Abstract
Cyanobacteria possess the simplest known circadian clock, which presents a unique opportunity to study how rhythms are generated and how input signals from the environment reset the clock time. The kaiABC locus forms the core of the oscillator, and the remarkable ability to reconstitute oscillations using purified KaiABC proteins has allowed researchers to study mechanism using the tools of quantitative biochemistry. Autotrophic cyanobacteria experience major shifts in metabolism following a light-dark transition, and recent work suggests that input mechanisms that couple the day-night cycle to the clock involve energy and redox metabolites acting directly on clock proteins. We offer a summary of the current state of knowledge in this system and present a perspective for future lines of investigation.
INTRODUCTION
Life near the surface of the Earth must contend with a rhythmically changing environment due to the rotation of the planet on its axis. Apparently in response to this challenge, many organisms have developed circadian clocks, endogenous timing systems that generate near-24 hour rhythms in behavior and physiology in anticipation of dusk and dawn. A defining property of a true circadian clock is that these endogenous rhythms are self-sustaining and will persist in constant laboratory environments.
Over the past decades, it has become increasingly clear that there are intimate connections between circadian rhythms and metabolism in plants, animals, and fungi. Strikingly, recent reports have identified a conserved link between circadian rhythmicity and the oxidation state of peroxiredoxins in organisms across the kingdoms, though the mechanistic nature of this link has yet to be elucidated [1]. In this review, we discuss the recent expansion of work on the simplest known model organism that exhibits circadian rhythms—photosynthetic bacteria—and what is known about the coupling between the clock and metabolism in these prokaryotes. Though quite sophisticated in function, the cyanobacterial clock has a relatively tractable genetic structure compared to plant and animal model systems.
Remarkably, the fundamental oscillatory mechanism in the cyanobacterial clock can be reconstituted using a mixture of three purified proteins in vitro [2]. These developments make it possible to bring the full power of both bacterial genetics and quantitative biochemistry to bear on the subject of biological rhythms, a topic once limited to phenomenological studies. We conclude by discussing recent work that has begun to study how the clock is coupled to metabolism in the context of the reconstituted oscillator—a minimal system formed by the KaiABC proteins that autonomously generates ~24 h rhythms. We offer a perspective for future studies analyzing how the circadian clock is integrated with bacterial physiology in vivo.
THE CYANOBACTERIAL CIRCADIAN CLOCK
Despite long-standing suspicions that regulatory mechanisms in bacteria might be too simple to support true circadian rhythms, or that the rapid (faster than a day) doubling times of many bacteria would somehow obviate the need for a circadian clock [3,4], researchers in the late 1980s discovered a bona fide circadian rhythm in unicellular cyanobacterial species [5,6]. Significantly, these initial discoveries centered on the relationship between rhythms and metabolism—unicellular cyanobacteria cannot simultaneously carry out oxygenic photosynthesis and nitrogen fixation, because the nitrogenases used by these organisms are poisoned by oxygen. An endogenous circadian rhythm allows a cyanobacterial cell to temporally separate photosynthesis from nitrogenase activity, even when constantly illuminated.
Progress into deciphering the molecular mechanisms that generate rhythms in cyanobacteria proceeded rapidly with the development of the genetically tractable isolate Synechococcus elongatus PCC 7942 (henceforth S. elongatus, notably not a nitrogen fixer) [3]. The introduction of bioluminescent and fluorescent reporters into this strain to permit automated detection of transcriptional rhythms has made it a powerful model organism to investigate the cyanobacterial circadian system.
Analysis of this organism has revealed many fundamental properties of the clock mechanism. Time-lapse microscopy studies have indicated that circadian rhythms are nearly or completely a cell-autonomous phenomenon, and that daughter cells inherit the clock phase of the mother with high fidelity [7]. Nevertheless, bulk cultures of S. elongatus can maintain high amplitude, coherent rhythms for many days, reflecting highly robust single-cell clocks that can preserve a stable phase on the timescale of weeks without further input from the environment [7,8].
