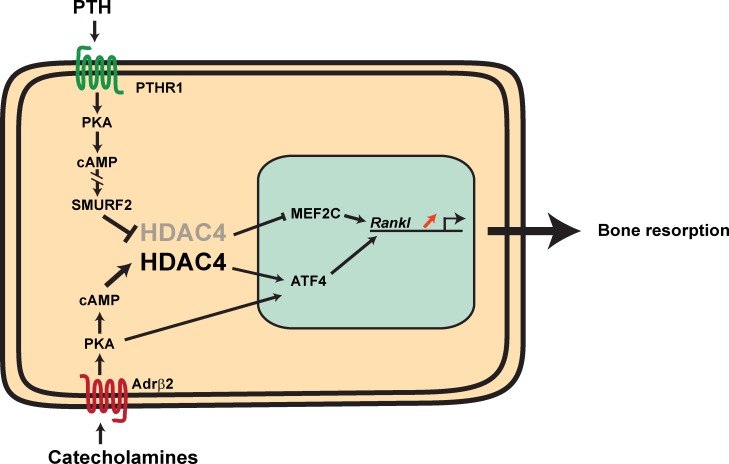
During osteoclast differentiation, two extracellular cues that induce cAMP production both converge on HDAC4 and induce Rankl expression but do so using distinct signaling pathways.
Abstract
Parathyroid hormone (PTH) and the sympathetic tone promote Rankl expression in osteoblasts and osteoclast differentiation by enhancing cyclic adenosine monophosphate production through an unidentified transcription factor for PTH and through ATF4 for the sympathetic tone. How two extracellular cues using the same second messenger in the same cell elicit different transcriptional events is unknown. In this paper, we show that PTH favors Rankl expression by triggering the ubiquitination of HDAC4, a class II histone deacetylase, via Smurf2. HDAC4 degradation releases MEF2c, which transactivates the Rankl promoter. Conversely, sympathetic signaling in osteoblasts favors the accumulation of HDAC4 in the nucleus and its association with ATF4. In this context, HDAC4 increases Rankl expression. Because of its ability to differentially connect two extracellular cues to the genome of osteoblasts, HDAC4 is a critical regulator of osteoclast differentiation.
Introduction
Osteoblasts are bone-specific, multifunctional cells that are responsible for bone formation through the synthesis of proteins of the bone extracellular matrix (Karsenty et al., 2009). Because osteoblasts express the osteoclast differentiation factor RANKL (receptor activator of nuclear factor κ–B ligand), they also favor bone resorption (Teitelbaum, 2000). In addition, osteoblasts are endocrine cells (Karsenty and Ferron, 2012). In agreement with the diversity of their functions, multiple extracellular cues can affect osteoblasts.
Parathyroid hormone (PTH) regulates Rankl expression through several mechanisms (Tawfeek et al., 2010). In one of them, PTH binds to its cognate receptor, PTH1R, a G protein–coupled receptor on osteoblasts, and enhances Rankl expression in a cAMP-dependent manner (Kondo et al., 2002). Ex vivo assays have suggested that CREB mediates the PTH regulation of Rankl expression (Fu et al., 2002), but in vivo, however, inactivation of Creb in osteoblasts does not affect Rankl expression or bone resorption. Thus, the transcriptional mechanisms whereby PTH signaling in osteoblasts affects Rankl expression are not elucidated (Kajimura et al., 2011). Another systemic cue affecting Rankl expression in osteoblasts is the sympathetic nervous system. Catecholamines, after their binding to another G protein–coupled receptor, the β2 adrenergic receptor, also use cAMP as a second messenger to enhance Rankl expression by recruiting the transcription factor ATF4 (Elefteriou et al., 2005). These observations raise the following question: How do two distinct regulatory signals that use the same second messenger elicit different transcriptional events in the same cell type?
Chromatin structure, which is influenced by posttranslational modifications of histone proteins around which the DNA is wrapped, is a major determinant of gene expression (Jenuwein and Allis, 2001; Allis et al., 2007). Histone acetylation promotes gene transcription by relaxing the chromatin structure, whereas deacetylation of histones by histone deacetylases (HDACs) induces chromatin condensation and transcriptional repression (Berger, 2002; Verdin et al., 2003; Allis et al., 2007). Class II HDACs contain a poorly active catalytic domain and a long N-terminal extension to which transcription factors can bind. The existence of this domain has suggested that class II HDACs can link extracellular cues to the genome of a given cell (Verdin et al., 2003; Haberland et al., 2009).
One class II HDAC, HDAC4, prevents chondrocyte hypertrophy in part by inhibiting the activity of the transcription factor Runx2, a master gene of skeletogenesis, in proliferating chondrocytes (Vega et al., 2004). Given the regulation of bone formation that Runx2 exerts through its expression in osteoblasts, this suggested that HDAC4 also regulates bone formation (Karsenty et al., 2009).
We show here that, in vivo, HDAC4 does not inhibit Runx2 functions in osteoblasts but instead integrates PTH and sympathetic signaling. PTH signaling in osteoblasts favors Rankl expression by inducing, via the E3 ubiquitin ligase Smurf2, HDAC4 ubiquitination; this releases MEF2c, which can then activate Rankl expression. Conversely, the sympathetic tone promotes HDAC4 accumulation in the nucleus of osteoblasts and its interaction with ATF4. Thus, under the control of the sympathetic tone, HDAC4 favors Rankl and osteoclast differentiation. Lastly, HDAC4 does not inhibit Runx2 functions in osteoblasts in vivo because catecholamines disrupt the physical interaction between HDAC4 and Runx2. This study identifies HDAC4 as a link between two extracellular signals and the genome of osteoblasts.
Results and discussion
HDAC4 inhibits Rankl expression in osteoblasts
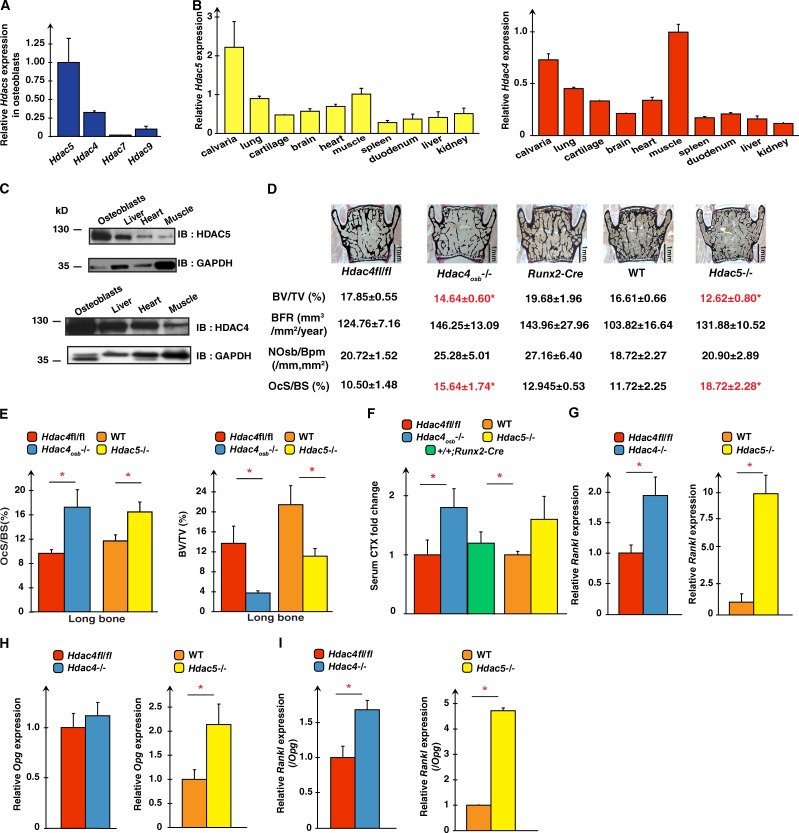
To study the functions of HDACs in osteoblasts, we asked which ones were expressed in these cells. Hdac5 expression was twofold higher in bone than in any other tissue (Fig. 1 B). Hdac4 expression was also higher in bone than in other tissues where it exerts important functions (Fig. 1, A and B; Zhang et al., 2002; Chang et al., 2004; Vega et al., 2004). Similar results were obtained at the protein level (Fig. 1 C).
