Abstract
Argonaute proteins and their small RNA co-factors short interfering RNAs (siRNAs) are known to inhibit gene expression at the transcriptional and post-transcriptional levels. In Caenorhabditis elegans, the Argonaute CSR-1 binds thousands of endogenous siRNAs (endo-siRNAs) antisense to germline transcripts and associates with chromatin in a siRNA-dependent manner. However, its role in gene expression regulation remains controversial. Here, we used a genome-wide profiling of nascent RNA transcripts to demonstrate that the CSR-1 RNAi pathway promotes sense-oriented Pol II transcription. Moreover, a loss of CSR-1 function resulted in global increase in antisense transcription and ectopic transcription of silent chromatin domains, which led to reduced chromatin incorporation of centromere-specific histone H3. Based on these findings, we propose that the CSR-1 pathway has a role in maintaining the directionality of active transcription thereby propagating the distinction between transcriptionally active and silent genomic regions.
INTRODUCTION
RNA interference (RNAi) is largely known as a negative regulator of gene expression through different mechanisms. Those include inhibition of mRNA translation, mRNA or pre-mRNA degradation, and inhibition of transcription by promoting heterochromatin assembly (reviewed in refs. 1,2). Short interfering RNAs (siRNAs) bound by conserved Argonaute proteins, some of which have an endonucleolytic capacity, select the complementary RNA targets regulated by RNAi (reviewed in ref. 3). However, neither RNA-RNA interaction nor Argonaute-mediated cleavage excludes the possibility that siRNAs may have a positive effect on gene expression. Here, we report that an RNAi pathway enhances transcription of genes targeted by endogenous siRNAs (endo-siRNAs) in C. elegans genome wide.
endo-siRNAs were initially discovered in C. elegans4, and later in Drosophila and mammals (reviewed in ref. 5). They can be antisense to protein-coding transcripts or can be derived from pseudogenes, transposons and intergenic regions that produce double-stranded RNA (dsRNA) (reviewed in ref. 2). Most known cases of gene regulation by endo-siRNAs include inhibition of their targets at the post-transcriptional level (reviewed in refs. 5-7). However, association of nuclear Argonaute proteins with transcriptionally-active gene loci has also been described in metazoans, including human cells8-12, (reviewed in ref. 13).
The network of endo-siRNA pathways in C. elegans is very extensive (reviewed in ref. 7). In the germline, endo-siRNAs that are produced by the activities of RNA-dependent RNA Polymerases (RdRPs) on target transcripts are termed 22G-RNAs. They belong to two major endo-RNAi pathways: 1) the worm-specific AGO (WAGO) pathway14, which silences pseudogenes, transposons, cryptic loci, and some protein-coding genes, and 2) the CSR-1 pathway, which targets more than 4,000 highly active genes preferentially expressed in the germline15,16. Surprisingly, CSR-1-bound 22G-RNAs do not inhibit expression of their targets15,16. However, the Argonaute CSR-1 has been shown to associate with chromatin in a 22G-RNA-dependent manner15, which suggests nuclear functions. Moreover, inactivation of the CSR-1 RNAi pathway components leads to sterility, abnormal P-granules, chromosome segregation defects, mislocalization of centromeric proteins, such as the variant histone CENP-A, and embryonic lethality15-23. Recently, the CSR-1 pathway has been shown to directly promote histone pre-mRNA processing24 and to positively regulate expression of some spermiogenesis-related genes25. Therefore, the key unanswered question is whether CSR-1-bound 22G-RNAs globally regulate expression of CSR-1 target genes in the nucleus.
To determine the effect of the CSR-1 pathway on RNA Polymerase II (Pol II) transcription we utilized the Global Run-On sequencing (GRO-seq) method26, which allows to map the position, amount and orientation of transcriptionally-engaged Pol II genome wide. We assayed transcription in a loss-of-function mutant of Dicer-related helicase, drh-3, which is depleted of most endo-siRNAs14 and in a csr-1 hypomorphic strain that has reduced expression of CSR-115. Our genomic analyses demonstrate that the CSR-1 pathway promotes Pol II transcription of its active target genes in a siRNA-dependent manner genome wide. Moreover, a loss of CSR-1 activity results in a global increase in antisense transcription and ectopic expression of silent genes, which affects chromatin organization. Therefore, our results suggest that CSR-1 and its associated 22G-RNAs participate in a positive-feedback mechanism that stimulates and restricts sense-oriented Pol II transcription, thus propagating the distinction between “active” and “silenced” chromatin domains.
RESULTS
Reduced transcription of CSR-1 targets in endo-RNAi mutants
Several studies have shown that endo-siRNA abundance negatively correlates with the expression of their target genes in Drosophila, C. elegans and mouse (reviewed in refs. 5-7). CSR-1-associated endo-siRNAs are antisense to more than 4,000 protein-coding genes (CSR-1 targets) expressed in the germline. However, RNA profiling of csr-1 mutant worms did not detect significant increases in steady-state RNA levels of CSR-1 target genes compared to wild type (WT)15. Moreover, a decrease in expression was observed in the csr-1 mutant for a significant number of CSR-1 targets (Supplementary Fig. 1a). Importantly, we have recently shown that CSR-1 binds to histone mRNAs (a prominent group of CSR-1 targets) and promotes their biogenesis24. Given the fact that CSR-1 is present in the nuclear fraction and associates with chromatin at its target genes in a siRNA-dependent manner15, we explored the possibility that CSR-1 can globally regulate its target genes at the transcriptional level. To study the relationship between CSR-1 22G-RNAs and Pol II transcription genome wide, we analyzed the distribution of transcriptionally-engaged Pol II in WT worms and mutants affecting the CSR-1 pathway by GRO-seq. Knowing that csr-1 null mutations cause sterility17, we utilized a previously described hypomorphic csr-1(tm892) mutant strain in which CSR-1 function was partially rescued by a transgenic CSR-1 protein expressed from the germline-specific promoter15. We confirmed that CSR-1 is present in the nuclear fraction and that the csr-1 hypomorphic strain expresses CSR-1 protein isoforms at a lower level compared to WT (Supplementary Fig. 1c, d). We also used a loss-of-function mutant of Dicer-related helicase, drh-3(ne4253) (ref. 14), which is depleted of most 22G-RNAs that belong to the WAGO and CSR-1 pathways. Importantly, the levels of histone proteins and their incorporation into chromatin in these two loss-of-function mutant strains were similar to those in WT (Supplementary Fig. 1b, e). Therefore, the possible indirect effect of histone depletion on transcription, which may occur after prominent down-regulation of CSR-1 by RNAi24, should not be prevalent in these homozygous viable strains used for GRO-seq. Also, we performed GRO-seq using late L3 and early L4 stage worms before the occurrence of germline defects at later stages that could compromise our transcriptional profiling.
To evaluate the global effect of the CSR-1 pathway on Pol II transcription, we divided the GRO-seq reads into groups corresponding to promoters, gene bodies and gene ends, as previously described27. We compared cumulative GRO-seq signals from CSR-1 targets and from all genes in csr-1 hypomorph and drh-3(ne4253) versus WT, and, notably, we found a substantial global reduction of Pol II transcription at CSR-1 target genes in both mutants (Fig. 1a-c). The effects on transcription that we observed in the drh-3 mutant were specific to the CSR-1 target genes, because transcription of the WAGO protein-coding target genes14 was not globally affected (Supplementary Fig. 2a, b). These results indicate that CSR-1 specifically promotes transcription of its target genes in a 22G-RNA-dependent manner. To confirm our analyses of GRO-seq data with a different statistical and normalization approach, we calculated the differential gene expression in csr-1 hypomorph and drh-3(ne4253) mutants compared to WT using the R package DESeq28. In agreement with the analysis using cumulative reads, we only found a significant enrichment of CSR-1 targets among the genes that are transcriptionally down-regulated in the mutants (Fig. 1d, e). Interestingly, we also found that the up-regulated genes were significantly depleted of CSR-1 targets (Fig. 1d, e). Therefore, we quantified the cumulative GRO-seq signals from all the non-target genes and found a global increase in Pol II transcription among the non-target genes in csr-1 hypomorph and drh-3(ne4253) mutant compared to WT (Fig. 1a, b). Finally, we compared our data with the published microarray results generated on csr-1(tm892) mutant and WT adult worms15 and found a significant overlap among genes misregulated in the mutant that were identified by the microarray and by the GRO-seq (Supplementary Fig. 2c, d). This indicates that the changes in nascent RNA levels that we detected in CSR-1 pathway mutants correlate with the changes in steady-state mRNA levels that were published earlier15. Collectively, these results demonstrate that the CSR-1 pathway has a profound impact on Pol II transcription genome wide.
