Abstract
The study of interactions between hepatitis C virus (HCV) with its mammalian host, along with the development of more effective therapeutics and vaccines has been delayed by the lack of a suitable small animal model. HCV readily infects only humans and chimpanzees, which poses logistic, economic and ethical challenges with analyzing HCV infection in vivo. Progress has been made in understanding the determinants that dictate HCV’s narrow host range providing a blueprint for constructing a mouse model with inheritable susceptibility to HCV infection. Indeed, genetically humanized mice were generated that support viral uptake, replication and production of infectious virions – albeit at low levels. These efforts are complemented with attempts to select for viral variants that are inherently more capable of replicating in non-human species. In parallel, engraftment of relevant human tissues into improved xenorecipients is being continuously refined. Incorporating advances in stem-cell-biology and tissue engineering may allow the generation of patient-specific humanized mice. Construction of such mouse “avatars” may allow analyzing functionally patient-specific differences with respect to susceptibility to infection, disease progression and responses to treatment. In this review, we discuss the three, before mentioned approaches to overcome current species barriers and generate a small animal model for HCV infection, i.e. genetic modification of mice to increase their susceptibility to the virus; genetic modification of HCV, to increase its pathogenicity for mice; and the introduction of human liver and immune cells into immunodeficient mice, to create “humanized” mice. Although in the foreseeable future there will not be a single model that perfectly mimics the natural course of HCV in humans there is reason for optimism. The spectrum of murine animal models for hepatitis C provides a broad arsenal for analyzing the disease. These models may play an important role by prioritizing vaccine candidates and possibly refining combination anti-viral drug therapies. This article forms part of a symposium in Anti-viral Research on “Hepatitis C: next steps toward global eradication.”
Keywords: Hepatitis C, Animal models, Species tropism, Anti-viral immunity, Drug development, Vaccines
1. Do we still need a small animal model for hepatitis C?
Hepatitis C remains a major medical problem affecting at least 150 million chronic carriers who are at risk of developing serious liver diseases, including fibrosis, cirrhosis and hepatocellular carcinoma. Until recently curative therapies were poorly tolerated and ineffective in the majority of patients (National institutes of health consensus development conference statement, 2002; Jesudian et al., 2013). However, therapeutic regimens are emerging that hold promise of reliably eradicating HCV in the majority of patients. This optimistic view is blurred by the risk of emerging resistance, unfavorable drug-drug interactions and treatment side effects. These potential complications paired with the projected high cost of future therapies will limit access to those in developed countries with adequate medical infrastructure. Additionally, even in developed countries, a significant number of patients is not diagnosed (Edlin, 2011). Thus, the impact of combinations of directly acting anti-virals (DAAs) and host targeting anti-virals (HTAs) on a global scale, including in developing countries remains to be seen.
Although proof of concept for numerous novel therapeutic modalities against HCV (Olsen et al., 2011; Lanford et al., 2010) has been shown in chimpanzees, most emerging therapeutics were not routinely assessed for treatment efficacy in preclinical animal models. Thus, it appears that HCV animal models are not essential for the process. However, development of novel therapeutics could have possibly been sped up and complications of initially very promising drug candidates (Lamarre et al., 2003) could have potentially been predicted (Vanwolleghem et al., 2007) if a predictive (small) animal model(s) had been available. The rapidly evolving field of anti-HCV DAAs and HTAs could still benefit from predictive animal models for refinements of future combination therapies guiding the design of costly clinical trials. Beyond evaluating drug efficacy, HCV animal models could also be employed to analyze potentially interfering interactions between anti-HCV DAAs and other commonly administered drugs and may even serve to predict immune mediated, dose-independent toxicities.
While the DAA/HTA pipeline looks promising, development of therapeutic or preventative vaccines, which may arguably have the broadest impact on reducing the global HCV burden, remains a formidable challenge. Few vaccination approaches are being tested in clinical trials (Feinstone et al., 2012). The complexity of the mammalian immune system, HCV’s astounding genetic and antigenic diversity, our limited understanding of correlates of protection, and immune exhaustion in chronic HCV carriers are confounding problems for developing an effective HCV vaccine. Presumably, a vast number of permutations of antigens, adjuvants, vaccine vectors and administration regimens can only be systematically evaluated in small animal models and results will likely provide important guiding cues for prioritizing candidates for clinical development.
Beyond the practical applications a small animal model would be of great utility for basic research applications. Study of HCV has been hampered by the lack of infection systems that mimic accurately the unique host environment of the liver (Sheahan et al., 2010). Only a few human cell lines support efficiently HCV replication but their transformed nature does not reflect normal hepatocyte physiology (reviewed in Sheahan et al., 2010). Primary human hepatocytes replicate HCV only inefficiently, in part due to the difficulty of maintaining their highly differentiated phenotype in cell culture. Study of HCV in the physiological three dimensional context of an intact liver would be highly desirable. This would be facilitated by the use of an animal model (Billerbeck et al., 2013).
2. Desired features of a mouse model for hepatitis C?
As holds true for any experimental system for a human disease a mouse model for hepatitis C should optimally mimic closely as many, if not all relevant clinical features observed in patients. A mouse model for HCV should be readily susceptible to all HCV genotypes resulting in persistent viremia in the majority of exposed animals. In order to dissect mechanistically correlates of protective immunity, persistence and immune-mediated pathogenesis the model should be fully immune-competent and amenable to genetic manipulations. Since it can often take decades in humans between the acute phase of infection and development of severe liver pathologies it is impractical, and perhaps even unrealistic, to aim to wait for the natural disease progression in laboratory mice whose life-span is limited to less than three years. To overcome this challenge mouse strains may be used that are more prone to liver disease development possibly accelerating and exacerbating development of clinically relevant symptoms. From a practical perspective an animal model for hepatitis C should be highly reproducible, easy to propagate, high in throughput and cheap in production.
The chimpanzee model fulfills many of these requirements. Research using chimpanzees has been instrumental in the discovery and analysis of HCV infection and it remains the gold standard for all other animal models. However, their very high costs and scarcity have limited cohort sizes in experimental studies, which – in combination with the inter-individual variability of an outbred species – limits the reproducibility and thus their practical utility. Furthermore, use of chimpanzee in biomedical research is banned in many countries and growing ethical concerns in the US have led to an NIH memorandum that severely restricts all federally-funded (HCV) research involving chimpanzees (NIH 2011, posting date. http://grants.nih.gov/grants/guide/notice-files/NOT-OD-12-025.html). This creates a pressing need for alternative models to bridge the gap between currently available in vitro models for HCV infection and the complex situation in an (chronically) infected patient.