Figure 1.
HDAC4 and HDAC5 inhibit Rankl expression in osteoblasts. (A) Expression of class IIa Hdacs in mouse osteoblasts. The expression of each gene is compared with the one of Hdac5. (B) Hdac4 (right) and Hdac5 (left) expression in various tissues. Expression of these genes in each tissue is compared with their expression in muscle. (C) HDAC4 and HDAC5 accumulation in different tissues. Anti-GAPDH was used as a loading control. (D and E) Histomorphometric analyses of vertebrae (D) and long bones (E) of Hdac4osb−/− (n = 7) and Hdac5−/− (n = 7) mice compared with Hdac4fl/fl (n = 8) or WT (n = 5) controls, respectively. BV/TV (%), percentage of bone volume over trabecular volume; NOsb/Bpm., number of osteoblasts per bone perimeter; BFR, bone formation rate; OcS/BS (%), percentage of osteoclast surface per bone surface. (F) Serum CTX levels in Hdac4fl/fl (n = 13), Hdac4 osb−/− (n = 12), WT (n = 10), Hdac5−/− (n = 9), and +/+Runx2-Cre (n = 4). Results are expressed as fold changes compared with levels seen in Hdac4fl/fl mice. (G) Rankl expression in Hdac4fl/fl (n = 4), Hdac4−/− (n = 4; left), WT (n = 4), and Hdac5−/− (n = 4; right) osteoblasts. (H and I) Opg expression (H) and Rankl/Opg ratio (I) in Hdac4fl/fl (n = 4), Hdac4−/− (n = 4), WT (n = 4), and Hdac5−/− (n = 4) osteoblasts. For G–I, results are expressed as fold changes compared with levels seen in Hdac4fl/fl or WT osteoblasts. Results are given as means ± SEM. *, P < 0.05 by Student’s t test. IB, immunoblot.
We used Hdac5−/− mice to study HDAC5 functions in osteoblasts and generated mice lacking Hdac4 in osteoblasts by crossing Hdac4fl/fl mice with mice expressing the Cre recombinase under the control of Runx2 regulatory elements (Rauch et al., 2010). Before using Hdac4osb−/− mice, because Runx2 expression is not restricted to osteoblasts, we verified that Hdac4 had been efficiently deleted from osteoblasts but not from other cell types and that there was no overexpression of either Hdac4 in the Hdac5−/− mice or of Hdac5 in the Hdac4osb−/− mice (Fig. S1, A and B).
Because HDAC4 inhibits Runx2 functions in chondrocytes, we expected that it would do the same in osteoblasts and that Hdac4osb−/− mice would display a high bone mass because of an increase in bone formation parameters. Instead, both Hdac4osb−/− and Hdac5−/− mice demonstrated a low bone mass phenotype affecting all bones tested. This was caused by a 50% increase in the bone surface covered by osteoclasts compared with control bones, whereas bone formation parameters were modestly affected (Fig. 1, D and E). This increase in bone resorption was caused by a marked increase in Rankl expression in Hdac4−/− and Hdac5−/− compared with control osteoblasts, whereas Opg expression was not changed to the same extent. As a result, the ratio of Rankl/Opg was increased over 50% in Hdac4−/− and Hdac5−/− compared with wild-type (WT) osteoblasts (Fig. 1, G–I). Expression of other regulatory genes and of type I collagen was normal in Hdac4−/− osteoblasts (Fig. S1 C). Thus, through their expression in osteoblasts, HDAC4 and HDAC5 inhibit Rankl expression and bone resorption. An explanation for the absence of HDAC4’s influence on bone formation is proposed below (see Fig. 4 H). For the rest of this work, because of the cell-specific nature of the gene deletion, we studied Hdac4osb−/− mice.
Figure 4.
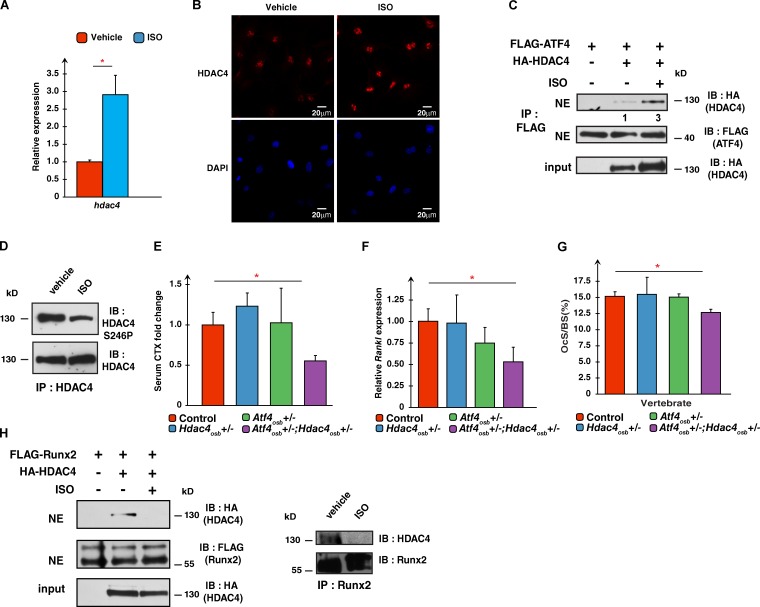
The sympathetic tone stabilizes HDAC4 to favor Rankl expression. (A) Expression of Hdac4 and in primary osteoblasts treated with vehicle or ISO for 2 h. Results are presented as fold changes compared with levels seen in vehicle-treated cells. (B) Immunofluorescence detection of HDAC4 in WT primary osteoblasts treated with vehicle or ISO. (C) Coimmunoprecipitation assay in ROS17/2.8 cells showing an interaction between FLAG-ATF4 and HA-HDAC4. Nuclear extracts (NE) were immunoprecipitated after ISO treatment with an anti-FLAG antibody, and proteins were detected by Western blotting with anti-HA or anti-FLAG antibodies. The numbers shown under each lane of the top blot represent the fold enhancement in intensity of the bands. (D) Immunoprecipitation of HDAC4 in mouse primary osteoblasts treated with vehicle or ISO. Proteins were analyzed by Western blot using anti-HDAC4 or anti–HDAC4-S246P antibodies. (E) Serum CTX levels in control (n = 8), Hdac4osb+/− (n = 3), Atf4osb+/− (n = 3), and Atf4osb+/−; Hdac4osb+/− (n = 3) mice, shown as fold changes compared with control levels. (F) Rankl expression in long bones of control (n = 8), Hdac4osb+/− (n = 3), Atf4osb+/− (n = 3), and Atf4osb+/−;Hdac4osb+/− (n = 3) mice. Results are presented as fold changes compared with levels seen in WT bones. (G) Histomorphometric quantification of the bone surface covered by osteoclasts in vertebrae of control (n = 8), Hdac4osb+/− (n = 3), Atf4osb+/− (n = 3), and Atf4osb+/−;Hdac4osb+/− (n = 3) mice. OcS/BS (%), percentage of osteoclast surface per bone surface. (H, left) Coimmunoprecipitation assay in ISO-treated ROS17/2.8 cells showing interaction between FLAG-Runx2 and HA-HDAC4. Nuclear extracts were immunoprecipitated with the anti-FLAG antibody. Purified proteins were detected by Western blotting with anti-HA or anti-FLAG antibodies. (right) Immunoprecipitation of Runx2 in mouse osteoblasts treated with vehicle or ISO. Proteins were analyzed by Western blotting using an anti-Runx2 or anti-HDAC4 antibodies. Results are given as means ± SEM. *, P < 0.05 by Student’s t test. IB, immunoblot; IP, immunoprecipitation.
