et al., 2004; Lehmann, 2011). We therefore tested whether the ubiquitination status of PCNA plays a role in stabilizing SIVA1. As shown in Fig. 2 E, depletion of RAD18, which abolished PCNA monoubiquitination after UV treatment, has no effect on SIVA1 protein stability, indicating that RAD18-mediated PCNA monoubiquitination is not required for maintaining the steady-state levels of SIVA1.
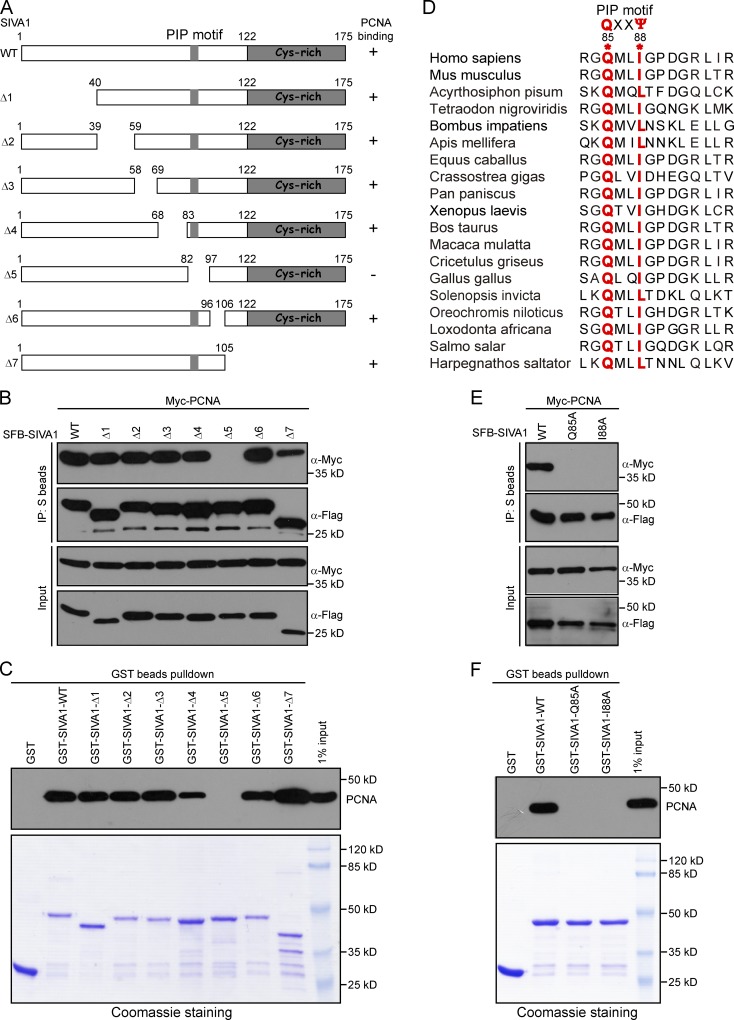
SIVA1 interacts with PCNA via a putative PCNA-interacting peptide (PIP) box
We next mapped the regions within SIVA1 responsible for its interaction with PCNA by using SFB-tagged wild-type SIVA1 and a series of SIVA1 deletion mutants (Fig. 3 A). Coimmunoprecipitation experiments revealed that a domain spanning amino acids 83–96 of SIVA1 contributes to its interaction with PCNA (Fig. 3 B). Consistently, whereas purified recombinant wild-type SIVA1 and the other mutants were able to pull-down endogenous PCNA from HEK293T cell extracts very efficiently, the SIVA1-Δ5 mutant failed to do so (Fig. 3 C). Interestingly, this region of SIVA1 (residues 83–96) has been highly conserved throughout evolution, suggesting that it may carry out an important function of SIVA1 (Fig. 3 D).
Figure 3.
The PIP box of SIVA1 is responsible for PCNA binding. (A) Schematic representation of wild-type and deletion mutants of SIVA1 used in this study. (B) Residues 83–96 of SIVA1 are responsible for PCNA binding. HEK293T cells were transiently transfected with plasmids encoding tagged PCNA and SIVA1. Cell lysates were immunoprecipitated with S beads, and Western blot analysis was performed with anti-Flag and anti-Myc antibodies. (C) Purified GST, GST-tagged wild-type SIVA1, or its deletion mutants immobilized on Sepharose beads were incubated with HEK293T cell lysates. Endogenous bound PCNA was analyzed by anti-PCNA immunoblotting. Input GST or GST-SIVA1 proteins were shown at the bottom. (D) Sequence alignment of the PCNA-binding region in SIVA1 from different species. (E and F) The PIP box of SIVA1 is required for SIVA1 to bind to PCNA. HEK293T cells were transfected with plasmids encoding SFB-tagged wild-type SIVA1 or two PIP box mutants (Q85A and I88A) together with plasmids encoding Myc-tagged PCNA. (E) Coprecipitation was performed using S protein beads, and immunoblotting was performed using antibodies as indicated. (F) Purified GST or GST-tagged wild-type SIVA1 or two PIP box mutants (Q85A and I88A) immobilized on Sepharose beads were incubated with HEK293T cell lysates. Endogenous bound PCNA was analyzed by anti-PCNA immunoblotting. IP, immunoprecipitation; WT, wild type.
Many PCNA-associated proteins have been shown to interact with PCNA via their corresponding PIP box, which has the consensus motif QXX(M/L/I)XX(F/Y)(F/Y). During visual inspection of the SIVA1 amino acid sequence, we identified a potentially divergent PIP box that coincided with the region required for its interaction with PCNA (Fig. 3 D). To test whether SIVA1 might similarly interact with PCNA via its putative PIP box, we generated two point mutants with mutations within the consensus PIP box (Q85A and I88A; Fig. 3 D). As shown in Fig. 3 E, whereas wild-type SIVA1 was able to interact with PCNA, both the Q85A and I88A mutants failed to do so. Furthermore, pull-down experiments consolidated the aforementioned notion that SIVA1 interacts with PCNA via its PIP box (Fig. 3 F).
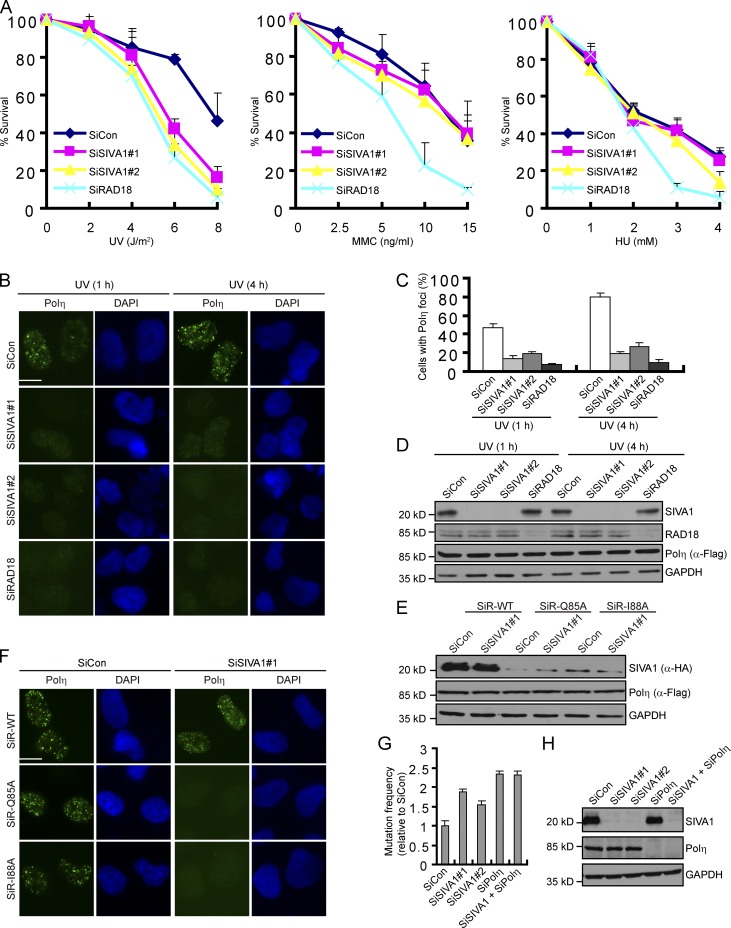
SIVA1 is required for Polη recruitment after UV damage
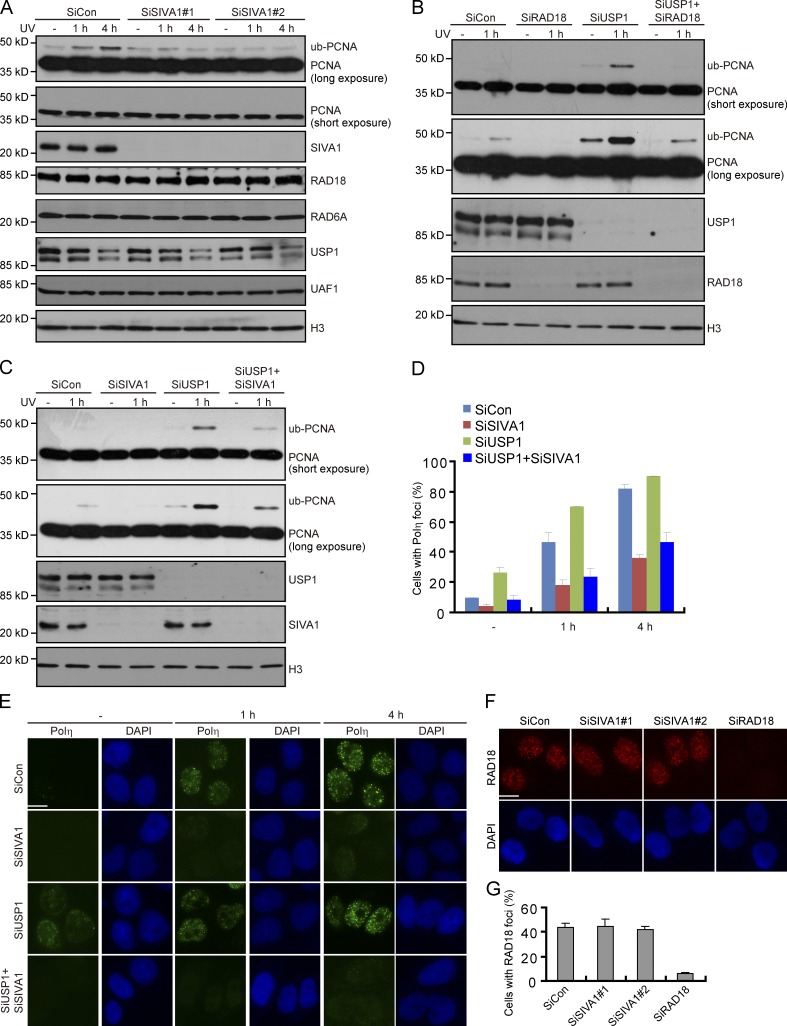
To investigate the cellular function of SIVA1, we knocked down its expression in human HeLa cells using two independent siRNAs specifically targeting SIVA1. Interestingly, SIVA1 knockdown cells showed significantly elevated sensitivity to UV as compared with cells treated with control siRNA (Fig. 4 A). SIVA1 knockdown cells, however, showed little or no hypersensitivity to mitomycin C (MMC) and hydroxyurea (HU), suggesting a specific involvement of SIVA1 in the response to UV-induced DNA damage (Fig. 4 A).
