ERM activation by SLK kinase promotes polarized association at the mitotic cortex of LGN and NuMA, a necessary step in proper spindle orientation.
Abstract
Mitotic spindle orientation relies on a complex dialog between the spindle microtubules and the cell cortex, in which F-actin has been recently implicated. Here, we report that the membrane–actin linkers ezrin/radixin/moesin (ERMs) are strongly and directly activated by the Ste20-like kinase at mitotic entry in mammalian cells. Using microfabricated adhesive substrates to control the axis of cell division, we found that the activation of ERMs plays a key role in guiding the orientation of the mitotic spindle. Accordingly, impairing ERM activation in apical progenitors of the mouse embryonic neocortex severely disturbed spindle orientation in vivo. At the molecular level, ERM activation promotes the polarized association at the mitotic cortex of leucine-glycine-asparagine repeat protein (LGN) and nuclear mitotic apparatus (NuMA) protein, two essential factors for spindle orientation. We propose that activated ERMs, together with Gαi, are critical for the correct localization of LGN–NuMA force generator complexes and hence for proper spindle orientation.
Introduction
Oriented cell divisions are crucial for embryogenesis and adult tissue homeostasis, and rely on the accurate control of spindle orientation during mitosis (Gonzalez, 2007; Knoblich, 2008; Minc and Piel, 2012). Spindle orientation depends on the polarized localization of force generator complexes linking the spindle microtubules to the cell cortex, notably the Gαi–leucine-glycine-asparagine repeat protein (LGN)–nuclear mitotic apparatus (NuMA) complex (Siller and Doe, 2009; Morin and Bellaïche, 2011). Intriguingly, it has also been shown that spindle orientation requires the integrity of cortical F-actin (Théry et al., 2005; Toyoshima and Nishida, 2007; Kunda and Baum, 2009; Fink et al., 2011; Luxenburg et al., 2011; Sandquist et al., 2011; Castanon et al., 2013). Thus deciphering the pathways involved in the organization of the mitotic F-actin cortex and their potential impact on force generators constitutes a major challenge to unravel the mechanisms governing oriented cell division.
Ezrin/radixin/moesin (ERM) proteins are key, regulated organizers of cortical F-actin–rich structures (Fehon et al., 2010). We and others previously reported that the sole ERM protein encoded in flies (dMoesin) is essential for maintaining cortical stability throughout mitosis and for spindle orientation in Drosophila melanogaster cells (Carreno et al., 2008; Kunda et al., 2008; Nakajima et al., 2013). However, mechanistically, it is not known whether rocking spindles observed upon dMoesin depletion resulted from the large cortical deformations associated with that depletion or from a more instructive role in properly localizing the force generator machinery. In mammalian cells, previous work reported mutant situations in which there is a correlation between a reduction in ERM activation and spindle orientation defects (Théry et al., 2005; Luxenburg et al., 2011). However, these situations correspond to either acute inhibition of the Src family tyrosine kinases or knockout of the broad range transcription factor Srf, leaving unclear whether ERM activation plays a specific role in spindle orientation.
Here, we report that the direct activation of the three mammalian ERMs by the Ste20-like kinase (SLK) is crucial for guiding the mitotic spindle toward the expected orientation in two mammalian models of oriented cell division: micropatterned cells and apical progenitors of the mouse neocortex. Importantly, we found that proper localization of LGN and NuMA at the cortex depends on ERM activation, thereby providing molecular insights on the role of ERMs in spindle orientation.
Results and discussion
SLK directly phosphorylates mammalian ERMs and controls their cortical activation in mitosis
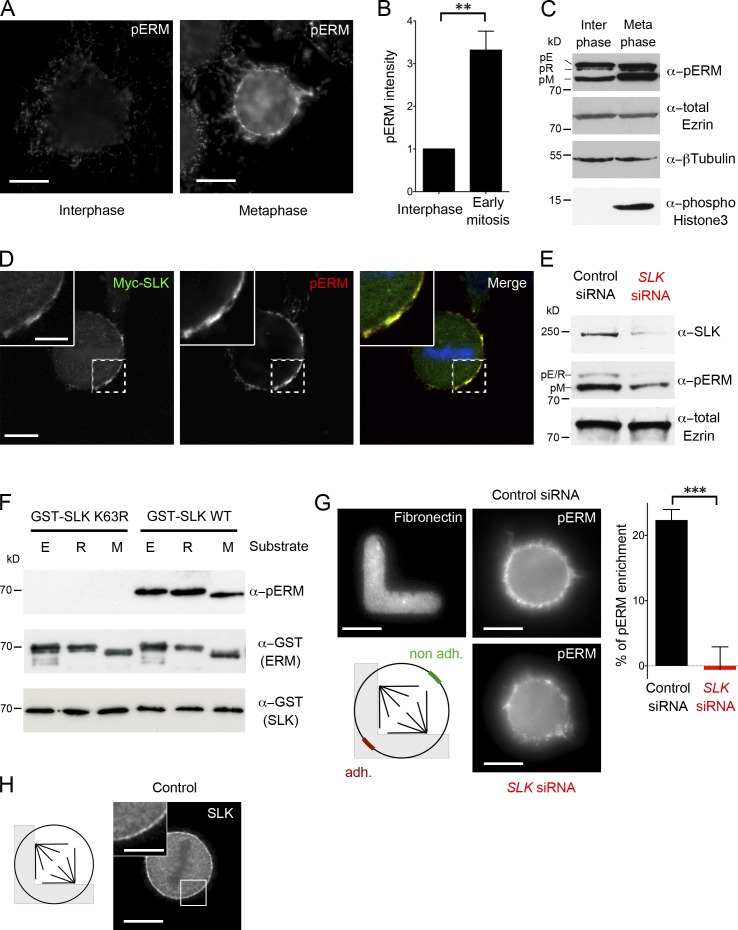
We first aimed to better characterize mammalian ERM activation through the cell cycle. Ezrin, radixin, and moesins are activated by phosphorylation at a conserved threonine residue (T567, T564, and T558, respectively; Matsui et al., 1998). Using an antibody that specifically detects this phosphorylation event (Fievet et al., 2004), we confirmed that activated ERMs (hereafter pERMs) predominantly localized at the metaphase cell cortex in HeLa cells (Fig. 1 A). We measured a threefold increase in pERM staining (Fig. 1 B), as well as increased activation of the three ERMs in metaphase, whereas total amounts of ERMs (e.g., total ezrin) remained stable (Fig. 1 C). Later, pERMs were found highly enriched in cleavage furrows (unpublished data), as previously reported (Kawano et al., 1999; Carreno et al., 2008; Kunda et al., 2008).
Figure 1.