HDAC4 inhibits osteoclast differentiation by preventing MEF2c to transactivate Rankl
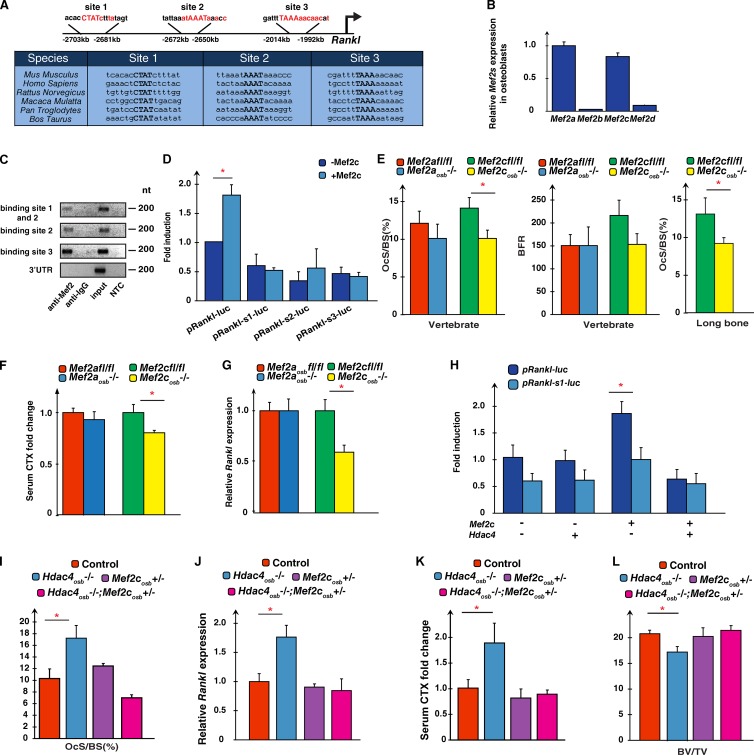
To elucidate how HDAC4 inhibits Rankl expression in osteoblasts, we asked whether members of the MEF2 family of transcription factors regulate Rankl expression. There were two reasons to ask this question. The first one is the well-documented interaction of HDAC4 with MEF2 proteins in other cell types (Zhang et al., 2002). Second, there are three putative MEF2 binding sites in the mouse Rankl promoter that are conserved in all vertebrate species analyzed, and Mef2a and Mef2c are highly expressed in osteoblasts (Fig. 2, A and B).
Figure 2.
HDAC4 prevents MEF2c to transactivate Rankl.(A) Putative MEF2 binding sites in the Rankl promoter of the mouse and other bony vertebrate species. Red letters indicate the core MEF2 binding sequence. (B) Mef2 gene expression in WT osteoblasts (n = 3). Expression of each gene is compared with the expression of Mef2a. (C) Chromatin immunoprecipitation assay in mouse osteoblasts analyzing MEF2 binding to the Rankl promoter. NTC, no template control. (D) DNA cotransfection assays in COS cells with a Rankl promoter luciferase construct (pRankl-luc) either WT or harboring a mutation in either one of the MEF2 binding sites alone or with a MEF2c expression vector. (E) Histomorphometric quantification the bone surface covered by osteoclasts and bone formation rate in vertebrae of Mef2afl/fl (n = 5), Mef2aosb−/− (n = 5), Mef2cfl/fl (n = 6), and Mef2cosb−/− (n = 8; left) mice and long bones of Mef2cfl/fl (n = 6) and Mef2cosb−/− (n = 8) mice (right). BFR, bone formation rate; OcS/BS (%), percentage of osteoclast surface per bone surface. (F) Serum CTX levels in Mef2afl/fl (n = 8), Mef2aosb−/− (n = 7), Mef2cfl/fl (n = 6), and Mef2cosb−/− (n = 7) mice. Results are represented as fold changes compared with levels seen in Mef2cfl/fl mice. (G) Rankl expression in Mef2afl/fl (n = 8), Mef2aosb−/− (n = 7), Mef2cfl/fl (n = 13), and Mef2cosb−/− (n = 14) mice. Results are expressed as fold changes compared with levels seen in Mef2cfl/fl mice. (H) DNA cotransfection assays in COS cells of pRankl-luc either WT or mutant for the MEF2 binding site 1, along with MEF2c and HDAC4 expression vectors. (I) Histomorphometric quantification of the bone surface covered by osteoclasts in the vertebrae of control (n = 5), Mef2cosb+/− (n = 4), Hdac4osb−/− (n = 4), and Hdac4osb−/−;Mef2cosb+/− (n = 3) mice. (J) Rankl expression in control (n = 10), Mef2cosb+/− (n = 4), Hdac4osb−/− (n = 4), and Hdac4osb−/−;Mef2cosb+/− (n = 5) calvaria. (K) Serum CTX levels of control (n = 6), Mef2cosb+/− (n = 3), Hdac4osb−/− (n = 4), and Hdac4osb−/−;Mef2cosb+/− (n = 3) mice. In I and K, results are presented as fold changes compared with levels seen in control mice. (L) Histomorphometric analyses of BV/TV percentage in vertebrae of control (n = 9), Mef2cosb+/− (n = 4), Hdac4osb−/− (n = 5), and Hdac4osb−/−;Mef2cosb+/− (n = 3) mice. Results are given as means ± SEM. *, P < 0.05 by Student’s t test.