Figure 4.
SIVA1 regulates localization of Polη. (A) Clonogenic survival assays in SIVA1-depleted HeLa cells after UV, MMC, or HU treatment. RAD18 depletion is used as a positive control. Cells were permitted to grow for 14 d before staining. Experiments were performed in triplicates. Results shown are means of three independent experiments and were presented as means ± SD. (B–D) SIVA1 is required for Polη foci formation. A XP30RO cell line to express SFB-tagged Polη (XP30RO-Polη) under the control of a tetracycline-inducible promoter was generated. The resulting cell line was transfected twice with the indicated siRNAs and was induced by 1 µg/ml doxycycline addition for 24 h before 50 J/m2 UV treatment. Cells were then fixed and processed for Polη immunofluorescence. (B) Representative Polη foci were shown. (C) Quantification results were the mean of three independent experiments and were presented as means ± SD. More than 100 cells were counted in each experiment. (D) Knockdown efficiency was confirmed by immunoblotting. (E and F) PIP box mutants of SIVA1 could not rescue Polη foci formation in SIVA1-depleted cells. XP30RO-Polη–derivative cell lines stably expressing HA-tagged siRNA#1-resistant wild-type SIVA1 or two PIP box mutants (Q85A and I88A) were generated. The resulting cell line was transfected twice with the indicated siRNAs and was induced by doxycycline addition for 24 h before 50 J/m2 UV treatment. 4 h later, cells were fixed and processed for Polη immunofluorescence. (F) Representative Polη foci were shown. (E) The exogenous SIVA1 expression was confirmed by immunoblotting using the anti-HA antibody. (G and H) HEK293T cells transfected with the indicated siRNAs for 48 h were treated with UV-damaged supF plasmid. After a further 48 h, the supF plasmid was recovered, and supF mutation frequencies were measured from 10,000 colonies. (G) Results were the mean of three independent experiments and were presented as means ± SD. (H) Knockdown efficiency was confirmed by immunoblotting. SiCon, control siRNA; SiR, siRNA resistant; WT, wild type. Bars, 10 µm.
A major universal tolerance mechanism is TLS, in which specialized low-fidelity DNA polymerases elongate the DNA across the lesion. To investigate how SIVA1 contributes to the repair and/or tolerance of UV damage, we engineered a Polη-deficient XP30RO cell line to express SFB-tagged Polη (XP30RO-Polη) under the control of a tetracycline-inducible promoter (Fig. S2, A and B) and tested the effect of SIVA1 depletion on the ability of Polη to form foci after UV irradiation. Remarkably, depletion of SIVA1 significantly reduced UV-induced Polη focus formation (Fig. 4, B–D). Similar results were also obtained with HeLa cells (Fig. S2, C–E). These results suggest that SIVA1 may confer cellular resistance to UV radiation by promoting recruitment of Polη to the sites of UV-induced DNA damage.
Previous studies have shown that genotoxin-induced CHK1 activation in TLS-deficient cells is exacerbated and that this may be a result of the persistence of unfilled postreplicative single-stranded gaps (Bi et al., 2005, 2006; Lopes et al., 2006; Callegari et al., 2010; Yang et al., 2013). Because SIVA1 is required for efficient Polη foci formation, we sought to examine whether SIVA1 depletion affects DNA damage-induced CHK1 phosphorylation. As shown in Fig. S3 A, SIVA1 depletion, like RAD18 depletion, resulted in prolonged phosphorylation of CHK1 after UV damage.
To further test whether the PIP motif is important for SIVA1 function in vivo, we generated a XP30RO-Polη cell line to stably express siRNA#1-resistant wild-type SIVA1 or its point mutants defective in PCNA binding and examined Polη focus formation. The expression of exogenous SIVA1 or SIVA1 mutants was confirmed by immunoblotting in the cells transfected with control siRNA or SIVA1 siRNA#1 (Fig. 4 E). Notably, the expression level of both the SIVA1 mutants defective in PCNA binding (Q85A and I88A) is significantly lower than that of wild type (Fig. 4 E). This finding further supports our conclusion that SIVA1 is normally stabilized in vivo by PCNA (Fig. 2 A). Accordingly, the defect of Polη foci formation in SIVA1-depleted cells was readily restored by the reexpression of wild-type SIVA1 but not the point mutants defective in PCNA binding (Fig. 4 F and Fig. S3 B).
Depletion of SIVA1 causes increased mutation frequency
Cells deficient in Polη exhibit hypersensitivity to UV damage and display elevated mutation frequency. Given that SIVA1 is required for efficient recruitment of Polη to sites of UV-induced DNA damage, we propose that SIVA1 may also be involved in mutagenesis suppression. To test this possibility, we measured the mutation frequency using the pZ189 shuttle vector system (Akasaka et al., 1992). As shown in Fig. 4 (G and H), the mutation frequency in SIVA1-depleted cells was dramatically elevated. Moreover, codepletion of SIVA1 and Polη did not cause a further increase in mutation frequency, suggesting that SIVA1 and Polη may operate in the same pathway (Fig. 4, G and H).
SIVA1 is required for efficient PCNA monoubiquitination
RAD18/RAD6-dependent monoubiquitination of PCNA at Lys164 (K164) is thought to promote direct lesion bypass by recruiting TLS polymerases to the stalled replication forks. The association of SIVA1 with PCNA and the requirement of SIVA1 for Polη recruitment raised the intriguing possibility that SIVA1 may play a critical role in PCNA monoubiquitination. In agreement with this idea, SIVA1 depletion results in a dramatic reduction in the level of PCNA monoubiquitination (Fig. 5 A). Importantly, suppression of SIVA1 expression did not markedly alter the cell cycle distribution (Fig. S3 C), suggesting that these observed phenotypes in SIVA1 knockdown cells were not caused by any change in cell cycle.
Figure 5.
SIVA1 is required for efficient PCNA monoubiquitination. (A) SIVA1 is required for UV-induced PCNA monoubiquitination. HeLa cells were transfected twice with control siRNA or siRNAs specific for SIVA1. 48 h after transfection, cells were untreated or treated with 50 J/m2 UV for the indicated times. Chromatin fractions were isolated and immunoblotted with the indicated antibodies. (B and C) HeLa cells were transfected twice with the indicated siRNAs. 48 h after transfection, cells were untreated or treated with 50 J/m2 UV for 1 h. Chromatin fractions were isolated and immunoblotted with the indicated antibodies. (D and E) A HeLa cell line to stably express HA-Flag–tagged Polη was generated. The resulting cell line was transfected twice with the indicated siRNAs. 48 h after transfection, cells were untreated or treated with 50 J/m2 UV for the indicated times before fixing and processed for Polη immunofluorescence. (E) Representative Polη foci were shown. (D) Quantification results were the mean of three independent experiments and were presented as means ± SD. More than 100 cells were counted in each experiment. (F and G) SIVA1 is not required for RAD18 foci formation. HeLa cells were transfected twice with control siRNA or siRNAs specific for SIVA1 or RAD18. 48 h after transfection, cells were treated with 50 J/m2 UV for 4 h before fixing and processed for RAD18 immunofluorescence. (F) Representative RAD18 foci were shown. (G) Quantification results were the mean of three independent experiments and were presented as mean ± SD. More than 100 cells were counted in each experiment. SiCon, control siRNA; ub, ubiquitin. Bars, 10 µm.