Figure 1. CSR-1 pathway positively regulates Pol II transcription.
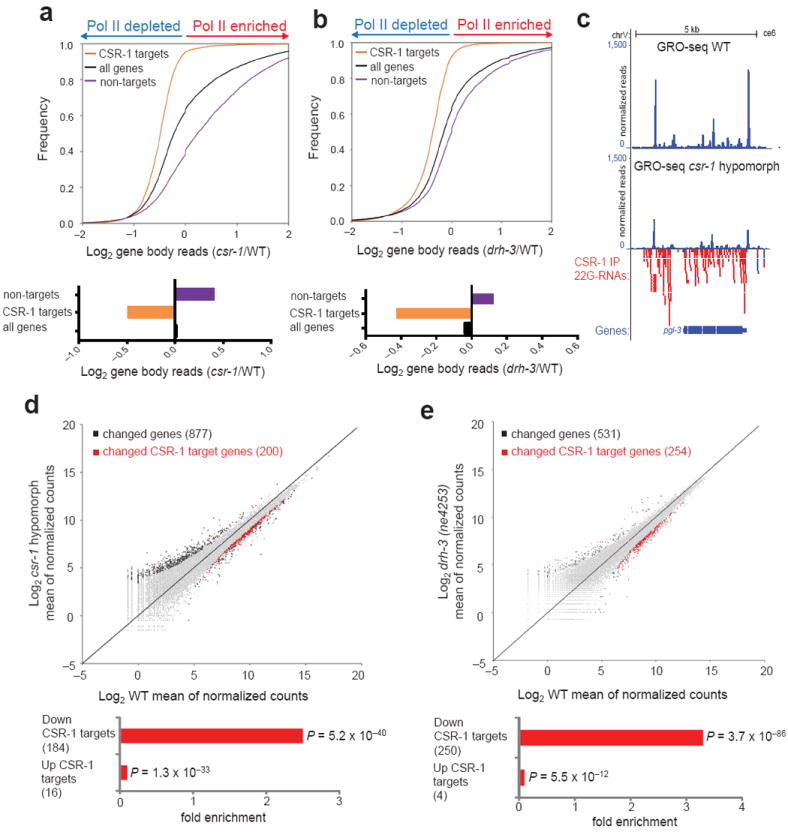
(a) Top, cumulative distribution plots of normalized GRO-seq gene body reads (log2 of the csr-1/WT read ratio): CSR-1 targets, as defined in ref. 15, (orange line), non-target genes (purple line) and total genes (black line). Bottom, the averages of the log2 ratio between csr-1 hypomorphic mutant and WT normalized gene body reads. (b) GRO-seq analysis performed as in a, considering drh-3(ne4253) and WT normalized GRO-seq gene body reads. (c) An example of CSR-1 target gene, pgl-3. Track listing from top to bottom: normalized GRO-seq reads in WT larvae, normalized GRO-seq reads in csr-1 hypomorphic mutant larvae, and CSR-1 22G-RNAs15. Gene models are based on UCSC Genome Browser (ce06). (d) Top, scatter plot of mean of normalized counts (log2) in csr-1 hypomorphic mutant or WT calculated using DESeq package28. The black squares represent genes with significant changes in Pol II transcription considering a False Discovery Rate (FDR) of < 0.05. The red squares represent CSR-1 target genes with significant changes in Pol II transcription (FDR < 0.05). The number of significantly changed genes is shown in parentheses. Bottom, significant enrichment or depletion of CSR-1 targets among, respectively, genes up-regulated and down-regulated by GRO-seq in csr-1 mutant versus WT, P values for the significance of enrichment or depletion were calculated using Fisher’s exact test. (e) Top, scatter plot, as in d, of mean of normalized counts (log2) in drh-3 (ne4352) or WT. Bottom, significant enrichment or depletion of CSR-1 targets as in d.
Next, we investigated which step of Pol II transcription was regulated by CSR-1 22G-RNAs. The decrease in transcription observed in CSR-1 pathway mutants occurred along the entire length of the genes, including promoter regions (Fig. 2a, b and Supplementary Fig. 3a, b). Consistently, we detected decreased levels of Pol II at the promoters of CSR-1 target genes in drh-3(ne4253) compared to WT by Pol II ChIP-qPCR experiments with an antibody that recognizes the non-phosphorylated isoform of Pol II (Supplementary Fig. 3c). In addition, we found an interaction between Pol II and CSR-1 in co-immunoprecipitation experiments (Supplementary Fig. 2d). Importantly, we found that the interaction between Pol II and CSR-1 was dependent on RNA and not on DNA (Fig. 2c). This suggests that CSR-1 and its associated 22G-RNAs interact with Pol II through nascent RNA transcripts. To prove this, we analyzed nuclear RNA that immunoprecipitated with FLAG–CSR-1 expressed from a transgene that rescued sterility of csr-1 null mutant33. Indeed, we found an enrichment of germline-specific nascent transcripts (CSR-1 targets) in FLAG–CSR-1 RNA-IP compared to control IP performed with non-transgenic worms (Fig. 2d). Treatment of nuclear extracts with DNase I did not affect the interaction between CSR-1 and nascent RNA (Fig. 2d). However, inhibition of drh-3 by RNAi caused a substantial reduction in the amount of immunoprecipitated RNA (Fig. 2d). These results indicate that the interaction between CSR-1 and nascent transcripts is mediated by 22G-RNAs and not dependent on DNA.
Figure 2. CSR-1 interacts with Pol II and nascent transcripts.
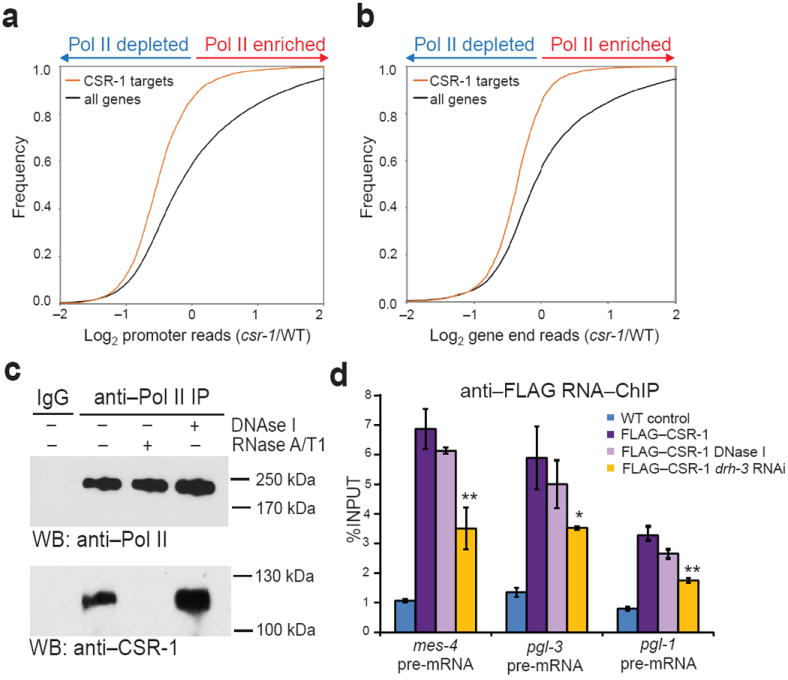
(a, b) Cumulative distribution plots of normalized GRO-seq promoter reads (log2 of the reads’ ratios: csr-1 hypomorph/WT or drh-3(ne4253)/WT): CSR-1 targets (orange line), and all genes (black line). (c) Co-immunopercipitation experiments with nuclear extracts treated or not with DNase I or RNase A/T1. The top panels show western blots with an antibody that recognizes the non-phosphorylated isoform of Pol II and the bottom panels show western blots with an antibody recognizing CSR-1. See Supplementary Fig. 8 for uncropped blot images. (d) RNA-ChIP-qPCR results obtained with an antibody against FLAG and nuclear extracts from non-transgenic worms (blue bars) or transgenic worms expressing FLAG–CSR-1 protein: without DNase I treatment (purple bars) or treated with DNase I (light purple bars). Yellow bars show results with FLAG–CSR-1 strain treated with RNAi against drh-3. Error bars, s.d. (n = 3 technical replicates). *P < 0.05; **P < 0.01 by one-tailed Student’s t test in comparison to the transgenic strain expressing FLAG–CSR-1 protein.