Chromatin immunoprecipitation assays verified that MEF2 proteins could bind to each putative MEF2 binding site present in the mouse Rankl promoter (Fig. 2 C). In DNA cotransfection assays performed in COS cells, which do not express Rankl, an expression vector for Mef2c transactivated a construct containing a 3-kb-long fragment of the Rankl promoter driving the luciferase gene (pRankl-luc); disrupting each of these three binding sites decreased the activity of this promoter fragment by ≥50% and abrogated MEF2c’s ability to activate pRankl-luc (Fig. 2 D). The results provided in vitro evidence suggesting that MEF2 proteins are important to regulate Rankl expression in osteoblasts.
To determine whether what was observed in vitro exists in vivo, we generated mutant mice lacking Mef2a or Mef2c in osteoblasts only and verified that this manipulation had efficiently deleted Mef2a or Mef2c from osteoblasts but not from other cell types (Fig. S2). Although mice lacking Mef2a in osteoblasts did not display any abnormalities of bone resorption, Mef2cosb−/− mice exhibited a significant decrease in the bone surface covered by osteoclasts in all bones analyzed (Fig. 2 E). Serum CTX levels were also significantly lower in Mef2cosb−/− than in Mef2cfl/fl mice, and Rankl expression was decreased nearly 50% in Mef2cosb−/− compared with control mice (Fig. 2, F and G).
The opposite influence of HDAC4 and MEF2c on Rankl expression along with the ability of HDAC4 to interact physically with MEF2c suggested that HDAC4 might inhibit Rankl expression by hampering the transactivating function of MEF2c. Indeed, in DNA cotransfection experiments, the ability of MEF2c to transactivate the Rankl-luc construct in COS cells was abrogated when an Hdac4 expression vector was cotransfected (Fig. 2 H). In vivo, Rankl expression, serum CTX values, and bone mass were normal in Hdac4osb−/− mice lacking one allele of Mef2c in osteoblasts only (Fig. 2, I–L). Collectively, these results suggest the existence of an HDAC4 ┤ MEF2c →Rankl regulatory loop in mouse osteoblasts regulating osteoclast differentiation.
PTH favors Rankl expression through MEF2c by promoting HDAC4 proteasomal degradation
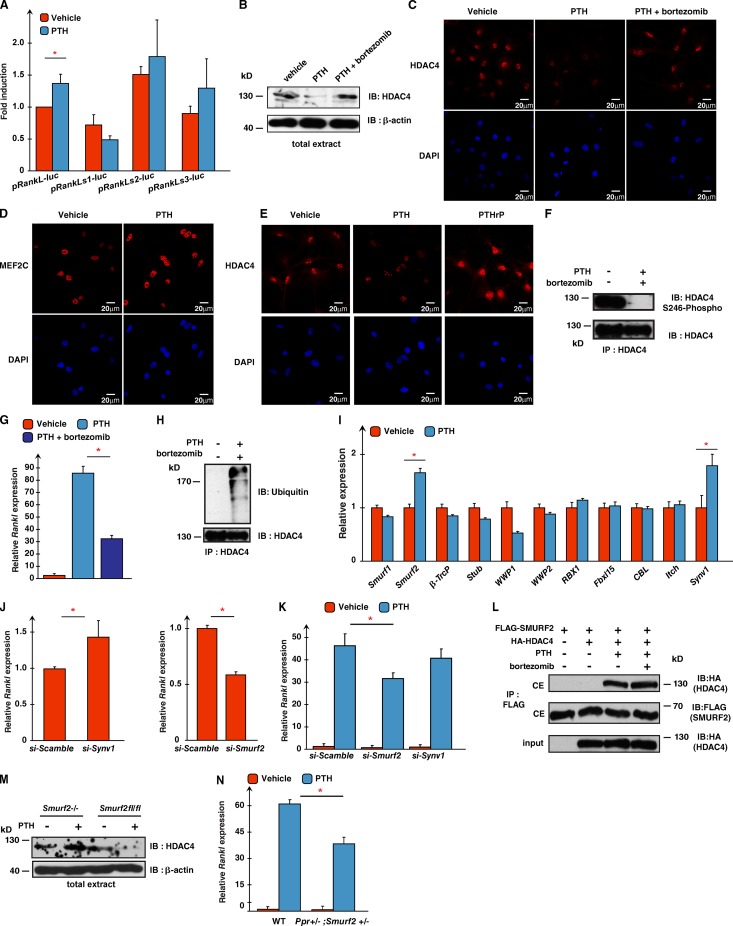
Next, we asked whether PTH signaling in osteoblasts promotes Rankl expression by disrupting this HDAC4 ┤ MEF2c →Rankl pathway. PTH increased the activity of pRankl-luc in ROS17/2.8 osteoblastic cells, and mutating the first MEF2 binding site in this promoter abrogated this effect of PTH (Fig. 3 A). Additionally, PTH triggered a marked decrease in HDAC4 accumulation in both the cytoplasm and the nucleus of mouse osteoblasts, without affecting MEF2c accumulation. This is different from what is observed after treatment of osteoblasts with PTH-related protein (PTHrP), a molecule that bears similarities to PTH and that signals through the same receptor (Fig. 3, B–E; Kronenberg, 2003), because PTHrP allows HDAC4 to move to the nucleus by inducing its dephosphorylation on S246 (Kozhemyakina et al., 2009). PTH induced the same dephosphorylation in HDAC4 in mouse osteoblasts (Fig. 3 F), but this could not explain our findings, as PTH induces a near complete disappearance of HDAC4 from osteoblast nuclei (Fig. 3 C).
Figure 3.