Despite the ability of the circadian rhythm to persist robustly through cell divisions, it is not true that the cell cycle and circadian clock are uncoupled in this organism. It has been observed in both turbidostatic bulk culture and in single cells that the circadian clock imposes an inhibitory control on cell division near dusk (so-called “circadian gating”), so that cells are discouraged from dividing at clock times corresponding to dusk, though DNA continues to be replicated [9,10]. Though this phenomenon appears as an isolated inhibitory window lasting a few hours per day under constant light laboratory growth conditions, in a cycling environment cell division is completely prevented in the dark, independent of clock time. Therefore the phenomenon of clock-gated cell division may instead by viewed as an anticipatory block on cell division at dusk that briefly precedes the growth-halting effects of darkness.
Some of the original transcriptional reporters used to measure rhythms in S. elongatus were fused to the promoters of photosystem genes (e.g. psbAI) which oscillate with a very high amplitude. Subsequent analysis using both promoter trap methods and microarray analyses have shown that many (if not all) of the genes in the S. elongatus genome have a rhythmic, circadian component to their expression [11–13]. These global rhythms in gene expression are likely related to changes in the supercoiling status of the cyanobacterial genome which has been shown to oscillate during the circadian cycle [13,14]. Though many temporal patterns of gene expression can be observed in S. elongatus, the vast majority of genes peak in expression either near subjective dusk or subjective dawn, when cells are kept in constant light [11]. As with the case with the circadian control of cell division discussion above, actual darkness forces a massive reduction in the transcriptional output of the cell, though a small number of genes are transiently induced in the dark in clock-specific manner [12,15].
Genetic analysis of the S. elongatus clock has been extremely successful at illuminating many of the molecular mechanisms that underlie rhythmicity in this organism. A cleverly designed genetic screen identified a locus of three genes, named kaiA, kaiB, and kaiC, all of which are required for rhythmic transcription and behavior [16]. In a sense we will make precise in the following section, the kai genes are the core of the oscillatory mechanism.
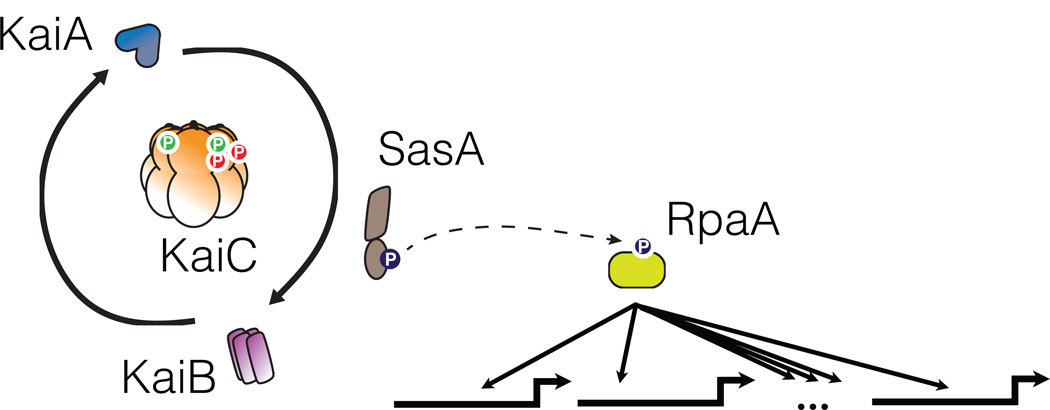
Further analysis has identified the major clock output pathway, which transduces timing information from the Kai proteins to control transcription and the rhythmic physiology described above. The histidine kinase SasA interacts with the Kai proteins and rhythmically phosphorylates the response regulator RpaA [17]. Phosphorylated RpaA has been shown to bind directly to a regulon of ~170 genes which include many of the high amplitude genes which peak at subjective dusk. In the absence of this signaling pathway, no noticeable rhythms in transcription remain [18]. Thus the Kai oscillator serves as a signaling platform that interfaces with a canonical bacterial two-component system to control transcription. Other genes that peak near subjective dawn and the global effects that cause rhythmic expression of even heterologous promoters may be downstream of this primary regulon. RpaA controls expression of multiple sigma factors, a modular subunit of RNA polymerase that may assist in generating global rhythms. Another possibility is that oscillatory transcription of the RpaA regulon results in rhythms of the metabolic state of the cell that could influence DNA supercoiling—DNA gyrase activity is known to be quite sensitive to the ATP/ADP ratio [19].
