Abstract
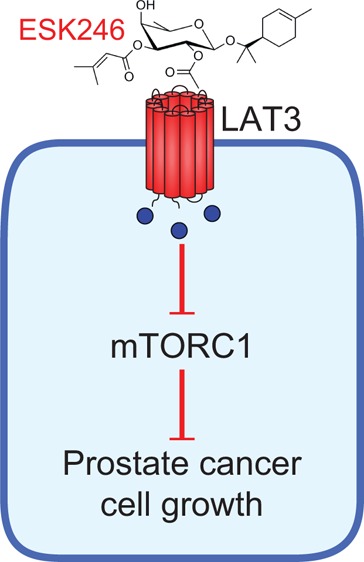
The l-type amino acid transporter (LAT) family consists of four members (LAT1–4) that mediate uptake of neutral amino acids including leucine. Leucine is not only important as a building block for proteins, but plays a critical role in mTORC1 signaling leading to protein translation. As such, LAT family members are commonly upregulated in cancer in order to fuel increased protein translation and cell growth. To identify potential LAT-specific inhibitors, we established a function-based high-throughput screen using a prefractionated natural product library. We identified and purified two novel monoterpene glycosides, ESK242 and ESK246, sourced from a Queensland collection of the plant Pittosporum venulosum. Using Xenopus laevis oocytes expressing individual LAT family members, we demonstrated that ESK246 preferentially inhibits leucine transport via LAT3, while ESK242 inhibits both LAT1 and LAT3. We further show in LNCaP prostate cancer cells that ESK246 is a potent (IC50 = 8.12 μM) inhibitor of leucine uptake, leading to reduced mTORC1 signaling, cell cycle protein expression and cell proliferation. Our study suggests that ESK246 is a LAT3 inhibitor that can be used to study LAT3 function and upon which new antiprostate cancer therapies may be based.
l-type amino acid transporters (LATs) mediate the Na+-independent uptake of neutral amino acids, including the essential branched chain amino acids (BCAAs) leucine, isoleucine and valine. LATs are composed of two distinct families, SLC7 (LAT1/SLC7A5 and LAT2/SLC7A8) and SLC43 (LAT3/SLC43A1 and LAT4/SLC43A2). LAT1 and LAT2 have a broad substrate range and associate with the 4F2hc glycoprotein (SLC3A2) to form a heterodimeric obligatory exchanger of high affinity.1−5 LAT3 and LAT4 have a narrower substrate range and utilize facilitated diffusion to transport neutral amino acids.6−8
Expression of LATs on mammalian cells is critical to mediate uptake of amino acids that can subsequently be used for energy production and as building blocks for protein production. Amino acids, especially leucine, are also a crucial component of the mTORC1 signaling pathway, which controls protein translation.9 Translation can only begin when sufficient amino acids, in particular leucine, are present within the cell. Recent data suggests that intracellular leucine levels are detected by a leucyl-tRNA synthetase (LRS),10,11 which is thought to activate the Rag GTPase complex, binding to Raptor and activating mTORC1 signaling on the surface of lysosomes.10−14 Therefore, changes in LAT expression and function can control intracellular amino acid levels and mTORC1 regulated protein translation.
LATs have been shown to be critical mediators of protein translation and cell growth in a variety of cancers.15−21 In prostate cancer, we have shown increased LAT3 expression in primary cancer and increased LAT1 expression in metastasis.16 Knockdown of either LAT3 or LAT1 expression in prostate cancer cell lines inhibits mTORC1 pathway activation, cell growth, and cell cycle both in vitro and in vivo.16,17
The leucine analogue 2-aminobicyclo[2.2.1]heptane-2-carboxylic acid (BCH) is commonly used to inhibit l-type amino acid transport in vitro. However, BCH targets all members of the LAT family and is used in millimolar quantities, making it unsuitable as a LAT inhibitor in vivo.22 A novel LAT1 inhibitor, JPH203/KYT-0353, has also been reported. This tyrosine analogue has only been tested on LAT1 and LAT2, showing selectivity for LAT1-mediated transport, as well as suppression of HT-29 colorectal adenocarcinoma cell growth in vitro and in vivo.23
In this study, we screened a prefractionated natural product library and identified two novel LAT inhibitors, ESK242 and ESK246. We further demonstrate that ESK246 preferentially inhibits LAT3, reducing LAT3-mediated leucine uptake in Xenopus laevis oocytes. We show that ESK246 is a more potent LAT inhibitor than ESK242, reducing leucine uptake, mTORC1 signaling, cell cycle protein expression, and proliferation in prostate cancer cell lines.
Results and Discussion
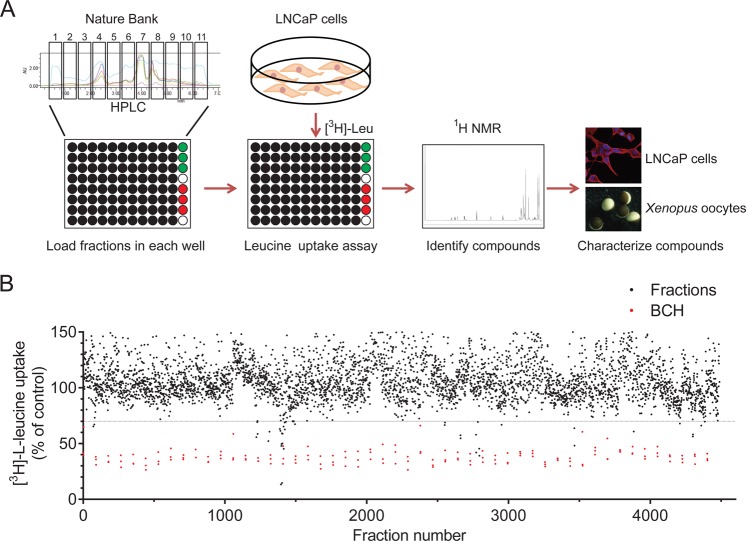
High-throughput Screening of the Prefractionated Nature Bank Library. To discover LAT3-specific inhibitors, we used a function-based strategy for high-throughput screening (HTS) of the Nature Bank prefractionated library (Figure 1). Our high-throughput screen incorporated a 15 min [3H]-l-leucine uptake assay using the androgen-responsive prostate cancer cell line, LNCaP (Figure 1A). The HTS screen was performed on a subset of the Nature Bank lead-like enhanced (LLE) fraction library. This library consists of over 200 000 semipurified fractions sourced from plants and marine invertebrates collected from Australia, China and Papua New Guinea.24,25
Figure 1.