PTH favors Rankl expression by disrupting the HDAC4–MEF2c interaction. (A) DNA transfection assays in ROS17/2.8 cells using a pRankl-luc reporter construct for either WT (pRankl-luc) or mutated for each MEF2 binding site. Cells were treated with PTH or vehicle for 16 h. (B) Western blot analysis of total extracts from primary osteoblasts treated with vehicle, PTH, or PTH plus bortezomib for 2 h using anti-HDAC4 and anti–β-actin antibodies. (C) Immunofluorescence showing HDAC4 localization in mouse primary osteoblasts treated with vehicle or PTH alone or in the presence of bortezomib. (D) Immunofluorescence assays showing MEF2c localization in mouse primary osteoblasts treated with vehicle or PTH. (E) Immunofluorescence assays showing HDAC4 localization in mouse primary osteoblasts treated with vehicle, PTH, or PTHrP. (F) Immunoprecipitation of HDAC4 in WT primary osteoblasts treated with vehicle or PTH plus bortezomib. Proteins were analyzed by Western blotting using anti-HDAC4 or anti-HDAC4 phospho-S246 antibodies. (G) Rankl expression in primary osteoblasts treated with vehicle, PTH, or PTH plus bortezomib. Results are presented as fold changes compared with levels seen in vehicle-treated osteoblasts. (H) Immunoprecipitation of HDAC4 in primary osteoblasts treated with vehicle or PTH plus bortezomib. Proteins were analyzed by Western blotting using anti-HDAC4 or antiubiquitin antibodies. (I) Analysis of expression of E3 ubiquitin ligases in WT primary osteoblasts treated with PTH. Results for each gene are presented as fold change compared with levels seen in vehicle-treated osteoblasts. (J) Rankl expression in scrambled-, Synv1-, or Smurf2 siRNA–transfected primary osteoblasts. Results are represented as fold changes compared with levels seen in scrambled siRNA–transfected cells. (K) Rankl expression in scrambled-, Smurf2-, or Synv1 siRNA–transfected primary osteoblasts treated with vehicle or PTH. Results are presented as fold changes compared with levels seen in vehicle-treated scrambled siRNA–transfected cells. (L) Coimmunoprecipitation assay in ROS17/2.8 cells showing interaction between FLAG-Smurf2 and HA-HDAC4. Cytoplasmic extracts (CE) were immunoprecipitated after treatment with PTH or vehicle with an anti-FLAG antibody, and proteins were detected by Western blotting with anti-HA or anti-FLAG antibodies. (M) Analysis of Rankl expression in WT or Ppr+/−;Smurf2+/− primary osteoblasts treated with vehicle or PTH. (N) Western blot analysis of total extracts from primary osteoblasts treated with vehicle or PTH using anti-HDAC4 and anti–β-actin antibodies. Results are given as means ± SEM. *, P < 0.05 by Student’s t test. IB, immunoblot; IP, immunoprecipitation.
Because PTH does not decrease Hdac4 expression in osteoblasts (Fig. S3 A), we asked whether it induces proteasomal degradation of HDAC4 in osteoblasts. In support of this idea, the PTH-induced decrease of HDAC4 accumulation in osteoblasts was prevented by the addition of an inhibitor of proteasomal degradation, bortezomib (Fig. 3, B and C). Bortezomib also significantly decreased the ability of PTH to up-regulate Rankl expression in osteoblasts, and treating osteoblasts with PTH and bortezomib induced ubiquitination of HDAC4 (Fig. 3, G and H).
PTH up-regulated the expression of two E3 ubiquitin ligases that are highly expressed in osteoblasts Smurf2 and Synv1 (Fig. 3 I; Sévère et al., 2013). We tested whether Smurf2 and Synv1 are involved in PTH signaling in osteoblasts through siRNA knockdown experiments. In each experiment, we verified that we had efficiently decreased either Smurf2 or Synv1 accumulation, while not affecting Hdac4 expression (Fig. S3, B and C). Decreasing Synv1 accumulation in osteoblasts increased Rankl expression and did not prevent PTH from inducing Rankl expression, whereas decreasing Smurf2 accumulation in mouse osteoblasts significantly lowered the expression of Rankl and hampered the ability of PTH to induce Rankl expression (Fig. 3, J and K). Three additional experimental evidences suggested that Smurf2 is a major target of PTH signaling in osteoblasts: first, Smurf2 interacts with HDAC4 only in the presence of PTH; second, PTH did not trigger HDAC4 ubiquitination in Smurf2−/− osteoblasts; and third, the PTH induction of Rankl expression was decreased nearly 50% in osteoblasts lacking one allele of Ppr (encoding the PTH receptor) and one allele of Smurf2 (Fig. 3, J–N). These results indicate that PTH favors Rankl expression in osteoblasts, in part, by recruiting Smurf2 to ubiquitinate HDAC4. This abrogates the physical interaction between HDAC4 and MEF2c and allows MEF2c to activate the Rankl promoter.
The sympathetic tone favors Rankl expression by stabilizing HDAC4
That PTH regulates Rankl expression in an HDAC4-dependent manner raised the hypothesis that another systemic regulator of Rankl expression in osteoblasts, the sympathetic nervous system (Elefteriou et al., 2005), might also recruit HDAC4 to achieve this function. To test whether this was the case, we treated mouse osteoblasts with isoproterenol (ISO), a surrogate of the sympathetic tone. Like PTH, ISO favored Hdac4 expression (Fig. 4 A), but unlike PTH, ISO increased HDAC4 accumulation in the nucleus of osteoblasts and promoted its interaction with ATF4 (Fig. 4, B and C). ISO also decreased the phosphorylation of HDAC4 on S246, an event that favors HDAC4 translocation to the nucleus (Fig. 4 D). In vivo, mice lacking one allele of Hdac4 and one allele of Atf4 only in osteoblasts demonstrated a significant decrease in Rankl expression, circulating CTX values, and the bone surface covered by osteoclasts (Fig. 4, E–G). Of note, although ISO treatment of ROS17/2.8 osteoblastic cells or mouse osteoblasts increased the interaction between HDAC4 and ATF4, it disrupted in tandem the interaction between HDAC4 and Runx2. This latter observation provides an explanation as to why in the living animal, in which sympathetic signaling is constantly present, disruption of HDAC4 in osteoblasts does not affect Runx2-regulated bone formation (Ducy et al., 1999).
This study reveals that Hdac4, through its expression in osteoblasts, prevents expression of Rankl, the major osteoclast differentiation factor, and serves as a bridge between two extracellular cues, regulating Rankl expression and the genome of osteoblasts (Fig. 5). On the one hand, PTH signaling in osteoblasts favors Rankl expression by inhibiting HDAC4 functions. To do that, PTH induces the ubiquitination of HDAC4 by enhancing the expression of an E3 ubiquitin ligase, Smurf2, in osteoblasts and promoting the interaction of Smurf2 and HDAC4. This frees MEF2c that can transactivate the Rankl promoter. Our results do not exclude in any way the fact that PTH also favors Rankl expression in osteoblasts by signaling in T lymphocytes (Gao et al., 2008; Tawfeek et al., 2010). On the other hand, the sympathetic tone favors the accumulation of HDAC4 in osteoblasts and its interaction with ATF4, the only known transcriptional mediator of the sympathetic regulation of Rankl expression (Elefteriou et al., 2005). This mechanism is different from the PKA-induced proteolysis described by Backs et al. (2011) in cardiomyocytes. Although we cannot exclude that such a proteolysis occurs in osteoblasts, we note that if it were the case, ISO would prevent the expression of Rankl instead of increasing it.
Figure 5.
Model of HDAC4 functions in osteoblasts. PTH signaling favors HDAC4 (in gray) degradation through ubiquitination; this releases MEF2c, which can transactivate the Rankl promoter and favor bone resorption. The sympathetic tone favors HDAC4 accumulation, its translocation to the nucleus, its association with ATF4, and thereby, Rankl expression. The broken lines indicate that additional proteins yet to be identified are involved.