BIOCHEMICAL RECONSTITUTION
The prevailing models for the generation of circadian rhythms in eukaryotes are based on transcriptional negative feedback loops, where a gene product ultimately induces the repression of its own promoter. Because kaiBC is a high-amplitude rhythmic target of the clock output, it was initially appealing to apply to eukaryotic transcription-centric models to the cyanobacterial clock [20]. However, circadian rhythms persist in S. elongatus even when kaiBC is expressed from constitutive or opposite-phase promoters [21]. An even stronger result is that circadian timing persists in the dark when gene expression is nearly abolished, and in the presence of inhibitors of transcription and translation [22]. Under these conditions, rhythmic phosphorylation of the core clock enzyme KaiC can still be observed.
This line of investigation culminated with the striking report that recombinantly expressed, purified KaiA, KaiB, and KaiC proteins generate a stable near-24 hour rhythm in a test tube in the presence of ATP—effectively a reconstituted clock [2]. This reconstituted system has many remarkable properties in common with the circadian rhythm in vivo. Despite being based on enzyme-catalyzed reactions, the period of the reconstituted rhythms are quite resistant to changes in temperature. Many point mutations that alter the period of the clock in vivo cause similar changes to the period in the reconstituted system [2]. These results established unequivocally that post-translational interactions between the Kai proteins are primarily responsible for determining the periodicity of the circadian rhythm, despite the fact that the kaiBC gene participates in a transcriptional feedback loop. Single-cell analyses have recently shown that this feedback loop is important for reducing noise in the circadian rhythm [23]. Taken together, these results raise the question of whether the importance of post-translational mechanisms in establishing timing in the eukaryotic circadian feedback loops may be underestimated.
Much biochemical and mathematical modeling work has gone into understanding the basis of oscillations in the reconstituted system. KaiC is a hexameric enzyme, and each subunit consists of two similar AAA+ ATPase-like domains, CI and CII [24]. The overall rate of ATP turnover from KaiC is very slow and reflects the circadian timescale of the oscillations [25]. Phosphorylation of KaiC occurs through an autokinase reaction at the subunit interfaces in CII; two residues, Ser431 and Thr432, are phosphorylated in a specific order due to strong kinetic preferences [26,27]. KaiC can also dephosphorylate itself using the same active site, acting on Ser431 and Thr432 with similar kinetic preferences, using ADP as a phosphate acceptor [28,29].
KaiC kinase activity is stimulated by interaction with KaiA [30]. At the transition from the phosphorylation phase of the oscillation to the dephosphorylation phase, increasing Ser431 phosphorylation promotes KaiB binding to KaiC. This KaiB•KaiC complex can then trap KaiA, leaving it unable to activate KaiC molecules and allowing dephosphorylation to occur [27].
The basic logic of an activator (KaiA) that is inhibited when KaiC reaches a critical phosphorylation state is a potential mechanism that couples many KaiC molecules together. This coupling ensures that KaiC molecules that begin to drift out of phase will be brought back in sync, making the oscillation a collective property of all of the molecules in the reaction. This collective behavior helps to explain the remarkable robustness of the rhythms in single cells—because there are many 1,000s of KaiC molecules coupled together in each cell, progression through a cycle involves thousands of elementary reactions as each KaiC hexamer becomes phosphorylated, essentially averaging over the fundamental stochasticity in each underlying chemical reaction. Thus, the Law of Large Numbers may effectively suppress stochasticity in the clock time [31].