High-throughput screening for LAT3 inhibitors. (A) Schematic representation of the function-based drug discovery process. Eleven HPLC fractions of each biota sample were aliquoted into 96-well plates, with 88 fractions on each plate. Triplicate wells of negative control (DMSO; green) and positive control (BCH; red) were also loaded. LNCaP cells (which express high levels of LAT3) and [3H]-l-leucine were added to each well for 15 min to identify any fractions that inhibit LAT3-mediated leucine uptake. Verified fractions were examined by 1H NMR to identify the structure of compounds. Novel compounds were characterized using amino acid uptake assays in Xenopus laevis oocytes and LNCaP prostate cancer cell based assays. (B) A leucine uptake assay was used to screen 4488 fractions from the Nature Bank library in LNCaP cells (n = 1 assay per fraction). Threshold for inhibition was set at 70% of control (dotted line) and BCH positive controls are indicated (red).
The HTS involved screening the Nature Bank library, initially analyzing 4488 fractions (51 plates × 88 fractions) for activity against LAT3-mediated [3H]-l-leucine uptake (Figure 1B). Each plate also contained 3 negative control wells (DMSO, 0.5% (v/v)) and 3 wells of the LAT family inhibitor BCH (10 mM) as a positive control. BCH consistently inhibited leucine uptake to 30–40% of control, while the negative control ranged from 90–110% on each plate (Figure 1B). In order to reduce the number of hits, we restricted our analysis from the initial screen to fractions that reduced leucine uptake to less than 70% of control, which resulted in a total of 31 fractions (0.7% of analyzed fractions). These fractions were retested in LNCaP cells using the [3H]-l-leucine uptake assay as well as a cell viability assay to determine whether they could inhibit prostate cancer cell viability. Fraction 11711.8-21-11 showed substantial inhibition of both leucine uptake and cell growth (Supporting Information (SI) Figure S1A). As some fractions may contain toxic compounds that rapidly damage cell membranes, such as detergents, leading to the low [3H]-l-leucine levels and cell growth, we also examined cell morphology after 48 h treatment, confirming that fraction 11711.8-21-11 did not damage the cell membranes (SI Figure S1B). Fraction 11711.8-21-11 originated from a collection of the plant Pittosporum venulosum (Queensland, Australia) and yielded two new LAT inhibitors, ESK242 and ESK246 (Figure 2A). The structures of these new natural products were identified by extensive spectroscopic and spectrometric analyses, confirmed by chemical synthesis, and characterized by a variety of biological methods.
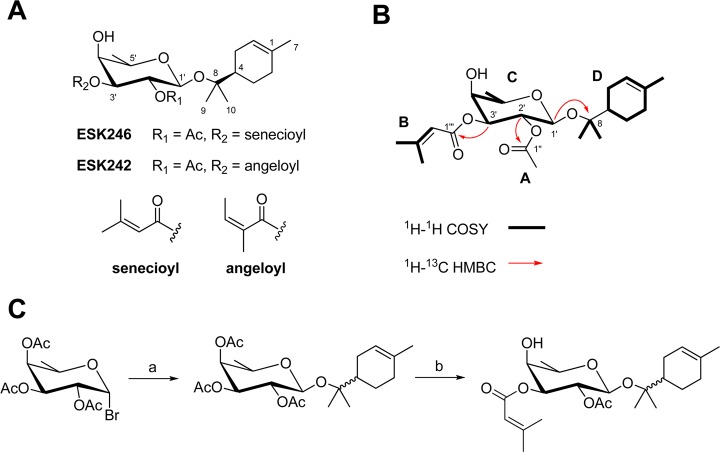
Figure 2.
Identification of compound structure. (A) Structures of two new natural products ESK242 and ESK246. (B) Crucial NMR correlations used to establish the planar structure of ESK246. Bold lines indicate 1H–1H COSY correlations that were used to identify the 1,1-dimethyl vinyl spin system (fragment B), fucopyranoside spin system (fragment C) and the terpineol spin system (fragment D). Red arrows depict the crucial 1H–13C HMBC correlations used to identify the position of the aglycone substituent and the esterification sites of the fucopyranoside residue. (C) Synthetic route to ESK246. Reagents used: (a) α-terpineol, Ag2CO3, CH2Cl2, rt, 48 h. (b) i. NaOMe, MeOH, rt, 3 h. ii. 3,3-Dimethylacryloyl chloride, py, CH2Cl2, 0 °C, 18 h. iii. Ac2O, py, CH2Cl2, 0 °C, 3 h. In addition to the synthesis of diastereomers of α-terpineol shown, an identical synthetic route was used with the (4S)-α-terpineol starting material.
Isolation and Identification of ESK242 and ESK246
ESK246 (trivial name venuloside A) was isolated as an optically active clear oil ([α]D +39, c 0.1, MeOH) with a molecular formula C23H36O7, as established from HRESIMS measurements. The 1H NMR spectrum of ESK 246 in C6D6 was well resolved and showed 21 resonances composed of 2 sp2-hybridized methines, 6 sp3-hybridized methines, 3 diastereotopic methylene pairs, and 7 sp3-hybridized methyls (SI Table S1). The 13C NMR spectrum showed two carbonyls (δC 168.9, 165.8), three other quaternary carbons (δC 157.7, 133.6 and 79.4), two olefinic resonances (δC 121.4 and 116.4), four oxymethines (δC 95.9, 73.8, 70.4, 70.3, and 70.0), one other methine (δC 44.4), three methylenes (δC 31.3, 26.96, and 24.0), and seven methyls (δC 27.02, 23.9, 23.53, 23.52, 20.7, 20.2, and 16.6). Interpretation of 2D NMR data allowed for the identification of four partial structures depicted in Figure 2B, namely an acetyl substituent (fragment A), a 3-methylbut-2-enoyl group (fragment B), a sugar moiety (fragment C), and a ten-carbon aglycone unit (fragment D). Characteristic chemical shifts for the methyl (δH 1.87, 3H, s, δC 20.7) and carbonyl resonances (δC 168.9, C-1″) were used to assign fragment A as the acetyl substituent. Fragment B had two methyl vinyl resonances (δH 2.14, d, J = 1.0 Hz, δC 27.02 and 1.45, d, J = 1.0 Hz; δC 20.2), coupled to each other and to an olefin proton (δH 5.77, δC 116.4), in addition to a quaternary carbon (δC 157.7) and an ester carbonyl resonance (δC 165.8) consistent with a senecioyl (3-methylbut-2-enoyl) moiety. Partial structure C contained an anomeric proton, four other oxymethines and a doublet methyl resonance, consistent with the presence of a six-deoxy sugar moiety. The 1H NMR chemical shifts and coupling constants for H-1′ (δH 4.47, d, J = 7.8 Hz), H-2′ (δH 5.66, dd, J = 10.3, 7.8 Hz), H-3′ (5.16, dd, J = 10.4, 3.2 Hz), H-4′ (3.72, br d, J = 3.2 Hz), H-5′ (3.10, br q, J = 6.3 Hz), and H-6′-Me (1.17, d, J = 6.3 Hz) protons, together with the ROESY data showed that the glycosyl moiety was β-fucopyranoside. The downfield shift of H-2′ and H-3′ resonances suggested that the C-2 and C-3 hydroxyl groups on the sugar moiety were esterified. Chemical shifts assigned to the acetyl and senecioyl substituents were positioned on C-2′ and C-3′ respectively following crucial 1H–13C HMBC correlations from the H-2′ glycosidic resonance to C-1″ carbonyl on the acetyl group, as well as the glycosidic H-3′ to C-1‴ carbonyl on the senecioyl group (Figure 2B). The sugar residue in ESK246 was therefore concluded to be 2′-acetyl-3′-senecioyl-β-fucopyranoside. Signals for the aglycone fragment D, C10H17 as suggested by the molecular formula, were composed of an olefinic multiplet, three diastereotopic methylene pairs, a single methine and three methyl resonances, consistent with the presence of an α-terpineol moiety. Crucial HMBC correlation from the anomeric H-1′ to the quaternary C-8 carbon (δC 79.4) (Figure 2B) connected substructure D to the sugar unit to complete the planar structure of the glycoside.