3. Approaches to generate a small animal model for HCV infection
Several independent but possibly complementary approaches have been taken to overcome current species barriers and to generate a small animal model for HCV infection, immunity and pathogenesis (reviewed in Billerbeck et al., 2013):
Expression of HCV proteins in transgenic mice.
Use of potential surrogates for HCV, i.e. viruses genetically related to HCV, which replicate more readily in non-human species. While original efforts focused on GB virus B transmission to new world monkeys (Simons et al., 1995; Lanford et al., 2003; Karayiannis et al., 1989; Schaluder et al., 1995; Bukh et al., 2001), more recently viruses even more closely related to HCV have been found in a number of other species, including dogs (Kapoor et al., 2011), horses (Burbelo et al., 2012) and rodents (Kapoor et al., 2013). However, it remains to be analyzed how similar the life-cycles of these viruses are to HCV’s and whether they would cause hepatitis in their respective hosts.
Humanization of the mouse liver and immune system by transplanting human hematopoietic stem cells and hepatocytes into the same murine recipient, thus allowing studies of pathology, immune correlates, and mechanisms of pathogen persistence.
Systematic screens to identify and overcome species restrictions allowing for genetic host adaptation and thus creating inbred murine models for HCV. Likewise, HCV could be adapted to infect hepatocytes of non-human origin, such as mice (Bitzegeio et al., 2010) and/or smaller non-human primates (Sourisseau et al., 2013).
Our discussions in this article will primarily focus on the latter two approaches as these focus on the virus that actually causes disease in humans and are aimed at modeling clinically relevant histopathology as a consequence of the possibly unique inflammatory milieu induced during the replicative cycle of HCV.
4. Genetically humanized mouse models for HCV infection – immunity and pathogenesis?
Mice are usually resistant to HCV infection but progress has been made towards a better understanding of the determinants restricting HCV’s replicative cycle to humans and chimpanzees. Mouse cells do not support HCV uptake, which creates a first barrier for a broader host range. HCV – complexed with host lipoproteins in lipoviro particles – engages a large number of cellular factors to enter human hepatocytes. The exact mechanism of HCV uptake has not been completely elucidated but is a multistep process (reviewed in Zeisel et al., 2013) initiated by attachment to glycosaminoglycans (Koutsoudakis et al., 2006; Barth et al., 2003), binding to low-density lipoprotein receptor (LDLR) (Molina et al., 2007; Agnello et al., 1999; Monazahian et al., 1999; Owen et al., 2009), the scavenger receptor class B type I (SCARB1; Scarselli et al., 2002) and the tetraspanin CD81 (Pileri et al., 1998) on the hepatocyte surface. HCV subsequently engages the tight junction proteins, claudin-1 (CLDN1; Evans et al., 2007) and occludin (OCLN; Liu et al., 2009; Ploss et al., 2009), ultimately resulting in uptake via receptor mediated endocytosis. Signaling through the receptor tyrosine kinases epidermal growth factor receptor (EGFR) and ephrin receptor A2 (EphA2; Lupberger et al., 2011) appears to lead to the formation of CD81–CLDN1 complexes required for HCV entry.
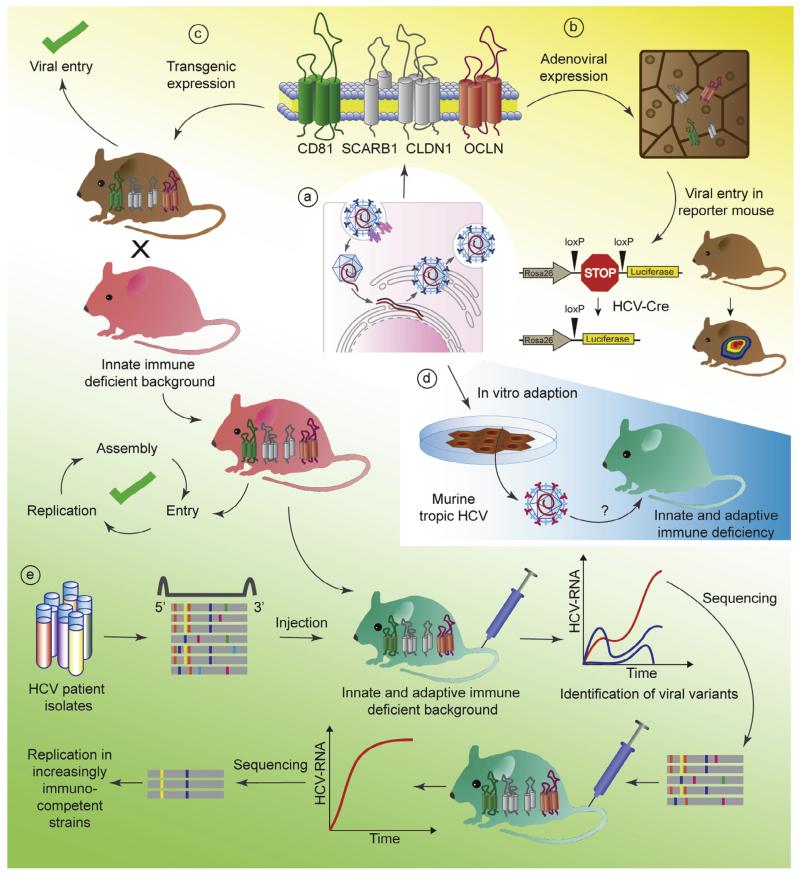
More recently the cholesterol uptake receptor Niemann Pick C1 like 1 (NPC1L1, Sainz et al., 2012) and transferrin receptor (TfR,Martin and Uprichard, 2013) have been implicated in HCV uptake, but their exact role has yet to be determined. Although there is sequence divergence between the murine and human orthologs of most of these molecules (reviewed in Sandmann and Ploss, 2013), only a few seem to account for the block at the level of entry. Expression of CD81, SCARB1, CLDN1 and OCLN are all required, but only CD81 and OCLN constitute the minimal set of human factors needed to facilitate viral uptake in mouse or hamster cells (Ploss et al., 2009) (Fig. 1a). These in vitro data also hold up in mice as adenoviral delivery (Dorner et al., 2011) or transgenic expression (Dorner et al., 2013) of human CD81 and OCLN in mice (Fig. 1b and c) is sufficient to allow for HCV entry into murine hepatocytes via an HCV glycoprotein mediated uptake process. Proof of concept was established that a mouse model for HCV uptake can be employed to assess the efficacy of entry inhibitors (Dorner et al., 2011; Giang et al., 2012; Anggakusuma et al., 2013) and also vaccine candidates (Dorner et al., 2011) which are based primarily on antibody-mediated protection.
Fig. 1.