It is well established that the level of PCNA monoubiquitination is regulated by both the RAD18–RAD6 ubiquitin ligase complex and the USP1–UAF1 deubiquitinase complex. The requirement of SIVA1 for the efficient monoubiquitination of PCNA suggests that it either positively regulates ubiquitin ligation or negatively regulates USP1–UAF1 via direct or indirect mechanisms. If SIVA1 positively regulates ubiquitin ligation, depletion of SIVA1 will compromise PCNA monoubiquitination even in the absence of USP1. On the other hand, if SIVA1 negatively regulates USP1–UAF1, depletion of SIVA1 will not affect PCNA monoubiquitination in the absence of USP1. To test these possibilities, we first analyzed the monoubiquitination of PCNA in SIVA1 and USP1 single and double knockdown cells. Consistent with previous findings (Huang et al., 2006; Cohn et al., 2007; Kim et al., 2009), depletion of USP1 by using siRNA results in increased levels of monoubiquitinated PCNA both in the absence and presence of UV damage (Fig. 5, B and C). Interestingly, like that of RAD18, depletion of SIVA1 compromised PCNA monoubiquitination even when USP1 was simultaneously depleted (Fig. 5, B and C). Consistently, depletion of SIVA1 compromised Polη focus formation when USP1 was simultaneously depleted (Fig. 5, D and E; and Fig. S3 D). These results, collectively with our observation that SIVA1 depletion did not affect the chromatin association of USP1 and UAF1 (Fig. 5 A), exclude the possibility that SIVA1 may act as an antagonist of PCNA deubiquitination by the USP1–UAF1 complex.
Next, we examined how SIVA1 facilitates the establishment of PCNA monoubiquitination. Because PCNA monoubiquitination occurs in a chromatin context, we asked whether SIVA1 regulates the chromatin association of RAD18 or RAD6, the ubiquitin-conjugating enzyme required for PCNA monoubiquitination. Surprisingly, however, depletion of SIVA1 had no effect on the levels of chromatin-bound RAD18 and RAD6 (Fig. 5 A). In addition, knockdown of SIVA1 did not affect the formation of RAD18 foci after UV treatment (Fig. 5, F and G). These results indicate that SIVA1 does not promote PCNA monoubiquitination by facilitating the binding and/or retention of RAD18 and RAD6 on chromatin.
SIVA1 associates with and recruits RAD18 to PCNA
Substrate-specific receptors dictate the specificity for the ubiquitination reaction by selectively recruiting substrates to the E3 ubiquitin ligase machinery. The observation that SIVA1 interacts with PCNA and is required for efficient PCNA monoubiquitination, despite normal RAD18 foci formation in SIVA1-depleted cells, suggested that SIVA1 may serve as a substrate receptor for RAD18 ubiquitin ligase. To test this possibility, we first examined whether SIVA1 physically interacts with RAD18. Coimmunoprecipitation experiments showed that SFB-tagged RAD18 interacted with Myc-tagged SIVA1 (Fig. 6 A). We further demonstrated that endogenous RAD18 interacts with SIVA1 and that the interaction between SIVA1 and RAD18 is enhanced after UV treatment (Fig. 6, A and B).
Figure 6.
SIVA1 associates with and recruits RAD18 to PCNA. (A) SIVA1 interacts with RAD18. HEK293T cells were transiently transfected with plasmids encoding SFB-tagged Morc3 or RAD18 together with plasmids encoding Myc-tagged SIVA1. 24 h after transfection, cells were left untreated or treated with 50 J/m2 UV for 1 h. Coprecipitation was performed using S protein beads, and immunoblotting was performed using antibodies as indicated. (B) Association of endogenous SIVA1 with RAD18 in HeLa cells was analyzed by immunoprecipitation using the anti-SIVA1 antibody. Cells transfected with the indicated siRNAs were treated with 50 J/m2 UV for 1 h or left untreated and then lysed with NETN buffer containing Benzonase. Cell lysates were incubated with protein A agarose beads conjugated with indicated antibodies, and Western blot analysis was performed as indicated. (C) Residues 40–58 of SIVA1 are responsible for RAD18 binding. HEK293T cells were transiently transfected with the indicated plasmids. 24 h after transfection, cells were treated with 50 J/m2 UV for 1 h. Coprecipitation was performed using S protein beads, and immunoblotting was performed using antibodies as indicated. (D) Direct in vitro binding between recombinant GST-SIVA1 and His-SUMO–RAD18 purified from E. coli. GST served as negative control for RAD18 binding. (top) RAD18 was detected by immunoblotting. (bottom) Purified proteins visualized by Coomassie staining. (E) Overexpression of SIVA1 enhances the interaction between RAD18 and PCNA. HEK293T cells were transiently transfected with the indicated plasmids. 24 h after transfection, cells were left untreated or treated with 50 J/m2 UV for 1 h. Coprecipitation was performed using S protein beads, and immunoblotting was performed using antibodies as indicated. (F) The RAD18–PCNA interaction is diminished in SIVA1-depleted cells. HeLa cells were transfected twice with control siRNA or siRNA specific for SIVA1. 48 h after transfection, cells were left untreated or treated with 50 J/m2 UV for 1 h. Cells were then Triton X-100 extracted and formaldehyde fixed, and cell lysates were immunoprecipitated with protein A agarose beads conjugated with anti-PCNA antibody. Western blot analysis was performed as indicated. (G) RAD18 depletion does not affect SIVA1–PCNA interaction. HeLa cells transfected with the indicated siRNAs were treated with 50 J/m2 UV for 1 h or left untreated and then lysed with NETN buffer containing Benzonase. Cell lysates were then incubated with protein A agarose beads conjugated with anti-SIVA1 antibody, and Western blot analysis was performed according to standard procedures. (H) SIVA1 facilitates RAD18–PCNA complex assembly in vitro. (top) RAD18 and SIVA1 were detected by immunoblotting. (bottom) Purified proteins were visualized by Coomassie staining. Con, control; IP, immunoprecipitation; ub, ubiquitin; WT, wild type.
To identify the regions within SIVA1 responsible for its interaction with RAD18, we performed coimmunoprecipitation experiments and found that a domain spanning amino acids 40–58 of SIVA1 is responsible for RAD18 binding (Fig. 6 C). Conversely, we generated a series of RAD18 deletion mutants to examine which domain of RAD18 might be required for its interaction with SIVA1. Interestingly, the RAD18 region that mediates its binding to SIVA1 is located at residues 331–375 and thus overlaps with the RAD18 binding region to RAD6 (Fig. S4; Bailly et al., 1997; Watanabe et al., 2004).
To test whether SIVA1 binding to RAD18 through a direct interaction, an in vitro GST pull-down assay was performed using GST-fused SIVA1 and His-small ubiquitin-like modifier (SUMO)–fused RAD18 recombinant proteins (Hibbert et al., 2011). Notably, His-SUMO–RAD18 could be pulled down by GST-SIVA1 but not by GST alone (Fig. 6 D), suggesting that SIVA1 directly interacts with RAD18. Moreover, consistent with the results of coimmunoprecipitation experiments, the GST–SIVA1-Δ2 fusion protein failed to pull down His-SUMO–RAD18 (Fig. 6 D).
The association of SIVA1 with RAD18 and PCNA through nonoverlapping regions and the similar effects of SIVA1 and RAD18 on PCNA monoubiquitination raised the intriguing possibility that SIVA1 might facilitate the RAD18–PCNA interaction in the cells, thus leading to PCNA monoubiquitination. In a mammalian overexpression system, RAD18 interacted with PCNA weakly, probably through endogenous SIVA1. This interaction was enhanced by ectopically expressed SIVA1 (Fig. 6 E). In untransfected cells, endogenous RAD18 interacted with PCNA in a UV-dependent manner, and this interaction was diminished by knockdown of SIVA1 (Fig. 6 F). Moreover, the SIVA1–PCNA interaction was not affected when RAD18 was ablated by siRNA (Fig. 6 G). These results suggest that SIVA1 is the central component of the RAD18–SIVA1–PCNA complex and physically connects RAD18 to PCNA in vivo. In agreement with this conclusion, the recombinant SIVA1 protein facilitated RAD18–PCNA complex assembly in vitro (Fig. 6 H).
SIVA1 promotes RAD18-mediated PCNA monoubiquitination in vivo and in vitro
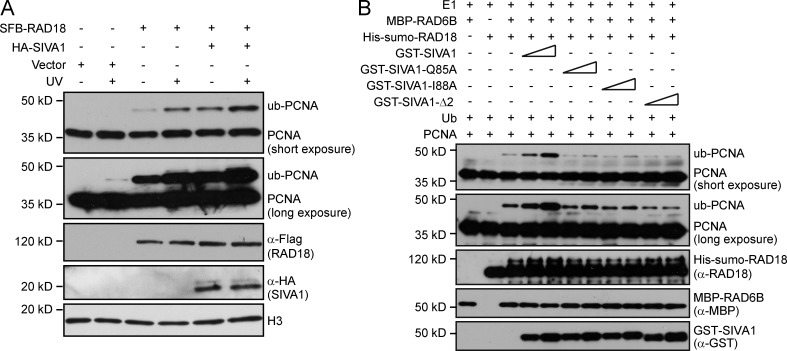
As shown in Fig. 5 A and Fig. 6 (E–H), SIVA1 mediates the interaction between RAD18 and PCNA and is required for efficient PCNA monoubiquitination. Therefore, it would be interesting to test whether SIVA1 can promote RAD18-mediated PCNA monoubiquitination. We first investigated RAD18-mediated monoubiquitination of PCNA in cells overexpressing SIVA1. As we expected, RAD18-mediated monoubiquitination of PCNA was significantly enhanced in SIVA1-overexpressed cells compared with that in control cells both in the presence and absence of UV treatment (Fig. 7 A).
Figure 7.