In summary, our findings suggest that CSR-1 has a direct effect on the Pol II complex and promotes its association with CSR-1 target loci through interactions of 22G-RNAs with nascent transcripts.
Increased antisense transcription in CSR-1 pathway mutants
Given that the GRO-seq method can distinguish the orientation of Pol II transcripts, we analyzed changes in antisense transcription. Surprisingly, we detected a global increase in antisense Pol II transcription in both csr-1 and drh-3 mutants (Supplementary Fig. 4a, b). To confirm these results with a different normalization method, we calculated the numbers of genes with marked up-regulated or down-regulated antisense transcription in CSR-1 pathway mutants compared to WT using DESeq28. Indeed, we found that the majority of genes with substantial changes in antisense transcriptions were up-regulated in CSR-1 pathway mutants (Fig. 3a, b). Interestingly, the increase in antisense transcription was also observed on CSR-1 target genes (Fig. 3c). Because CSR-1 primarily associates with 22G-RNAs antisense to mRNA transcripts15 and we found that it interacted with nascent RNA Pol II transcripts (Fig. 2d), these results are consistent with the possibility that CSR-1-associated 22G-RNAs stabilize sense-oriented Pol II machinery through their interaction with nascent sense RNA. This, in turn, may reduce the frequency of Pol II initiation in the antisense orientation.
Figure 3. Increased antisense Pol II transcription in CSR-1 pathway mutants.
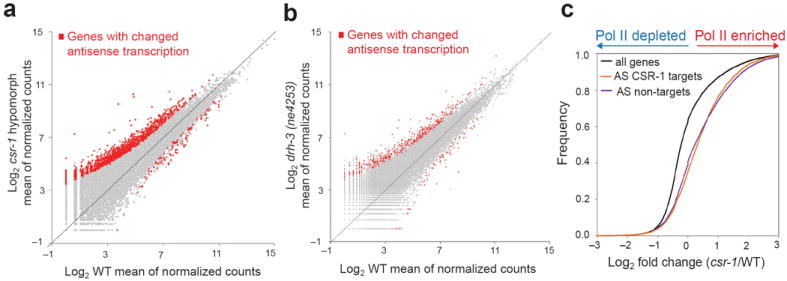
(a, b) Scatter plots, as in Fig. 1d, e, of mean of normalized counts (log2) calculated using DESeq package28 and showing genes with statistically significant changes in antisense Pol II transcription in csr-1 hypomorph or drh-3(ne4253) mutants versus WT, respectively, (FDR < 0.05). (c) Cumulative distribution plots of normalized GRO-seq reads showing CSR-1 targets (antisense (AS) reads, orange line) and non-targets (AS reads, purple line) compared to the sense reads of all genes (black line). Only reads from the gene body were considered for the analyses shown in c.
Functional analysis of gene expression changes
An interesting feature of our GRO-seq findings is the global increase in transcription of non-CSR-1-target genes in csr-1 and drh-3 mutants (Fig. 1a, b, d, e). To further explore the nature of this increased transcription, we performed functional analysis of GRO-seq data using categories of genes that have been previously defined by their expression profiles29-32. We found that genes that were highly expressed at the developmental stage used for our analysis (late L3 and early L4), including ubiquitous and germline-enriched genes, showed a significant global decrease in Pol II transcription in csr-1 hypomorph compared to WT (Supplementary Fig. 5a). Conversely, genes that are low-expressed or not expected to be expressed at the studied developmental stage, including the serpentine gene family and spermatogenesis-related genes, showed a significant global increase in Pol II transcription (Supplementary Fig. 5a). This analysis suggests that the CSR-1 pathway differentially regulates highly expressed genes and low-expressed or silent genes. Indeed, when we analyzed cumulative GRO-seq signals from the top 20% most highly transcribed genes and the lowest 20% transcribed genes (defined by GRO-seq) we found that highly expressed genes showed a global depletion in Pol II transcription in the studied mutants, and that low-expressed or silent genes showed a global increase in transcription (Fig. 4a and Supplementary Fig. 5d). The changes in Pol II transcription in these categories of genes were correlated with the enrichment or depletion of CSR-1 targets (Supplementary Fig. 5b, c). Similar results were obtained in GRO-seq analyses when genes were divided based on quartile of expression (Fig. 4b and Supplementary Fig. 5e). Finally, we evaluated the distribution of Pol II transcription along the chromosomes. Strikingly, changes in Pol II transcription on each chromosome were in accordance with the chromosomal enrichment of CSR-1 targets (Fig. 4c, d). Collectively, these results are consistent with a model of genome-wide redistribution of Pol II from highly transcribed genes to low-transcribed or silenced genes in CSR-1 pathway mutants. Therefore, we propose that CSR-1 and its associated 22G-RNAs have a role in restricting the localization of Pol II to highly transcribed genes.
Figure 4. Categories and chromosomal distributions of genes affected by CSR-1 pathway mutants.
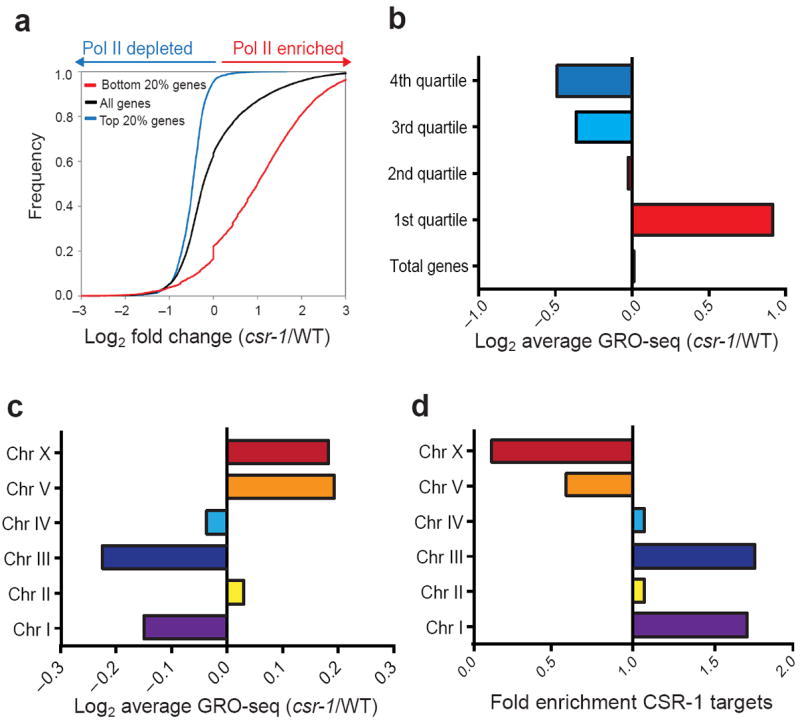
(a) Cumulative distribution plots of normalized GRO-seq reads showing changes in transcription in csr-1 hypomorph compared to WT: top 20% highly-transcribed genes (blue line), bottom 20% genes (red line) and all genes (black line). Only reads from the gene body were considered for this analysis. (b) The average of the log2 ratios between the csr-1 hypomorphic mutant and WT normalized gene body reads quantified for genes grouped by quartiles of expression, based on GRO-seq. (c) The averages of the log2 ratios between the csr-1 hypomorphic mutant and WT reads quantified for genes present on each autosome. (d) Enrichment or depletion of CSR-1 target genes among C. elegans chromosomes, see the formula used to calculate enrichment or depletion in online methods section.