GENETICS OF CLOCK INPUT
Though the Kai proteins alone are capable of generating a highly robust rhythm, there must be additional mechanisms in the cell that cause this rhythm to synchronize appropriately with light-dark cycles in the environment. Genetic screens based on random mutagenesis or transposon insertions to identify mutants with clocks that did not respond appropriately to perturbation by a 4–5 hour dark pulse yielded two results: a point mutation in kaiC (A422V) [32] and the deletion of a histidine kinase named cikA (circadian input kinase) [33].
Although CikA has phytochrome-like properties by sequence analysis, it does not appear to function as a direct light sensor [34]. Recent genetic and biochemical analysis has indicated that CikA plays a role in the clock output mechanism and modulates RpaA phosphorylation [35,36]. Thus, the basis of the clock input phenotype of cikA-null remains opaque. Further, even the clock of cikA-null cells can be effectively reset by a 12 hour dark pulse, implying that there must be cikA-independent input mechanisms.
METABOLIC INPUT IN VITRO
An alternative hypothesis is that information about the time of day can be communicated to the Kai oscillator through photosynthesis directly. This idea is attractive since, as a phototroph, the metabolic state of S. elongatus is highly dependent on light availability. Further, it is known that the phase of the circadian rhythm can be reset through prolonged treatment with a photosynthesis inhibitor, similar to the effect of a dark pulse [37].
This led us and others to look for direct interactions between the Kai proteins and metabolites that change in response to a loss of photosynthetic activity. Because ATP is a required cofactor for KaiC phosphorylation, ATP levels were a natural target. Although ATP is present at saturating concentrations for KaiC binding in vivo, the ATP/ADP ratio is a potent regulator of KaiC’s autokinase activity [38]. By simulating the changes in ATP/ADP that occur during a dark pulse, we were able to recreate phase shifts in the purified oscillator that mimic those seen in vivo [38] (Fig. 2). Despite the impact of ATP/ADP on KaiC kinase activity, the period of the purified oscillator is remarkably insensitive to sustained changes in ATP/ADP. We recently proposed a mechanism to account for this “nutrient compensation” of the oscillator period based on an invariant delay in KaiB binding set by slow CI ATPase activity [39].
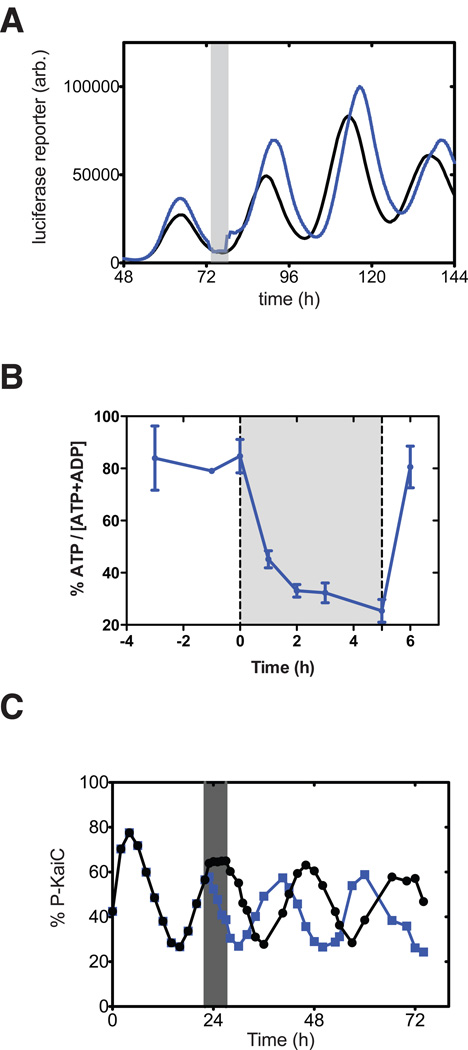
Figure 2.