SIVA1 promotes PCNA monoubiquitination in vivo and in vitro. (A) SIVA1 promotes RAD18-mediated PCNA monoubiquitination in vivo. HeLa cells were transiently transfected with the indicated plasmids. 48 h after transfection, cells were left untreated or treated with 50 J/m2 UV for 1 h. Chromatin fractions were isolated and immunoblotted with the indicated antibodies. (B) SIVA1 promotes RAD18-mediated PCNA monoubiquitination in vitro. 0.4 µM PCNA was incubated in the absence or presence of various combinations of 50 µM ubiquitin, 50 nM Uba1, 0.2 µM RAD6, 0.2 µM RAD18, and SIVA1 (0.5 µM or 1 µM) as indicated. The assay was performed in reaction buffer containing 25 mM Tris, pH 8.0, 50 mM NaCl, and 0.1 mM DTT. The reactions were activated with 2 mM ATP and incubated at 30°C for 1 h. Proteins were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. ub, ubiquitin.
Next, we performed in vitro ubiquitination assays using PCNA as the substrate in the presence of wild-type or mutant SIVA1 along with the RAD18–RAD6 complex to examine whether SIVA1 can promote RAD18-mediated PCNA monoubiquitination in vitro. As expected, the recombinant RAD18–RAD6 complex was able to monoubiquitinate PCNA as shown by the appearance of an electrophoretically shifted form of PCNA. Interestingly, the ubiquitin-modified PCNA was significantly enhanced in the presence of recombinant wild-type SIVA1 but not the mutants defective in PCNA or RAD18 binding (Fig. 7 B).
SIVA1 is functionally linked to RAD18
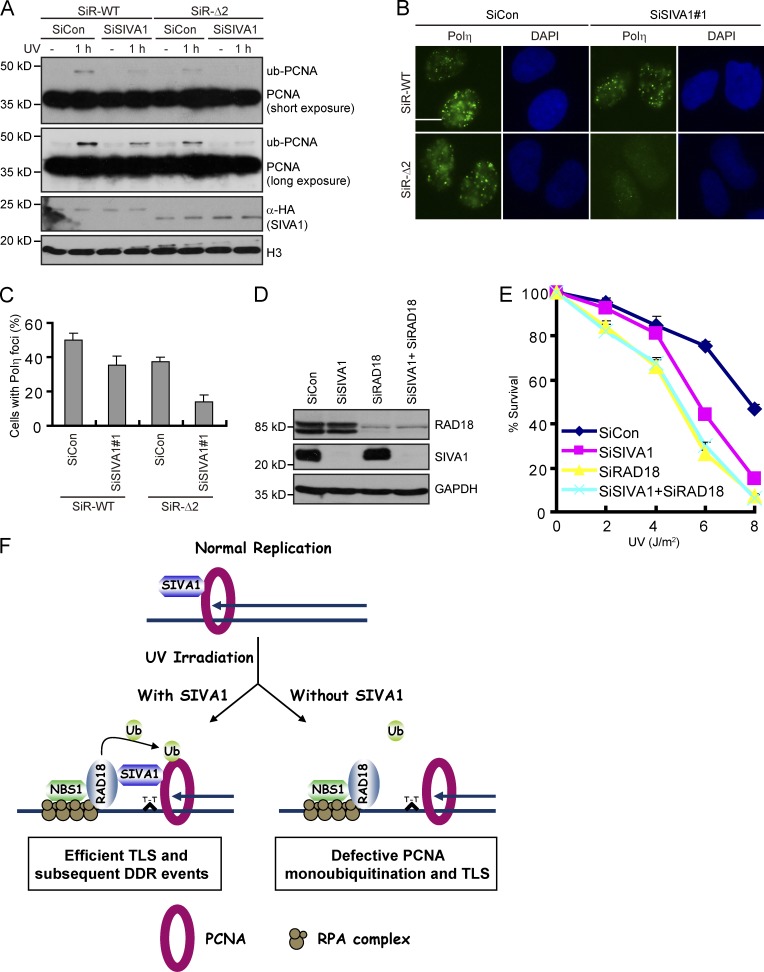
To investigate the biological significance of the interaction between SIVA1 and RAD18, we performed rescue experiments to examine whether the ability of SIVA1 to bind RAD18 is important for its function in PCNA monoubiquitination and Polη foci formation. As shown in Fig. 8 (A–C) and Fig. S5, siRNA-resistant wild-type SIVA1, but not the deletion mutants defective in RAD18 binding, was able to restore PCNA monoubiquitination and Polη foci formation in SIVA1-depleted cells. These results indicate that the RAD18-binding activity is required for SIVA1 to function in vivo.
Figure 8.
SIVA1 is functionally linked to RAD18. (A) The mutant defective in RAD18 binding failed to rescue PCNA monoubiquitination in cells with SIVA1 depletion. A HeLa cell line stably expressing HA-tagged siRNA#1-resistant wild-type SIVA1 (SiR-WT) or its deletion mutant defective in RAD18 binding (SiR-Δ2) was generated. The resulting cell lines were transfected twice with control siRNA or SIVA1 siRNA#1. 48 h after the second transfection, cells were left untreated or treated with 50 J/m2 UV for 1 h. Chromatin fractions were isolated and immunoblotted with the indicated antibodies. (B and C) The mutant defective in RAD18 binding failed to rescue Polη foci formation in cells with SIVA1 depletion. A XP30RO-Polη–derivative cell line stably expressing HA-tagged siRNA#1-resistant wild-type SIVA1 or the deletion mutant defective in RAD18 binding was generated. The resulting cell line was transfected twice with the indicated siRNAs and was induced by doxycycline addition for 24 h before 50 J/m2 UV treatment. 1 h later, cells were fixed and processed for Polη immunofluorescence. (B) Representative Polη foci were shown. Bar, 10 µm. (C) Quantification results were the mean of three independent experiments and were presented as means ± SD. More than 100 cells were counted in each experiment. (D and E) SIVA1 and RAD18 function in the same pathway. HeLa cells transfected with the indicated siRNAs were treated with increasing doses of UV. Survival curves are shown for the indicated cell lines. Data are presented as means ± SD from three different experiments. (F) Model depicting the molecular function of SIVA1 in regulating TLS as described under the Discussion section. SiCon, siRNA control; ub, ubiquitin.
To further define the in vivo functional link between SIVA1 and RAD18, we performed an epistasis analysis of the two proteins. As shown in Fig. 8 (D and E), the UV hypersensitivity of cells depleted of both RAD18 and SIVA1 was not greater than when each protein was depleted individually, indicating that SIVA1 and RAD18 function in the same pathway.
Discussion
In this study, we have provided several lines of evidence to show that SIVA1 is a critical regulator of PCNA monoubiquitination in response to UV damage. First, SIVA1 constitutively interacts with PCNA through a highly conserved putative PIP box. Second, cells depleted of SIVA1 showed marked increases in cellular sensitivity to UV treatment. Third, SIVA1 depletion largely prevented the formation of Polη foci after UV treatment. Fourth, SIVA1 promotes RAD18-mediated PCNA monoubiquitination in vivo and in vitro. Finally, depletion of SIVA1 resulted in a marked decrease in PCNA monoubiquitination, which was accompanied by reduced RAD18–PCNA complex formation. Our data are consistent with a model in which SIVA1 serves as a molecular bridge between RAD18 and PCNA, thus targeting the E3 ligase activity of RAD18 onto PCNA (Fig. 8 F).
A key event in the regulation of TLS is the monoubiquitination of PCNA, a homotrimeric protein that acts as an auxiliary factor for DNA polymerases. Upon UV damage, PCNA is monoubiquitinated by the RAD18–RAD6 complex. This modification thus provides a platform for recruiting repair factors as well as specialized translesion DNA polymerases, which operate to bypass bulky DNA adducts during DNA replication. Besides the RAD18–RAD6 complex, many other proteins, including RPA (Davies et al., 2008), Claspin (Yang et al., 2008), CHK1 (Yang et al., 2008), C1orf124/Spartan/DVC1 (Centore et al., 2012; Davis et al., 2012; Ghosal et al., 2012; Juhasz et al., 2012; Machida et al., 2012; Mosbech et al., 2012), NBS1 (Yanagihara et al., 2011), and Polη (Durando et al., 2013), are also required for efficient PCNA monoubiquitination. Most of these proteins are involved directly or indirectly in recruiting RAD18 to stalled replication forks. In contrast, SIVA1 serves as an adaptor that bridges RAD18 to PCNA and is not required for RAD18 nuclear foci formation and chromatin recruitment. Based on these findings, we propose that, in response to UV damage, single-stranded DNA regions are exposed and are coated with RPA at stalled replication forks. Through its interaction with RPA, NBS1, and/or other auxiliary factors, RAD18 is recruited to these stalled replication forks. SIVA1 then serves as a molecular bridge between RAD18 and PCNA, thus targeting the E3 ligase activity of RAD18 onto PCNA (Fig. 8 F). However, in the absence of SIVA1, although RAD18 is still efficiently recruited to UV damage sites, it fails to interact efficiently with PCNA, leading to defects in PCNA monoubiquitination and subsequent Polη recruitment and TLS activation (Fig. 8 F).