The absolute configuration of the natural product was determined via comparison of chiro-optical and NMR spectral data with that of the synthetically prepared analogues. Hydrolysis of ESK246 in 5% HCl in MeOH afforded a mixture of methyl-3-O-senecioyl-β-fucopyranoside, methyl-3-O-senecioyl-α-fucopyranoside and methyl-4-O-senecioyl-α-fucopyranoside (see Supporting Information). Comparison of the optical rotation values of the ESK246 hydrolysis-sourced α- and β-anomers of methyl-3-O-senecioyl-fucopyranoside with that of the synthetic α- and β-anomers of methyl-3-O-senecioyl-d-fucopyranoside established the absolute configuration of fucose residue as d (see Supporting Information). The d-configuration of the fucose sugar is typical of plant-sourced glycosides, whereas l has been reported from bacteria- and plant-sourced polysaccharides, as well as glycolipids and glycoproteins of animal origin.26 Resolving the absolute configuration of the α-terpineol proved to be a challenge as the acid hydrolysis work-up and isolation did not yield any terpineol product. The absolute configuration of the natural product was therefore resolved with total synthesis of the racemic and (4S)-α-terpineol diastereomers of ESK246. Starting from d-fucose and the readily available (4rac)- and (4S)-α-terpineols, a four-step synthesis was achieved (Figure 2C and Supporting Information text). Koenigs–Knorr glycosylation of bromo-fucosyl donor with α-terpineol afforded the fully protected α-terpineol fucoside, which was subsequently deprotected under basic conditions (NaOMe/MeOH) to give the α-terpineol fucoside. Utilizing the inherent difference in reactivity of the fucose hydroxyl groups,27 selective acylation at the 3′O position of α-terpineol fucoside with 3,3-dimethylacryloyl chloride was achieved.28 Introduction of the acetate moiety at the 2′O position was carried out under basic conditions using acetic anhydride to give the final product.
In the 1H NMR spectrum of the C-4 epimeric mixure of ESK246, the greatest signal dispersion between the two isomers was observed with the H-9 and H-10 resonances (SI Figure S2). These two sets of resonances were therefore used as a point of differentiation in the determination of the absolute configuration of ESK246. In an 1H NMR-based titration experiment depicted in SI Figure S3, addition of the naturally occurring ESK246 to the C-4 epimeric mixture of α-terpineol-8-O-β-d-(2′-acetyl, 3′-senecioyl) fucopyranoside resulted in the enhancement of the resonances associated with the (4R)-α-terpineol epimer, therefore securing the stereochemistry of the natural product as (4R)-α-terpineol-8-O-β-d-(2′-acetyl, 3′-senecioyl) fucopyranoside.
ESK242 (trivial name venuloside B) was isolated as an optically active clear oil ([α]D +20, c 0.1, MeOH) with a molecular formula C23H36O7, as established from HRESIMS measurements. The compound was isomeric with ESK246 and showed an identical number of 1H and 13C resonances in the NMR spectra. The most significant differences between the NMR spectra of the two constitutional isomers were centered on the two methyl vinyl resonances H-4‴ (δH 2.05, m; δC 16.0) and H-5‴ (δH 1.94, m; δC 20.8) as well as an olefinic methine resonance at H-3‴ (δH 5.79, m; δC 139.3). In contrast to ESK246, which had a senecioyl group at C-3, ESK242 showed two methyl vinyl resonances each coupled to an olefin proton, consistent with a 1,2-dimethyl vinyl group. A ROESY experiment (τmix = 400 ms) assigned the geometry of the double bond to be cis, based on the observation of a correlation between H-3‴ and H-5‴ methyl resonances, consistent with the presence of an angeloyl group. Crucial 1H–13C HMBC correlations from the glycosidic H-3′ to C-1‴ placed the angeloyl group on C-3′ of the sugar moiety. Interpretation of 1D and 2D NMR data (SI Table S1) established the structure of ESK242 as α-terpineol-2′-acetate-3′-angeloyl-β-fucopyranoside. Based on the structural homology with ESK246 and the close correspondence of the NMR chemical shifts (SI Table S1) the absolute configuration of ESK242 is proposed to be (4R)-α-terpineol-8-O-β-d-(2′-acetyl, 3′-angeloyl) fucopyranoside.
Effects of ESK242 and ESK246 on Leucine Uptake in Prostate Cancer Cell Lines
To examine the inhibitory effects of ESK242 and ESK246, we next investigated the impact of these new compounds on LNCaP and PC-3 prostate cancer cell lines. We have previously shown that LAT3 is the dominant leucine transporter in LNCaP cells, while LAT1 plays a more important role in PC-3 cells (SI Figure S4A).16 In the presence of compounds ESK242 and ESK246, leucine uptake was decreased in a dose-dependent manner in LNCaP cells (Figure 3A). The IC50 of ESK242 is 29.6 ± 1.2 μM, while the IC50 of ESK246 is 8.12 ± 1.2 μM. Both compounds had an IC50 more than 2 orders of magnitude lower than the universal LAT inhibitor, BCH, whose IC50 is 4060 ± 1.1 μM (Figure 3A). In contrast, neither compound substantially inhibited leucine uptake at the same concentration in PC-3 cells, in which LAT1 is more abundantly expressed (SI Figure S4B).