SLK directly phosphorylates mammalian ERM proteins and controls their cortical activation in mitosis. (A) Staining of pERMs in interphase and metaphase HeLa cells (single plane, same settings). (B) FACS quantification of pERM levels (mean ± SEM; arbitrary units) in early mitosis (MPM2-positive cells) and interphase (MPM2-negative cells). n = 4; **, P < 2 × 10−3 (Student t test). (C) Western blot of total lysates from interphase and metaphase cells, using antibodies against pERMs, total ezrin, β-tubulin (loading control), and phospho-Histone3 (mitotic marker). (D) Confocal slice of a metaphase cell expressing Myc-SLK (green) and stained for pERMs (red). (E) Western blot of total lysates from metaphase cells treated with control siRNA (black) or SLK siRNA (red), using antibodies against SLK, pERMs, and total ezrin. (F) In vitro kinase assay using recombinant wild-type (WT) or catalytically dead (K63R) kinase domain of SLK (aa 1–344) and GST-ezrinC-ter, GST-radixinC-ter, or GST-moesinC-ter, as substrates, in the presence of ATP. ERM phosphorylation was detected by Western blot with pERM antibodies. (G) Staining of pERMs in mitotic cells plated on L-shaped micropatterns, after control or SLK depletion, as indicated. (top left) Fibronectin staining showing the micropattern shape. The bias [100 × (adh. − nonadh.)/(nonadh.)] of pERM staining at the cortex facing the adhesive versus nonadhesive substrate is presented on the right. Mean ± SEM; n > 85 cells; ***, P < 10−3 (Student t test). (H) Staining of endogenous SLK in a metaphase micropatterned cell. Bars: (main) 10 µm; (insets) 5 µm.
In vertebrates, the identity of the ERM activators has been a matter of debate (Fehon et al., 2010); however, recent evidence pointed out a role for kinases of the SLK family in interphase cells (Belkina et al., 2009; Viswanatha et al., 2012). In addition, we and others previously reported that ERM activation in mitosis depended on the dSlik kinase in D. melanogaster S2 cells (Hipfner et al., 2004; Carreno et al., 2008; Kunda et al., 2008). In the present study, we thus focused our attention on the localization and function of the human orthologue SLK. First, a Myc-tagged version of SLK localized at the metaphase cortex where it colocalized with pERMs in HeLa cells (Fig. 1 D). Importantly, upon siRNA-mediated depletion of SLK (Fig. S1 A), the phosphorylation of ERMs was reduced in metaphase cells (Fig. 1 E), which was confirmed quantitatively by FACS analysis (46.5 ± 5.8% reduction, n = 3 experiments). To investigate a direct involvement of SLK in ERM activation, we performed an in vitro kinase assay using recombinant proteins, which demonstrated that SLK could efficiently phosphorylate the three ERMs (Fig. 1 F). This phosphorylation was abolished when ATP was omitted (Fig. S1 B) or when a critical catalytical residue (K63) of SLK was mutated (Fig. 1 F).
Importantly, the localization of activated ERMs at the metaphase cell cortex was strongly reduced in SLK-depleted cells cultured either directly on glass (not depicted) or on fibronectin-coated micropatterns (Fig. 1 G). In control cells cultured on L-shaped micropatterns, activated ERM were 22 ± 3% more enriched in the part of the cortex facing the adhesive area of the micropattern (Fig.1 G), as previously reported (Théry et al., 2005). Interestingly, this bias was severely reduced after SLK depletion (Fig.1 G and Fig. S1 C), indicating that both the amount and the precise distribution of pERMs at the mitotic cortex depend on SLK. To determine whether this bias was caused by uneven localization of SLK, we examined its endogenous distribution and found that it is uniformly detected at the cortex (1.7 ± 0.5% enrichment, n = 120 cells; Fig. 1 H). Thus a possible explanation is that the activation of the kinase may be uneven, which would be consistent with previous work indicating that the activity of the kinase is under tight regulation: SLK activity has been shown to rise at the G2/M transition (O’Reilly et al., 2005), whereas we detected no change in protein levels (Fig. S1 D). Alternatively, SLK may have additional substrates that contribute to pERM polarization. Finally, phosphatidylinositol 4,5-bisphosphate levels and dephosphorylation events, which are also important determinants for ERM activation, could regulate pERM levels and distribution in mitosis in concert with SLK (Roubinet et al., 2011; Kunda et al., 2012). Altogether, our results indicate that SLK directly phosphorylates the three mammalian ERM proteins and is critical for ERM activation at the mitotic cell cortex. Furthermore, perturbing SLK expression constitutes an efficient means to impact ERM activation.
ERM activation is required for correct reorientation of the mitotic spindle
To test the role of ERM activation in oriented cell division, we used L-shaped fibronectin-coated micropatterns, which sets a predictable spindle orientation in the X-Y plane along a 45° angle (Théry et al., 2005; Fink et al., 2011; Fig. 2 A). This angle (hereafter “final angle”) was measured within 10 min after the metaphase–anaphase transition, and thus defined the orientation of the cell division axis. When micropatterned cells were recorded from the beginning of chromosome congression in prometaphase to anaphase onset, we observed that most spindles initially started from positions far from the optimal angle. However, spindles quickly rotated and reoriented toward the optimal angle within 15 min, and then remained close to this position (Fig. 2 B).
Figure 2.
ERM activation is required for guiding the mitotic spindle to its final correct orientation. (A) Control siRNA-treated Histone2B-expressing cells were cultured on L-shaped fibronectin micropattern and recorded by time-lapse microscopy. The distribution of the angle of cell division axis (α) at anaphase onset (black curve) is shown (mean ± SD on five independent experiments; n > 1,200 cells). Bar, 5 µm. (B) Evolution of the spindle angle as a function of time starting at late prometaphase, plotted for a dozen cells described in A. (C, E, and G) Same analysis as in A after SLK depletion (C, red curve), after simultaneous depletion of the three ERMs (E, blue curve), and after ezrin T567D overexpression (G, green curve). 500–1,000 cells from at least three independent experiments. In C and E, the black curve corresponds to cells transfected with control siRNA. In G, the gray curve corresponds to cells transfected with a control plasmid. In all cases, the angle distributions were different from controls, with P < 10−5 (Kolmogorov-Smirnov test). (D, F, and H) Same analysis as in B for cells described in C, E, and G, respectively.
In marked contrast to control cells, the number of cells with their spindles oriented at the final 45° optimal angle was severely reduced and the angle distribution was more widespread in SLK-depleted cells (Fig. 2 C and Fig. S1, A and E, for a second, independent siRNA). Remarkably, the initial distribution of spindle angles was as broad as in control cells (not depicted), but the process of spindle reorientation toward the optimal angle was compromised (Fig. 2 D).
Simultaneous depletion of the three ERMs resulted in very similar spindle orientation defects. The distribution of the final angle was close to the one measured in SLK-depleted cells, and spindles were unable to reorient toward the optimal angle (Fig. 2, E and F). Of note, ERM appeared to play redundant functions in spindle orientation, as single protein depletions did not lead to the aforementioned defects (Fig. S1 F).