Equally important as PCNA monoubiquitylation is PCNA deubiquitylation, which is performed by USP1 in a heterodimeric complex with its cofactor UAF1 (Huang et al., 2006; Cohn et al., 2007; Kim et al., 2009). Depletion of USP1 or UAF1 results in increased levels of monoubiquitinated PCNA both in the presence and absence of DNA damage. PCNA deubiquitylation also requires the PCNA-binding protein hELG1 (Lee et al., 2010; Parnas et al., 2010; Yang et al., 2011). Several studies have demonstrated that hELG1 specifically directs the USP1–UAF1 complex to PCNA at the damage site and in doing so initiates the switch from the TLS polymerases to the replicative polymerases (Lee et al., 2010; Parnas et al., 2010; Yang et al., 2011). It is apparent that the two scaffold proteins SIVA1 and hELG1 have opposing functions in the regulation of PCNA monoubiquitination and are important for determining the ultimate ubiquitination status of PCNA. The mechanisms that regulate this balance require further investigation.
Although most known factors involved in TLS are conserved from yeast to humans, the counterpart of SIVA1 protein is only found in higher eukaryotes. The reason for this difference between yeast and human cells is not yet clear but is likely to reflect a need for additional levels of control in mammalian cells. Previous studies have shown that limited PCNA monoubiquitination could be achieved in the absence of DNA, and significant PCNA monoubiquitination occurred only when PCNA is loaded onto DNA by RFC (Watanabe et al., 2004; Garg and Burgers, 2005; Haracska et al., 2006; Unk et al., 2006, 2008; Hibbert et al., 2011). In an in vitro ubiquitination assay using purified proteins, we showed that SIVA1 could promote RAD18-dependent PCNA monoubiquitination in the absence of DNA. Given that SIVA1 facilitates RAD18–PCNA complex assembly in vivo and in vitro, we speculate that SIVA1 may also enhance RAD18-dependent monoubiquitination of RFC-loaded PCNA. Further studies will be conducted to consolidate this working model.
It has been previously reported that SIVA1 regulates cell proliferation and participates in p53-dependent apoptosis (Du et al., 2009; Resch et al., 2009; Iorio-Morin et al., 2012; Wang et al., 2013). In contrast, we found that SIVA1 depletion does not significantly affect cell proliferation and cell cycle distribution (Fig. S3, A and C; and Fig. S5). The basis for these discrepancies is not clear, and additional experiments will be required to clarify the issue.
Using the SupF suppressor tRNA gene as the mutagenesis reporter gene, we found that SIVA1 depletion resulted in a hypermutator phenotype. Although it is not currently clear how SIVA1 suppresses mutagenesis, given that both SIVA1 and Polη (Durando et al., 2013) forms a complex with RAD18, we speculate that SIVA1 may bind directly or indirectly to Polη and may favor the recruitment of Polη to monoubiquitinated PCNA, thereby facilitating the error-free bypass of UV lesions. However, we cannot exclude the possibility that SIVA1 may have other roles in suppressing mutagenesis.
In summary, we have identified a function for SIVA1 as an adaptor for the E3 ligase RAD18, showing that collaboration of SIVA1 and RAD18 controls the level of PCNA monoubiquitination. These findings will provide new insights into the molecular mechanisms of the replicative bypass of base damage to DNA by TLS, which is critically important for the maintenance of genomic integrity and tumor suppression.
Materials and methods
Antibodies
Rabbit polyclonal anti-SIVA1, anti-USP1, and anti-UAF1 antibodies were generated by immunizing rabbits with MBP-SIVA1 (residues 1–175), MBP-USP1 (residues 351–785), or MBP-UAF1 (residues 400–677) fusion proteins expressed and purified from E. coli, respectively. Antisera were affinity purified using an immobilization and purification kit (AminoLink Plus; Thermo Fisher Scientific). Anti-RAD18 and anti-RAD6A antibodies were purchased from Bethyl Laboratories, Inc. Anti-PCNA and anti-Myc (9E10) antibodies were purchased from Santa Cruz Biotechnology, Inc. and Covance, respectively. Anti-GAPDH, anti-H3, and anti-HA antibodies were purchased from EMD Millipore. The anti-Flag (M2) antibody was purchased from Sigma-Aldrich.
Cell culture and transfection
HEK293T and HeLa cells were maintained in DMEM supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. Cell transfection was performed using Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol.
Constructs
SIVA1, PCNA, and RAD18 cDNAs were obtained from Thermo Fisher Scientific. The PCR-amplified DNA fragment containing wild-type SIVA1, RAD18, or PCNA was subcloned into the pDONR201 vector using Gateway Technology (Invitrogen). Site-directed mutagenesis was performed according to standard procedures to obtain the SIVA1 mutants. For transient expression of PCNA, RAD18, SIVA1, or its mutants, the corresponding fragment in the entry vector was transferred into a Gateway-compatible destination vector (Invitrogen), which harbors either an N-terminal triple-epitope tag (S protein tag, Flag epitope tag, and Streptavidin-binding peptide tag) or an N-terminal Myc tag or HA tag. All clones were sequenced to verify desired mutations.
The establishment of stable cell lines and affinity purification of SFB-tagged protein complexes
HEK293T cells were transfected with plasmids encoding SFB-tagged SIVA1 or PCNA. Cell lines stably expressing tagged proteins were selected by culturing in medium containing 2 µg/ml puromycin and confirmed by immunoblotting and immunostaining. For affinity purification, HEK293T cells stably expressing tagged proteins were lysed with NETN buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40) containing Benzonase (EMD Millipore) for 20 min. The supernatants were cleared at 14,000 rpm to remove debris and then incubated with streptavidin-conjugated beads (GE Healthcare) for 2 h at 4°C. The beads were washed three times with NETN buffer, and then, bead-bound proteins were eluted with NETN buffer containing 1 mg/ml biotin (Sigma-Aldrich). The elutes were incubated with S protein beads (EMD Millipore). The beads were again washed three times with NETN buffer and subjected to SDS-PAGE. Protein bands were excised and digested, and the peptides were analyzed by mass spectrometry.
Protein purification
SIVA1 wild type and mutants were cloned into pDONR201 as entry clones and were then transferred to pDEST15 destination vector (Invitrogen) for the expression of GST-tagged fusion protein in E. coli. Cells were grown at 37°C until log phase and were induced with 0.2 mM IPTG at 18°C for 16 h. Cells were harvested and resuspended in lysis buffer (20 mM Tris-HCl, 300 mM NaCl, 1% Triton X-100, 2 mM DTT, and 1 µg/ml each of leupeptin, aprotinin, and pepstatin) and then sonicated. The extract was centrifuged at 18,000 rpm for 40 min. The supernatant was collected and incubated with glutathione–Sepharose resin for 4 h at 4°C. After washing the beads with washing buffer (20 mM Tris-HCl, 500 mM NaCl, 0.5% NP-40, 2 mM DTT, and 1 µg/ml each of leupeptin, aprotinin, and pepstatin), the bound protein was used for pull-down assays or eluted with 20 mM of reduced glutathione for in vitro ubiquitination assays. Full-length PCNA or RAD6B was cloned into pDONR201 as an entry clone and was then transferred to the Gateway-compatible destination vector for the expression of MBP-tagged fusion protein in E. coli. Cells were harvested and resuspended in lysis buffer (20 mM Tris-HCl, 300 mM NaCl, 1% Triton X-100, 1 mM EDTA, 10 mM 2-mercaptoethanol, and 1 µg/ml each of leupeptin, aprotinin, and pepstatin). After sonicating, the extract was centrifuged at 18,000 rpm for 40 min. The supernatant was collected and incubated with amylose resins for 2 h at 4°C. The bound protein was eluted with lysis buffer containing 10 mM maltose. Full-length RAD18 was cloned into the pET28-N-His-SUMO vector (EMD Millipore) for the expression of His-SUMO–tagged RAD18 in E. coli. The SUMO tag on RAD18 was able to improve its solubility to allow expression and purification in the absence of RAD6 (Hibbert et al., 2011). Cells were harvested and resuspended in lysis buffer (20 mM Hepes, 300 mM NaCl, 1% Triton X-100, and 1 µg/ml each of leupeptin, aprotinin, and pepstatin). After sonicating, the extract was centrifuged at 18,000 rpm for 40 min. The supernatant was collected and incubated with cobalt agarose for 2 h at 4°C. After washing the beads with washing buffer (20 mM Hepes, 500 mM NaCl, 1% Triton X-100, 5 mM imidazole, and 1 µg/ml each of leupeptin, aprotinin, and pepstatin), the bound protein was eluted with lysis buffer containing 200 mM imidazole.
Coimmunoprecipitation and Western blotting
For whole-cell extracts, the cells were solubilized in NETN lysis buffer supplemented with 50 U/µl Benzonase, protease inhibitors, and phosphatase inhibitors. After removal of cell debris by centrifugation, the soluble fractions were collected. For Flag immunoprecipitations, a 0.8-ml aliquot of lysate was incubated with 1 µg of the Flag monoclonal antibody and 25 µl of a 1:1 slurry of protein A–Sepharose for 2 h at 4°C. For endogenous immunoprecipitations, 1 mg of the whole-cell extract was incubated with 25 µl of a 1:1 slurry of protein A–Sepharose coupled with 2 µl of the indicated antibodies for 2 h at 4°C. The Sepharose beads were washed three times with NTEN buffer, boiled in 2× SDS loading buffer, and resolved on SDS-PAGE. Membranes were blocked in 5% milk in TBST (TBS with Tween) buffer and then probed with antibodies as indicated. To detect endogenous interaction between PCNA and RAD18, coimmunoprecipitation was conducted as described previously with modification (Kannouche et al., 2004). HeLa cells treated with the indicated siRNA were left untreated or treated with 50 J/m2 UV and recovered for 1 h. Cells were then rinsed twice in cold PBS, incubated for 5 min on ice with gentle shaking in buffer A (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM Pipes, pH 6.8, 1 mM EGTA, 0.2% Triton X-100, and protease inhibitors). After Triton X-100 extraction, the adhering cellular material was fixed by shaking with 1% formaldehyde in PBS for 10 min at 4°C. Glycine was added to a final concentration of 0.125 M for 5 min to quench the cross-linking. Cellular material were then washed twice with cold PBS, harvested, centrifuged, and dissolved in 200 µl buffer B (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 0.1% SDS). After 10-min incubation on ice, the samples were sonicated to shear the DNA to 200–1,500 bp and diluted five times with dilution buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM EDTA, and 0.2% Triton X-100). Redissolved proteins were incubated with monoclonal mouse anti-PCNA antibody overnight at 4°C and then with 30 µl of protein A–Sepharose. 2 h later, the beads were washed three times with dilution buffer and boiled in 2× SDS loading buffer, and the bound proteins were analyzed by electrophoresis and immunoblotting.