Figure 3.
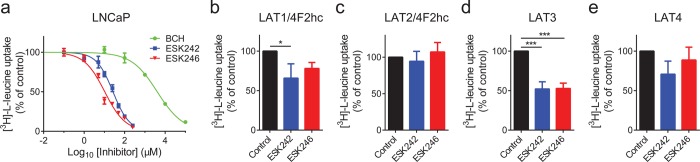
(A) Inhibition of LAT3-mediated [3H]-l-leucine uptake in LNCaP cells. The IC50 of BCH, ESK242 and ESK246 was calculated to be 4060 ± 1.1 μM, 29.6 ± 1.2 μM, and 8.12 ± 1.2 μM, respectively. (B–E) [3H]-l-leucine uptake assay in the presence or absence of 50 μM ESK242 or ESK246 in oocytes expressing LAT1/4F2hc (B), LAT2/4F2hc (C), LAT3 (D), or LAT4 (E). Data show mean ± SEM (n = 3), *P < 0.05, **P < 0.01, ***P < 0.001.
Characterization of ESK242 and ESK246 by Amino Acid Uptake Assay in Oocytes
The differential effects on leucine uptake in LNCaP and PC-3 cells suggested these compounds selectively inhibit individual LAT family members. To test the specificity of the purified compounds, we expressed LAT1/4F2hc, LAT2/4F2hc, LAT3 or LAT4 in Xenopus laevis oocytes and [3H]-l-leucine uptake assays were performed. After 4 days, transporters were expressed on the plasma membrane of oocytes, as shown by an increase in [3H]-l-leucine uptake compared to uninjected oocytes (SI Figure S5A). To demonstrate expression of the LAT3 transporter on the surface of oocytes, we performed a surface biotinylation assay, which confirmed LAT3 expression in injected oocytes only (SI Figure S5B). The optimal concentration of leucine, and duration of leucine uptake, was determined by dose response and time course experiments, showing 1 mM l-leucine for 30 min to be optimal (SI Figures S5C and D). After treatment with 50 μM ESK242 or ESK246 for 30 min, LAT1/4F2hc transport activity was significantly inhibited by ESK242 (34.2%; Figure 3B). LAT3-mediated leucine transport was significantly inhibited by both ESK242 (47.9%) and ESK246 (47.3%; Figure 3D). Neither compound significantly inhibited leucine uptake mediated by LAT2/4F2hc (Figure 3C) or LAT4 (Figure 3E). These results suggest that ESK246 is a preferential LAT3 inhibitor and ESK242 inhibits both LAT1 and LAT3. We next determined the IC50 of both compounds for LAT3 in oocytes. The IC50 of ESK246 was calculated at 146.7 ± 2.4 μM, which is lower than the IC50 of ESK242 at 281.8 ± 1.3 μM (Figure 4A), suggesting that ESK246 has a higher affinity for LAT3 than ESK242.
Figure 4.
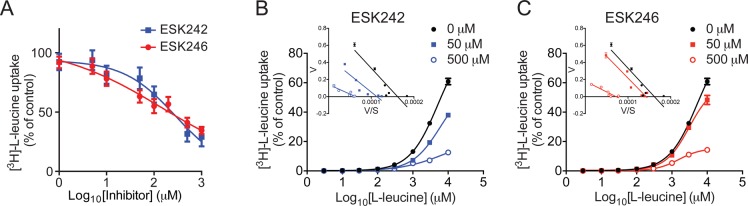
ESK242 and ESK246 inhibit LAT3 via a mixed mode of inhibition. (A) Increasing doses of ESK242 and ESK246 inhibit [3H]-l-leucine transport in oocytes expressing LAT3 with IC50 values of 281.8 ± 1.3 μM and 146.7 ± 2.4 μM, respectively. (B–C) [3H]-l-leucine dose response in the absence or presence of 50 μM or 500 μM ESK242 (B) or ESK246 (C). Data are fitted to the Michaelis–Menten equation and normalized to the maximal rate of transport of [3H]-l-leucine alone. An Eadie–Hofstee transformation was performed (inset, B–C). Data show mean ± SEM (n = 5).
In order to investigate the nature of inhibition of LAT3 by ESK242 and ESK246, leucine dose responses were performed in the absence or presence of 50 μM and 500 μM of each inhibitor. As the concentration of inhibitor increases, the Km of leucine for LAT3 (6.3 ± 0.5 mM) increases slightly in the presence of 50 μM ESK242 (7.5 ± 0.5 mM) but decreases to 2.7 ± 0.5 mM in the presence of 500 μM ESK242 (Figure 4B). In the presence of 50 μM and 500 μM ESK246, the Km of leucine for LAT3 decreases to 4.7 ± 0.6 mM and 2.1 ± 0.3 mM, respectively (Figure 4C) suggesting a competitive mechanism of action for these compounds. The maximal rate of uptake also decreases as the inhibitor concentration increases, which would not be expected for a purely competitive inhibitor suggesting that these compounds may be acting in a mixed competitive/noncompetitive manner. An Eadie–Hofstee transformation of the data (Figure 4B and C, inset) also demonstrates that both ESK242 and ESK246 are acting as mixed inhibitors.