Reconstitution of metabolic clock input. (A) Clock resetting in vivo. A luminescence reporter of clock-driven transcription is recorded while cells grow in constant light. The experimental culture (blue points) is exposed to a 5 hour dark pulse (gray bar) which shifts the phase of the subsequent rhythm relative to a control (black points). (B) Energy charge drops in darkness. Measurement of [ATP]/([ATP] + [ADP]) during a similar dark pulse (gray bar). (C) Simulating changes in ATP/ADP ratio to match those seen in vivo using a buffer exchange protocol recapitulates phase shifts in the purified system. ATP/ADP is lowered for the experimental reaction (blue points) relative to a control (black points) during the gray bar.
Similarly, it has been recently shown that KaiA reversibly aggregates in the presence of oxidized quinone, and that this metabolic input can also be used to create phase shifts in the purified oscillator [40,41]. Because flow through the electron chain ceases on a faster timescale than ATP depletion in the dark, it has been suggested that redox signals through quinone and energy signals through ATP/ADP work together during a dark pulse to appropriately reset the clock [41].
CONCLUSIONS
The reconstituted KaiABC oscillator presents a uniquely powerful tool for unraveling the mechanisms responsible for clock input, because various effects can be studied in isolation. We expect that combining this biochemical approach more closely with genetics will lead to an integrated understanding of how the oscillator functions in its in vivo context.
A major question that remains to be answered involves the relationship between the cikA-null and kaiC (A422V) mutant phenotypes and the metabolic mechanisms that have been characterized biochemically. One possibility is that CikA is involved in a separate input mechanism with the ability to partially override metabolic signals. Another is that these mutants might alter the state of the cell so that the metabolic contrast between light and dark is less severe.
We currently lack a complete structural understanding of how ATP/ADP and quinone act on the Kai proteins to alter their activity. Similarly, we do not know how changes in abundance of the Kai proteins might alters the system’s response to metabolic cues. Relatedly, it should be noted that darkness halts the transcription of the kai genes while allowing post-translational modifications to continue, and that this likely has implications for clock input [42]—the suppression of transcription by darkness can itself be thought of as a metabolic input.
Finally, it has become clear that metabolism is intimately involved with clock input, but does the clock itself generate important rhythms in metabolism? Many genes involved in central carbon metabolism are under clock control [13]. There is certainly a precedent for this in the circadian field, particularly in plants [43,44], but clear evidence for metabolic rhythms has been scant so far in cyanobacteria. If indeed a major function of the circadian clock is to prepare the metabolic state of the cell for changes in the environment, it may point the way to uncovering the basis of clock-dependent fitness advantages [45].
Figure 1.
Schematic of cyanobacterial clock. The core oscillatory mechanism consists of post-translational modification of KaiC and interactions with KaiA and KaiB. Rhythmic interactions between SasA and KaiC lead to phosphotransfer to the response regulator RpaA. RpaA targets many genes, including kaiBC.
HIGHLIGHTS.
We discuss the role of cyanobacterial clock in regulating cell physiology
We summarize the biochemistry of the Kai proteins
We discuss recent results connecting clock input to metabolism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451.. The authors show that purified KaiA, KaiB, and KaiC proteins are sufficient to generate a near-24 hour rhythm in KaiC phosphorylation in a test tube. They showed that this in vitro oscillator is temperature-compensated, and that several in vivo period mutants in KaiC recapitulate the altered period in vitro. This work paved the way for the biochemical study of circadian clock mechanism.