RNAi
All siRNAs were synthesized by Thermo Fisher Scientific. The siRNAs were 21 bp, and sequences are as follows: SIVA1 siRNA#1, 5′-CCCUGUGUGGCCUCGUGGAdTdT-3′; SIVA1 siRNA#2, 5′-GGCGGUCUGCGGUCAGUGUdTdT-3′; RAD18 siRNA, 5′-ACUCAGUGUCCAACUUGCUdTdT-3′; USP1 siRNA, 5′-GAAAUACACAGCCAAGUAAUU-3′; and control siRNA, 5′-UUCAAUAAAUUCUUGAGGUUU-3′. The siRNA-resistant wild-type and mutant SIVA1 constructs were generated by changing six nucleotides in the SIVA1 siRNA#1-targeting region (C453T, G456A, T459C, C462T, C465A, and G468A substitutions). The siRNA transfection was performed with 100 nM siRNA duplexes using Lipofectamine RNAiMAX (Invitrogen) following the manufacturer’s instructions. Transfection was repeated twice with an interval of 24 h to achieve the maximal RNAi effect. Lentiviral nonsilencing control shRNA and shRNA target sets were purchased from Thermo Fisher Scientific. The PCNA targeting sequences are #1, 5′-AGACAAGTAATGTCGATAA-3′, and #2, 5′-TCAGTATGTCTGCAGATGT-3′. The shRNAs were packaged into lentiviruses by cotransfecting with packaging plasmids pMD2G and pSPAX2 (provided by S. Zhou, Baylor College of Medicine, Houston, TX) into HEK293T cells. 48 h after transfection, the supernatant was collected for infection of HeLa cells. Infection was repeated twice with an interval of 24 h to achieve maximal infection efficiency.
Immunofluorescence staining
Indirect immunofluorescence was performed as previously described (Huang et al., 2009; Liu et al., 2010; Wan et al., 2013). HeLa or XP30RO (gift from C. Guo, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing, China) cells cultured on coverslips were treated with 50 J/m2 UV and recovered for the indicated times. Cells were then washed with PBS, preextracted with buffer containing 0.5% Triton X-100 for 5 min, and fixed with 3% paraformaldehyde for 10 min at room temperature. For PCNA and SIVA1 coimmunofluorescence staining, HeLa cells were denatured by 2.5 M HCl for 1 h at room temperature after 3% paraformaldehyde fixation. Cells were then incubated in primary antibody for 30 min at room temperature. After three 5-min washes with PBS, secondary antibody was added at room temperature for 30 min. Cells were then stained with DAPI to visualize nuclear DNA. Images were captured with use of a fluorescence microscope (Eclipse 80i; Nikon) equipped with a Plan Fluor 60× oil objective lens (NA 0.5–1.25; Nikon) and a camera (CoolSNAP HQ2; Photometrics). Images were captured using NIS-Elements basic research imaging software (Nikon) and analyzed using Photoshop CS3 (Adobe).
Mutation frequency
HEK293T cells were transfected twice with control siRNA or siRNAs targeting SIVA1. 24 h after the second transfection, cells were introduced with the SupF tRNA gene containing shuttle plasmid pZ189 irradiated with 300 J/m2 UV. 48 h later, the plasmid was extracted from cells using a purification kit (AxyPrep Plasmid Miniprep; Axygen). To exclude the methylated template, the purified plasmid was digested with DpnI, and then, E. coli strain MBM7070 was transformed with the processed plasmid and selected for 100 µg/ml ampicillin resistance on Luria–Bertani plates containing 100 µg/ml X-gal and 1 mM IPTG. In Fig. 4 G, white (mutant) and blue (wild type) colonies were counted, and the mutation frequency was calculated as the ratio of white colonies to total colonies. The pZ189 plasmid and MBM7070 strain were gifts from J. Shao (Zhejiang University, Hangzhou, China).
Cell survival assays
HeLa cells were transfected twice with control siRNA or siRNAs specifically targeting SIVA1 or RAD18. 24 h after transfection, 103 cells were split and transferred into 60-mm dishes. Cells were incubated for 24 h before they were treated with UV, MMC, or HU as indicated. The medium was replaced 24 h later, and cells were then incubated for 14 d. Resulting colonies were fixed and stained with Coomassie blue.
Chromatin fractionation
Preparation of chromatin fractions was as described previously (Huang et al., 2009; Liu et al., 2010) with modifications. In brief, 1 or 4 h after treatment with 50 J/m2 UV, cells were collected and washed with PBS. Cell pellets were subsequently resuspended in NETN buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40) and incubated on ice for 15 min. Nuclei were then recovered and resuspended in 0.2 M HCl. The soluble fraction was neutralized with 1 M Tris-HCl, pH 8.0, for further analysis.
Lentivirus packaging and infection
Tet-On–inducible SFB-tagged lentiviral vector and packaging plasmids (pMD2G and pSPAX2) were provided by S. Zhou (Baylor College of Medicine, Houston, TX). Polη entry constructs were transferred into the Gateway-compatible SFB-tagged lentiviral vector. Virus supernatant was collected 48 h after the cotransfection of lentiviral vectors and packaging plasmids (pMD2G and pSPAX2) into HEK293T cells. XP30RO cells were infected with viral supernatants with the addition of 8 µg/ml polybrene (Sigma-Aldrich), and stable pools were selected with medium containing 500 µg/ml G418 (EMD Millipore). The expression of the indicated genes in the stable pools was induced by the addition of 1 µg/ml doxycycline (Sigma-Aldrich) for 24 h for the experiments presented in this study.
Retrovirus production and infection
pDONR201 derivative constructs containing SIVA1 or Polη were transferred into a Gateway-compatible pEF1A-HA or pEF1A-HA-Flag retroviral vector, respectively. Virus supernatant was collected 48 h after the cotransfection of retroviral vectors and pCL-Eco and VSV-G into HEK293T cells. HeLa cells were infected with viral supernatant in the presence of 8 µg/ml polybrene (Sigma-Aldrich) and were then selected in growth medium containing 2 µg/ml puromycin (EMD Millipore).
In vitro ubiquitination assay
A standard in vitro ubiquitination reaction was performed at a final volume of 10 µl including ubiquitin at 50 µM, Uba1 at 50 nM, RAD6B at 0.2 µM, RAD18 at 0.2 µM, PCNA at 0.4 µM, and SIVA1 at 0.5 µM or 1 µM. The assay was performed in reaction buffer containing 25 mM Tris, pH 8.0, 50 mM NaCl, and 0.1 mM DTT. The reactions were activated with 2 mM ATP and incubated at 30°C for 1 h. Proteins were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
BrdU incorporation assays
HeLa cells were transfected twice with the indicated siRNAs for an internal 24 h. 48 h after the second transfection, 100 µM BrdU was added into the medium for 1 h. Cells were then harvested, washed with PBS, and fixed with ice-cold 70% ethanol overnight. Once centrifuged, cells were washed with PBS. DNA was denatured with 2.5 M HCl for 1 h at room temperature. After washing three times with PBS, cells were incubated in mouse anti-BrdU antibody (Roche) diluted 1:100 in blocking buffer (PBS + 0.1% Triton X-100 + 5% BSA) for 12 h followed by 3× wash with blocking buffer containing 500 mM NaCl. FITC-conjugated goat anti–mouse IgG (1:100; Jackson ImmunoResearch Laboratories, Inc.) was added and incubated for 4 h. After washing with blocking buffer containing 500 mM NaCl, cells were resuspended in PBS containing 20 µg/ml propidium iodide and 200 µg/ml RNase A at 37°C for 20 min. Cell cycle distribution was analyzed on a flow cytometer (FACScan; Beckman Coulter).
Quantitative RT-PCR
Total RNA was extracted using the GenElute Mammalian Total RNA Miniprep kit (Sigma-Aldrich). Purified RNA was then reverse transcribed to cDNA by using SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). Quantitative PCR was performed on an RT-PCR system (7500; Applied Biosystems) using the Power SYBR Green PCR Master Mix detection system (Applied Biosystems) with the following program: 50°C for 2 min, 95°C for 10 min, 95°C for 15 s, and 60°C for 1 min for 40 cycles. GAPDH was chosen as an internal control for the normalization of the total cDNA. The primers used were as follows: GAPDH forward, 5′-CGACCACTTTGTCAAGCTCA-3′, and reverse, 5′-TTACTCCTTGGAGGCCATGT-3′; SIVA1 forward, 5′-CAGATGCTGATTGGACCAGA-3′, and reverse, 5′-CATGAGGAACAGGCAATGG-3′; and PCNA forward, 5′-TTGCACTGAGGTACCTGAACTT-3′; and reverse, 5′-TGTCCCATATCCGCAATTTT-3′.