Effects of ESK242 and ESK246 on mTORC1 Signaling and Cell Growth in Prostate Cancer Cell Lines
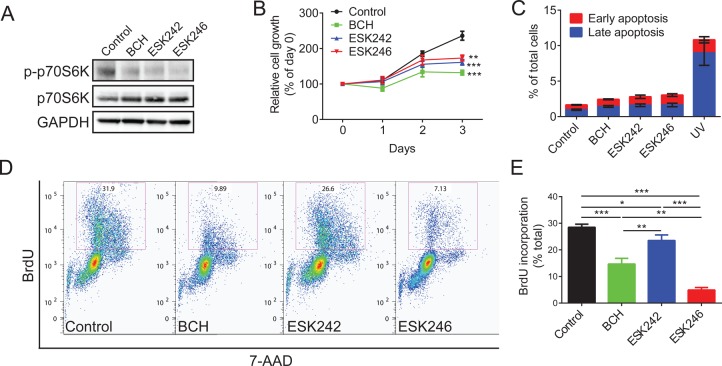
Having established that ESK242 and ESK246 inhibit leucine uptake in LNCaP cells with an IC50 between 8–30 μM (Figure 3A), we next assessed the downstream effects of leucine deprivation on signaling and cell growth. Leucine uptake mediated by LAT3 has been shown to regulate mTORC1 activation in prostate cancer cells.16 Both ESK242 and ESK246 suppressed phosphorylation (activation) of the mTORC1 target protein p70S6K (Figure 5A), consistent with a mechanism consequent to inhibition of LAT3-mediated leucine transport in LNCaP cells. We next examined cell viability in the presence of compounds using an MTT assay in prostate cancer cells. Both ESK242 and ESK246 led to lower cell viability in LNCaP cells after 3 days treatment (Figure 5B). Only compound ESK242, but not ESK246, significantly inhibited cell viability in PC-3 cells, (SI Figure S6A), which may be due to ESK242 inhibition of both LAT1 and LAT3. To determine whether decreased cell viability is due to activation of apoptosis or suppression of proliferation, we first examined apoptosis using flow cytometry to detect “flipped” Annexin-V protein levels in the plasma membrane. Cells exposed to UV showed increased Annexin-V flipping indicative of apoptosis (Figure 5C). Neither ESK242 nor ESK246 induced apoptosis, consistent with previous studies examining BCH (Figure 5C).16 Therefore, it is likely that the effects on viability are mediated through inhibition of cell cycle and proliferation. To test this, we next examined BrdU (bromodeoxyuridine, a thymidine analogue) incorporation in DNA using flow cytometry and showed the BrdU incorporation rate was decreased after BCH, ESK242, or ESK246 treatment (Figure 5D,E). Compound ESK246 showed the highest level of inhibition of BrdU incorporation (4.9%; 17.2% of control), which was significantly lower than either BCH (14.7%; 51.6% of control) or ESK242 treatment (23.5%; 82.7% of control; Figure 5E). Our previous studies have shown that reduced extracellular amino acid levels inhibit expression of the cell cycle regulator CDK1 and the ubiquitination enzyme UBE2C in prostate cancer and melanoma.17,29 Therefore, we examined expression of CDK1 and UBE2C in the presence of BCH, ESK242, and ESK246, showing expression of both proteins was down regulated by ESK246 (SI Figure S6B).
Figure 5.
Effects of ESK242 and ESK246 in LNCaP cells. (A) Representative Western blots (from n = 3) of p70S6K phosphorylation after BCH (10 mM), ESK242 (50 μM), and ESK246 (50 μM) inhibition. GAPDH was used as the loading control. (B) MTT cell viability assay (n = 3) in LNCaP cells incubated with BCH (10 mM), ESK242 (50 μM) and ESK246 (50 μM). Two-way ANOVA test was performed, **P < 0.01, ***P < 0.001. (C) Analysis of apoptosis (n = 3) in LNCaP cells using Annexin-V and PI staining after inhibition with BCH (10 mM), ESK242 (50 μM), and ESK246 (50 μM). (D,E) Analysis of cell proliferation using BrdU incorporation in LNCaP cells inhibited with BCH (10 mM), ESK242 (50 μM) and ESK246 (50 μM). Representative flow cytometry analysis (D) and quantification (E) from 3 separate experiments are shown. One-way ANOVA test was performed, *P < 0.05, **P < 0.01, ***P < 0.001.
Conclusions
We have utilized a function-based drug discovery platform that enabled high-throughput screening of LAT3-specific natural products from the Nature Bank fraction library. Using this platform, we identified two novel monoterpene glycosides, ESK242, a dual LAT1 and LAT3 inhibitor, and ESK246, which is more selective for inhibition of LAT3 in both oocytes and LNCaP cells. Notably, ESK246, inhibits LAT-mediated leucine transport in LNCaP cells with a ∼500-fold lower IC50 compared to the leucine analogue BCH. We anticipate that derivatives of these inhibitors could be used as prostate cancer therapeutics as well as to investigate the biological function of LAT1 and LAT3 during development and in disease.
Methods
General Chemical Experimental Procedures
UV spectra were recorded on a Jasco V650 UV/vis spectrophotometer. NMR spectra were recorded at 30 °C on either a Varian 500 or 600 MHz Unity INOVA spectrometer. The latter spectrometer was equipped with a triple resonance cold probe. The 1H and 13C NMR chemical shifts were referenced to the solvent peaks for benzene-d6 at δH 7.20 and δC 128.0, respectively, and for DMSO-d6 at δH 2.50 and δC 39.43, respectively. HRESIMS were recorded on a Bruker Daltronics Apex III 4.7e Fourier-transform mass spectrometer. Silica gel chromatography was performed using Merck silica gel 60 (0.015–0.040 mm). A Waters 600 pump equipped with a Waters 996 PDA detector and a Waters 717 autosampler were used for HPLC. A Phenomenex Onyx monolythic column [100 × 10 mm] was used for semipreparative HPLC separations. All solvents used for chromatography, UV, and MS were Lab-Scan HPLC grade (RCI Lab-Scan, Bangkok, Thailand), and the H2O was Millipore Milli-Q PF filtered. All starting materials and reagents for the synthesis were obtained from commercial suppliers, Sigma-Aldrich, Carbosynth, Alfa Aesar, Merck Millipore, and used without further purification. High-performance liquid chromatography (HPLC) grade solvents were obtained from Labscan, and purified using a PureSolv MD 5 solvent purification system from Innovative Technology. Reactions were performed in flame-dried glassware under positive N2 pressure, with magnetic stirring. Rubber septa and syringes were used for liquid transfers. Thin layer chromatography (TLC) was performed on 0.25 mm Merck silica gel 60 F254 plates and plates visualized by staining with Seebach’s stain; phosphomolybdic acid/cerium(IV) sulfate 2.5:1 in H2O/H2SO4 94:6 (mL).
Plant Material
Pittosporum venulosum (F. Muell) (Nature Bank code 11711.8) was collected in July 1995 from State Forest 144, Mt. Windsor Tableland, Queensland, Australia. The plant was identified by P.I. Forster and S.J. Figg. A voucher specimen (PIF17245) has been lodged with the Queensland Herbarium.
Isolation of ESK246 and ESK242
Pittosporum venulosum (10 g) was dried, ground, and sequentially extracted in hexane (250 mL), CH2Cl2 (250 mL) and MeOH (2 × 250 mL). All extracts were combined and reduced under pressure to yield a dark green oil (1.62 g). The combined organic extract was then subjected to a solvent–solvent partition with the compounds of interest concentrated in the hexane fraction. The hexane fraction (370 mg) was subjected to flash silica oxide chromatography (10 cm × 4 cm) eluting with a gradient from 100% hexane to 100% ethyl acetate (EtOAc). Fraction eluting with 8:2 (hexane/EtOAc) yielded ESK242 (149 mg, 1.49% dry weight), while the fraction eluting with 6:4 (hexane/EtOAc) yielded ESK246 (58 mg, 0.58% dry weight). The purity of ESK242 and ESK246 was confirmed by 1H NMR and 13C NMR (SI Figures S7–S10) to be >95% (SI Table S2).