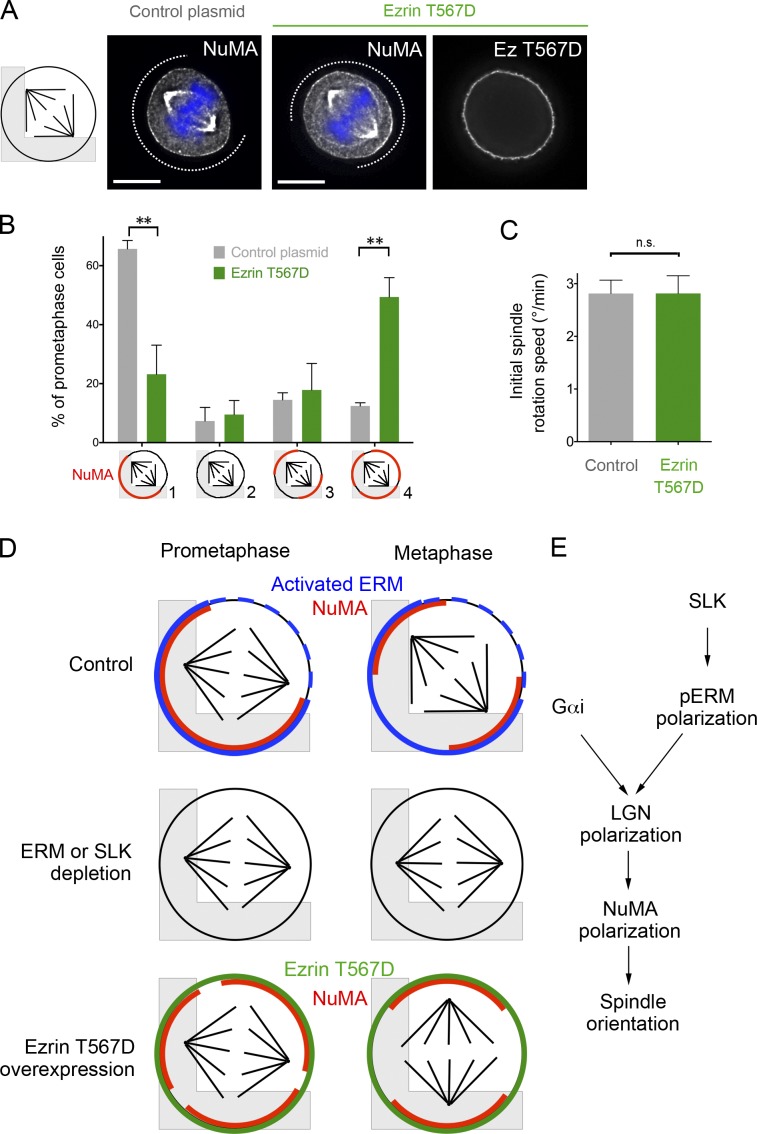
We next wondered whether constitutive ERM activation would also perturb spindle orientation. The hyperactive, phosphomimetic mutant of ezrin (T567D) uniformly localized to the mitotic cell cortex when overexpressed in micropatterned HeLa cells (see Fig. 5 A). As expected, the localization of this mutant did not require the presence of SLK (Fig. S1 G). Interestingly, as in SLK- or ERM-depleted cells, cells expressing ezrin T567D showed spindle orientation defects caused by a lack of reorientation (Fig. 2, G and H). This effect was specific to ezrin T567D, as overexpression of the wild-type or the nonphosphorylable ezrin T567A did not perturb spindle orientation (Fig. S1 H). Consistently, ezrin T567A did not change ERM activation or NuMA association to the cortex (unpublished data).
Figure 5.
ERM hyperactivation perturbs spindle orientation and NuMA localization, and a proposed model showing the role of activated ERMs in the polarized localization of NuMA and spindle orientation. (A) NuMA staining in prometaphase in control or ezrin T567D–expressing cells cultured on micropatterns. (B) Same quantification as in Fig. 4 D in control (gray) or ezrin T567D–expressing cells (green). (C) Same quantification as in Fig. 4 E in control or ezrin T567D–expressing cells. Phenotypic summary (D) and proposed model (E). Bars, 10 µm.
In contrast to D. melanogaster cells (Carreno et al., 2008; Kunda et al., 2008), mammalian cells depleted of ERMs or SLK did not display detectable spindle defects (Fig. S1 I), lack of cell rounding, or large cell shape changes (Fig. 2). However, the cortical F-actin was likely less associated with the plasma membrane, as occasional limited and transient blebs were detected in a fraction of cells depleted of ERMs (58 ± 6%) or SLK (40 ± 6% vs. 8 ± 4% in controls, n > 500). It is thus likely that additional cross-linkers play a role in cortical stability in mitotic mammalian cells. Importantly, we did not observe any correlation between the presence of blebs and the occurrence of spindle orientation defects. In addition, as observed in control cells, mitotic ERM- or SLK-depleted cells and ezrin T567D–expressing cells were connected to the fibronectin substrate through retraction fibers (Fig. S1 J), indicating normal contacts with the adhesive micropattern.
These data show that ERMs and the kinase controlling their activation play a key role in spindle orientation. Our results further indicate that proper levels and localization of activated ERMs are critical for the guidance of the spindle in an in vitro model of oriented cell division.
SLK depletion and ERM hyperactivation perturb spindle orientation in vivo
Apical progenitors of the embryonic mouse neocortex express ERMs (Gimeno et al., 2004) and constitute a well-established model of oriented cell division in vivo because spindles are oriented parallel to the apical ventricular surface (Konno et al., 2008; Fietz and Huttner, 2011; Morin and Bellaïche, 2011).