Online supplemental material
Fig. S1 shows that SIVA1 interacts with PCNA and demonstrates the specificity of the anti-SIVA1 antibody. Fig. S2 shows that SIVA1 depletion affects Polη foci formation. Fig. S3 shows that SIVA1 depletion does not markedly affect cell proliferation. Fig. S4 shows that residues 331–375 of RAD18 are responsible for SIVA1 binding. Fig. S5 shows that SIVA1-depleted cells complemented with wild type or Δ2 mutant of SIVA1 have comparable cell cycle profiles. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201311007/DC1.
Supplementary Material
Acknowledgments
We thank all our colleagues in the Huang laboratory for insightful discussions, Drs. Lee Zou and Titia Sixma for helpful comments, Dr. Caixia Guo for the XP30RO cell line, and Drs. Zongping Xia and Sheng Ye for technical help.
This work was supported in part by the National Program for Special Support of Eminent Professionals, National Basic Research Program of China grants 2012CB944402 and 2013CB911003, National Natural Science Funds for Distinguished Young Scholars, National Natural Science Foundation of China grant 31071243, Natural Science Foundation of Zhejiang Province grant R2110569, Zhejiang University K.P. Chao’s High Technology Development Foundation, and China’s Fundamental Research Funds for the Central Universities. T. Liu is a member of Feng laboratory and supported by National Natural Science Foundation of China grants 31171347 and 31090360 and the Ministry of Science and Technology grants 2012CB966600 and 2013CB945303.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- HU
- hydroxyurea
- MBP
- maltose-binding protein
- MMC
- mitomycin C
- PCNA
- proliferating cell nuclear antigen
- PIP
- PCNA-interacting peptide
- RFC
- replication factor C
- SUMO
- small ubiquitin-like modifier
- TLS
- translesion DNA synthesis
References
- Akasaka S., Takimoto K., Yamamoto K. 1992. G:C—>T:A and G:C—>C:G transversions are the predominant spontaneous mutations in the Escherichia coli supF gene: an improved lacZ(am) E. coli host designed for assaying pZ189 supF mutational specificity. Mol. Gen. Genet. 235:173–178 10.1007/BF00279358 [DOI] [PubMed] [Google Scholar]
- Bailly V., Prakash S., Prakash L. 1997. Domains required for dimerization of yeast Rad6 ubiquitin-conjugating enzyme and Rad18 DNA binding protein. Mol. Cell. Biol. 17:4536–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Slater D.M., Ohmori H., Vaziri C. 2005. DNA polymerase kappa is specifically required for recovery from the benzo[a]pyrene-dihydrodiol epoxide (BPDE)-induced S-phase checkpoint. J. Biol. Chem. 280:22343–22355 10.1074/jbc.M501562200 [DOI] [PubMed] [Google Scholar]
- Bi X., Barkley L.R., Slater D.M., Tateishi S., Yamaizumi M., Ohmori H., Vaziri C. 2006. Rad18 regulates DNA polymerase kappa and is required for recovery from S-phase checkpoint-mediated arrest. Mol. Cell. Biol. 26:3527–3540 10.1128/MCB.26.9.3527-3540.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M., Green C.M., Crosetto N., Rudolf F., Zapart G., Coull B., Kannouche P., Wider G., Peter M., Lehmann A.R., et al. 2005. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 310:1821–1824 10.1126/science.1120615 [DOI] [PubMed] [Google Scholar]
- Bienko M., Green C.M., Sabbioneda S., Crosetto N., Matic I., Hibbert R.G., Begovic T., Niimi A., Mann M., Lehmann A.R., Dikic I. 2010. Regulation of translesion synthesis DNA polymerase eta by monoubiquitination. Mol. Cell. 37:396–407 10.1016/j.molcel.2009.12.039 [DOI] [PubMed] [Google Scholar]
- Callegari A.J., Clark E., Pneuman A., Kelly T.J. 2010. Postreplication gaps at UV lesions are signals for checkpoint activation. Proc. Natl. Acad. Sci. USA. 107:8219–8224 10.1073/pnas.1003449107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centore R.C., Yazinski S.A., Tse A., Zou L. 2012. Spartan/C1orf124, a reader of PCNA ubiquitylation and a regulator of UV-induced DNA damage response. Mol. Cell. 46:625–635 10.1016/j.molcel.2012.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A., Elledge S.J. 2010. The DNA damage response: making it safe to play with knives. Mol. Cell. 40:179–204 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M.A., Kowal P., Yang K., Haas W., Huang T.T., Gygi S.P., D’Andrea A.D. 2007. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell. 28:786–797 10.1016/j.molcel.2007.09.031 [DOI] [PubMed] [Google Scholar]
- Davies A.A., Huttner D., Daigaku Y., Chen S., Ulrich H.D. 2008. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol. Cell. 29:625–636 10.1016/j.molcel.2007.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E.J., Lachaud C., Appleton P., Macartney T.J., Näthke I., Rouse J. 2012. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat. Struct. Mol. Biol. 19:1093–1100 10.1038/nsmb.2394 [DOI] [PubMed] [Google Scholar]
- Du W., Jiang P., Li N., Mei Y., Wang X., Wen L., Yang X., Wu M. 2009. Suppression of p53 activity by Siva1. Cell Death Differ. 16:1493–1504 10.1038/cdd.2009.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durando M., Tateishi S., Vaziri C. 2013. A non-catalytic role of DNA polymerase η in recruiting Rad18 and promoting PCNA monoubiquitination at stalled replication forks. Nucleic Acids Res. 41:3079–3093 10.1093/nar/gkt016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele M.J., Ciccia A., Elia A.E., Elledge S.J. 2011. Proliferating cell nuclear antigen (PCNA)-associated KIAA0101/PAF15 protein is a cell cycle-regulated anaphase-promoting complex/cyclosome substrate. Proc. Natl. Acad. Sci. USA. 108:9845–9850 10.1073/pnas.1106136108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E.C. 2005. Suffering in silence: the tolerance of DNA damage. Nat. Rev. Mol. Cell Biol. 6:943–953 10.1038/nrm1781 [DOI] [PubMed] [Google Scholar]
- Garg P., Burgers P.M. 2005. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1. Proc. Natl. Acad. Sci. USA. 102:18361–18366 10.1073/pnas.0505949102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal G., Leung J.W., Nair B.C., Fong K.W., Chen J. 2012. Proliferating cell nuclear antigen (PCNA)-binding protein C1orf124 is a regulator of translesion synthesis. J. Biol. Chem. 287:34225–34233 10.1074/jbc.M112.400135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Johnson R.E., Unk I., Phillips B., Hurwitz J., Prakash L., Prakash S. 2001a. Physical and functional interactions of human DNA polymerase eta with PCNA. Mol. Cell. Biol. 21:7199–7206 10.1128/MCB.21.21.7199-7206.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Johnson R.E., Unk I., Phillips B.B., Hurwitz J., Prakash L., Prakash S. 2001b. Targeting of human DNA polymerase iota to the replication machinery via interaction with PCNA. Proc. Natl. Acad. Sci. USA. 98:14256–14261 10.1073/pnas.261560798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Unk I., Johnson R.E., Phillips B.B., Hurwitz J., Prakash L., Prakash S. 2002. Stimulation of DNA synthesis activity of human DNA polymerase kappa by PCNA. Mol. Cell. Biol. 22:784–791 10.1128/MCB.22.3.784-791.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Unk I., Prakash L., Prakash S. 2006. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proc. Natl. Acad. Sci. USA. 103:6477–6482 10.1073/pnas.0510924103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbert R.G., Huang A., Boelens R., Sixma T.K. 2011. E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc. Natl. Acad. Sci. USA. 108:5590–5595 10.1073/pnas.1017516108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 419:135–141 10.1038/nature00991 [DOI] [PubMed] [Google Scholar]
- Huang J., Gong Z., Ghosal G., Chen J. 2009. SOSS complexes participate in the maintenance of genomic stability. Mol. Cell. 35:384–393 10.1016/j.molcel.2009.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T.T., Nijman S.M., Mirchandani K.D., Galardy P.J., Cohn M.A., Haas W., Gygi S.P., Ploegh H.L., Bernards R., D’Andrea A.D. 2006. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 8:339–347 10.1038/ncb1378 [DOI] [PubMed] [Google Scholar]
- Iorio-Morin C., Germain P., Roy S., Génier S., Labrecque P., Parent J.L. 2012. Thromboxane A2 modulates cisplatin-induced apoptosis through a Siva1-dependent mechanism. Cell Death Differ. 19:1347–1357 10.1038/cdd.2012.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.P., Bartek J. 2009. The DNA-damage response in human biology and disease. Nature. 461:1071–1078 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.E., Kondratick C.M., Prakash S., Prakash L. 1999. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 285:263–265 10.1126/science.285.5425.263 [DOI] [PubMed] [Google Scholar]
- Juhasz S., Balogh D., Hajdu I., Burkovics P., Villamil M.A., Zhuang Z., Haracska L. 2012. Characterization of human Spartan/C1orf124, an ubiquitin-PCNA interacting regulator of DNA damage tolerance. Nucleic Acids Res. 40:10795–10808 10.1093/nar/gks850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche P.L., Wing J., Lehmann A.R. 2004. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 14:491–500 10.1016/S1097-2765(04)00259-X [DOI] [PubMed] [Google Scholar]