ESK246
Clear oil; [α]D = +39 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 202 (3.59), 218 (3.72) nm; IR (KBr film) 3435, 2934, 1752, 1721, 1370, 1227, 1132, 1069 cm–1; 1H NMR (600 MHz, C6D6) and 13C NMR (150 MHz, C6D6). See SI Table S1. HRESIFTMS m/z [M + Na]+ 447.235553 (calcd for C23H36O7Na, 447.235325).
ESK242
Clear oil; [α]D = +20 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 202 (3.85), 218 (3.88) nm; IR (KBr film) 3487, 2976, 2936, 1752, 1718, 1371, 1232, 1148, 1069 cm–1; 1H NMR (600 MHz, C6D6) and 13C NMR (150 MHz, C6D6). See SI Table S1. HRESIFTMS m/z [M + Na]+ 447.235517 (calcd for C23H36O7Na, 447.235325).
Cell Lines
Human prostate cancer cell lines LNCaP-FGC and PC-3 were purchased from ATCC (Rockville, MD). LNCaP cells have been passaged directly from original low-passage stocks (2009), and we confirmed PC-3 cell identity by STR profiling in 2010 (Cellbank). Cells were cultured in RPMI 1640 medium (Life Technologies) containing 10% (v/v) fetal bovine serum (FBS), penicillin–streptomycin solution (Sigma-Aldrich), and 1 mM sodium pyruvate (Life Technologies). Cells were maintained at 37 °C in a fully humidified atmosphere containing 5% CO2.
Leucine Uptake Assay
The [3H]-l-leucine uptake was performed as detailed previously.16 Briefly, cells were cultured in 6-well plates in RPMI media. After collecting and counting, cells (3 × 104/well) were incubated with 0.3 μCi [3H]-l-leucine (200 nM; PerkinElmer) in leucine-free RPMI media (Life Technologies) with 10% (v/v) dialyzed FBS for 15 min at 37 °C. For high-throughput screening of Nature Bank fractions, LNCaP cells (104/well) were incubated with 0.3 μCi [3H]-l-leucine (200 nM) in HBSS with 10% (v/v) dialyzed FBS and 50 mM l-glutamine. Cells were directly added into 96-well plates containing preloaded fractions from Nature Bank. DMSO 0.5% (v/v) was used as the negative control and 10 mM BCH was used as the positive control. Cells were collected, transferred to filter paper using a 96-well plate harvester (Wallac PerkinElmer), dried, exposed to scintillation fluid, and counts measured using a liquid scintillation counter (PerkinElmer).
Cell Viability Assay
Cells in exponential growth phase were harvested and seeded (1 × 104/well) in a flat-bottomed 96-well plate. The cells were incubated overnight in RPMI media, prior to culture with or without each inhibitor. MTT solution (10 μL; 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrasodium bromide; Millipore) was added to each well for 4 h, prior to addition of 100 μL of isopropanol/HCl solution and mixed thoroughly. The plates were immediately read at 570 nm/630 nm in a PolarStar plate reader (BMG). Results were plotted as percentages of the absorbance observed in control wells (vehicle/DMSO).
Oocyte Uptake Assay
Human LAT1, LAT2, LAT3, LAT4, and 4F2hc cDNAs were subcloned into the oocyte transcription vector pOTV, respectively. The resulting transporter cDNAs were linearized with SpeI. Complementary RNA (cRNA) was transcribed with T7 RNA polymerase and capped with 5′-7-methylguanosine using the mMessage mMachine kit (Ambion). All cRNAs were purified using a NucAway Spin Column (Ambion).
Stage V oocytes were harvested from Xenopus laevis as described previously,30 and all surgical procedures followed a protocol approved under the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes. Oocytes were injected with 23 nL of cRNA mix containing 1:1 LAT1/4F2hc, LAT2/4F2hc, LAT3, or LAT4 cRNA, respectively (4.6 ng of cRNA in total), and incubated in standard frog Ringer’s solution (ND96:96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 5 mM HEPES, pH 7.5) supplemented with 50 μg/mL gentamycin, 2.5 mM sodium pyruvate, and 0.5 mM theophylline at 16–18 °C. Four days after injection, 5 oocytes per group were incubated in 500 μL of uptake solution (standard frog Ringer’s solution, ND96) containing 500 nM [3H]-l-leucine in a final concentration of 1 mM l-leucine for 30 min at RT. The LAT inhibitor BCH was used at 10 mM and ESK242 and ESK246 were used at various concentrations, as indicated in the text. Uptake was terminated by three rapid washes in ice-cold ND96 followed by lysis in 50 mM NaOH and 50% SDS. [3H]-l-leucine uptake was measured by scintillation counting using a Trilux β-counter (PerkinElmer Life Science).
Biotinylation Assay
Surface proteins on Xenopus laevis oocytes were isolated using the Cell Surface Protein Isolation Kit (Thermo Scientific Pierce). The oocytes were incubated for 30 min at 4 °C on an orbital shaker with Sulfo-NHS-SS-biotin. After three washes with PBS, the oocytes were sonicated in 500 μL of Lysis buffer containing protease inhibitors. The lysate was incubated on ice for 30 min and spun for 2 min at 10 000g. The labeled proteins were isolated from the supernatant using NeutrAvidin agarose and eluted with SDS-PAGE sample buffer containing 50 mM DTT. The eluted proteins were then analyzed by Western blotting as detailed below.
BrdU Incorporation Assay
Cells were seeded at a density of 2 × 105 in 6-well plates and allowed to adhere overnight. After serum starvation, cells were incubated with either DMSO, 10 mM BCH, 50 μM ESK242, or 50 μM ESK246 for 22 h. At the end of the treatment, BrdU (150 μg/mL) was added to culture media and incubated for another 2 h, followed by detachment using Tryple (Life Technologies). Cells were fixed and stained using the Becton Dickinson APC BrdU flow cytometry kit (BD). The BrdU antibody was diluted 1 in 50. Nuclei were counterstained by 7-AAD (7-aminoactinomycin). The cells were analyzed on a Canto II flow cytometer (BD) with postanalysis performed using FlowJo software (Tree Star Inc.).