Ectopic transcription of silent chromatin domains
Two distinct histone marks characterize transcriptionally silent and active chromatin domains in the C. elegans germline: the trimethylation of lysine 27 (H3K27me3) and lysine 36 (H3K36me3), respectively; these mutually exclusive domains are epigenetically propagated in the embryo31,33,34. Consistent with these results, we found that the genome-wide distribution of Pol II transcription in larvae, as defined by GRO-seq, positively correlated with the presence of H3K36me3 and negatively correlated with the presence of H3K27me3 (Fig. 5a, c). The centromeric histone H3 variant CENP-A is incorporated at regions devoid of transcription and co-localizes with H3K27me3 (ref. 35) (Fig. 5a-c). In contrast, GRO-seq signals were enriched at genomic regions that produce CSR-1-associated 22G-RNAs (Fig. 5b). Therefore, there is an inverse correlation between CENP-A domains and transcriptionally active 22G-RNA-producing regions, which was noted earlier35. Given the fact that normally silent genes were ectopically transcribed in CSR-1 pathway mutants (Fig. 4a, b and Supplementary Fig. 5a, d, e), we analyzed cumulative GRO-seq reads from CENP-A-enriched domains and H3K27me3-enriched genes and indeed detected an increase in transcription in csr-1 hypomorphic mutant compared to WT (Fig. 5d-f). These results are consistent with a loss of robust distinction between transcriptionally active and silent genomic regions in the studied mutants.
Figure 5. Relationship between silent and active chromatin domains in CSR-1 pathway mutants.
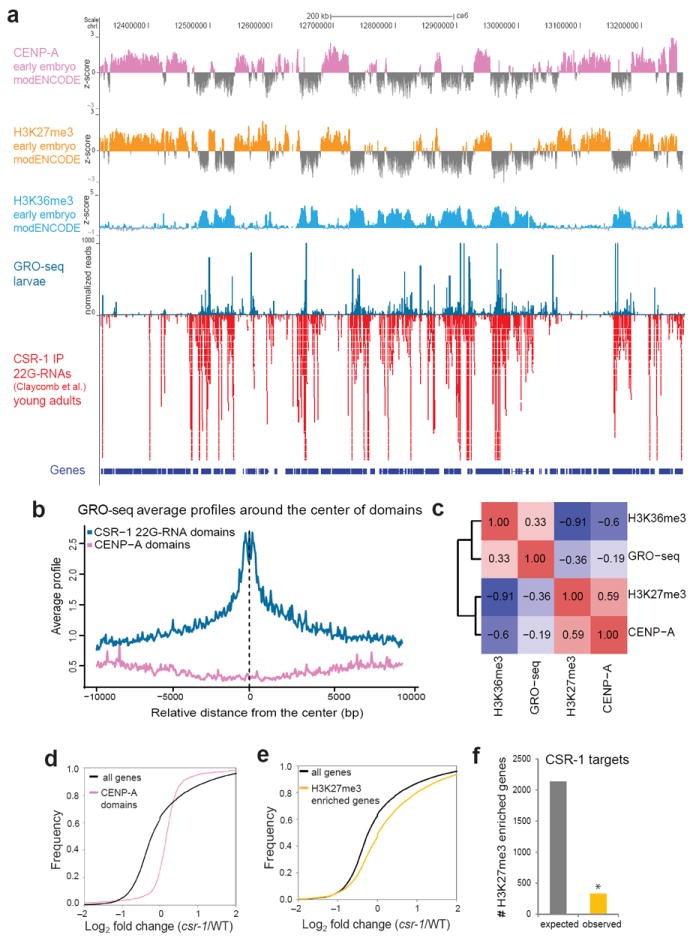
(a) A map of a portion of chromosome I, track listing from top to bottom: ChIP-chip peaks for CENP-A, H3K27me3 and H3K36me3 (early embryo data from modENCODE), GRO-seq reads (this study) and CSR-1-bound endo-siRNA reads15. Gene models are based on UCSC Genome Browser (ce06). (b) Average GRO-seq profiles at CSR-1-associated endo-siRNA-enriched genomic domains (blue line) and at CENP-A-enriched domains (pink line). (c) A heatmap showing the genome-wide correlation coefficient values between GRO-seq reads and H3K36me3 ChIP-chip, CENP-A ChIP-chip and H3K27me3 ChIP-chip peaks (all ChIP-chip data are from modENCODE). Positive correlations are shown in red and negative correlations in blue. (d) Cumulative distribution plots of normalized GRO-seq reads showing changes in transcription in csr-1 hypomorph compared to WT at CENP-A-enriched domains (red line) compared to all genes (black line). (e) Cumulative distribution plots, as in d, at genes enriched in H3K27me3 (yellow line) compared to all genes (black line). (f) Expected (grey bar) and observed (orange bar) numbers of CSR-1 target genes among the genes enriched in H3K27me3. Asterisk indicates significance of less than expected by chance calculated using hypergeometric test, P < 0.0001.
Altered CENP-A incorporation into chromatin
Mutants of the CSR-1 pathway show chromatin organization defects, including mislocalization of centromeric proteins, such as CENP-A15-17,19,23. Depletion of canonical histone proteins upon strong down-regulation of CSR-1 by RNAi is predicted to have a significant impact on chromatin integrity24. However, we thought of the possibility that ectopic transcription seen in the csr-1 and drh-3 hypomorphic mutants may also affect chromatin organization, because CENP-A incorporation at the holocentric C. elegans chromosomes is inhibited at the genomic loci transcribed in the germline35.
To test this hypothesis, we selected genomic regions that contain well-known active germline-specific CSR-1 target genes adjacent to silent non-target genes. We confirmed that in csr-1 hypomorphic mutant these regions showed a decreased Pol II transcription of active germline-specific target genes and increased transcription of silent non-target genes (Supplementary Fig. 6). Notably, the increased ectopic transcription of silent regions was still low compared to the transcription of active germline-specific target genes (Supplementary Fig. 6); nonetheless, it may affect chromatin organization. In agreement with previous data34,35, in WT embryos, we found that genes with low germline expression maintained higher levels of H3K27me3 and CENP-A compared to the adjacent active germline genes. Strikingly, this enrichment of H3K27me3 and CENP-A at non-germline genes was markedly reduced in CSR-1 pathway mutants (Fig. 6a-d). These findings are consistent with the prediction that ectopic transcription interferes with CENP-A deposition. Importantly, there was no increase in the levels of H3K27me3 and CENP-A on CSR-1 target genes in the mutant embryos (Fig. 4a-d). Moreover, no decrease in H3K36me3 levels and MES-4 binding at CSR-1 target genes was detected in mutant larvae (Supplementary Fig. 7). Thus, the reduced levels of H3K27me3 and CENP-A observed in CSR-1 pathway mutants were not the consequence of redistribution of H3H27me3, as observed upon reduction of mes-4 expression34. In addition, these results also exclude the possibility that reduced transcription of CSR-1 target genes observed in csr-1 and drh-3 mutants was due to ectopic silencing or reduced activity of chromatin factors that promote transcription, such as MES-4.
Figure 6. Decrease in H3K27me3 and CENP-A chromatin localization in CSR-1 pathway mutant embryos.