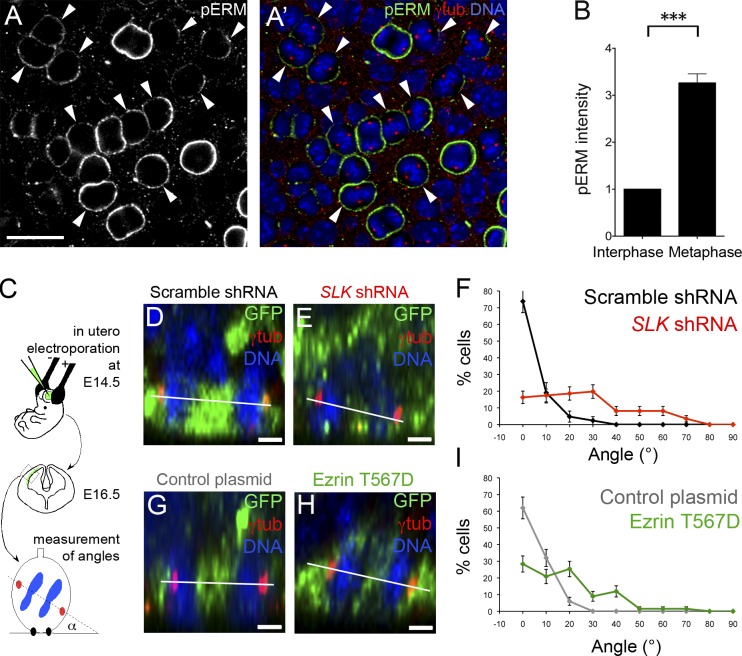
Using en face views of whole-mount neocortical vesicles from embryonic day 14.5 (E14.5) to E16.5 wild-type embryos, we found a strong activation of ERMs in mitotic apical progenitors (Fig. 3, A and A′). Quantifications of pERM immunostaining revealed a threefold increase at the cortex of metaphase cells (measured in the plane containing spindle poles), as compared with adjacent interphase cells (Fig. 3 B). To explore the role of ERM activation, we electroporated in utero control and SLK shRNA in the embryonic neocortex at E14.5 and analyzed cell division axis orientation at E16.5 (Fig. 3 C). In controls, the vast majority of spindles were parallel to the apical surface (Fig. 3, D and F), as expected (Konno et al., 2008). In contrast, Slk depletion led to a severe decrease in the proportion of spindles parallel to the ventricular surface and to a broadening of the angle distribution (Fig. 3, E and F; and Fig. S1 K, for a second, independent shRNA). Similarly, increasing ERM activation by the overexpression of the cortically localized ezrin T567D mutant (Fig. S1 L) led to profound misorientation of mitotic spindles (Fig. 3, G–I). Whether such defects could directly impact on brain organization or cell fate determination of post-mitotic neurons was not addressable because ERMs are additionally involved in cell migration (Belkina et al., 2009). Nevertheless, our results show that both ERM hyperactivation and depletion of the ERM activating kinase Slk strongly perturb the orientation of the cell division axis in mammalian neocortical progenitors in vivo.
Figure 3.
ERM activation is essential for proper spindle orientation in mouse apical progenitors. (A and A’) En face confocal view of apical progenitors at E16.5 stained for pERMs (green), γ-tubulin (red), and DAPI (blue). Focus has been made at the plane containing the spindle poles of mitotic progenitors. Arrowheads point to prometaphase and metaphase cells. Bar, 20 µm. (B) Quantification of cortical pERM intensity (mean ± SEM; arbitrary units) at the cortex in metaphase and adjacent interphase cells. n = 85 cells; ***, P < 10−5. (C) Schematic representation of the experimental procedure using in utero electroporation in mouse embryos at E14.5 and analyzed at E16.5. Transfected cells expressed GFP. (D, E, G, and H) Z-views obtained from en face confocal images of the cerebral neocortex electroporated with scramble (D) or Slk-shRNA–encoding plasmids (E) or with empty (G) or ezrin T567D–encoding vectors (H). The scramble shRNA panel (D) is shown again in Fig. S1 K. (F and I) Percentage of cells dividing at a specific angle α, using the measurement conventions indicated in C (mean ± SD; n = 50–85 cells from three to six independent embryos). The angle distributions were different from controls, with P < 10−5 (Kolmogorov-Smirnov test). Bars, 1 µm.
Localized LGN and NuMA association with the cell cortex relies on ERM activation
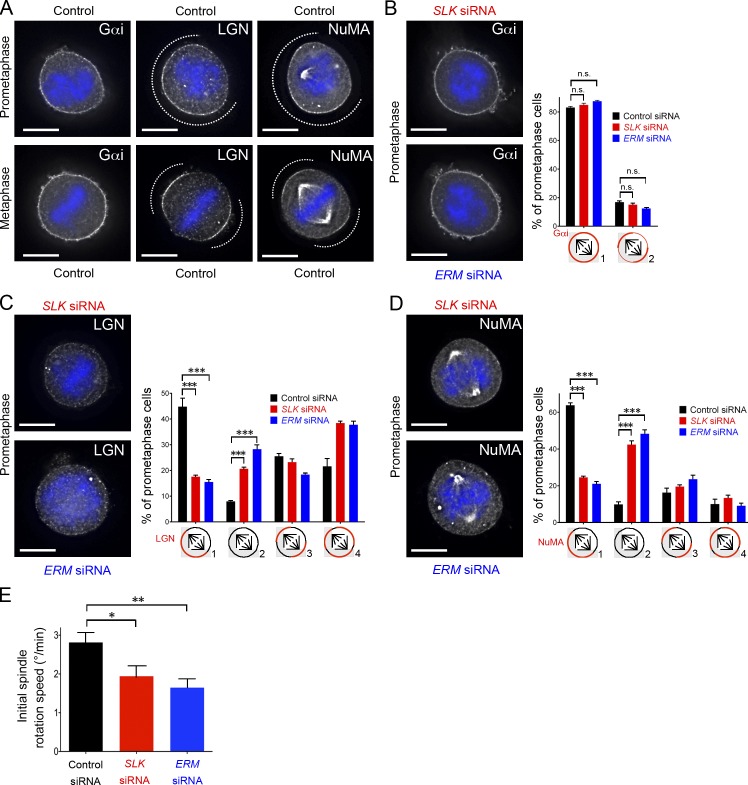
To explore how pERMs impact oriented cell divisions, we next investigated the endogenous localization of the Gαi–LGN–NuMA complex in control micropatterned cells. As in nonpatterned cells (Du and Macara, 2004; Woodard et al., 2010; Kiyomitsu and Cheeseman, 2012, 2013; Kotak et al., 2012), LGN and NuMA were found enriched as two “brackets” at the cortex facing the spindle poles in metaphase (Fig. 4 A, bottom). In addition, NuMA strongly associated with the spindle poles, as expected. In contrast, Gαi localized at the entire plasma membrane with a slight increased staining at the polar cortex (Fig. 4 A, bottom). Interestingly, in prometaphase, we observed that LGN and NuMA localized preferentially at the cortex facing the adhesive zone of the pattern, whereas Gαi already localized evenly at the plasma membrane (Fig. 4 A, top). This novel observation is of importance because it experimentally supports a theoretical model explaining spindle orientation in micropatterned cells. Indeed, it has been calculated that spindles would preferentially orient at the observed 45° angle if force generators are more enriched at the adhesive cortex (Théry et al., 2007). As expected, this biased localization of LGN–NuMA occurs in early mitosis, when spindle reorientation is maximal (Fig. 2 B).
Figure 4.
Localized LGN and NuMA association with the cell cortex relies on ERM activation. (A–D) Endogenous Gαi, LGN, and NuMA staining in prometaphase and metaphase cells cultured on L-shaped micropatterns. The micropatterns are oriented as in Fig. 1 G. Dotted lines indicate LGN–NuMA extent at the cortex. Metaphase corresponds to cells with fully congressed chromosomes and promometaphase corresponds to cells after nuclear envelope breakdown but without fully congressed chromosomes. DAPI staining is displayed in blue. The three prometaphase control panels are shown again in Fig. S2 A, and the NuMA metaphase panel is shown again in Fig. S2 B. Endogenous Gαi (B), LGN (C), and NuMA (D) staining in prometaphase micropatterned cells after SLK or ERM depletion. For each marker, the percentage of prometaphase cells (mean ± SEM) displaying the indicated localization (categories 1–4) is quantified after control (black), SLK (red), and ERM (blue) depletion. n > 50 cells, n = 2 (B); n > 125 cells, n = 3 (C); n = >150 cells, n = 6 (D); ***, P < 3 × 10−4 (Student t test). (E) Initial speed of spindle rotation (degree/min) in the first 5 min of chromosome congression, after control (black), SLK (red), and ERM (blue) depletion. Mean ± SEM; n > 60 cells; n = 3. n.s., not significant; *, P < 0.05; **, P < 2 × 10−2 (Student t test). Bars, 10 µm.