Apoptosis Assay
Cells were seeded at a density of 2 × 105 in 6-well plates, allowed to adhere overnight, before incubation with DMSO, 10 mM BCH, 50 μM ESK242, or 50 μM ESK246 for 48 h, respectively. Positive control group cells were irradiated in a UV Stratalinker 2400 (Stratagene) with a 400 000 μJ dosage and incubated in fresh media for 16 h. Cells were detached using Tryple and resuspended in 1 mL of binding buffer (HEPES-buffered PBS supplemented with 2.5 mM calcium chloride) containing antiannexin V-APC (BD) and incubated for 15 min in the dark at RT. PI solution (20 μg/mL) was added, and the cells were analyzed on a Canto II flow cytometer (BD) with postanalysis performed using FlowJo software.
Western Blots
Cells were seeded at a density of 2 × 105 in 6-well plates, allowed to adhere overnight, before incubation with DMSO, 10 mM BCH, 50 μM ESK242, or 50 μM ESK246 for 6 h or 3 d. Cells were lysed by the addition of lysis buffer (200 μL) with protease inhibitor Cocktail III (Bioprocessing Biochemical) and 1 mM Na3VO4 (Sigma). Equal protein (micro-BCA method; Pierce, IL) was loaded on 4–12% gradient gels (Life Technologies), electrophoresed, and transferred to PVDF membrane. The membrane was blocked with 2.5% (w/v) BSA in PBS-Tween20, and incubated with the primary and secondary antibodies. The secondary HRP-labeled antibodies were detected using enhanced chemiluminescence reagents (Pierce) on a Kodak Imager (Kodak). Antibodies used in this study were against LAT1 (Cosmo Bio), LAT3 (a kind gift from Kunimasa Yan, Kyorin University, Tokyo, Japan), α-tubulin (Santa Cruz), p-p70S6K, p70S6K, (Cell Signaling), UBE2C (Boston Biochem), CDK1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Abcam). Horseradish peroxidase-conjugated donkey antimouse IgG, donkey antirabbit IgG, and goat antimouse IgM were used as secondary antibodies (Millipore).
Statistical Analysis
Data are expressed as mean ± SEM. Experiments were performed in triplicate, except where noted in the Figure Legend. All data were analyzed using a one-way ANOVA test, apart from MTT assays that used a two-way ANOVA test in GraphPad Prism v6 (GraphPad Software, Inc.).
Acknowledgments
This research was supported by Movember through Prostate Cancer Foundation of Australia (YI0813 to Q.W.; PG2910 to M.J. and J.H.; YI0707 to J.H.; and the Australian Movember Revolutionary Team Award Targeting Advanced Prostate Cancer, J.H., R.J.Q., Q.W., and T.G); National Breast Cancer Foundation (ECF-12-05 J.H.); National Health and Medical Research Council (1051820 to J.H.; 1035693 to M.J.; 1048784 and 571093 to R.M.R.); Cure the Future and an anonymous foundation (J.E.J.R); Tour de Cure Fellowship (C.G.B); Queensland Government Smart Futures NIRAP program (R.J.Q.), Australian Research Council for NMR and MS equipment (LE0668477 and LE0237908 to R.J.Q.). The authors acknowledge the support of Compounds Australia, which is a recipient of funding from the Queensland Government Smart State Research Facilities Fund and Australian Government funding provided under the Super Science Initiative and financed from the Education Investment Fund. We thank the Queensland Herbarium for plant collection and identification and H. Vu for the HRESIMS data acquisition.
Supporting Information Available
This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
R.J.Q. and J.H. designed the experiments. Q.W. and J.H. screened the Nature Bank library in LNCaP cells. T.G. isolated and identified the structure of the natural products. S.B. and R.H.P. synthesized the natural products. C.G.B. and J.F. cloned the cDNA of transporters. J.F. performed the oocyte assays. Q.W. and A.M. examined the compounds in LNCaP cells. Q.W., T.G., S.B., R.H.P., J.F., R.M.R., J.E.J.R., M.J., R.J.Q., and J.H. interpreted results and edited the manuscript. Q.W., T.G., S.B., R.H.P., J.F., C.G.B, R.J.Q., and J.H. wrote the paper.
The authors declare no competing financial interest.
Supplementary Material
References
- Kanai Y.; Segawa H.; Miyamoto K.; Uchino H.; Takeda E.; Endou H. (1998) Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98). J. Biol. Chem. 273, 23629–23632. [DOI] [PubMed] [Google Scholar]
- Mastroberardino L.; Spindler B.; Pfeiffer R.; Skelly P. J.; Loffing J.; Shoemaker C. B.; Verrey F. (1998) Amino-acid transport by heterodimers of 4F2hc/CD98 and members of a permease family. Nature 395, 288–291. [DOI] [PubMed] [Google Scholar]
- Pineda M.; Fernandez E.; Torrents D.; Estevez R.; Lopez C.; Camps M.; Lloberas J.; Zorzano A.; Palacin M. (1999) Identification of a membrane protein, LAT-2, that co-expresses with 4F2 heavy chain, an l-type amino acid transport activity with broad specificity for small and large zwitterionic amino acids. J. Biol. Chem. 274, 19738–19744. [DOI] [PubMed] [Google Scholar]
- Rossier G.; Meier C.; Bauch C.; Summa V.; Sordat B.; Verrey F.; Kuhn L. C. (1999) LAT2, a new basolateral 4F2hc/CD98-associated amino acid transporter of kidney and intestine. J. Biol. Chem. 274, 34948–34954. [DOI] [PubMed] [Google Scholar]
- Segawa H.; Fukasawa Y.; Miyamoto K.; Takeda E.; Endou H.; Kanai Y. (1999) Identification and functional characterization of a Na+-independent neutral amino acid transporter with broad substrate selectivity. J. Biol. Chem. 274, 19745–19751. [DOI] [PubMed] [Google Scholar]
- Babu E.; Kanai Y.; Chairoungdua A.; Kim D. K.; Iribe Y.; Tangtrongsup S.; Jutabha P.; Li Y.; Ahmed N.; Sakamoto S.; Anzai N.; Nagamori S.; Endou H. (2003) Identification of a novel system l-amino acid transporter structurally distinct from heterodimeric amino acid transporters. J. Biol. Chem. 278, 43838–43845. [DOI] [PubMed] [Google Scholar]
- Bodoy S.; Martin L.; Zorzano A.; Palacin M.; Estevez R.; Bertran J. (2005) Identification of LAT4, a novel amino acid transporter with system l-activity. J. Biol. Chem. 280, 12002–12011. [DOI] [PubMed] [Google Scholar]