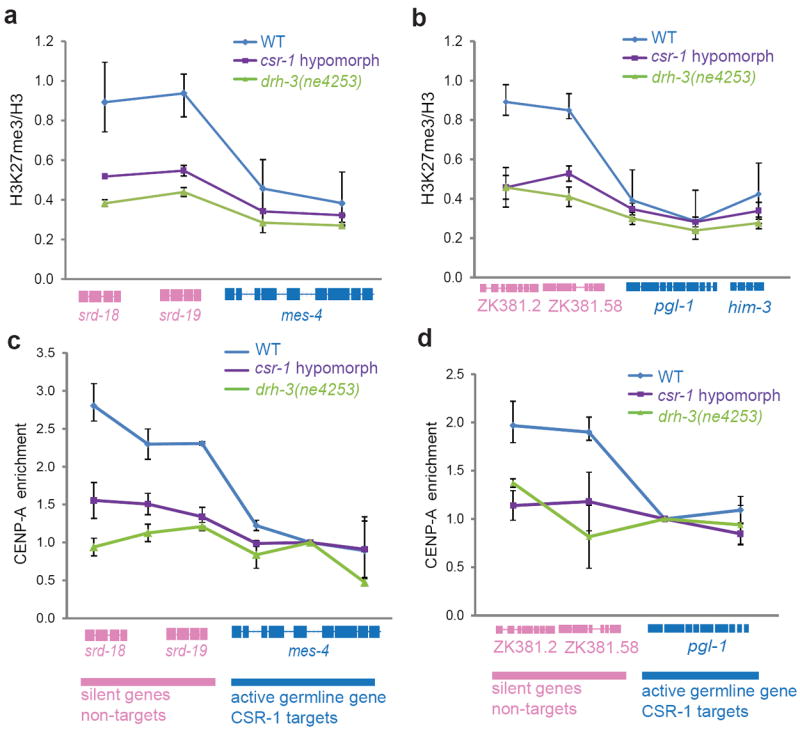
(a, b) ChIP-qPCR results showing the levels of H3K27me3 normalized to the total levels of histone H3 at two genomic loci in WT, csr-1 hypomorph and drh-3(ne4253) mutants. Genes colored in pink correspond to the non-target silent genes that increase in transcription in mutant larvae and genes colored in blue are active germline-specific CSR-1 target genes that decrease in transcription in CSR-1 pathway mutant larvae. Bars indicate ranges, (n = 2 biological replicates). Gene models are based on UCSC Genome Browser (ce06). (c, d) ChIP-qPCR as in a, b, showing the enrichment of CENP-A on chromatin in WT and csr-1 hypomorph and drh-3(ne4253) mutants. The enrichment of CENP-A has been calculated relative to a region in the CSR-1 target genes for each locus considered (circles). Bars indicate ranges, (n = 2 biological replicates).
The above results suggest that the global effect of the CSR-1 pathway on transcription is linked to the maintenance of distinct active and silent chromatin domains genome wide.
DISCUSSION
RNAi has been defined as a gene silencing mechanism. In metazoans, the discoveries of endo-siRNAs antisense to mRNA transcripts and transposons and their ability to repress their target RNAs further supported the paradigm that endogenous RNAi-related pathways negatively regulate gene expression (reviewed in refs. 5-7,13). In C. elegans, one of the two major germline endo-siRNA pathways, the WAGO pathway, has been implicated in silencing of transposons, pseudogenes, cryptic loci and some protein-coding genes14. Recently, a connection between the PIWI-interacting RNA (piRNA) pathway and the WAGO pathway has also been described36-40. piRNAs can initiate the production of endo-siRNAs that are loaded onto some WAGO Argonaute proteins to silence the pre-mRNAs of their targets and to promote the methylation of lysine 9 on histone H3 (H3K9me), which is a chromatin mark associated with gene silencing36,40.
The Argonaute CSR-1 is distinct among the many C. elegans Argonaute proteins in that it binds 22G-RNAs antisense to thousands of protein-coding genes15, and the CSR-1 pathway has been associated with the most dramatic developmental phenotypes15-23. These findings suggest that CSR-1 is involved in gene expression regulation on a global scale. Since CSR-1 localizes to chromatin15, we tested the possibility that it regulates gene expression at the transcriptional level. To investigate the transcriptional changes in mutants of the CSR-1 pathway we used the GRO-seq method, which allows the most sensitive quantitative view of the distribution of transcriptional-engaged Pol II in a genome-wide fashion26. Unexpectedly, we have discovered a global positive role of the CSR-1 pathway in transcriptional regulation in C. elegans. We found that CSR-1 associated with Pol II machinery to promote the high level of sense-oriented Pol II transcription on its target germline-expressed genes in a siRNA-dependent manner. Thus, it is likely that the multiple developmental abnormalities in CSR-1 pathway mutants may result from decreased transcription of germline genes.
Does CSR-1 play a direct role in promoting transcription?
Despite the lack of a clear mechanistic understanding of how CSR-1 regulates Pol II transcription, several lines of evidence support the idea that CSR-1 has a direct effect on its target genes. First, CSR-1 physically localizes to the target loci in the nucleus15. Second, we found that CSR-1 interacted with Pol II machinery and that this interaction was mediated by RNA. Third, we showed that CSR-1 interacted with nascent transcripts from the target loci in 22G-RNA dependent manner. Fourth, our GRO-seq analyses demonstrated down-regulation of Pol II transcription in CSR-1 pathway mutants specifically at genes targeted by antisense CSR-1-bound 22G-RNAs. Fifth, we observed similar transcriptional changes in drh-3(ne4253) and csr-1 hypomorphic mutants, indicating that sequence-specificity of DRH-3-dependent 22G-RNAs must play a role. Moreover, a decrease in transcription observed in drh-3(ne4253) mutant occurred specifically at the CSR-1 target genes and not at other 22G-RNA targets, such as WAGO protein-coding target genes.
In addition, we can also exclude several possible indirect effects of CSR-1 on transcription. It is unlikely that a decrease in canonical histone levels led to transcriptional changes observed here. Our GRO-seq analyses were performed on homozygous viable partial loss-of-function mutant strains, which did not show histone depletion seen in csr-1 RNAi-treated worms24. This allowed us to separate the indirect consequences of histone depletion from the apparent direct role of CSR-1 in transcription. Also, it is not likely that reduced transcription of CSR-1 target genes was due to ectopic silencing mediated by Polycomb group proteins or by the piRNA–WAGO system since we have not detected an increase in H3K27me3 on CSR-1 target genes in CSR-1 pathway mutants and we have shown that the occupancy of MES-4 and H3K36me3 was not reduced under these conditions. Importantly, similar changes in transcription were observed in both csr-1 and drh-3 mutants, although DRH-3 helicase is also required for WAGO 22G-RNA production14 and gene misregulation profile in drh-3 mutant would have been distinct from that of csr-1, if WAGO-bound 22G-RNAs were causing ectopic silencing in the absence of CSR-1. Although we cannot exclude all possible indirect effects, based on the above arguments, it is most likely that CSR-1 and its 22G-RNA cofactors have a direct role in promoting expression of their target genes.
Indirect effects of CSR-1 pathway on transcription
Another important aspect of CSR-1-mediated transcriptional regulation, which has been revealed by our GRO-seq analyses, is the global increase in transcription at the genomic regions that do not include CSR-1 target genes. We have shown that those regions correspond to the silent part of the genome and that the increased Pol II transcription occurred in sense and antisense orientation. Because CSR-1 does not bind 22G-RNAs corresponding to those silent regions, it is unlikely that CSR-1 participates in controlling their transcription directly. One possibility is that CSR-1-bound antisense 22G-RNAs interact with nascent sense transcripts and directly reinforce sense-oriented Pol II complexes to prevent premature termination. Indeed, we found that CSR-1 associated with nascent transcripts in 22G-RNA-dependent manner and that RNA mediated the interaction between CSR-1 and Pol II machinery. Therefore, in the CSR-1 pathway mutants there may be an increased availability of Pol II to initiate non-specific transcriptional events all over the genome, including antisense transcription at CSR-1 targets themselves (Fig. 7).
Figure 7. Model illustrating the proposed role of the CSR-1 pathway in regulation of Pol II transcription and chromatin organization.