Genetic approaches to overcome HCV species barriers. (a) Identification and expression of human specific factors and/or ablation of murine inhibitory pathways. (b) adenoviral expression of human CD81 (green) and OCLN (red) allows to visualize HCV entry using a sensitive Cre-activatable reporter. (c) Transgenic mice support HCV entry and when crossed to a mouse background blunted in innate anti-viral responses support the entire HCV life cycle. (d) Adaptation of HCV to infection of non-human cells in vitro. (e) Identification and characterization adaptive mutations within (patient-derived) viral populations capable of replicating mice increasingly or fully immunocompetent mouse strains.
Alternatively to the genetic host adaptation approach species barriers at the level of entry can also be overcome through viral adaptation. Using an in vitro selection approach, mutations within the HCV envelope proteins E1 and E2 were identified which allow HCV to enter cell lines expressing only mouse CD81, SCARB1, CLDN1 and OCLN (Bitzegeio et al., 2010) (Fig. 1d). Whether this murine tropic strain of HCV is indeed capable of entering mouse primary hepatocytes in vitro or in vivo has yet to be shown.
Previously, it was demonstrated that HCV replicons can be propagated in mouse cell lines albeit at low efficiency (Uprichard et al., 2006; Zhu et al., 2003). These data and subsequent work in human-mouse heterokaryons (Frentzen et al., 2011) suggested that dominant negative inhibitors putatively interfering with HCV RNA replication do not exist. Mutations were identified during replication of antibiotic-resistant HCV replicons in mouse cells, but those seemed to be random rather than adaptive (Uprichard et al., 2006), as none substantially enhanced replication efficiency in the murine cell environment when reintroduced into the parental subgenome (Zhu et al., 2003). This suggests that, although all essential cellular factors that support HCV replication are probably present in mouse cells, their mouse orthologs might not optimally interact with virally encoded components of HCV replication machinery. Consequently, expression of human replication co-factors may increase HCV RNA replication in mouse cells. In search for a mouse cellular environment that is more conducive to HCV’s replicative cycle, mouse embryonic fibroblasts (MEFs) with targeted deletions in genes critical for type I interferon (IFN) signaling were tested for their ability to replicate subgenomic and full-length HCV replicons (Nandakumar et al., 2013; Lin et al., 2010; Chang et al., 2006).
Cumulatively, these studies demonstrated that type I IFN dependent and independent pathways restrict HCV replication in mouse cells. Follow-up studies in cell lines derived from mice with defective innate immune signaling that were engineered to express human CD81 and OCLN and were supplemented with the liver specific microRNA 122 provided evidence that the entire HCV life-cycle can be recapitulated in a murine cellular environment (Vogt et al., 2013; Frentzen et al., 2014). MiR-122 forms a complex at the very 5′-end of the HCV genome, which affects HCV RNA replication and translation (reviewed in Conrad and Niepmann, 2013). The mature form of mouse miR-122 is identical to the human sequence, and is abundantly expressed in mouse and human hepatocytes in vivo (Chang et al., 2004). In contrast, many immortalized hepatocyte derived lines irrespective of species origin dedifferentiate during culture resulting in a loss of endogenous expression of miR-122 and conceivably other hepatocyte specific factors and thus have to be ectopically supplied (Frentzen et al., 2014 and reviewed in Sheahan et al., 2010). This work in engineered mouse cell lines extended previous studies that showed mouse cells can produce infectious HCV particles (Long et al., 2011).
These lines of investigations culminated in the construction of a genetically humanized mouse model. Mice transgenically expressing human CD81 and OCLN are capable of sustaining all steps of the viral life-cycle when crossed to e.g. STAT1 or IFNαβ receptor deficient backgrounds, which are profoundly impaired in type I and III IFN signaling (Dorner et al., 2013) (Fig. 1c). It is unclear why HCV’s sophisticated arsenal of anti-viral response evasion mechanisms does not seem to suffice to establish chronicity in human entry factor transgenic mice with intact anti-viral defenses. Conceivably, more rapid induction, greater magnitude and/or simply a different quality of anti-viral immunity in mice as compared to man may control more efficient HCV infection.
This inbred model already shows utility for dissecting genetically HCV infection (Dorner et al., 2013) offering the additional advantage of studying the virus in the 3D context of the liver, which e.g. takes better into account hepatocyte heterogeneity influenced by liver zonation or circadian rhythms. Hepatocytes along the porto-central axis of the liver are heterogenous dictated by environmental factors such as oxygen and nutrients levels, which form zonal gradients around the arteries and veins. These translate at the transcriptional level to specific pericentral versus periportal transcriptomic profiles (reviewed in Torre et al, 2010). Liver-specific transcription is further governed by organ-specific circadian rhythms (reviewed in Tong and Yin, 2013). The influence of these spatio-temporal expression patterns on physiological processes such as liver metabolism is well appreciated but the impact on hepatic inflammation, specifically HCV infection is not understood. Furthermore, additional precedence was established for assessing preclinically the efficacy of anti-HCV drug candidates in genetically humanized mice (Dorner et al., 2013).
To study unperturbed host responses to HCV it will be necessary to establish persistent HCV infection in fully immunocompetent mice. Efforts are ongoing to select for viral variants that replicate more robustly in sufficiently immunocompromised rodent strains harnessing the remarkable genetic plasticity of HCV (Fig. 1e). Passage of high titer sera through progressively more immunocompetent hosts may produce this outcome. Different, genetically diverse HCV isolates and genotypes may be distinct in their ability to establish chronicity in genetically humanized mice. A fully immunocompetent, inbred mouse model would enable testing and prioritizing vaccine candidates and may present with liver pathologies reminiscent of the clinical phenotype in patients which could be exacerbated on different genetic backgrounds.
5. Humanized mouse avatars to dissect patient-specific (immune) responses to HCV infection
Inbred mice are the most widely used species in infectious disease research and reflect many aspects of human biology remarkably well, which is partly rooted in the considerable genetic similarity between the species. However, humans and mice, whose lineages have diverged approximately 70 million years ago, differ tremendously in size and life-span and have evolved in distinct ecological niches which translates into differences with respect to immune activation and response to challenge (Mestas and Hughes, 2004). Thus, although genetically humanized mice offer tremendous opportunities for studying HCV infection in vivo, caution is warranted when extrapolating data to humans. Consequently, and given the fact, that the opportunity of conducting clinical studies in patients is limited, alternative animal models need to be considered optimally allowing the study of human host (immune) responses to HCV replicating in human hepatocytes.