We then tested the influence of ERM activation on LGN–NuMA polarization in prometaphase, as a similar bias in pERM localization had also been observed (Fig. 1 G). Although depletion of either SLK or the three ERMs did not change the uniform membrane association of Gαi (Fig. 4 B), this treatment strongly impaired LGN–NuMA polarization (Fig. 4, C and D, category 1; and Fig. S2 A, bottom right) and markedly increased the number of cells without LGN–NuMA at the cortex (Fig. 4, C and D, category 2; and Fig. S2 A for second siRNA). Remarkably, the cortical reduction of the force generator complex LGN–NuMA in prometaphase cells depleted for SLK or ERMs correlated with a reduction of the speed of spindle rotation, as compared with controls (Fig. 4 E). LGN and NuMA association with the cortex was also reduced in SLK- and ERM-depleted cells later in metaphase (not depicted and Fig. S2 B). Of note, depletion of either ezrin, radixin, or moesin only mildly impacted NuMA association with the cortex (Fig. S2 C), in line with our aforementioned observation that single ERM depletion has no impact on spindle orientation (Fig. S1 F).
Increasing activated ERM levels by the overexpression of ezrin T567D mutant, which localized in a uniform manner (Fig. 5 A), also perturbed the polarized NuMA localization at the cortex in prometaphase. However, in contrast to ERM- or SLK-depleted cells, NuMA was still associated with the cell cortex (Fig. 5, A and B). This correlated with the fact that the speed of spindle rotation was unchanged in ezrin T567D–expressing cells (Fig. 5 C). Remarkably, NuMA was additionally localized at the nonadhesive cortex in 49.4 ± 8.1% of the ezrin T567D–expressing cells in prometaphase (11.7 ± 5.2% in control cells; Fig. 5 B, category 4), likely accounting for the observed defects of spindle reorientation (Fig. 2, G and H). In metaphase, NuMA brackets eventually formed at the cortex facing the poles of the mispositioned spindles (Fig. S2 D).
Oriented cell divisions require that force generators are properly localized at the mitotic cortex, such that spindles align along the expected axis. It is well established that Gαi is essential for LGN–NuMA association with the cortex, but a key question is to understand how and when the localization of LGN–NuMA and thus force generators is biased while Gαi is uniformly distributed. In the present study, we found that (a) LGN and NuMA localize in a polarized manner at the cell cortex, which provides a molecular explanation for the preferential spindle orientation; (b) the biased localization of LGN–NuMA occurs early in mitosis, in prometaphase; and (c) the biased activation of ERMs by the SLK kinase is required for the LGN–NuMA polarization in prometaphase (Fig. 5 D). Thus spindle orientation relies on two essential inputs that control LGN–NuMA localization: SLK/pERMs and Gαi (Fig. 5 E).
The novel role of the actin organizers ERMs in force generator localization raises two interesting questions: first, whether activated ERMs may directly recruit LGN–NuMA (because we observed that ERM hyperactivation led to aberrant NuMA localization), and second, the role of actin in this process. We found that Gαi impairment by pertussis toxin impaired NuMA localization, even after ERM hyperactivation (ezrin T567D–expressing cells; Fig. S3 A). This demonstrates that activated ERMs are not sufficient to recruit force generator complexes at the cortex and that the SLK/ERMs input cooperate with the Gαi input. Regarding the role of F-actin, we found that NuMA localization at the cortex is lost after Latrunculin A treatment (Fig. S3 B). This is consistent with F-actin integrity being critical for Gαi and LGN association with the cortex (Luxenburg et al., 2011; Zheng et al., 2013). In addition, the F-actin requirement for NuMA localization cannot be bypassed by forced ERM activation (Fig. S3 C). Importantly, the amount of F-actin at the cell cortex was unchanged in SLK- or ERM-depleted cells (Fig. S3 D) and in ezrin T567D–expressing cells (Fig. S3 E), whereas LGN–NuMA localization is perturbed. Altogether, our results show that both the presence of F-actin and its organization by activated ERMs are critical for the correct localization of the LGN–NuMA complex.
In conclusion, we propose that ERM activation orchestrates the organization of the actin cortex and the biased recruitment of force generators, and possibly microtubule dynamics (Solinet et al., 2013), thereby promoting instructive interactions between the mitotic spindle and the cell periphery. In addition, our study reveals that ERM activation represents a major integration event linking extrinsic cues to the proper localization of force generators, thereby providing new insights into the integrated cascades governing spindle orientation.
Materials and methods
Plasmids, siRNA, and shRNA
Recombinant proteins were purified using GST fusion encoding plasmids (pGEX backbone; GE Healthcare): pGEX-2T-SLK kinase domain (aa 1–344), pGEX-2T-SLK kinase domain K63R (aa 1–344), pGEX-2T-ezrin C-terminal (aa 310–586), pGEX-2T-radixin C-terminal (aa 310–583), and pGEX-2T-moesin C-terminal (aa 310–577). We used pcDNA3 (Invitrogen), pEGFP (Takara Bio Inc.), and pCB6 (http://www.addgene.org/vector-database/2070/; a gift from M. Arpin, Institut Curie, Paris, France) backbone vectors for expression in mammalian cells (CMV promotor): peGFP-N1-ezrin T567D, peGFP-N1 empty, pCB6-ezrin T567D and pCB6 empty, and pcDNA3-myc SLK full length. Human SLK cDNA was purchased from Source BioScience and cloned into vectors using the Gateway technology (Invitrogen). Point mutations were introduced using QuickChange (Invitrogen). ERMs and SLK siRNAs were purchased from Thermo Fisher Scientific: EZR (5′-GCGCGGAGCUGUCUAGUGAUU-3′), RDX (5′-GGCAUUAAGUUCAGAAUUA-3′), MOE (5′-UCGCAAGCCUGAUACCAUU-3′), and SLK#7 (5′-GGTAGAGATTGACATATTA-3′; Figs. S1–S3) and SLK#10 (5′-GGAACATAGCCAAGAATTA-3′; Figs. 1, 2, 4, and S1–S3). Scramble and shRNA-encoding plasmids were purchased from Origene: SLK#1 (5′-GGAGTTAGATGAGGAACATAGCCAAGAAT-3′; Fig. 3) and SLK#2 (5′-GAAGACAGTGCTGAGGATACGCAGAGTAA-3′; Fig. S1).