- 3.Johnson CH, Golden SS, Ishiura M, Kondo T. Circadian clocks in prokaryotes. Mol Microbiol. 1996;21:5–11. doi: 10.1046/j.1365-2958.1996.00613.x. [DOI] [PubMed] [Google Scholar]
- 4.Winfree AT. The Geometry of Biological Time. edn 2nd. New York: Springer-Verlag; 2000. [Google Scholar]
- 5.Grobbelaar N, Huang TC, Lin HY, Chow TJ. Dinitrogen-Fixing Endogenous Rhythm in Synechococcus Rf-1. FEMS Microbiol Lett. 1986;37:173–177. [Google Scholar]
- 6.Mitsui A, Kumazawa S, Takahashi A, Ikemoto H, Cao S, Arai T. Strategy by Which Nitrogen-Fixing Unicellular Cyanobacteria Grow Photoautotrophically. Nature. 1986;323:720–722. [Google Scholar]
- 7.Mihalcescu I, Hsing W, Leibler S. Resilient circadian oscillator revealed in individual cyanobacteria. Nature. 2004;430:81–85. doi: 10.1038/nature02533. [DOI] [PubMed] [Google Scholar]
- 8.Teng SW, Mukherji S, Moffitt JR, de Buyl S, O'Shea EK. Robust circadian oscillations in growing cyanobacteria require transcriptional feedback. Science. 2013;340:737–740. doi: 10.1126/science.1230996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori T, Binder B, Johnson CH. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:10183–10188. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Q, Pando BF, Dong G, Golden SS, van Oudenaarden A. Circadian gating of the cell cycle revealed in single cyanobacterial cells. Science. 2010;327:1522–1526. doi: 10.1126/science.1181759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Golden SS, Kondo T, Ishiura M, Johnson CH. Bacterial luciferase as a reporter of circadian gene expression in cyanobacteria. J Bacteriol. 1995;177:2080–2086. doi: 10.1128/jb.177.8.2080-2086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito H, Mutsuda M, Murayama Y, Tomita J, Hosokawa N, Terauchi K, Sugita C, Sugita M, Kondo T, Iwasaki H. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc Natl Acad Sci U S A. 2009;106:14168–14173. doi: 10.1073/pnas.0902587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vijayan V, Zuzow R, O'Shea EK. Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc Natl Acad Sci U S A. 2009;106:22564–22568. doi: 10.1073/pnas.0912673106.. The authors used a microarray approach to analyze rhythmic transcription in Synechococcus. Consistent with previous indications of a rhythm in DNA supercoiling, this study shows that supercoiling changes exert a phase-dependent causal influence on genome-wide transcription by using drugs to inhibit DNA gyrase.
- 14.Smith RM, Williams SB. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci U S A. 2006;103:8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosokawa N, Hatakeyama TS, Kojima T, Kikuchi Y, Ito H, Iwasaki H. Circadian transcriptional regulation by the posttranslational oscillator without de novo clock gene expression in Synechococcus. Proc Natl Acad Sci U S A. 2011;108:15396–15401. doi: 10.1073/pnas.1019612108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo T, Tsinoremas NF, Golden SS, Johnson CH, Kutsuna S, Ishiura M. Circadian clock mutants of cyanobacteria. Science. 1994;266:1233–1236. doi: 10.1126/science.7973706. [DOI] [PubMed] [Google Scholar]
- 17.Takai N, Nakajima M, Oyama T, Kito R, Sugita C, Sugita M, Kondo T, Iwasaki H. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc Natl Acad Sci U S A. 2006;103:12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markson JS, Piechura JR, Puszynska AM, O'Shea EK. Circadian control of global gene expression by the cyanobacterial master regulator RpaA. Cell. 2013;155:1396–1408. doi: 10.1016/j.cell.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Workum M, van Dooren SJ, Oldenburg N, Molenaar D, Jensen PR, Snoep JL, Westerhoff HV. DNA supercoiling depends on the phosphorylation potential in Escherichia coli. Mol Microbiol. 1996;20:351–360. doi: 10.1111/j.1365-2958.1996.tb02622.x. [DOI] [PubMed] [Google Scholar]
- 20.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 21.Ditty JL, Canales SR, Anderson BE, Williams SB, Golden SS. Stability of the Synechococcus elongatus PCC 7942 circadian clock under directed antiphase expression of the kai genes. Microbiology. 2005;151:2605–2613. doi: 10.1099/mic.0.28030-0. [DOI] [PubMed] [Google Scholar]