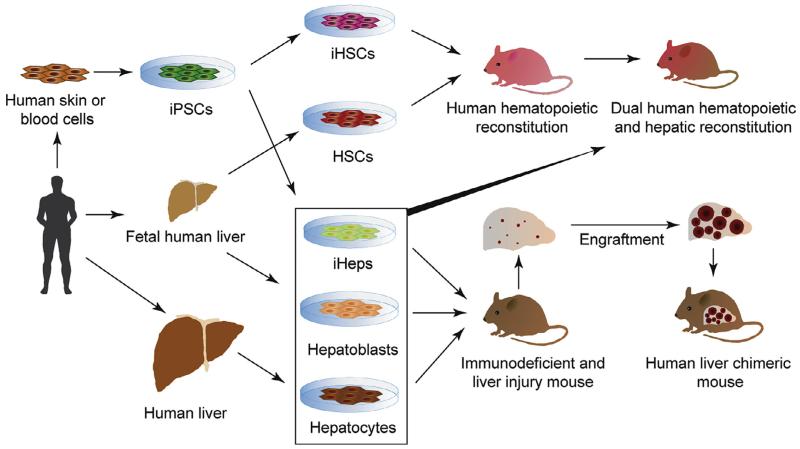
Engraftment of human hepatocytes into suitable xenorecipients has proven to be one solution to study HCV in its native environment (reviewed in Billerbeck et al., 2013, Meuleman and Leroux-Roels, 2008, de Jong et al., 2010). To facilitate engraftment, human hepatocytes are usually injected into immunodeficient recipients suffering from liver injury (Fig. 2). Suppression of the murine immune system is necessary to prevent graft rejection. The liver injury provides the expansion stimulus to the usually quiescent hepatocytes and gives the transplanted human liver cells a competitive growth advantage over mouse hepatocytes. Liver injury can be inflicted surgically, e.g. by partial hepatectomy, by treatment with hepatotoxins, such as retrorsine or carbon tetrachloride, or with genetic approaches. The latter are most widely used for stable engraftment as they provide a more selective control over the severity of the liver injury, and hepatotoxicity is usually limited to mouse hepatocytes.
Fig. 2.
Current and future approaches for the generation of humanized xenotransplantation models engrafted with patient-specific cells. Depicted are sources of primary human hepatocytes and hematopoetic stem cells isolated directly from fetal or adult organs or generated via directed differentiation of human pluripotent stem cells. Human cells are used to humanized liver and immune system in the same xenorecipient. iPSC = induced pluripotent stem cell, HSC = hematopoietic stem cell, iHSC = HSC generated through directed differentiation from iPSCs, iHeps = hepatocyte-like cells derived from iPSCs.
Susceptibility to HCV upon engraftment of human hepatocytes was shown in a number of immunodeficient liver injury models, including Alb-uPA (Meuleman et al., 2005; Mercer et al., 2001), FAH−/− (Bissig et al., 2010), AFC8 (Washburn et al., 2011) and more recently MUP-uPA (Tesfaye et al., 2013) and HSV-TK (Kosaka et al., 2013) mice. Human liver chimeric mice have proven their utility for analyzing HCV infection and testing the efficacy of various treatment modalities (reviewed in Meuleman and Leroux-Roels, 2008). The human hepatic graft offers the additional benefit of obtaining simultaneously human-like metabolic and toxicologic profiles.
Robust human hepatic chimerism can routinely be achieved with adult hepatocytes, which limits the analysis of host responses to few, often randomly selected donor lots. To enable the systematic investigation of human host genetics on HCV infection specifically or human liver disease in general it would be desirable to utilize patient specific hepatocytes. Induced pluripotent stem cells (iPSCs) can be routinely generated from any patient starting off with easily accessible cells such as lymphocytes or skin fibroblasts in a process of cellular reprogramming (Takahashi et al., 2007; Takahashi and Yamanaka, 2006). iPSCs are also amenable to genome engineering thereby allowing the creation of basically any desired genotype and control cells on an isogenic background (Hockemeyer et al., 2011). iPSCs can be differentiated efficiently towards the hepatocyte lineage (Touboul et al., 2010; Si-Tayeb et al., 2010), yielding hepatocyte-like cells (iHeps), that not only express hepatocyte-specific markers but also have hepatocyte function and are susceptible to human hepatotropic pathogens (Schwartz et al., 2012; Wu et al., 2012; Roelandt et al., 2012) (Fig. 2). However, iHeps, similar to fetal hepatoblasts (Haridass et al., 2009), do not to seem to respond to the proliferative signals in the injured liver in contrast to adult hepatocytes. Efforts are ongoing to systematically define the parameters influencing the engraftment of human iHeps with the goal of generating iHep-derived liver chimeric mice. Improvements in differentiation protocols leading to a more faithful hepatocyte phenotype (Shan et al., 2013) may need to be coupled with alternative engraftment strategies including the use of engineered tissue organoids, i.e. ex vivo produced structures mimicking liver function and architecture (Stevens et al., 2013; Chen et al., in press) implanted into improved xenorecipient strains.
A considerable shortcoming of currently available human liver chimeric mouse models is their lack of cellular and humoral immune response due to their highly immunocompromized status which is necessary to prevent graft rejection. To study HCV-specific immune responses, which counteract the infection but are also thought to contribute to the progression of liver pathogenesis, attempts have been made to co-engraft mice with both human hepatocytes and components of a human immune system in a single recipient (Fig. 2). Proof of concept for this approach was established in a study demonstrating that co-injection with a mixture of human fetal hepatoblasts, non-parenchymal cells and hematopoietic stem cells yielded dually engrafted animals. Although the hepatic chimerism was very low, mice were susceptible to HCV infection, mounted antigen-specific immune responses and developed signs of liver fibrosis (Washburn et al., 2011) additional refinements will be necessary: To increase the utility of the system, first, protocols need to be devised that reliably produce a more robust, i.e. high level dual chimerism. Furthermore, a considerable logistical challenge for such dually engrafted animals will be sourcing of donor-matched hepatocytes, non-parenchymal cells and HSCs. These populations can be isolated from fetal livers but given the usually small organ size only small cohorts of humanized cells could be generated for a single donor.
Here, progress in the generation of engraftable iHSCs from iPSCs (reviewed in Chou et al., 2013) in combination with iHeps derived from the same iPSC donor may solve this issue. Additionally, since human immune responses are generally weak in HSC-transplanted mice, further modifications will be needed to improve both the cellular complexity and functionality of the engrafted human immune system. Those include but are not limited to the expression of human MHC in the absence of mouse MHC to ensure faithful presentation of self- and virally derived peptides to human T cells and to reduced graft-versus-host-disease; co-transplantation of HSC donor-matched human thymic cortical epithelium to facilitate proper T cell selection; the expression of human orthologs of non-redundant cytokines which exhibit limited biological cross-reactivity to foster development of underrepresented human immune – in particular erythro-myeloid – cell lineages; the improvement the organization of lymphoid architecture, especially in spleen and lymphnodes, to allow for adequate T and B cell priming; genetically replacing non-compatible immune cell receptors and chemokines expressed on non-hematopoietically derived cells to improve e.g. immune cell trafficking; the introduction of a human microbiome to account for effects of species-specific commensals on the immune system (reviewed in Rongvaux et al., 2013).