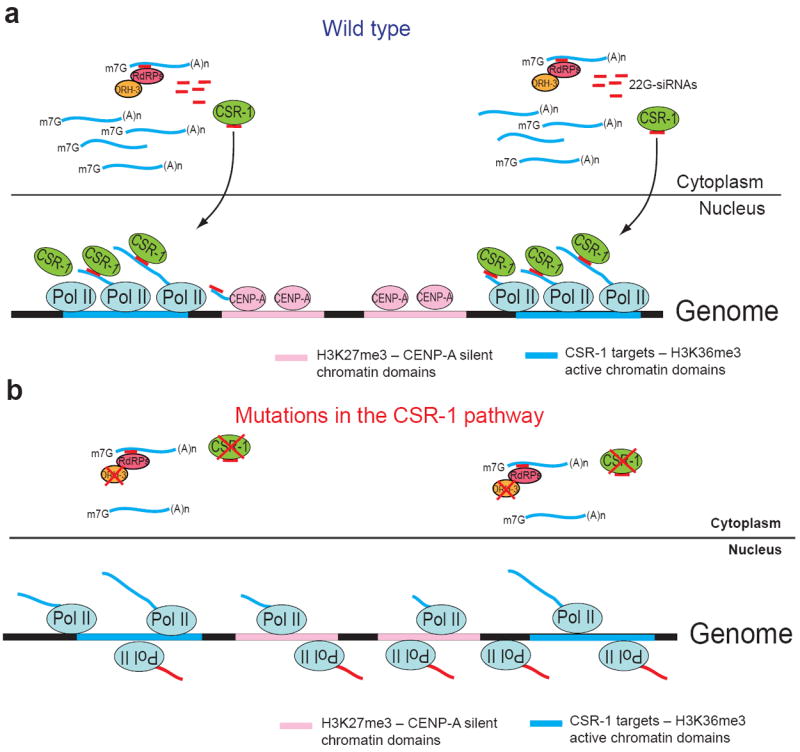
(a) Cytoplasmic mRNAs derived from actively transcribed regions are used as templates by RdRP complexes containing DRH-3 to produce antisense 22G-RNAs that are loaded onto CSR-1 Argonaute15. CSR-1 and its associated siRNAs translocate into the nucleus where they interact with complementary nascent RNA transcripts to promote and localize Pol II transcription on active genes and to inhibit sense and antisense transcription at the silent chromatin regions. This leads to the reinforcement of the distinction between active and silent chromatin domains. (b) Impaired function of the components of the CSR-1 pathway (such as CSR-1 or DRH-3) leads to the reduction in the levels of Pol II transcription on active genes and results in the ectopic sense and antisense transcription. Ectopic transcription at normally silent chromatin domains leads to reduced CENP-A incorporation and to the loss of robust differences between active and silent chromatin.
In different systems, Argonaute proteins have been shown to participate in several other nuclear processes, including splicing10,12 and three-dimensional genome organization8,41. Also, we have described the role of the CSR-1 pathway in histone pre-mRNA processing24. Here we found that transcription of histone genes, along with other CSR-1 targets, was reduced in csr-1 and drh-3 mutants. Therefore, there is another possibility for explaining our results: it is conceivable that CSR-1 globally affects pre-mRNA processing of its target genes, which is linked to transcriptional re-initiation. In this regard, RNA processing factors have been implicated in promoting gene loop formation on active genes42, and there is evidence that gene looping reinforces transcription re-initiation and promotes the directionality of Pol II transcription by favoring sense over antisense transcription initiation43,44.
In addition to histone mRNA processing, CSR-1 has also been implicated in translational regulation in the germline45. Also, an inhibitory effect of CSR-1-bound 22G-RNAs on some target mRNAs has been suggested46. Therefore, future investigation of direct and indirect inputs of CSR-1 on gene expression, possibly by multiple mechanisms, and their interconnections will be quite challenging, but it also promises to be very exciting.
Transcriptional regulation and chromatin organization
In addition to the challenge of understanding the mechanism by which CSR-1 regulates Pol II transcription, another important question is why an RNAi-based mechanism has evolved to potentiate transcription of active genes. In this study, we have accumulated evidence suggesting that the germline CSR-1 pathway is important for the propagation of the distinction between active and silent transcriptional domains in the progeny. In S. pombe, siRNAs generated from the peri-centromeric transcripts promote H3K9 methylation and heterochromatin formation in cis-, which facilitates centromere function (reviewed in ref. 1). Holocentric chromosomes in C. elegans incorporate centromeric proteins along the entire length of chromosomes. Moreover, CENP-A protein does not co-localize with H3K9 methylated regions, which are present on chromosomal arms35,47. Instead, CENP-A is incorporated on transcriptionally silenced chromatin domains enriched in H3K27me3, whereas CSR-1 22G-RNAs are produced from active chromatin domains that are depleted of CENP-A35. Therefore, there is an inverse correlation between CENP-A domains and CSR-1 22G-RNAs35. Here, we have shown that CSR-1 22G-RNAs generated from actively transcribed genomic regions promoted the maintenance of Pol II transcription from these target genomic loci in cis-. Moreover, we have demonstrated that decreased activity of CSR-1 led to increased transcription along the CENP-A-enriched domains, which was sufficient to reduce the incorporation of CENP-A in the embryos. Thus, we propose that the effect of CSR-1 on transcription is also important for proper CENP-A incorporation at the regions devoid of transcription (Fig. 7).
Interestingly, a recent study has shown that CSR-1 and two additional Argonaute proteins, ALG-3 and ALG-4, promoted expression of some spermatogenesis-specific CSR-1 target genes and suggested that heritable 22G-RNAs transmit gene-activating information from one generation to the next25. Also, CSR-1-bound 22G-RNAs are thought to promote expression of artificial transgenes in an epigenetic manner48,49. These observations are consistent with the global positive effect of CSR-1 on transcription revealed by our studies. It is possible that heritable CSR-1 22G-RNAs potentiate germline transcription genome wide, which then leads to definition of chromatin domains that are marked by distinct chromatin modifications and persist in the zygote and developing embryos.
Since the association of nuclear Argonautes with euchromatin has been shown in other animals, including human cells8-12, and gene activation has been described upon targeting human Argonaute proteins to promoter regions using siRNAs50-53, positive regulation of transcription by endogenous RNAi may be common in metazoans.
ONLINE METHODS
Strains
Strains were maintained at 20°C unless otherwise noted, using standard methods54. Bristol N2 was the wild type strain used. Strains used in this study were: drh-3(ne4253), csr-1(tm892); neIs19[pie-1∷3xflag∷csr-1, unc-119(+)], LGII: neSi1[cb-unc-119(+) 3xflag∷csr-1]. For the GRO-seq experiments, synchronous populations of worms were grown for approximately 40 hr post-hatching at 20°C on OP-50 E. coli at a density of ~100,000 animals per 15 cm Petri dish. For early embryo preparations, worms were grown at 20°C on OP-50 E. coli at a density of ~40,000 animals per 15 cm Petri dish until gravid adults contained five embryos or fewer per worm, then embryos were extracted by hypochlorite treatment.
Cytoplasmic and nuclear fractionation
Nuclear and cytoplasmic extracts were prepared as follows: worms were washed off the plates with cold M9 and washed 3-4 times. The worm pellet was resuspended in 2ml of cold Nuclear Run-On (NRO) lysis buffer (10 mM Tris pH 7.5, 2 mM MgCl2, 3 mM CaCl2, 0.5% IGEPAL and 10% glycerol), transferred to a steel dounce (on ice) and stroked 30 times. The worm lysate was centrifuged for 2 min at 40 × g and the supernatant was centrifuged again for 5 min at 1,000 × g to pellet the nuclei. Next, the supernatant was stored as a cytoplasmic fraction and the nuclei pellet was washed three times with 1 ml of cold NRO lysis buffer and then resuspended in extraction buffer (50 mM Tris, 300 mM NaCl, 1 mM EDTA, 0.1% NP40, protease inhibitor cocktails (Fermentas)) and sonicated with 30 sec pulses 4 times using Branson microtip sonicator at 10% power in the cold room. The resulting material was used for western blotting and co-immunoprecipitation experiments or washed an additional time with 1ml of cold Freezing Buffer (50 mM Tris-Cl pH 8.3, 40% glycerol, 5 mM MgCl2, 0.1 mM EDTA) and then resuspended in 100 μl of Freezing Buffer and used for the GRO-seq experiments.