- 22.Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 23.Tseng R, Chang YG, Bravo I, Latham R, Chaudhary A, Liwang A. KaiA Assists the KaiB-KaiC Interaction and KaiB/SasA Competition in the Circadian Clock of Cyanobacteria. J Mol Biol. 2013 doi: 10.1016/j.jmb.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi F, Itoh N, Uzumaki T, Iwase R, Tsuchiya Y, Yamakawa H, Morishita M, Onai K, Itoh S, Ishiura M. Roles of two ATPase-motif-containing domains in cyanobacterial circadian clock protein KaiC. J Biol Chem. 2004;279:52331–52337. doi: 10.1074/jbc.M406604200. [DOI] [PubMed] [Google Scholar]
- 25. Terauchi K, Kitayama Y, Nishiwaki T, Miwa K, Murayama Y, Oyama T, Kondo T. ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc Natl Acad Sci U S A. 2007;104:16377–16381. doi: 10.1073/pnas.0706292104.. The authors provide biochemical evidence that the period of the circadian rhythm is closely linked to the rate of catalytic turnover for the core enzyme KaiC. This study shows that each KaiC molecule consumes less than 1 ATP molecule every hour, and that the rate of ATP turnover is correlated with the oscillator period in many mutants. Further, they show that the rate of ATP turnover is nearly insensitive to temperature over the funcitonal range of the oscillator.
- 26.Nishiwaki T, Satomi Y, Kitayama Y, Terauchi K, Kiyohara R, Takao T, Kondo T. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 2007;26:4029–4037. doi: 10.1038/sj.emboj.7601832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rust MJ, Markson JS, Lane WS, Fisher DS, O'Shea EK. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318:809–812. doi: 10.1126/science.1148596.. The authors show that phosphorylation and dephosphorylation of KaiC occur at two residues (Ser431 and Thr432) in an ordered fashion. Further, the study shows that measured rate constants for phosphorylation at each site along with the assumption that P~Ser431 triggers formation of a complex that traps the activator KaiA can produce stable oscillations.
- 28.Egli M, Mori T, Pattanayek R, Xu Y, Qin X, Johnson CH. Dephosphorylation of the core clock protein KaiC in the cyanobacterial KaiABC circadian oscillator proceeds via an ATP synthase mechanism. Biochemistry. 2012;51:1547–1558. doi: 10.1021/bi201525n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishiwaki T, Kondo T. Circadian autodephosphorylation of cyanobacterial clock protein KaiC occurs via formation of ATP as intermediate. J Biol Chem. 2012;287:18030–18035. doi: 10.1074/jbc.M112.350660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci U S A. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22:2127–2134. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiyohara YB, Katayama M, Kondo T. A novel mutation in kaiC affects resetting of the cyanobacterial circadian clock. J Bacteriol. 2005;187:2559–2564. doi: 10.1128/JB.187.8.2559-2564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitz O, Katayama M, Williams SB, Kondo T, Golden SS. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science. 2000;289:765–768. doi: 10.1126/science.289.5480.765. [DOI] [PubMed] [Google Scholar]
- 34.Mutsuda M, Michel KP, Zhang X, Montgomery BL, Golden SS. Biochemical properties of CikA, an unusual phytochrome-like histidine protein kinase that resets the circadian clock in Synechococcus elongatus PCC 7942. J Biol Chem. 2003;278:19102–19110. doi: 10.1074/jbc.M213255200. [DOI] [PubMed] [Google Scholar]
- 35. Gutu A, O'Shea EK. Two antagonistic clock-regulated histidine kinases time the activation of circadian gene expression. Mol Cell. 2013;50:288–294. doi: 10.1016/j.molcel.2013.02.022.. The authors show that the histidine kinase CikA, discovered in a screen for clock input phenotypes, acts on the output transcription factor RpaA. The study further establishes that CikA is a bifunctional enzyme (i.e. can both phosphotranfer to RpaA and act to dephosphorylate RpaA) and that the phosphatase activity of CikA depends on the state of the KaiABC oscillator.