6. A multipronged approach towards the most suitable animal model for hepatitis C
To gain a mechanistic understanding of HCV biology and host-responses in the physiological context of the liver and to address the medical need for more effective interventions, in particular for vaccine development, animal models are needed. The issue is particularly pressing in light of the now highly restricted access to the chimpanzee model, which – is the only non-human species readily permissive to HCV infection.
To create more tractable animal models a number of distinct but likely complementary approaches are being pursued. The identification of new viruses, which are genetically related to HCV in a variety of species may shed light on the evolutionary origins of HCV but may also open opportunities to establish surrogate models. Over the last few years we have gained a better understanding about the determinants restricting HCV’s host tropism which has allowed to overcome species barriers through genetic host humanization and possibly also through viral adaptation. In current models the entire HCV life-cycle can be recapitulated opening unprecedented opportunities to dissect an HCV infection in vivo. However, in order to increase the utility of the system, particularly to study HCV immunity and pathogenesis it will be necessary to establish a more robust HCV RNA replication, optimally in fully immunocompetent mice.
In parallel, humanized xenotransplantation models are continuously being refined. Human liver chimeric mice engrafted with human adult hepatocytes have a proven track record for studying HCV infection. With continuous advances in stem cell biology and improvements in directed differentiation protocols it may become possible to create humanized mouse avatars engrafted with patient specific cells. Such models would have great utility to dissect the hepatocyte-intrinsic host genetic factors influencing HCV infection. Infection with HCV can cause clinical phenotypes ranging from apparent resistance to fulminant infection. Undoubtedly, host genetics have a dramatic impact on the outcome of infection. For example, genome-wide association studies have revealed a strong association between single nucleotide polymorphisms (SNP) located within the interleukin 28B (IL28B) locus and clearance of HCV infection and response to interferon-based therapy (Ge et al., 2009; Suppiah et al., 2009; Tanaka et al., 2009; Thomas et al., 2009). Subsequently, it was demonstrated that some of these variants upstream of IL28B create a new interferon gene IFNL4, which is associated with impaired HCV clearance (Prokunina-Olsson et al., 2013). However, the functional consequences of the presence of less favorable alleles have been impossible to decipher due to the lack of adequate model systems. Likewise, some individuals exhibit hyper-, hypo- or null alleles of genes implicated in the HCV life cycle (van Zelm et al., 2010; Hadj-Rabia et al., 2004; Feldmeyer et al., 2006; Vergeer et al., 2011; Innerarity et al., 1987; Mahley, 1988; Zhang et al., 2008).
Infection of humanized mice engrafted with stem cell derived hepatocytes, into which any specific gene alteration could be introduced, would allow to characterize functionally the impact of these variants on an isogenic background. Beyond hepatitis C humanized avatar mice could also be employed to dissect genetic variations affecting for example the life-cycles of other hepatotropic pathogens, non-virally associated liver inflammation (e.g. non-alcoholic fatty liver disease (Romeo et al., 2008), metabolic disorders (e.g. a1-antitrypsin deficiency, familial hypercholesterolemia, or glycogen storage diseases)) or hepatocellular carcinoma (Zhang, 2012).
Co-engraftment of a donor-matched human liver and a hematopoietic system in a single xenorecipient will provide means to study human immune response, possibly enabling analysis of HCV associated pathogenesis and to study clinically relevant co-infections with hepatitis B virus and/or human immunodeficiency virus.
This multipronged approach will yield a plethora of complementary models each with their own unique strengths and weaknesses and utility for specific applications when studying HCV vivo. To ensure relevance, the development efforts need to ensure that new and refined models accurately reflect important hallmarks of HCV infection observed in patients.
Acknowledgements
The authors thank Brigitte Heller and Benjamin Winer for edits and critical discussion of the manuscript. Work in the laboratory is in part supported by grants from the National Institutes of Health (2 R01 AI079031-05A1, 1 R01 AI107301-01, 1 R56 AI106005-01), the Walter Reed Army Institute of Research and Bill and Melinda Gates Foundation. M.v.S. is a recipient of a fellowship from the German Research Foundation (Deutsche Forschungsgemeinschaft).
References
- National institutes of health consensus development conference statement: management of hepatitis C 2002 (June 10–12, 2002), 02, National Institutes of Health Consensus Development Conference Statement. Gastroenterology. 123:2082–2099. doi: 10.1053/gast.2002.1232082. [DOI] [PubMed] [Google Scholar]
- Agnello V, Abel G, Elfahal M, Knight GB, Zhang Q-X. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. U.S.A. 1999;96:12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anggakusuma, Colpitts CC, Schang LM, Rachmawati H, Frentzen A, Pfaender S, Behrendt P, Brown RJ, Bankwitz D, Steinmann J, Ott M, Meuleman P, Rice CM, Ploss A, Pietschmann T, Steinmann E. Turmeric curcumin inhibits entry of all hepatitis C virus genotypes into human liver cells. Gut. 2013 doi: 10.1136/gutjnl-2012-304299. [DOI] [PubMed] [Google Scholar]
- Barth H, Schafer C, Adah MI, Zhang F, Linhardt RJ, Toyoda H, Kinoshita-Toyoda A, Toida T, Van Kuppevelt TH, Depla E, Von Weizsacker F, Blum HE, Baumert TF. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 2003;278:41003–41012. doi: 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- Billerbeck E, de Jong Y, Dorner M, de la Fuente C, Ploss A. Animal models for hepatitis C. Curr. Top. Microbiol. Immunol. 2013;369:49–86. doi: 10.1007/978-3-642-27340-7_3. [DOI] [PubMed] [Google Scholar]