GRO-seq
GRO-seq experiments were performed as described in (ref. 27). Two biological replicates were generated for the csr-1 hypomorphic mutant and the corresponding WT samples using ~300,000 worms for each experiment, and two biological replicates were prepared for the drh-3(ne4253) mutant and the corresponding WT samples using ~100,000 worms for each experiment.
Data Analysis
Two independent biological replicates of GRO-seq were generated using csr-1 hypomorphic strain and the corresponding N2 wild type (WT) (called Mut_CSR-1 and WT_CSR-1, respectively); ~300,000 worms were used for each replicate. Since the drh-3(ne4253) mutant strain has a reduced number of progeny we included two biological replicates of drh-3(ne4253) using ~100,000 worms and compared them with two biological replicates of N2 (WT) generated using an equal number of worms (Mut_DRH-3 and WT_DRH-3, respectively). Sequencing was performed to a length of 50 nucleotides. Since cloned RNA ranged from 16 to 180 nucleotides in length, the sequencing reads were post-processed to eliminate the 3′ linker sequence, if present, so that resulting reads ranged from 16 to 50 nucleotides in length. The reads were then aligned using “bwa 0.6.1” to the C. elegans genome (ce6 assembly), and rRNA reads were removed based on genomic locations:
| chrI | 15060000 | 15072421 | Ce_18S_rDNA 0 + |
| chrV | 17115500 | 17132700 | Ce_5S_rDNA 0 + |
| chrV | 17428800 | 17429040 | Ce_5S_rDNA-singleton 0 + |
| chrX | 16929621 | 16940441 | Ce_repetitiveLocus 0 + |
After filtering, we obtained 44,155,515 reads from csr-1 hypomorphic mutant [26,584,972 reads from the first replicate (called Mut_CSR-1_Rep1) and 17,570,543 reads from the second replicate (called Mut_CSR-1_Rep2)] and 36,133,711 reads from the corresponding WT [16,052,870 reads from the first replicate (called WT_CSR-1_Rep1) and 20,080,841 reads from the second replicate (called WT_CSR-1_Rep2)]. From drh-3(ne4253) mutant, we obtained after filtering 34,588,133 reads [17,492,181 reads from the first replicate (called Mut_DRH-3_Rep1) and 17,095,952 reads from the second replicate (called Mut_DRH-3_Rep2)], and 63,000,071 reads from the corresponding WT [21,660,645 reads from the first replicate (called WT_DRH-3_Rep1) and 41,339,426 reads from the second replicate (called WT_DRH-3_Rep2)]. We further filtered the data by strand (±) to analyze sense and antisense Pol II transcription. After ascertaining concordant results with the individual replicate pairs (WT vs. mutant), replicate reads for each WT and mutant were pooled, using downsampling on the replicate with higher read count to ensure equal representation of either replicate, and the pooled reads were processed together to increase statistical power.
For each annotated C. elegans transcript, the aligned reads were binned based on the annotated transcriptional start site (TSS) and transcriptional termination site (TTS) into 3 regions as follows: ‘promoter’ (from 600bp upstream of the annotated TSS to 200bp downstream), ‘gene body’ (from 200bp downstream of the annotated TSS to 200bp upstream of the annotated TTS), and ‘gene end’ (from 200bp upstream of the annotated TTS to 400bp downstream). We then calculated region-specific RPKM values (# of mapped reads per region / ([region length in KB] × [# of total mapped reads in million]). We also calculated statistically significant transcriptional changes in csr-1 hypomorph and drh-3(ne4253) mutants versus WT using DESeq package28 with a FDR of < 0.05.
For the enrichment or depletion of gene sets, we first calculated the number of genes expected by chance according to r = (n1 × n2)/N, with n1 = number genes in set 1, n2 = number of genes in set 2, N = total number of C. elegans genes considered, and then the enrichment was calculated by dividing the number of genes in common to the two datasets by r.
The average data profiles in Figure 5B were calculated using Cistrome–Galaxy55 with the CEAS tool (SitePro: Aggregation plot tool for signal profiling (version 1.0.0)), and the heatmap showing the genome-wide correlation coefficient values in Figure 5C was calculated using Multiple wiggle files correlation tool (version 1.0.0).
The overlaps between gene lists were calculated using the online tool at http://www.nemates.org. Representation factors “r” to quantify the enrichment or depletion of gene sets were calculated according to r = (n1,2)/[(n1 × n2)/N], with n1,2 = number of genes common to set 1 and 2, n1 = number genes in set 1, n2 = number of genes in set 2, N = total number of genes considered.
Genes enriched in H3K27me3 were calculated using H3K27me3 ChIP-chip data sets generated with early WT embryos in ref. 34. We determined the number of genes enriched in H3K27me3 as described ref. 34.
The genomic coordinates of CENP–A enriched domains were taken from CENP–A ChIP–chip data set described in ref. 35 and available at http://www.modencode.org/. MES–4-enriched domains were taken from MES-4–FLAG ChIP-chip available at http://www.modencode.org/ The data sets of endo-siRNA cloned from CSR-1 IP complexes and from csr-1(tm892) and ego-1(om97) mutants that were used for our analyses are from ref. 15.
The genes sets identified in this study and published gene sets used for comparisons are listed in Supplementary Tables 1 and 2.
Western blotting
The protein concentration in nuclear and cytoplasmic extracts was quantified by the Bradford assay and equal amounts of proteins were then resolved on precast NuPAGE Novex 4-12% Bis-Tris gels (Invitrogen) for 1h at 4°C and transferred to a nylon membrane via semidry transfer (BioRad) at a constant current of 0.12 A for 1h. The antibodies used are as follows: anti-Pol II 8WG16 (Covance, MMS-126R, 1:1,000 dilution), anti-CSR-1 (a gift from J. Claycomb and the Mello lab, validated in ref. 15, 1:2,000 dilution), anti-H2B (Abcam, ab1790, 1:2,000 dilution), anti-actin (Millipore, MAB1501R, 1:2,000 dilution), anti-H2A (Abcam, ab13923, 1:2,000 dilution), anti-rabbit IgG HRP-labeled (PerkinElmer, NEF812001EA, 1:10,000 dilution), anti-mouse IgG HRP-labeled (PerkinElmer, NEF822001EA, 1:10,000 dilution). Validation information for the commercial antibodies is included at the manufacturers’ websites. Original images of blots used in this study can be found in Supplementary Figure 8.
Co-immunoprecipitation
C. elegans nuclear protein extracts from L4 staged worms were used for immunoprecipitation with specific antibodies as described in ref. 27, followed by western blotting analysis. The antibodies used are as follows: 10 μg of anti-Pol II 8WG16 (Covance, MMS-126R), 10 μg of anti-CSR-1 (a gift from J. Claycomb and the Mello lab).
Chromatin Immunoprecipitation
Chromatin Immunoprecipitation (ChIP) experiments were performed as described in ref. 27 using early embryos or L4 worms. The antibodies used are as follows: 5 μg of anti-H3 (Millipore, 05-928), 2.5 μg of anti-H3K27me3 (Diagenode, pAb-195-050), 10 μg of anti-H3K36me3 (Abcam, ab9050), 10 μg of anti-MES-4 (Novus, 29400002), 10 μg of anti-CeCENP-A (a gift from the Desai lab, validated in ref. 35), 10 μg of anti-Pol II 8WG16 (Covance, MMS-126R). Validation information for the commercial antibodies is included at the manufacturers’ websites.