- Fukuhara D.; Kanai Y.; Chairoungdua A.; Babu E.; Bessho F.; Kawano T.; Akimoto Y.; Endou H.; Yan K. (2007) Protein characterization of NA+-independent system l-amino acid transporter 3 in mice: A potential role in supply of branched-chain amino acids under nutrient starvation. Am. J. Pathol. 170, 888–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball S. R.; Shantz L. M.; Horetsky R. L.; Jefferson L. S. (1999) Leucine regulates translation of specific mRNAs in L6 myoblasts through mTOR-mediated changes in availability of eIF4E and phosphorylation of ribosomal protein S6. J. Biol. Chem. 274, 11647–11652. [DOI] [PubMed] [Google Scholar]
- Han J. M.; Jeong S. J.; Park M. C.; Kim G.; Kwon N. H.; Kim H. K.; Ha S. H.; Ryu S. H.; Kim S. (2012) Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 149, 410–424. [DOI] [PubMed] [Google Scholar]
- Bonfils G.; Jaquenoud M.; Bontron S.; Ostrowicz C.; Ungermann C.; De Virgilio C. (2012) Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol. Cell 46, 105–110. [DOI] [PubMed] [Google Scholar]
- Zoncu R.; Bar-Peled L.; Efeyan A.; Wang S.; Sancak Y.; Sabatini D. M. (2011) mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 334, 678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y.; Bar-Peled L.; Zoncu R.; Markhard A. L.; Nada S.; Sabatini D. M. (2010) Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.; Goraksha-Hicks P.; Li L.; Neufeld T. P.; Guan K. L. (2008) Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata T.; Ferdous G.; Tsuruta T.; Satoh T.; Baba S.; Muto T.; Ueno A.; Kanai Y.; Endou H.; Okayasu I. (2009) l-Type amino-acid transporter 1 as a novel biomarker for high-grade malignancy in prostate cancer. Pathol. Int. 59, 7–18. [DOI] [PubMed] [Google Scholar]
- Wang Q.; Bailey C. G.; Ng C.; Tiffen J.; Thoeng A.; Minhas V.; Lehman M. L.; Hendy S. C.; Buchanan G.; Nelson C. C.; Rasko J. E.; Holst J. (2011) Androgen receptor and nutrient signaling pathways coordinate the demand for increased amino acid transport during prostate cancer progression. Cancer Res. 71, 7525–7536. [DOI] [PubMed] [Google Scholar]
- Wang Q.; Tiffen J.; Bailey C. G.; Lehman M. L.; Ritchie W.; Fazli L.; Metierre C.; Feng Y. J.; Li E.; Gleave M.; Buchanan G.; Nelson C. C.; Rasko J. E.; Holst J. (2013) Targeting amino acid transport in metastatic castration-resistant prostate cancer: Effects on cell cycle, cell growth, and tumor development. J. Natl. Cancer Inst. 105, 1463–1473. [DOI] [PubMed] [Google Scholar]
- Kobayashi K.; Ohnishi A.; Promsuk J.; Shimizu S.; Kanai Y.; Shiokawa Y.; Nagane M. (2008) Enhanced tumor growth elicited by l-type amino acid transporter 1 in human malignant glioma cells. Neurosurgery 62, 493–503. [DOI] [PubMed] [Google Scholar]
- Kim C. S.; Cho S. H.; Chun H. S.; Lee S. Y.; Endou H.; Kanai Y.; Kim do K. (2008) BCH, an inhibitor of system l-amino acid transporters, induces apoptosis in cancer cells. Biol. Pharm. Bull. 31, 1096–1100. [DOI] [PubMed] [Google Scholar]
- Kaira K.; Oriuchi N.; Imai H.; Shimizu K.; Yanagitani N.; Sunaga N.; Hisada T.; Tanaka S.; Ishizuka T.; Kanai Y.; Endou H.; Nakajima T.; Mori M. (2008) Prognostic significance of l-type amino acid transporter 1 expression in resectable stage I–III non-small-cell lung cancer. Br. J. Cancer 98, 742–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shennan D. B.; Thomson J.; Gow I. F.; Travers M. T.; Barber M. C. (2004) l-Leucine transport in human breast cancer cells (MCF-7 and MDA-MB-231): Kinetics, regulation by estrogen and molecular identity of the transporter. Biochim. Biophys. Acta 1664, 206–216. [DOI] [PubMed] [Google Scholar]
- Matthews R. H.; Sardovia M.; Lewis N. J.; Zand R. (1975) Biphasic kinetic plots and specific analogs distinguishing and describing amino acid transport sites in S37 ascites tumor cells. Biochim. Biophys. Acta 394, 182–192. [DOI] [PubMed] [Google Scholar]
- Oda K.; Hosoda N.; Endo H.; Saito K.; Tsujihara K.; Yamamura M.; Sakata T.; Anzai N.; Wempe M. F.; Kanai Y.; Endou H. (2010) l-Type amino acid transporter 1 inhibitors inhibit tumor cell growth. Cancer Sci. 101, 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp D.; Davis R. A.; Campitelli M.; Ebdon J.; Quinn R. J. (2012) Drug-like properties: Guiding principles for the design of natural product libraries. J. Nat. Prod. 75, 72–81. [DOI] [PubMed] [Google Scholar]
- Camp D.; Campitelli M.; Carroll A. R.; Davis R. A.; Quinn R. J. (2013) Front-loading natural-product-screening libraries for log P: Background, development, and implementation. Chem. Biodiversity 10, 524–537. [DOI] [PubMed] [Google Scholar]
- Takahashi S.; Kuzuhara H. (1997) Simple syntheses of l-fucopyranose andfucosidase inhibitors utilizing the highly stereoselective methylationof an arabinofuranoside 5-urose derivative. J. Chem. Soc., Perkin Trans. 1 5, 607–612. [Google Scholar]
- Lindhorst T. K. (2000) Essentials of Carbohydrate Chemistry and Biochemistry; Wiley-VCH, Weinheim/New York, pp xiii, 217. [Google Scholar]
- Bartlett C. J.; Day D. P.; Chan Y.; Allin S. M.; McKenzie M. J.; Slawin A. M.; Page P. C. (2012) Enantioselective total synthesis of (+)-scuteflorin A using organocatalytic asymmetric epoxidation. J. Org. Chem. 77, 772–774. [DOI] [PubMed] [Google Scholar]
- Wang Q.; Beaumont K. A.; Otte N. J.; Font J.; Bailey C. G.; van Geldermalsen M.; Sharp D. M.; Tiffen J. C.; Ryan R. M.; Jormakka M.; Haass N. K.; Rasko J. E. J.; Holst J. (2014) Targeting glutamine transport to suppress melanoma cell growth. Int. J. Cancer 10.1002/ijc.28749. [DOI] [PubMed] [Google Scholar]
- Poulsen M. V.; Vandenberg R. J. (2001) Niflumic acid modulates uncoupled substrate-gated conductances in the human glutamate transporter EAAT4. J. Physiol. 534, 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.