- Bissig KD, Wieland SF, Tran P, Isogawa M, Le TT, Chisari FV, Verma IM. Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. J. Clin. Invest. 2010;120:924–930. doi: 10.1172/JCI40094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitzegeio J, Bankwitz D, Hueging K, Haid S, Brohm C, Zeisel MB, Herrmann E, Iken M, Ott M, Baumert TF, Pietschmann T. Adaptation of hepatitis C virus to mouse CD81 permits infection of mouse cells in the absence of human entry factors. PLoS Pathog. 2010;6:e1000978. doi: 10.1371/journal.ppat.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukh J, Apgar CL, Govindarajan S, Purcell RH. Host range studies of GB virus-B hepatitis agent, the closest relative of hepatitis C virus, in New World monkeys and chimpanzees. J. Med. Virol. 2001;65:694–697. doi: 10.1002/jmv.2092. [DOI] [PubMed] [Google Scholar]
- Burbelo PD, Dubovi EJ, Simmonds P, Medina JL, Henriquez JA, Mishra N, Wagner J, Tokarz R, Cullen JM, Iadarola MJ, Rice CM, Lipkin WI, Kapoor A. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J. Virol. 2012;86:6171–6178. doi: 10.1128/JVI.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, Zaret KS, Taylor JM. MiR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- Chang KS, Cai Z, Zhang C, Sen GC, Williams BR, Luo G. Replication of hepatitis C virus (HCV) RNA in mouse embryonic fibroblasts: protein kinase R (PKR)-dependent and PKR-independent mechanisms for controlling HCV RNA replication and mediating interferon activities. J. Virol. 2006;80:7364–7374. doi: 10.1128/JVI.00586-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AA, Thomas DK, Ong LL, Schwartz RE, Golub TR, Bhatia SN. Humanized mice with ectopic artificial liver tissues. Proc. Natl. Acad. Sci. U. S. A. 108:11842–118427. doi: 10.1073/pnas.1101791108. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou BK, Ye Z, Cheng L. Generation and homing of iPSC-derived hematopoietic cells in vivo. Mol. Ther. 2013;21:1292–1293. doi: 10.1038/mt.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad KD, Niepmann M. The role of microRNAs in hepatitis C virus RNA replication. Arch. Virol. 2013 doi: 10.1007/s00705-013-1883-4. [DOI] [PubMed] [Google Scholar]
- de Jong YP, Rice CM, Ploss A. New horizons for studying human hepatotropic infections. J. Clin. Invest. 2010;120:650–653. doi: 10.1172/JCI42338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner M, Horwitz JA, Robbins JB, Barry WT, Feng Q, Mu K, Jones CT, Schoggins JW, Catanese MT, Burton DR, Law M, Rice CM, Ploss A. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–211. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner M, Horwitz JA, Donovan BM, Labitt RN, Budell WC, Friling T, Vogt A, Catanese MT, Satoh T, Kawai T, Akira S, Law M, Rice CM, Ploss A. Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature. 2013;501:237–241. doi: 10.1038/nature12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlin BR. Perspective: test and treat this silent killer. Nature. 2011;474:S18–S19. doi: 10.1038/474S18a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- Feinstone SM, Hu DJ, Major ME. Prospects for prophylactic and therapeutic vaccines against hepatitis C virus. Clin. Infect. Dis. 2012;55(Suppl 1):S25–S32. doi: 10.1093/cid/cis362. [DOI] [PubMed] [Google Scholar]
- Feldmeyer L, Huber M, Fellmann F, Beckmann JS, Frenk E, Hohl D. Confirmation of the origin of NISCH syndrome. Hum. Mutat. 2006;27:408–410. doi: 10.1002/humu.20333. [DOI] [PubMed] [Google Scholar]
- Frentzen A, Huging K, Bitzegeio J, Friesland M, Haid S, Gentzsch J, Hoffmann M, Lindemann D, Zimmer G, Zielecki F, Weber F, Steinmann E, Pietschmann T. Completion of hepatitis C virus replication cycle in heterokaryons excludes dominant restrictions in human non-liver and mouse liver cell lines. PLoS Pathog. 2011;7:e1002029. doi: 10.1371/journal.ppat.1002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentzen A, Anggakusuma, Gurlevik E, Hueging K, Knocke S, Ginkel C, Brown RJ, Heim M, Dill MT, Kroger A, Kalinke U, Kaderali L, Kuehnel F, Pietschmann T. Cell entry, efficient RNA replication, and production of infectious hepatitis C virus progeny in mouse liver-derived cells. Hepatology. 2014;59:78–88. doi: 10.1002/hep.26626. [DOI] [PubMed] [Google Scholar]
- Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- Giang E, Dorner M, Prentoe JC, Dreux M, Evans MJ, Bukh J, Rice CM, Ploss A, Burton DR, Law M. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc. Natl. Acad. Sci. U.S.A. 2012;109:6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadj-Rabia S, Baala L, Vabres P, Hamel-Teillac D, Jacquemin E, Fabre M, Lyonnet S, De Prost Y, Munnich A, Hadchouel M, Smahi A. Claudin-1 gene mutations in neonatal sclerosing cholangitis associated with ichthyosis: a tight junction disease. Gastroenterology. 2004;127:1386–1390. doi: 10.1053/j.gastro.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Haridass D, Yuan Q, Becker PD, Cantz T, Iken M, Rothe M, Narain N, Bock M, Norder M, Legrand N, Wedemeyer H, Weijer K, Spits H, Manns MP, Cai J, Deng H, Di Santo JP, Guzman CA, Ott M. Repopulation efficiencies of adult hepatocytes, fetal liver progenitor cells, and embryonic stem cell-derived hepatic cells in albumin-promoter-enhancer urokinase-type plasminogen activator mice. Am. J. Pathol. 2009;175:1483–1492. doi: 10.2353/ajpath.2009.090117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innerarity TL, Weisgraber KH, Arnold KS, Mahley RW, Krauss RM, Vega GL, Grundy SM. Familial defective apolipoprotein B-100: low density lipoproteins with abnormal receptor binding. Proc. Natl. Acad. Sci. U.S.A. 1987;84:6919–6923. doi: 10.1073/pnas.84.19.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesudian AB, de Jong YP, Jacobson IM. Emerging therapeutic targets for hepatitis C virus infection. Clin. Gastroenterol. Hepatol. 2013;11(612-619):e611. doi: 10.1016/j.cgh.2013.04.003. [DOI] [PubMed] [Google Scholar]
- Kapoor A, Simmonds P, Gerold G, Qaisar N, Jain K, Henriquez JA, Firth C, Hirschberg DL, Rice CM, Shields S, Lipkin WI. Characterization of a canine homolog of hepatitis C virus. Proc. Natl. Acad. Sci. U.S.A. 2011;108:11608–11613. doi: 10.1073/pnas.1101794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Simmonds P, Scheel TK, Hjelle B, Cullen JM, Burbelo PD, Chauhan LV, Duraisamy R, Sanchez Leon M, Jain K, Vandegrift KJ, Calisher CH, Rice CM, Lipkin WI. Identification of rodent homologs of hepatitis C virus and pegiviruses. mBio. 2013;4(e00216-e00213) doi: 10.1128/mBio.00216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiannis P, Petrovic LM, Fry M, Moore D, Enticott M, McGarvey MJ, Scheuer PJ, Thomas HC. Studies of GB hepatitis agent in tamarins. Hepatology. 1989;9:186–192. doi: 10.1002/hep.1840090204. [DOI] [PubMed] [Google Scholar]