Cell culture, transfection, and drug treatment
Wild-type and stably expressing mCherry-Histone2B HeLa cells were cultured as described in Fink et al. (2011) and seeded 24 h before treatment. Cell synchronization was achieved by double thymidine (Sigma-Aldrich) block and mitotic shake (Théry et al., 2005). For actin depolymerization experiments, cells were treated for 1 h with 2.4 µM Latrunculin A (Molecular Probes). Pertussis toxin (Sigma-Aldrich) was used for 4 h at 400 ng/ml, as described previously (Woodard et al., 2010). For transfections, cells were treated following the manufacturer’s instructions using either LTX (Invitrogen) for 24 h or HiPerFect (QIAGEN) for 72 h, for plasmids or siRNA, respectively. Coverslips with L-shaped fibronectin micropatterns were prepared and used as described previously (Fink et al., 2011). In brief, coveslips were first covered with poly-l-lysine-g-polyethyleneglycol to passivate the surface. After UV illumination through a mask destroying the poly-l-lysine-g-polyethyleneglycol in the unprotected areas, fibronectin was added at 50 µg/ml for 1 h. Cells in suspension were eventually added on the rinsed coverslips. Fibronectin (Sigma-Aldrich) and Fibrinogen coupled to Cy5 (Molecular Probes) were used for these experiments.
Immunofluorescence and FACS
For pERMs, SLK, Gαi, LGN, NuMA, and actin staining, HeLa cells were fixed for 20 min in 10% ice-cold TCA, rinsed with PBS, and permeabilized with 0.1% Triton X-100 for 5 min. For other stainings, cells were fixed with 4% PFA for 10 min at 37°C. Cells were then processed for immunofluorescence microscopy as previously described (Miserey-Lenkei et al., 2010) and mounted in Mowiol (EMD Millipore). The following primary antibodies were used: rabbit anti-pERMs (1:400; #3141; Cell Signaling Technology), mouse anti-ezrin (1:100; 610602; BD; a gift from A. Alcover, Institut Pasteur, Paris, France), mouse anti-Gαi (1:20; Sc-56536; Santa Cruz Biotechnology, Inc.), mouse anti-MPM2 (1:100; ab14581-50; Abcam), mouse anti–β-tubulin (1:1,000, T5168; Sigma-Aldrich), mouse anti–γ-tubulin (1:100; T6557; Sigma-Aldrich), rabbit anti-LGN (1:500; a gift of F. Matsuzaki, RIKEN Center for Developmental Biology, Kobe, Japan), mouse anti-Myc (1: 1,000; a gift from S. Moutel, Institut Curie, Paris, France), rabbit or mouse anti-VSVG (1:200; gifts from M. Arpin and S. Moutel, respectively), rabbit anti-NuMA (1:500; 36999; AbCam), mouse anti-actin (1:200; MAB1501R; EMD Millipore), and mouse anti-SLK (1:200, for Western blots; W4009748; Sigma-Aldrich). Rabbit anti-SLK antibodies have been raised by Agrobio against recombinant SLK (aa 328–822) and were used for immunofluorescence experiments (1:200). A FACScalibur (BD) was used for FACS analysis as in Chesneau et al. (2012). Early mitotic cells were gated based on high levels of the marker MPM2.
In utero mouse electroporation and immunostaining
Plasmids were purified with the Endofree plasmid maxi kit (Sigma-Aldrich), injected in the lateral ventricle of E14.5 embryos in utero, and electroporated as previously described (Mire et al., 2012) using an electroporator (CUY21 Edit; Nepagene). Embryos were harvested in L15-glucose 48 h after electroporation. Brains were dissected and open or cut in the vibratome; the electroporated parts or the 100-µm sections were fixed in 10% ice-cold TCA for 20 min for pERM staining or in 4% PFA for 2 h for other stainings and processed for immunofluorescence as described previously (Deck et al., 2013).
Microscope image acquisition, processing, and quantification
Immunofluorescence analysis.
For Fig. 1 D, confocal images were acquired with a confocal microscope (LSM700; Carl Zeiss) equipped with a 100× oil immersion objective (1.4 NA Plan Apochromat). For all other images and quantitative analysis, cells were examined under an inverted microscope (Ti; Nikon) equipped with a 100× oil immersion objective (1.4 NA Plan Apochromat). In each experiment, all images were acquired with the same exposure time and settings using Metamorph software (Molecular Devices) driving a charge coupled device camera (Coolsnap HQ; Photometrics). 16-bit images were converted into 8-bit images using ImageJ software (National Institutes of Health). Brightness and contrast of representative images were then adjusted to display equivalent background levels using Photoshop CS4 (Adobe) without changing the gamma. Intensity quantifications were all done on untreated images using ImageJ software. For endogenous SLK, Gαi, LGN, and NuMA staining, images were deconvolved using Huygens software (Scientific Volume Imaging) with the microscope PFS (10 iterations, threshold 200, and signal-to-noise ratio of 40).
Time-lapse microscopy and measurement of spindle orientation.
HeLa cells expressing mCherry-Histone2B plated on L-shaped micropatterns were placed in chambers (Chamlide; Live Cell Instrument) equilibrated with 5% CO2 and maintained at 37°C. Single cells were imaged every 5 min using an inverted microscope (Ti), equipped with a 10× air objective (0.3 NA Plan Fluor; Nikon). Chromosome plates were followed throughout mitosis using the cherry channel. Spindle orientation was calculated in early anaphase as depicted in Fig. 2, based on the angle measured between separated chromosomes and the pattern, using the angle tool of ImageJ.
Measurement of spindle orientation in mouse cerebral cortex.
Samples were acquired from the tissue apical surface with a 40× oil immersion objective (HCX PL APO NA 1.25; Leica) plus 3× zoom, on an inverted confocal microscope (TCS SP5; Leica). Z-views were generated in ImageJ. Spindle orientation measurements were performed using the angle tool of ImageJ on the Z-view images, taking the apical membrane as a reference plane.
Western blots
Cells in 6-well plates were lysed by addition of Laemmli buffer and detached by scratching. Western blot experiments were performed as described previously (Dambournet et al., 2011), using rabbit anti-pERMs (1:1,000), rabbit anti-SLK (1:1,000), mouse anti–β-tubulin (1:1,000), mouse anti-ezrin (1:5,000; a gift from M. Arpin), rabbit anti-phosphoHistone3 (1:1,000; #06-570; EMD Millipore), and mouse anti-GST (1:1,000; 554805; BD) antibodies.