RNA Chromatin Immuniprecipitation (RNA-ChIP)
C. elegans nuclear extracts from L4 staged worms, previously fixed with 2% paraformaldehyde, were used for immunoprecipitation using 30 μl of anti-FLAG M2 affinity gel (Sigma, A2220). Chromatin Immunoprecipitations were performed as described in ref. 27 except that the protein-RNA complexes were eluted from the beads using FLAG peptide (Sigma, F4799) and treated with 20 μg of proteinase K at 45°C for 1 h and then de-crosslinked at 65°C for 1h. The RNA was isolated using TRI Reagent (Ambion, AM9738) according to the manufacturer’s instructions. After treatment with TURBO™ DNase (Ambion, AM2238) the RNA was subjected to RT-qPCR using primers to detect pre-mRNAs. The sequence of primers is available upon request. Where indicated, the DNase treatment of the chromatin extract has been performed at 25°C for 30min using 20 U of TURBO™ DNase (Ambion, AM2238).
Supplementary Material
Acknowledgments
We thank A. Leshinsky, R.F. Cook, and C.A. Whittaker from the Swanson Biotechnology Center at the Massachusetts Institute of Technology (Koch Institute for Integrative Cancer Research) for the sequencing of nascent transcript libraries, W. Gu (University of Massachusetts, Worcester) for providing siRNA sequencing datasets, and members of the Grishok, Maniatis and Hobert labs at the Columbia University for discussions. Reagents were kindly provided by C. Mello (University of Massachusetts, Worcester) and A. Desai (University of California, San Diego). This work was supported by US National Institutes of Health grant 1DP2OD006412-01 (A.G.).
Footnotes
AUTHOR CONTRIBUTIONS
G.C. designed and performed the experiments. G.C., S.H., S.O. and R.S. performed bioinformatic analyses of generated and published data. G.C., S.H., S.O., R.S. and A.G. analyzed the data, and G.C. and A.G. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Accession codes
All sequencing data have been deposited in GEO with accession GSE49946.
References
- 1.Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev. 2010;20:134–41. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 3.Hammond SM. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–9. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–18. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- 5.Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673–8. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lau NC. Small RNAs in the animal gonad: guarding genomes and guiding development. Int J Biochem Cell Biol. 2010;42:1334–47. doi: 10.1016/j.biocel.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grishok A. Biology and Mechanisms of Short RNAs in Caenorhabditis elegans. Adv Genet. 2013;83:1–69. doi: 10.1016/B978-0-12-407675-4.00001-8. [DOI] [PubMed] [Google Scholar]
- 8.Moshkovich N, et al. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev. 2011;25:1686–701. doi: 10.1101/gad.16651211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cernilogar FM, et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–5. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taliaferro JM, et al. Two new and distinct roles for Drosophila Argonaute-2 in the nucleus: alternative pre-mRNA splicing and transcriptional repression. Genes Dev. 2013;27:378–89. doi: 10.1101/gad.210708.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang V, et al. Ago1 Interacts with RNA polymerase II and binds to the promoters of actively transcribed genes in human cancer cells. PLoS Genet. 2013;9:e1003821. doi: 10.1371/journal.pgen.1003821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ameyar-Zazoua M, et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol. 2012;19:998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- 13.Cecere G, Grishok AA. Nuclear Perspective on RNAi Pathways in Metazoans. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbagrm.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu W, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–44. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Claycomb JM, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–34. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Wolfswinkel JC, et al. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–48. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 17.Yigit E, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–57. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 18.She X, Xu X, Fedotov A, Kelly WG, Maine EM. Regulation of heterochromatin assembly on unpaired chromosomes during Caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet. 2009;5:e1000624. doi: 10.1371/journal.pgen.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duchaine TF, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–54. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 20.Maine EM, et al. EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired dna during C. elegans meiosis. Curr Biol. 2005;15:1972–8. doi: 10.1016/j.cub.2005.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vought VE, Ohmachi M, Lee MH, Maine EM. EGO-1, a putative RNA-directed RNA polymerase, promotes germline proliferation in parallel with GLP-1/notch signaling and regulates the spatial organization of nuclear pore complexes and germline P granules in Caenorhabditis elegans. Genetics. 2005;170:1121–32. doi: 10.1534/genetics.105.042135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Updike DL, Strome S. A genomewide RNAi screen for genes that affect the stability, distribution and function of P granules in Caenorhabditis elegans. Genetics. 2009;183:1397–419. doi: 10.1534/genetics.109.110171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura M, et al. Dicer-related drh-3 gene functions in germ-line development by maintenance of chromosomal integrity in Caenorhabditis elegans. Genes Cells. 2007;12:997–1010. doi: 10.1111/j.1365-2443.2007.01111.x. [DOI] [PubMed] [Google Scholar]
- 24.Avgousti DC, Palani S, Sherman Y, Grishok A. CSR-1 RNAi pathway positively regulates histone expression in C. elegans. EMBO J. 2012;31:3821–32. doi: 10.1038/emboj.2012.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conine CC, et al. Argonautes Promote Male Fertility and Provide a Paternal Memory of Germline Gene Expression in C. elegans. Cell. 2013;155:1532–44. doi: 10.1016/j.cell.2013.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cecere G, Hoersch S, Jensen MB, Dixit S, Grishok A. The ZFP-1(AF10)/DOT-1 Complex Opposes H2B Ubiquitination to Reduce Pol II Transcription. Mol Cell. 2013;50:894–907. doi: 10.1016/j.molcel.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinke V, et al. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6:605–16. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, et al. Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics. 2009;10:213. doi: 10.1186/1471-2164-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rechtsteiner A, et al. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 2010;6:e1001091. doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–81. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furuhashi H, et al. Trans-generational epigenetic regulation of C. elegans primordial germ cells. Epigenetics Chromatin. 2010;3:15. doi: 10.1186/1756-8935-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaydos LJ, Rechtsteiner A, Egelhofer TA, Carroll CR, Strome S. Antagonism between MES-4 and Polycomb repressive complex 2 promotes appropriate gene expression in C. elegans germ cells. Cell Rep. 2012;2:1169–77. doi: 10.1016/j.celrep.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gassmann R, et al. An inverse relationship to germline transcription defines centromeric chromatin in C. elegans. Nature. 2012;484:534–7. doi: 10.1038/nature10973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee HC, et al. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bagijn MP, et al. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science. 2012;337:574–8. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shirayama M, et al. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashe A, et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buckley BA, et al. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–51. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lei EP, Corces VG. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet. 2006;38:936–41. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- 42.Perkins KJ, Lusic M, Mitar I, Giacca M, Proudfoot NJ. Transcription-dependent gene looping of the HIV-1 provirus is dictated by recognition of pre-mRNA processing signals. Mol Cell. 2008;29:56–68. doi: 10.1016/j.molcel.2007.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH. Crosstalk between mRNA 3’ end processing and transcription initiation. Mol Cell. 2010;40:410–22. doi: 10.1016/j.molcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 44.Tan-Wong SM, et al. Gene loops enhance transcriptional directionality. Science. 2012;338:671–5. doi: 10.1126/science.1224350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friend K, et al. A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat Struct Mol Biol. 2012;19:176–83. doi: 10.1038/nsmb.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maniar JM, Fire AZ. EGO-1, a C. elegans RdRP, modulates gene expression via production of mRNA-templated short antisense RNAs. Curr Biol. 2011;21:449–59. doi: 10.1016/j.cub.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu T, et al. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 2011;21:227–36. doi: 10.1101/gr.115519.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wedeles CJ, Wu MZ, Claycomb JM. Protection of Germline Gene Expression by the C. elegans Argonaute CSR-1. Dev Cell. 2013 doi: 10.1016/j.devcel.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 49.Seth M, et al. The C. elegans CSR-1 Argonaute Pathway Counteracts Epigenetic Silencing to Promote Germline Gene Expression. Dev Cell. 2013 doi: 10.1016/j.devcel.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li LC, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A. 2006;103:17337–42. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwartz JC, et al. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15:842–8. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chu Y, Yue X, Younger ST, Janowski BA, Corey DR. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38:7736–48. doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu J, et al. Promoter-associated small double-stranded RNA interacts with heterogeneous nuclear ribonucleoprotein A2/B1 to induce transcriptional activation. Biochem J. 2012;447:407–16. doi: 10.1042/BJ20120256. [DOI] [PubMed] [Google Scholar]
- 54.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu T, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