- 36.Taniguchi Y, Takai N, Katayama M, Kondo T, Oyama T. Three major output pathways from the KaiABC-based oscillator cooperate to generate robust circadian kaiBC expression in cyanobacteria. Proc Natl Acad Sci U S A. 2010;107:3263–3268. doi: 10.1073/pnas.0909924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katayama M, Kondo T, Xiong J, Golden SS. ldpA encodes an iron-sulfur protein involved in light-dependent modulation of the circadian period in the cyanobacterium Synechococcus elongatus PCC 7942. J Bacteriol. 2003;185:1415–1422. doi: 10.1128/JB.185.4.1415-1422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rust MJ, Golden SS, O'Shea EK. Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science. 2011;331:220–223. doi: 10.1126/science.1197243.. The authors show that during dark pulses that cause phase shifts in the cyanobacterial clock, the energy charge of the cell (ATP/ADP ratio) falls dramatically. Simultaing these changes in ATP/ADP in the purified KaiABC oscillator causes similar phase shifts in vitro. Thus metabolic cues can act directly on the Kai proteins to adjust the circadian rhythm. The authors identify a potential mechanism based on the sensitivity of KaiC's kinase activity to ATP/ADP.
- 39. Phong C, Markson JS, Wilhoite CM, Rust MJ. Robust and tunable circadian rhythms from differentially sensitive catalytic domains. Proc Natl Acad Sci U S A. 2013;110:1124–1129. doi: 10.1073/pnas.1212113110.. The authors show that the function of N-terminal ATPase domain of KaiC is required for the negative feedback that switches the oscillator kinase mode to phosphatase mode. Further, this study shows that the activity of the N-terminal domain is only weakly sensitive to the ATP/ADP metabolic cue relative to the kinase activity of the C-terminal domain. Mathematical modeling suggests a mechanism where a sequence of metabolism-sensitive steps and insensitive steps can create an oscillation that can be phase-shifted by metabolic cues but maintain a homeostatic period near 24 hours.
- 40.Wood TL, Bridwell-Rabb J, Kim YI, Gao T, Chang YG, LiWang A, Barondeau DP, Golden SS. The KaiA protein of the cyanobacterial circadian oscillator is modulated by a redox-active cofactor. Proc Natl Acad Sci U S A. 2010;107:5804–5809. doi: 10.1073/pnas.0910141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim YI, Vinyard DJ, Ananyev GM, Dismukes GC, Golden SS. Oxidized quinones signal onset of darkness directly to the cyanobacterial circadian oscillator. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1216401109.. This study extends previous work from the same lab showing that the clock components KaiA and CikA interact with quinone in a redox-dependent fashion. In these experiments, the authors show that it is possible to reversibly inactivate KaiA in the in vitro oscillator by manipulating redox, and that these effects cause phase shifts in the rhythm. The authors also determine that the plastoquinone pool becomes rapidly oxidized in vivo following a light-dark transition. This study opens the door to the study of the role of redox in modulating the cyanobacterial clock.
- 42.Hosokawa N, Kushige H, Iwasaki H. Attenuation of the posttranslational oscillator via transcription-translation feedback enhances circadianphase shifts in Synechococcus. Proc Natl Acad Sci U S A. 2013;110:14486–14491. doi: 10.1073/pnas.1302243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Espinoza C, Degenkolbe T, Caldana C, Zuther E, Leisse A, Willmitzer L, Hincha DK, Hannah MA. Interaction with diurnal and circadian regulation results in dynamic metabolic and transcriptional changes during cold acclimation in Arabidopsis. PLoS One. 2010;5:e14101. doi: 10.1371/journal.pone.0014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graf A, Schlereth A, Stitt M, Smith AM. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci U S A. 2010;107:9458–9463. doi: 10.1073/pnas.0914299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci U S A. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]