Lastly, that compound heterozygous mice lacking one allele of Ppr in osteoblasts and one allele of Mef2c also in osteoblasts have normal bone resorption parameters (unpublished data) argues against the existence of a cross talk between the two arrows of the current models. The different mechanisms whereby PTH and the sympathetic tone use HDAC4 as an intermediary step to fulfill their functions in osteoblasts suggest that these two extracellular cues must recruit different intracellular signaling molecules. Further biochemical studies will help clarify this aspect of PTH and sympathetic signaling in osteoblasts.
Materials and methods
Mouse generation
As previously reported, Hdac5−/− mice were generated by fusing in frame the lacZ cDNA and neomycin resistance cassette under the control of the phosphoglycerate kinase promoter to the 5′ region of exon 3, therefore placing a β-galactosidase (β-gal) reporter gene under the control of endogenous Hdac5 promoter (provided by E. Olson and R. Bassel-Duby, University of Texas Southwestern Medical Center, Dallas, TX; Chang et al., 2004). Hdac4 floxed mice were generated by the insertion of loxP sites, flanking exon 6, which results in an out-of-frame mutation in the Hdac4 allele (provided by E. Olson and R. Bassel-Duby; Potthoff et al., 2007). Hdac4osb−/− mice were generated by intercrossing Hdac4fl/fl mice with Runx2-Cre transgenic mice (Rauch et al., 2010). Mef2a and Mef2c conditional alleles were generated by flanking exon 2 with loxP sites of each gene (provided by E. Olson and R. Bassel-Duby; Arnold et al., 2007; Akhtar et al., 2012). Mef2aosb−/− and Mef2cosb−/− mice (C57BL/6J) were generated by intercrossing Mef2afl/fl or Mef2cfl/fl mice and Runx2-Cre transgenic mice. Hdac4osb−/−;Mef2cosb+/− mice were obtained from crosses between F2 generation Hdac4osb+/−;Mef2cosb+/− and Hdac4fl/fl or Hdac4osb+/− and Hdac4osb+/−;Mef2cosb+/− mice. The Atf4 conditional allele was generated by placing a floxed neomycin cassette upstream of exon 2 and a loxP site downstream of exon 3 (Yoshizawa et al., 2009). Hdac4osb+/−;Atf4osb+/− mice were generated by crossing Atf4fl/fl or Atf4fl/+ mice with Hdac4osb+/− mice. Hdac4fl/fl, Mef2afl/fl, or Mef2cfl/fl mice littermates were used for the control mice for Hdac4osb−/−, Mef2aosb−/−, and Mef2cosb−/− mice, respectively. Floxed mice and Cre-expressing mice littermates (control in the figures) were used as control mice for Hdac4osb−/−;Mef2cosb+/− and Hdac4osb+/−;Atf4osb+/−. Pprfl/fl mice were provided by H. Kronenberg (Massachusetts General Hospital, Boston, MA). Mice genotypes were determined by PCR. All animal procedures were approved by the Columbia University Medical Center Institutional Animal Care and Use Committee and conform to the relevant regulatory standards.
Bone histomorphometry
Lumbar vertebrae or tibia dissected from 2- or 3-mo-old male mice were fixed for 24 h, dehydrated with graded concentrations of ethanol, and embedded in methyl methacrylate resin according to standard protocols. Von Kossa/Van Gieson, toluidine blue, and tartrate-resistant acid phosphatase stainings were used to measure bone volume over tissue volume (BV/TV) and osteoblast and osteoclast numbers and surface, respectively. Bone formation rates were assessed after calcein double labeling. Calcein (Sigma-Aldrich) was dissolved in calcein buffer (0.15 M NaCl and 2% NaHCO3) and injected twice at 0.125 mg/g body weight on days 1 and 4, and mice were killed on day 6. Vertebrate pictures for Von Kossa/Van Gieson were obtained using a microscope (DM4000B; Leica) equipped with a camera (DFC300 FX; Leica) using a 2.5× magnification. Images were acquired with Fire software (Leica), and BV/TV was analyzed using ImageJ software (National Institutes of Health). All quantifications were acquired with a fluorescent microscope (DMLB; Leica) equipped with a charge-coupled device camera (DXC-390; Sony) at 40× magnification. Analysis of the parameters was performed using OsteoMeasure Analysis system (OsteoMetrics).
Cell-based assays
Primary osteoblasts were isolated from calvaria of 2-d-old mice as described previously (Ducy and Karsenty, 1995; Ferron et al., 2010). In brief, calvaria were sequentially digested for 5 and 60 min at 37°C in α-modified MEM (α-MEM; Life Technologies) containing 1 mg/ml collagenase type 2 (Worthington Biochemical Corporation) and 0.25% trypsin-EDTA (Life Technologies). Cells of the first digest were discarded, whereas cells released from the second digestion were plated in α-MEM and 10% fetal bovine serum. For in vitro gene inactivation, floxed osteoblasts were divided into two groups and infected with either GFP- or Cre-expressing adenovirus (University of Iowa). For siRNA transfections, osteoblasts were transfected with siRNA pools (On-TARGETplus; Thermo Fisher Scientific) according to the manufacturer’s instructions. For immunofluorescence, cells were trypsinized, replated after 2 d of culture, and left in regular media conditions for 1 d. On experiment day, cells were washed twice with PBS and treated with indicated amounts of PTH (1–34) (10 nM; ProSpec), PTHrP (1–36) (10 nM; Bachem), ISO (10 µM; Sigma-Aldrich), and bortezomib (25 nM; Selleckchem) for 2 h. Immunofluorescence was performed using a standard protocol. Pictures were obtained on a confocal microscope (LSM 710; Carl Zeiss) at 63× magnification. For gene expression, osteoblasts were cultured in α-MEM containing 10% FBS supplemented with 5 mM β-glycerophosphate and 100 µg/ml ascorbic acid for 10 d. Cells were fasted in serum-free media for 16 h and treated with 10 nM PTH, 10 µM ISO, or 25 nM bortezomib for 2 h. Cells were then collected in TRIZOL (Invitrogen), and RNA isolation, cDNA preparation, and real-time PCR analysis were performed following standard protocols. In brief, total RNA was extracted, DNase I treated, and reverse transcribed with random primers using the cDNA synthesis kit (SuperScript III; Invitrogen). The cDNA samples were then used as templates for quantitative PCR analysis that was performed using the Taq SYBR Green Supermix with ROX (Bio-Rad Laboratories) and specific primers (Table S1) on a CFX Connect apparatus (Bio-Rad Laboratories). Expression levels of all quantitative PCR reactions were normalized using hprt expression levels as an internal control for each sample.