In vitro kinase assay
Recombinant proteins were purified from Escherichia coli as described in Dambournet et al. (2011). In brief, BL21(DE3) bacteria were induced for 3 h at 37°C with 1 mM IPTG. Cells were lysed by sonication in Lysis buffer (50 mM Tris, pH 8, 2 mM EDTA, 1 mM DTT, and protease inhibitors; Roche) with 1 mg/ml Lysosyme (Sigma-Aldrich). GST-tagged proteins were purified on GST-agarose beads (GE Healthcare) following the manufacturer’s instructions. 60 nM of recombinant GST-SLK kinase domain (aa 1–344) were mixed with 20 µM of recombinant GST-ezrin C-terminal (aa 310–586), GST-radixin C-terminal (aa 310–583), or GST-moesin C-terminal (aa 310–577) in kinase buffer (60 mM Hepes, pH 7.4, 10 mM MgCl2, 1 mM NaVO4, 1 mM DTT, and 0.5 mM EDTA) with 100 µM ATP. After 1 h at 37°C, Laemmli buffer addition stopped the reaction and products were analyzed by Western blot with antibodies against pERMs and GST.
Statistics
Data were analyzed, graphs were drawn, and statistics were done using Excel (Microsoft) and GraphPad prism (GraphPad Software). Data were compared with Student t test (Fig. 1, B and G; Fig. 3 B; Fig. 4, B–E; Fig. 5, B and C; Fig. S1 C; Fig. S2; and Fig. S3) or with Kolmogorov-Smirnov test (Fig. 2; Fig. 3, F and I; and Fig. S1, E and K).
Online supplemental material
Fig. S1 shows spindle orientation defects in micropatterned cells and apical progenitor cells treated with a second, independent SLK siRNA or shRNA; it also shows the absence of visible defects on retraction fibers or spindle morphology after SLK or ERM depletion. Fig. S2 shows the localization of Gαi, LGN, and NuMA in SLK#7- or ERM-depleted cells and in ezrin T567D–expressing cells. Fig. S3 shows NuMA and F-actin localization in control cells, in Latrunculin A–, or pertussis toxin–treated cells, and in SLK- or ERM-depleted mitotic cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201401049/DC1.
Supplementary Material
Acknowledgments
We thank A. Alcover, M. Arpin, I. Cheeseman, F. Matsuzaki, and S. Moutel for reagents or protocols; and M. Arpin, R. Basto, Y. Bellaiche, G. Hickson, J. Mathieu, X. Morin, M. Théry, and F. Schweisguth for critical reading and discussions. We thank the Imagopole Institut Pasteur, the Nikon Imaging Center at Institut Curie-Centre National de la Recherche Scientifique, the Imaging facility of the Institut de Biologie de L’Ecole Normale Supérieure, and K. Klinkert for image processing.
This work has been supported by the Institut Pasteur, the Centre National de la Recherche Scientifique, the Schlumberger Foundation for Education and Research, and the Fondation pour la Recherche Médicale (Equipe FRM DEQ20120323707) to A. Echard; the Fondation Association pour la Recherche sur le Cancer to M. Machicoane and J. Fink; European Young Investigator award, Institut National de la Santé et de la Recherche Médicale, European Molecular Biology Organization Young Investigator Program to S. Garel; and Institut National du Cancer grant 2011-1-PL BIO-11-IC to M. Piel. C.A. de Frutos was supported by a Marie Curie postdoctoral fellowship and the Investissements d’Avenir (ANR-10-LABX-54 MEMO LIFE and ANR-11-IDEX-0001-02 PSL* Research University).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- E
- embryonic day
- ERM
- ezrin/radixin/moesin
- LGN
- leucine-glycine-asparagine repeat protein
- NuMA
- nuclear mitotic apparatus
- SLK
- Ste20-like kinase
References
- Belkina N.V., Liu Y., Hao J.J., Karasuyama H., Shaw S. 2009. LOK is a major ERM kinase in resting lymphocytes and regulates cytoskeletal rearrangement through ERM phosphorylation. Proc. Natl. Acad. Sci. USA. 106:4707–4712 10.1073/pnas.0805963106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno S., Kouranti I., Glusman E.S., Fuller M.T., Echard A., Payre F. 2008. Moesin and its activating kinase Slik are required for cortical stability and microtubule organization in mitotic cells. J. Cell Biol. 180:739–746 10.1083/jcb.200709161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanon I., Abrami L., Holtzer L., Heisenberg C.P., van der Goot F.G., Gonzalez-Gaitan M. 2013. Anthrax toxin receptor 2a controls mitotic spindle positioning. Nat. Cell Biol. 15:28–39 10.1038/ncb2632 [DOI] [PubMed] [Google Scholar]
- Chesneau L., Dambournet D., Machicoane M., Kouranti I., Fukuda M., Goud B., Echard A. 2012. An ARF6/Rab35 GTPase cascade for endocytic recycling and successful cytokinesis. Curr. Biol. 22:147–153 10.1016/j.cub.2011.11.058 [DOI] [PubMed] [Google Scholar]
- Dambournet D., Machicoane M., Chesneau L., Sachse M., Rocancourt M., El Marjou A., Formstecher E., Salomon R., Goud B., Echard A. 2011. Rab35 GTPase and OCRL phosphatase remodel lipids and F-actin for successful cytokinesis. Nat. Cell Biol. 13:981–988 10.1038/ncb2279 [DOI] [PubMed] [Google Scholar]
- Deck M., Lokmane L., Chauvet S., Mailhes C., Keita M., Niquille M., Yoshida M., Yoshida Y., Lebrand C., Mann F., et al. 2013. Pathfinding of corticothalamic axons relies on a rendezvous with thalamic projections. Neuron. 77:472–484 10.1016/j.neuron.2012.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q., Macara I.G. 2004. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 119:503–516 10.1016/j.cell.2004.10.028 [DOI] [PubMed] [Google Scholar]
- Fehon R.G., McClatchey A.I., Bretscher A. 2010. Organizing the cell cortex: the role of ERM proteins. Nat. Rev. Mol. Cell Biol. 11:276–287 10.1038/nrm2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fietz S.A., Huttner W.B. 2011. Cortical progenitor expansion, self-renewal and neurogenesis—a polarized perspective. Curr. Opin. Neurobiol. 21:23–35 10.1016/j.conb.2010.10.002 [DOI] [PubMed] [Google Scholar]
- Fievet B.T., Gautreau A., Roy C., Del Maestro L., Mangeat P., Louvard D., Arpin M. 2004. Phosphoinositide binding and phosphorylation act sequentially in the activation mechanism of ezrin. J. Cell Biol. 164:653–659 10.1083/jcb.200307032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink J., Carpi N., Betz T., Bétard A., Chebah M., Azioune A., Bornens M., Sykes C., Fetler L., Cuvelier D., Piel M. 2011. External forces control mitotic spindle positioning. Nat. Cell Biol. 13:771–778 10.1038/ncb2269 [DOI] [PubMed] [Google Scholar]
- Gimeno L., Corradi A., Cobos I., Consalez G.G., Martinez S. 2004. Ezrin gene, coding for a membrane-cytoskeleton linker protein, is regionally expressed in the developing mouse neuroepithelium. Gene Expr. Patterns. 4:749–754 10.1016/j.modgep.2004.03.007 [DOI] [PubMed] [Google Scholar]