- Kosaka K, Hiraga N, Imamura M, Yoshimi S, Murakami E, Nakahara T, Honda Y, Ono A, Kawaoka T, Tsuge M, Abe H, Hayes CN, Miki D, Aikata H, Ochi H, Ishida Y, Tateno C, Yoshizato K, Sasaki T, Chayama K. A novel TK-NOG based humanized mouse model for the study of HBV and HCV infections. Biochem. Biophys. Res. Commun. 2013;441:230–235. doi: 10.1016/j.bbrc.2013.10.040. [DOI] [PubMed] [Google Scholar]
- Koutsoudakis G, Kaul A, Steinmann E, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 2006;80:5308–5320. doi: 10.1128/JVI.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarre D, Anderson PC, Bailey M, Beaulieu P, Bolger G, Bonneau P, Bos M, Cameron DR, Cartier M, Cordingley MG, Faucher AM, Goudreau N, Kawai SH, Kukolj G, Lagace L, LaPlante SR, Narjes H, Poupart MA, Rancourt J, Sentjens RE, St George R, Simoneau B, Steinmann G, Thibeault D, Tsantrizos YS, Weldon SM, Yong CL, Llinas-Brunet M. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature. 2003;426:186–189. doi: 10.1038/nature02099. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Chavez D, Notvall L, Brasky KM. Comparison of tamarins and marmosets as hosts for GBV-B infections and the effect of immunosuppression on duration of viremia. Virology. 2003;311:72–80. doi: 10.1016/s0042-6822(03)00193-4. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LT, Noyce RS, Pham TN, Wilson JA, Sisson GR, Michalak TI, Mossman KL, Richardson CD. Replication of subgenomic hepatitis C virus replicons in mouse fibroblasts is facilitated by deletion of interferon regulatory factor 3 and expression of liver-specific microRNA 122. J. Virol. 2010;84:9170–9180. doi: 10.1128/JVI.00559-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Yang W, Shen L, Turner JR, Coyne CB, Wang T. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J. Virol. 2009;83:2011–2014. doi: 10.1128/JVI.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long G, Hiet MS, Windisch MP, Lee JY, Lohmann V, Bartenschlager R. Mouse hepatic cells support assembly of infectious hepatitis C virus particles. Gastroenterology. 2011;141:1057–1066. doi: 10.1053/j.gastro.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S, Royer C, Fischer B, Zahid MN, Lavillette D, Fresquet J, Cosset FL, Rothenberg SM, Pietschmann T, Patel AH, Pessaux P, Doffoel M, Raffelsberger W, Poch O, McKeating JA, Brino L, Baumert TF. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat. Med. 2011;17:589–595. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- Martin DN, Uprichard SL. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc. Natl. Acad. Sci. U.S.A. 2013;110:10777–10782. doi: 10.1073/pnas.1301764110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, Addison WR, Fischer KP, Churchill TA, Lakey JR, Tyrrell DL, Kneteman NM. Hepatitis C virus replication in mice with chimeric human livers. Nat. Med. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J. Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- Meuleman P, Leroux-Roels G. The human liver-uPA-SCID mouse: a model for the evaluation of antiviral compounds against HBV and HCV. Antiviral Res. 2008;80:231–238. doi: 10.1016/j.antiviral.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Meuleman P, Libbrecht L, De Vos R, de Hemptinne B, Gevaert K, Vandekerckhove J, Roskams T, Leroux-Roels G. Morphological and biochemical characterization of a human liver in a uPA-SCID mouse chimera. Hepatology. 2005;41:847–856. doi: 10.1002/hep.20657. [DOI] [PubMed] [Google Scholar]
- Molina S, Castet V, Fournier-Wirth C, Pichard-Garcia L, Avner R, Harats D, Roitelman J, Barbaras R, Graber P, Ghersa P, Smolarsky M, Funaro A, Malavasi F, Larrey D, Coste J, Fabre JM, Sa-Cunha A, Maurel P. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J. Hepatol. 2007;46:411–419. doi: 10.1016/j.jhep.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Monazahian M, Bohme I, Bonk S, Koch A, Scholz C, Grethe S, Thomssen R. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. J. Med. Virol. 1999;57:223–229. doi: 10.1002/(sici)1096-9071(199903)57:3<223::aid-jmv2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Nandakumar R, Finsterbusch K, Lipps C, Neumann B, Grashoff M, Nair S, Hochnadel I, Lienenklaus S, Wappler I, Steinmann E, Hauser H, Pietschmann T, Kroger A. Hepatitis C virus replication in mouse cells is restricted by IFN-dependent and -independent mechanisms. Gastroenterology. 2013;145(1414-1423):e1411. doi: 10.1053/j.gastro.2013.08.037. [DOI] [PubMed] [Google Scholar]
- Olsen DB, Davies ME, Handt L, Koeplinger K, Zhang NR, Ludmerer SW, Graham D, Liverton N, MacCoss M, Hazuda D, Carroll SS. Sustained viral response in a hepatitis C virus-infected chimpanzee via a combination of direct-acting antiviral agents. Antimicrob. Agents Chemother. 2011;55:937–939. doi: 10.1128/AAC.00990-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DM, Huang H, Ye J, Gale M., Jr. Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology. 2009;394:99–108. doi: 10.1016/j.virol.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, Porter-Gill P, Mumy A, Kohaar I, Chen S, Brand N, Tarway M, Liu L, Sheikh F, Astemborski J, Bonkovsky HL, Edlin BR, Howell CD, Morgan TR, Thomas DL, Rehermann B, Donnelly RP, O’Brien TR. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelandt P, Obeid S, Paeshuyse J, Vanhove J, Van Lommel A, Nahmias Y, Nevens F, Neyts J, Verfaillie CM. Human pluripotent stem cell-derived hepatocytes support complete replication of hepatitis C virus. J. Hepatol. 2012;57:246–251. doi: 10.1016/j.jhep.2012.03.030. [DOI] [PubMed] [Google Scholar]
- Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux A, Takizawa H, Strowig T, Willinger T, Eynon EE, Flavell RA, Manz MG. Human hemato-lymphoid system mice: current use and future potential for medicine. Annu. Rev. Immunol. 2013;31:635–674. doi: 10.1146/annurev-immunol-032712-095921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz B, Jr., Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA, Uprichard SL. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat. Med. 2012;18:281–285. doi: 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann L, Ploss A. Barriers of hepatitis C virus interspecies transmission. Virology. 2013;435:70–80. doi: 10.1016/j.virol.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaluder GG, Dawson GJ, Simons JN, Pilot-Matias TJ, Gutierrez RA, Heynen CA, Knigge MF, Kurpiewski GS, Buijk SL, Leary TP, et al. Molecular and serologic analysis in the transmission of the GB hepatitis agents. J. Med. Virol. 1995;46:81–90. doi: 10.1002/jmv.1890460117. [DOI] [PubMed] [Google Scholar]