Molecular biology and biochemistry
For DNA cotransfection assays, COS cells were cultured in DMEM supplemented with 10% FBS. Cells were transfected with 100 ng pCMV14-FLAG-MEF2c or 10 ng pCDNA3.1-HA-HDAC4 expression vectors, pGL3-Basic-Rankl (pRankl-luc), pGL3-Basic-Rankls1 (pRankls1-luc), pGL3-Basic-Rankls2 (pRankls2-luc), or pGL3-Basic-Rankls3 (pRankls3-luc; 5 ng), and pCMV–β-gal reporter vectors. DNA transfections were performed in ROS17/2.8 cells cultured in DMEM F12 (1:1) supplemented with 10% FBS. Cells were transfected with 5 ng pRankl-luc or 5 ng pRankls1-luc and 200 ng pCMV–β-gal reporter vectors. 32 h after transfection, cells were washed with PBS and treated with serum-free media containing either 10 nM PTH or vehicle for the indicated times. Luciferase and β-gal assays were performed using standard procedures. For coimmunoprecipitation assays, ROS17/2.8 cells were transfected with 6 µg pCMV5-FLAG-ATF4, 6 µg pCMV5-FLAG-Runx2, 6 µg pCMV5-Smurf2, or pCDNA3.1-HA-HDAC4 (2 µg). 48 h later, cells were washed with PBS and treated with media containing 0.1% FBS and 10 nM PTH, 25 nM bortezomib, or 10 µM ISO for 2 h. Cells were lysed in hypotonic buffer and disrupted by Dounce homogenizer. The cytosolic fraction was separated from the pellet by centrifugation at 4°C. The nuclear-soluble fraction was obtained by incubation of the pellet in high-salt buffer to get a final NaCl concentration of 300 mM. Tagged proteins from each fraction were immunoprecipitated with anti-Flag M2 overnight. The next day, protein G–agarose beads were added and incubated for 2 h at 4°C. Each immunoprecipitation was washed five times with TGEN 150 buffer (20 mM Tris, pH 7.65, 150 mM NaCl, 3 mM MgCl2, 0.1 mM EDTA, 10% glycerol, and 0.01% NP-40), and proteins were eluted with Laemmli buffer and boiled for 10 min. For immunoprecipitation in primary osteoblasts, cells were lysed with 20 mM Tris, pH 7.65, 150 mM NaCl, and 1% NP-40 in and disrupted by Dounce homogenizer. The soluble fraction was incubated with indicated antibodies overnight at 4°C. The next day, protein G–agarose beads were added and incubated for 2 h at 4°C. Each immunoprecipitation was washed five times with TGEN 150 buffer (20 mM Tris, pH 7.65, 150 mM NaCl, 3 mM MgCl2, 0.1 mM EDTA, 10% glycerol, and 0.01% NP-40), and proteins were eluted with Laemmli buffer and boiled for 10 min.
Antibodies used in this study are mouse monoclonal anti-FLAG (F3165; Sigma-Aldrich), rabbit polyclonal anti-MEF2c (ab79436; Abcam), rabbit polyclonal anti-MEF2 (sc-313X; Santa Cruz Biotechnology, Inc.), rabbit polyclonal anti-HDAC4 (sc-11418; Santa Cruz Biotechnology, Inc.), rabbit monoclonal anti-HA (C29F4; Cell Signaling Technology), rabbit polyclonal anti-HDAC5 (2082; Cell Signaling Technology), rabbit polyclonal anti–phospho-HDAC4S-246 (provided by T.P. Yao, Duke University School of Medicine, Durham, NC; Cohen et al., 2007), rabbit monoclonal anti-GAPDH (14C10; Cell Signaling Technology), and mouse monoclonal antiubiquitin (3936; Cell Signaling Technology). DAPI was included in the mounting media (Electron Microscopy Sciences).
Chromatin immunoprecipitation
Mouse osteoblasts were washed twice with PBS and cross-linked with 1.1% formaldehyde for 10 min at 37°C. Cross-linking was stopped by 125 mM glycine with gentle rocking. Cells were washed with PBS containing 0.25 mM PMSF, nuclear extracts were prepared, and each sample was sonicated to obtain sheared fragments of 200–800 bp. Samples were precleared with normal rabbit IgG with incubation for 1 h and agarose G beads for 2 h at 4°C. An anti-MEF2 antibody was added to cleared lysates and incubated overnight at 4°C. The next day, agarose protein G beads were blocked with 200 µg/ml salmon sperm DNA and 1 mg/ml BSA for 1 h at 4°C. Lysates were incubated with blocked beads for 2 h at 4°C and washed once in low salt buffer, twice in high salt, twice in LiCl buffer, and twice in Tris EDTA buffer. Washed beads were eluted in elution buffer by incubating at 37°C for 2 h. The DNA was reverse cross-linked by incubating all samples at 65°C for 6 h. After DNA extraction, PCR analysis was performed on chromatin immunoprecipitated material with specific primers (for binding sites 1 and 2, 5′-GGGTCCTCTTAGCACCTTGAC-3′ and 5′-GACATGCTTAGATGAAAAATG-3′; for binding site 2, 5′-GTGGCATGGGTTTATTTATT-3′ and 5′-TCCTGAGTGCTGGGATTAAA-3′; for binding site 3, 5′-AGGCTCTCTTGTGACCTAGAG-3′ and 5′-CACACTTCCCACTTTGCCCAC-3′; for 3′UTR, 5′-CTAGAATTTCCCCAAGTCTTC-3′ and 5′-GGCTGGCCCCTATCCTTTGC-3′) designed to detect the Mef2 binding sites within the mouse Rankl promoter.
Statistical analyses
Results are given as means ± SEM. Statistical analyses were performed using unpaired, two-tailed Student’s t test. In all figures, error bars represent SEM, and asterisks represent P < 0.05.
Online supplemental material
Fig. S1 shows osteoblast-specific deletion of Hdac4. Fig. S2 shows osteoblast-specific deletion of Mef2a and Mef2c. Fig. S3 shows that PTH regulation of Hdac4 expression is independent of Smurf2 or Synv1 inactivation. Table S1 shows primers used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201403138/DC1.
Supplementary Material
Acknowledgments
We thank Dr. P. Ducy for valuable suggestions and critical reading of the manuscript, Dr. H. Kronenberg for generously providing the Pprfl/fl mice, Dr. T.-P. Yao for kindly providing us with the anti–HDAC4-S246 phosphoantibody, and Dr. R. Bassel-Duby and Dr. E. Olson for generously providing the Hdac4fl/fl, Hdac5−/−, Mef2afl/fl, and Mef2cfl/fl mice.