- Gonzalez C. 2007. Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat. Rev. Genet. 8:462–472 10.1038/nrg2103 [DOI] [PubMed] [Google Scholar]
- Hipfner D.R., Keller N., Cohen S.M. 2004. Slik Sterile-20 kinase regulates Moesin activity to promote epithelial integrity during tissue growth. Genes Dev. 18:2243–2248 10.1101/gad.303304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y., Fukata Y., Oshiro N., Amano M., Nakamura T., Ito M., Matsumura F., Inagaki M., Kaibuchi K. 1999. Phosphorylation of myosin-binding subunit (MBS) of myosin phosphatase by Rho-kinase in vivo. J. Cell Biol. 147:1023–1038 10.1083/jcb.147.5.1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., Cheeseman I.M. 2012. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 14:311–317 10.1038/ncb2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., Cheeseman I.M. 2013. Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell. 154:391–402 10.1016/j.cell.2013.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J.A. 2008. Mechanisms of asymmetric stem cell division. Cell. 132:583–597 10.1016/j.cell.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Konno D., Shioi G., Shitamukai A., Mori A., Kiyonari H., Miyata T., Matsuzaki F. 2008. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat. Cell Biol. 10:93–101 10.1038/ncb1673 [DOI] [PubMed] [Google Scholar]
- Kotak S., Busso C., Gönczy P. 2012. Cortical dynein is critical for proper spindle positioning in human cells. J. Cell Biol. 199:97–110 10.1083/jcb.201203166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P., Baum B. 2009. The actin cytoskeleton in spindle assembly and positioning. Trends Cell Biol. 19:174–179 10.1016/j.tcb.2009.01.006 [DOI] [PubMed] [Google Scholar]
- Kunda P., Pelling A.E., Liu T., Baum B. 2008. Moesin controls cortical rigidity, cell rounding, and spindle morphogenesis during mitosis. Curr. Biol. 18:91–101 10.1016/j.cub.2007.12.051 [DOI] [PubMed] [Google Scholar]
- Kunda P., Rodrigues N.T., Moeendarbary E., Liu T., Ivetic A., Charras G., Baum B. 2012. PP1-mediated moesin dephosphorylation couples polar relaxation to mitotic exit. Curr. Biol. 22:231–236 10.1016/j.cub.2011.12.016 [DOI] [PubMed] [Google Scholar]
- Luxenburg C., Pasolli H.A., Williams S.E., Fuchs E. 2011. Developmental roles for Srf, cortical cytoskeleton and cell shape in epidermal spindle orientation. Nat. Cell Biol. 13:203–214 10.1038/ncb2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Maeda M., Doi Y., Yonemura S., Amano M., Kaibuchi K., Tsukita S., Tsukita S. 1998. Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J. Cell Biol. 140:647–657 10.1083/jcb.140.3.647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minc N., Piel M. 2012. Predicting division plane position and orientation. Trends Cell Biol. 22:193–200 10.1016/j.tcb.2012.01.003 [DOI] [PubMed] [Google Scholar]
- Mire E., Mezzera C., Leyva-Díaz E., Paternain A.V., Squarzoni P., Bluy L., Castillo-Paterna M., López M.J., Peregrín S., Tessier-Lavigne M., et al. 2012. Spontaneous activity regulates Robo1 transcription to mediate a switch in thalamocortical axon growth. Nat. Neurosci. 15:1134–1143 10.1038/nn.3160 [DOI] [PubMed] [Google Scholar]
- Miserey-Lenkei S., Chalancon G., Bardin S., Formstecher E., Goud B., Echard A. 2010. Rab and actomyosin-dependent fission of transport vesicles at the Golgi complex. Nat. Cell Biol. 12:645–654 10.1038/ncb2067 [DOI] [PubMed] [Google Scholar]
- Morin X., Bellaïche Y. 2011. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev. Cell. 21:102–119 10.1016/j.devcel.2011.06.012 [DOI] [PubMed] [Google Scholar]
- Nakajima Y., Meyer E.J., Kroesen A., McKinney S.A., Gibson M.C. 2013. Epithelial junctions maintain tissue architecture by directing planar spindle orientation. Nature. 500:359–362 10.1038/nature12335 [DOI] [PubMed] [Google Scholar]
- O’Reilly P.G., Wagner S., Franks D.J., Cailliau K., Browaeys E., Dissous C., Sabourin L.A. 2005. The Ste20-like kinase SLK is required for cell cycle progression through G2. J. Biol. Chem. 280:42383–42390 10.1074/jbc.M510763200 [DOI] [PubMed] [Google Scholar]
- Roubinet C., Decelle B., Chicanne G., Dorn J.F., Payrastre B., Payre F., Carreno S. 2011. Molecular networks linked by Moesin drive remodeling of the cell cortex during mitosis. J. Cell Biol. 195:99–112 10.1083/jcb.201106048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandquist J.C., Kita A.M., Bement W.M. 2011. And the dead shall rise: actin and myosin return to the spindle. Dev. Cell. 21:410–419 10.1016/j.devcel.2011.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller K.H., Doe C.Q. 2009. Spindle orientation during asymmetric cell division. Nat. Cell Biol. 11:365–374 10.1038/ncb0409-365 [DOI] [PubMed] [Google Scholar]
- Solinet S., Mahmud K., Stewman S.F., Ben El Kadhi K., Decelle B., Talje L., Ma A., Kwok B.H., Carreno S. 2013. The actin-binding ERM protein Moesin binds to and stabilizes microtubules at the cell cortex. J. Cell Biol. 202:251–260 10.1083/jcb.201304052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry M., Racine V., Pépin A., Piel M., Chen Y., Sibarita J.B., Bornens M. 2005. The extracellular matrix guides the orientation of the cell division axis. Nat. Cell Biol. 7:947–953 10.1038/ncb1307 [DOI] [PubMed] [Google Scholar]
- Théry M., Jiménez-Dalmaroni A., Racine V., Bornens M., Jülicher F. 2007. Experimental and theoretical study of mitotic spindle orientation. Nature. 447:493–496 10.1038/nature05786 [DOI] [PubMed] [Google Scholar]
- Toyoshima F., Nishida E. 2007. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 26:1487–1498 10.1038/sj.emboj.7601599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanatha R., Ohouo P.Y., Smolka M.B., Bretscher A. 2012. Local phosphocycling mediated by LOK/SLK restricts ezrin function to the apical aspect of epithelial cells. J. Cell Biol. 199:969–984 10.1083/jcb.201207047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodard G.E., Huang N.N., Cho H., Miki T., Tall G.G., Kehrl J.H. 2010. Ric-8A and Giα recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. Mol. Cell. Biol. 30:3519–3530 10.1128/MCB.00394-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Wan Q., Liu J., Zhu H., Chu X., Du Q. 2013. Evidence for dynein and astral microtubule–mediated cortical release and transport of Gαi/LGN/NuMA complex in mitotic cells. Mol. Biol. Cell. 24:901–913 10.1091/mbc.E12-06-0458 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.