- Schwartz RE, Trehan K, Andrus L, Sheahan TP, Ploss A, Duncan SA, Rice CM, Bhatia SN. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proc. Natl. Acad. Sci. U.S.A. 2012;109:2544–2548. doi: 10.1073/pnas.1121400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J, Schwartz RE, Ross NT, Logan DJ, Thomas D, Duncan SA, North TE, Goessling W, Carpenter AE, Bhatia SN. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat. Chem. Biol. 2013;9:514–520. doi: 10.1038/nchembio.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T, Jones CT, Ploss A. Advances and challenges in studying hepatitis C virus in its native environment. Expert Rev. Gastroenterol. Hepatol. 2010;4:541–550. doi: 10.1586/egh.10.53. [DOI] [PubMed] [Google Scholar]
- Simons JN, Leary TP, Dawson GJ, Pilot-Matias TJ, Muerhoff AS, Schlauder GG, Desai SM, Mushahwar IK. Isolation of novel virus-like sequences associated with human hepatitis. Nat. Med. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, North PE, Dalton S, Duncan SA. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourisseau M, Goldman O, He W, Gori JL, Kiem HP, Gouon-Evans V, Evans MJ. Hepatic cells derived from induced pluripotent stem cells of pigtail macaques support hepatitis C virus infection. Gastroenterology. 2013;145(966-969):e967. doi: 10.1053/j.gastro.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens KR, Ungrin MD, Schwartz RE, Ng S, Carvalho B, Christine KS, Chaturvedi RR, Li CY, Zandstra PW, Chen CS, Bhatia SN. InVERT molding for scalable control of tissue microarchitecture. Nat. Commun. 2013;4:1847. doi: 10.1038/ncomms2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E, Riordan S, Sheridan D, Smedile A, Fragomeli V, Muller T, Bahlo M, Stewart GJ, Booth DR, George J. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat. Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, Ito Y, Mita E, Tanaka E, Mochida S, Murawaki Y, Honda M, Sakai A, Hiasa Y, Nishiguchi S, Koike A, Sakaida I, Imamura M, Ito K, Yano K, Masaki N, Sugauchi F, Izumi N, Tokunaga K, Mizokami M. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat. Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- Tesfaye A, Stift J, Maric D, Cui Q, Dienes HP, Feinstone SM. Chimeric mouse model for the infection of hepatitis B and C viruses. PLoS One. 2013;8:e77298. doi: 10.1371/journal.pone.0077298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, Goedert JJ, Kirk GD, Donfield SM, Rosen HR, Tobler LH, Busch MP, McHutchison JG, Goldstein DB, Carrington M. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Yin L. Circadian rhythms in liver physiology and liver diseases. Compr. Physiol. 2013;3:917–940. doi: 10.1002/cphy.c120017. [DOI] [PubMed] [Google Scholar]
- Torre C, Perret C, Colnot S. Molecular determinants of liver zonation. Prog. Mol. Biol. Transl. Sci. 2010;97:127–150. doi: 10.1016/B978-0-12-385233-5.00005-2. [DOI] [PubMed] [Google Scholar]
- Touboul T, Hannan NR, Corbineau S, Martinez A, Martinet C, Branchereau S, Mainot S, Strick-Marchand H, Pedersen R, Di Santo J, Weber A, Vallier L. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology. 2010;51:1754–1765. doi: 10.1002/hep.23506. [DOI] [PubMed] [Google Scholar]
- Uprichard SL, Chung J, Chisari FV, Wakita T. Replication of a hepatitis C virus replicon clone in mouse cells. Virol. J. 2006;3:89. doi: 10.1186/1743-422X-3-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zelm MC, Smet J, Adams B, Mascart F, Schandene L, Janssen F, Ferster A, Kuo CC, Levy S, van Dongen JJ, van der Burg M. CD81 gene defect in humans disrupts CD19 complex formation and leads to antibody deficiency. J. Clin. Invest. 2010;120:1265–1274. doi: 10.1172/JCI39748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanwolleghem T, Meuleman P, Libbrecht L, Roskams T, De Vos R, Leroux-Roels G. Ultra-rapid cardiotoxicity of the hepatitis C virus protease inhibitor BILN 2061 in the urokinase-type plasminogen activator mouse. Gastroenterology. 2007;133:1144–1155. doi: 10.1053/j.gastro.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Vergeer M, Korporaal SJ, Franssen R, Meurs I, Out R, Hovingh GK, Hoekstra M, Sierts JA, Dallinga-Thie GM, Motazacker MM, Holleboom AG, Van Berkel TJ, Kastelein JJ, Van Eck M, Kuivenhoven JA. Genetic variant of the scavenger receptor BI in humans. N. Engl. J. Med. 2011;364:136–145. doi: 10.1056/NEJMoa0907687. [DOI] [PubMed] [Google Scholar]
- Vogt A, Scull MA, Friling T, Horwitz JA, Donovan BM, Dorner M, Gerold G, Labitt RN, Rice CM, Ploss A. Recapitulation of the hepatitis C virus life-cycle in engineered murine cell lines. Virology. 2013;444:1–11. doi: 10.1016/j.virol.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn ML, Bility MT, Zhang L, Kovalev GI, Buntzman A, Frelinger JA, Barry W, Ploss A, Rice CM, Su L. A humanized mouse model to study hepatitis C virus infection, immune response, and liver disease. Gastroenterology. 2011;140:1334–1344. doi: 10.1053/j.gastro.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Robotham JM, Lee E, Dalton S, Kneteman NM, Gilbert DM, Tang H. Productive hepatitis C virus infection of stem cell-derived hepatocytes reveals a critical transition to viral permissiveness during differentiation. PLoS pathog. 2012;8:e1002617. doi: 10.1371/journal.ppat.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel MB, Felmlee DJ, Baumert TF. Hepatitis C virus entry. Curr. Top. Microbiol. Immunol. 2013;369:87–112. doi: 10.1007/978-3-642-27340-7_4. [DOI] [PubMed] [Google Scholar]
- Zhang Z. Genomic landscape of liver cancer. Nat.Genet. 2012;44:1075–1077. doi: 10.1038/ng.2412. [DOI] [PubMed] [Google Scholar]
- Zhang SY, Boisson-Dupuis S, Chapgier A, Yang K, Bustamante J, Puel A, Picard C, Abel L, Jouanguy E, Casanova JL. Inborn errors of interferon (IFN)-mediated immunity in humans: insights into the respective roles of IFN-alpha/beta, IFN-gamma, and IFN-lambda in host defense. Immunol. Rev. 2008;226:29–40. doi: 10.1111/j.1600-065X.2008.00698.x. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Guo JT, Seeger C. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J. Virol. 2003;77:9204–9210. doi: 10.1128/JVI.77.17.9204-9210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]