- Kim J.M., Parmar K., Huang M., Weinstock D.M., Ruit C.A., Kutok J.L., D’Andrea A.D. 2009. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev. Cell. 16:314–320 10.1016/j.devcel.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.Y., Yang K., Cohn M.A., Sikdar N., D’Andrea A.D., Myung K. 2010. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through Its interactions with PCNA and USP1. J. Biol. Chem. 285:10362–10369 10.1074/jbc.M109.092544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A.R. 2011. Ubiquitin-family modifications in the replication of DNA damage. FEBS Lett. 585:2772–2779 10.1016/j.febslet.2011.06.005 [DOI] [PubMed] [Google Scholar]
- Lehmann A.R., Kirk-Bell S., Arlett C.F., Paterson M.C., Lohman P.H., de Weerd-Kastelein E.A., Bootsma D. 1975. Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proc. Natl. Acad. Sci. USA. 72:219–223 10.1073/pnas.72.1.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A.R., Niimi A., Ogi T., Brown S., Sabbioneda S., Wing J.F., Kannouche P.L., Green C.M. 2007. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst.). 6:891–899 10.1016/j.dnarep.2007.02.003 [DOI] [PubMed] [Google Scholar]
- Liu T., Ghosal G., Yuan J., Chen J., Huang J. 2010. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 329:693–696 10.1126/science.1192656 [DOI] [PubMed] [Google Scholar]
- Lopes M., Foiani M., Sogo J.M. 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 21:15–27 10.1016/j.molcel.2005.11.015 [DOI] [PubMed] [Google Scholar]
- Machida Y., Kim M.S., Machida Y.J. 2012. Spartan/C1orf124 is important to prevent UV-induced mutagenesis. Cell Cycle. 11:3395–3402 10.4161/cc.21694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher V.M., Ouellette L.M., Curren R.D., McCormick J.J. 1976. Frequency of ultraviolet light-induced mutations is higher in xeroderma pigmentosum variant cells than in normal human cells. Nature. 261:593–595 10.1038/261593a0 [DOI] [PubMed] [Google Scholar]
- Masutani C., Araki M., Yamada A., Kusumoto R., Nogimori T., Maekawa T., Iwai S., Hanaoka F. 1999a. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 18:3491–3501 10.1093/emboj/18.12.3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C., Kusumoto R., Yamada A., Dohmae N., Yokoi M., Yuasa M., Araki M., Iwai S., Takio K., Hanaoka F. 1999b. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 399:700–704 10.1038/21447 [DOI] [PubMed] [Google Scholar]
- McCulloch S.D., Kokoska R.J., Masutani C., Iwai S., Hanaoka F., Kunkel T.A. 2004. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 428:97–100 10.1038/nature02352 [DOI] [PubMed] [Google Scholar]
- Moldovan G.L., Pfander B., Jentsch S. 2007. PCNA, the maestro of the replication fork. Cell. 129:665–679 10.1016/j.cell.2007.05.003 [DOI] [PubMed] [Google Scholar]
- Mosbech A., Gibbs-Seymour I., Kagias K., Thorslund T., Beli P., Povlsen L., Nielsen S.V., Smedegaard S., Sedgwick G., Lukas C., et al. 2012. DVC1 (C1orf124) is a DNA damage-targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nat. Struct. Mol. Biol. 19:1084–1092 10.1038/nsmb.2395 [DOI] [PubMed] [Google Scholar]
- Ohmori H., Friedberg E.C., Fuchs R.P., Goodman M.F., Hanaoka F., Hinkle D., Kunkel T.A., Lawrence C.W., Livneh Z., Nohmi T., et al. 2001. The Y-family of DNA polymerases. Mol. Cell. 8:7–8 10.1016/S1097-2765(01)00278-7 [DOI] [PubMed] [Google Scholar]
- Parnas O., Zipin-Roitman A., Pfander B., Liefshitz B., Mazor Y., Ben-Aroya S., Jentsch S., Kupiec M. 2010. Elg1, an alternative subunit of the RFC clamp loader, preferentially interacts with SUMOylated PCNA. EMBO J. 29:2611–2622 10.1038/emboj.2010.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plosky B.S., Vidal A.E., Fernández de Henestrosa A.R., McLenigan M.P., McDonald J.P., Mead S., Woodgate R. 2006. Controlling the subcellular localization of DNA polymerases iota and eta via interactions with ubiquitin. EMBO J. 25:2847–2855 10.1038/sj.emboj.7601178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povlsen L.K., Beli P., Wagner S.A., Poulsen S.L., Sylvestersen K.B., Poulsen J.W., Nielsen M.L., Bekker-Jensen S., Mailand N., Choudhary C. 2012. Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nat. Cell Biol. 14:1089–1098 10.1038/ncb2579 [DOI] [PubMed] [Google Scholar]
- Prakash S., Johnson R.E., Prakash L. 2005. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74:317–353 10.1146/annurev.biochem.74.082803.133250 [DOI] [PubMed] [Google Scholar]
- Prasad K.V., Ao Z., Yoon Y., Wu M.X., Rizk M., Jacquot S., Schlossman S.F. 1997. CD27, a member of the tumor necrosis factor receptor family, induces apoptosis and binds to Siva, a proapoptotic protein. Proc. Natl. Acad. Sci. USA. 94:6346–6351 10.1073/pnas.94.12.6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch U., Schichl Y.M., Winsauer G., Gudi R., Prasad K., de Martin R. 2009. Siva1 is a XIAP-interacting protein that balances NFκB and JNK signalling to promote apoptosis. J. Cell Sci. 122:2651–2661 10.1242/jcs.049940 [DOI] [PubMed] [Google Scholar]
- Sale J.E., Lehmann A.R., Woodgate R. 2012. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 13:141–152 10.1038/nrm3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz V., Janel-Bintz R., Wagner J., Biard D., Shiomi N., Fuchs R.P., Cordonnier A.M. 2010. Role of the ubiquitin-binding domain of Polη in Rad18-independent translesion DNA synthesis in human cell extracts. Nucleic Acids Res. 38:6456–6465 10.1093/nar/gkq403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P., Ulrich H.D. 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 425:188–191 10.1038/nature01965 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yoshida N., Murakami N., Kawata K., Ishizaki H., Tanaka-Okamoto M., Miyoshi J., Zinn A.R., Shime H., Inoue N. 2007. Dynamic regulation of p53 subnuclear localization and senescence by MORC3. Mol. Biol. Cell. 18:1701–1709 10.1091/mbc.E06-08-0747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai K., Abbas T., Jazaeri A.A., Dutta A. 2010. CRL4(Cdt2) E3 ubiquitin ligase monoubiquitinates PCNA to promote translesion DNA synthesis. Mol. Cell. 37:143–149 10.1016/j.molcel.2009.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unk I., Hajdú I., Fátyol K., Szakál B., Blastyák A., Bermudez V., Hurwitz J., Prakash L., Prakash S., Haracska L. 2006. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc. Natl. Acad. Sci. USA. 103:18107–18112 10.1073/pnas.0608595103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unk I., Hajdú I., Fátyol K., Hurwitz J., Yoon J.H., Prakash L., Prakash S., Haracska L. 2008. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc. Natl. Acad. Sci. USA. 105:3768–3773 10.1073/pnas.0800563105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L., Han J., Liu T., Dong S., Xie F., Chen H., Huang J. 2013. Scaffolding protein SPIDR/KIAA0146 connects the Bloom syndrome helicase with homologous recombination repair. Proc. Natl. Acad. Sci. USA. 110:10646–10651 10.1073/pnas.1220921110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zha M., Zhao X., Jiang P., Du W., Tam A.Y., Mei Y., Wu M. 2013. Siva1 inhibits p53 function by acting as an ARF E3 ubiquitin ligase. Nat Commun. 4:1551 10.1038/ncomms2533 [DOI] [PubMed] [Google Scholar]
- Watanabe K., Tateishi S., Kawasuji M., Tsurimoto T., Inoue H., Yamaizumi M. 2004. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 23:3886–3896 10.1038/sj.emboj.7600383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L., Chu F., Cheng Y., Sun X., Borthakur A., Ramarao M., Pandey P., Wu M., Schlossman S.F., Prasad K.V. 2002. Siva-1 binds to and inhibits BCL-X(L)-mediated protection against UV radiation-induced apoptosis. Proc. Natl. Acad. Sci. USA. 99:6925–6930 10.1073/pnas.102182299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagihara H., Kobayashi J., Tateishi S., Kato A., Matsuura S., Tauchi H., Yamada K., Takezawa J., Sugasawa K., Masutani C., et al. 2011. NBS1 recruits RAD18 via a RAD6-like domain and regulates Pol η-dependent translesion DNA synthesis. Mol. Cell. 43:788–797 10.1016/j.molcel.2011.07.026 [DOI] [PubMed] [Google Scholar]
- Yang K., Moldovan G.L., Vinciguerra P., Murai J., Takeda S., D’Andrea A.D. 2011. Regulation of the Fanconi anemia pathway by a SUMO-like delivery network. Genes Dev. 25:1847–1858 10.1101/gad.17020911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.H., Shiotani B., Classon M., Zou L. 2008. Chk1 and Claspin potentiate PCNA ubiquitination. Genes Dev. 22:1147–1152 10.1101/gad.1632808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Durando M., Smith-Roe S.L., Sproul C., Greenwalt A.M., Kaufmann W., Oh S., Hendrickson E.A., Vaziri C. 2013. Cell cycle stage-specific roles of Rad18 in tolerance and repair of oxidative DNA damage. Nucleic Acids Res. 41:2296–2312 10.1093/nar/gks1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.