This work was supported by grant AR045548 from the National Institutes of Health (G. Karsenty). A. Obri acknowledges the Fondation pour la Recherche Médical for financial support.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- BV/TV
- bone volume over tissue volume
- HDAC
- histone deacetylase
- ISO
- isoproterenol
- PTH
- parathyroid hormone
- PTHrP
- PTH-related protein
- WT
- wild type
References
- Akhtar M.W., Kim M.S., Adachi M., Morris M.J., Qi X., Richardson J.A., Bassel-Duby R., Olson E.N., Kavalali E.T., Monteggia L.M. 2012. In vivo analysis of MEF2 transcription factors in synapse regulation and neuronal survival. PLoS ONE. 7:e34863 10.1371/journal.pone.0034863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis C.D., Jenuwein T., Reinberg D., Caparros M.-L., 2007. Epigenetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 502 pp [Google Scholar]
- Arnold M.A., Kim Y., Czubryt M.P., Phan D., McAnally J., Qi X., Shelton J.M., Richardson J.A., Bassel-Duby R., Olson E.N. 2007. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev. Cell. 12:377–389 10.1016/j.devcel.2007.02.004 [DOI] [PubMed] [Google Scholar]
- Backs J., Worst B.C., Lehmann L.H., Patrick D.M., Jebessa Z., Kreusser M.M., Sun Q., Chen L., Heft C., Katus H.A., Olson E.N. 2011. Selective repression of MEF2 activity by PKA-dependent proteolysis of HDAC4. J. Cell Biol. 195:403–415 10.1083/jcb.201105063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S.L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142–148 10.1016/S0959-437X(02)00279-4 [DOI] [PubMed] [Google Scholar]
- Chang S., McKinsey T.A., Zhang C.L., Richardson J.A., Hill J.A., Olson E.N. 2004. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 24:8467–8476 10.1128/MCB.24.19.8467-8476.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen T.J., Waddell D.S., Barrientos T., Lu Z., Feng G., Cox G.A., Bodine S.C., Yao T.P. 2007. The histone deacetylase HDAC4 connects neural activity to muscle transcriptional reprogramming. J. Biol. Chem. 282:33752–33759 10.1074/jbc.M706268200 [DOI] [PubMed] [Google Scholar]
- Ducy P., Karsenty G. 1995. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol. Cell. Biol. 15:1858–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P., Starbuck M., Priemel M., Shen J., Pinero G., Geoffroy V., Amling M., Karsenty G. 1999. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 13:1025–1036 10.1101/gad.13.8.1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elefteriou F., Ahn J.D., Takeda S., Starbuck M., Yang X., Liu X., Kondo H., Richards W.G., Bannon T.W., Noda M., et al. 2005. Leptin regulation of bone resorption by the sympathetic nervous system and CART. Nature. 434:514–520 10.1038/nature03398 [DOI] [PubMed] [Google Scholar]
- Ferron M., Wei J., Yoshizawa T., Del Fattore A., DePinho R.A., Teti A., Ducy P., Karsenty G. 2010. Insulin signaling in osteoblasts integrates bone remodeling and energy metabolism. Cell. 142:296–308 10.1016/j.cell.2010.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q., Jilka R.L., Manolagas S.C., O’Brien C.A. 2002. Parathyroid hormone stimulates receptor activator of NFkappa B ligand and inhibits osteoprotegerin expression via protein kinase A activation of cAMP-response element-binding protein. J. Biol. Chem. 277:48868–48875 10.1074/jbc.M208494200 [DOI] [PubMed] [Google Scholar]
- Gao Y., Wu X., Terauchi M., Li J.Y., Grassi F., Galley S., Yang X., Weitzmann M.N., Pacifici R. 2008. T cells potentiate PTH-induced cortical bone loss through CD40L signaling. Cell Metab. 8:132–145 10.1016/j.cmet.2008.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M., Montgomery R.L., Olson E.N. 2009. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 10:32–42 10.1038/nrg2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D. 2001. Translating the histone code. Science. 293:1074–1080 10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- Kajimura D., Hinoi E., Ferron M., Kode A., Riley K.J., Zhou B., Guo X.E., Karsenty G. 2011. Genetic determination of the cellular basis of the sympathetic regulation of bone mass accrual. J. Exp. Med. 208:841–851 10.1084/jem.20102608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G., Ferron M. 2012. The contribution of bone to whole-organism physiology. Nature. 481:314–320 10.1038/nature10763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G., Kronenberg H.M., Settembre C. 2009. Genetic control of bone formation. Annu. Rev. Cell Dev. Biol. 25:629–648 10.1146/annurev.cellbio.042308.113308 [DOI] [PubMed] [Google Scholar]
- Kondo H., Guo J., Bringhurst F.R. 2002. Cyclic adenosine monophosphate/protein kinase A mediates parathyroid hormone/parathyroid hormone-related protein receptor regulation of osteoclastogenesis and expression of RANKL and osteoprotegerin mRNAs by marrow stromal cells. J. Bone Miner. Res. 17:1667–1679 10.1359/jbmr.2002.17.9.1667 [DOI] [PubMed] [Google Scholar]
- Kozhemyakina E., Cohen T., Yao T.-P., Lassar A.B. 2009. Parathyroid hormone-related peptide represses chondrocyte hypertrophy through a protein phosphatase 2A/histone deacetylase 4/MEF2 pathway. Mol. Cell. Biol. 29:5751–5762 10.1128/MCB.00415-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg H.M. 2003. Developmental regulation of the growth plate. Nature. 423:332–336 10.1038/nature01657 [DOI] [PubMed] [Google Scholar]
- Potthoff M.J., Wu H., Arnold M.A., Shelton J.M., Backs J., McAnally J., Richardson J.A., Bassel-Duby R., Olson E.N. 2007. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J. Clin. Invest. 117:2459–2467 10.1172/JCI31960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A., Seitz S., Baschant U., Schilling A.F., Illing A., Stride B., Kirilov M., Mandic V., Takacz A., Schmidt-Ullrich R., et al. 2010. Glucocorticoids suppress bone formation by attenuating osteoblast differentiation via the monomeric glucocorticoid receptor. Cell Metab. 11:517–531 10.1016/j.cmet.2010.05.005 [DOI] [PubMed] [Google Scholar]
- Sévère N., Dieudonné F.X., Marie P.J. 2013. E3 ubiquitin ligase-mediated regulation of bone formation and tumorigenesis. Cell Death Dis. 4:e463 10.1038/cddis.2012.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawfeek H., Bedi B., Li J.Y., Adams J., Kobayashi T., Weitzmann M.N., Kronenberg H.M., Pacifici R. 2010. Disruption of PTH receptor 1 in T cells protects against PTH-induced bone loss. PLoS ONE. 5:e12290 10.1371/journal.pone.0012290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitelbaum S.L. 2000. Bone resorption by osteoclasts. Science. 289:1504–1508 10.1126/science.289.5484.1504 [DOI] [PubMed] [Google Scholar]
- Vega R.B., Matsuda K., Oh J., Barbosa A.C., Yang X., Meadows E., McAnally J., Pomajzl C., Shelton J.M., Richardson J.A., et al. 2004. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 119:555–566 10.1016/j.cell.2004.10.024 [DOI] [PubMed] [Google Scholar]
- Verdin E., Dequiedt F., Kasler H.G. 2003. Class II histone deacetylases: versatile regulators. Trends Genet. 19:286–293 10.1016/S0168-9525(03)00073-8 [DOI] [PubMed] [Google Scholar]
- Yoshizawa T., Hinoi E., Jung D.Y., Kajimura D., Ferron M., Seo J., Graff J.M., Kim J.K., Karsenty G. 2009. The transcription factor ATF4 regulates glucose metabolism in mice through its expression in osteoblasts. J. Clin. Invest. 119:2807–2817 10.1172/JCI39366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.L., McKinsey T.A., Chang S., Antos C.L., Hill J.A., Olson E.N. 2002. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 110:479–488 10.1016/S0092-8674(02)00861-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.