ABSTRACT
Viruses have a profound influence on the ecology and evolution of plankton, but our understanding of the composition of the aquatic viral communities is still rudimentary. This is especially true of those viruses having RNA genomes. The limited data that have been published suggest that the RNA virioplankton is dominated by viruses with positive-sense, single-stranded (+ss) genomes that have features in common with those of eukaryote-infecting viruses in the order Picornavirales (picornavirads). In this study, we investigated the diversity of the RNA virus assemblages in tropical coastal seawater samples using targeted PCR and metagenomics. Amplification of RNA-dependent RNA polymerase (RdRp) genes from fractions of a buoyant density gradient suggested that the distribution of two major subclades of the marine picornavirads was largely congruent with the distribution of total virus-like RNA, a finding consistent with their proposed dominance. Analyses of the RdRp sequences in the library revealed the presence of many diverse phylotypes, most of which were related only distantly to those of cultivated viruses. Phylogenetic analysis suggests that there were hundreds of unique picornavirad-like phylotypes in one 35-liter sample that differed from one another by at least as much as the differences among currently recognized species. Assembly of the sequences in the metagenome resulted in the reconstruction of six essentially complete viral genomes that had features similar to viruses in the families Bacillarna-, Dicistro-, and Marnaviridae. Comparison of the tropical seawater metagenomes with those from other habitats suggests that +ssRNA viruses are generally the most common types of RNA viruses in aquatic environments, but biases in library preparation remain a possible explanation for this observation.
IMPORTANCE
Marine plankton account for much of the photosynthesis and respiration on our planet, and they influence the cycling of carbon and the distribution of nutrients on a global scale. Despite the fundamental importance of viruses to plankton ecology and evolution, most of the viruses in the sea, and the identities of their hosts, are unknown. This report is one of very few that delves into the genetic diversity within RNA-containing viruses in the ocean. The data expand the known range of viral diversity and shed new light on the physical properties and genetic composition of RNA viruses in the ocean.
INTRODUCTION
Viruses are integral to life in the ocean, contributing to the disease and mortality of their hosts, catalyzing evolution by mediating gene exchange, and influencing the partitioning of nutrients among trophic levels (1). The genetic material of viruses may consist of single-stranded DNA (ssDNA) or double-stranded DNA (dsDNA) or ssRNA or dsRNA, depending on the virus. Most studies of marine virioplankton over the past two decades have focused on DNA-containing viruses, which appear to be predominantly bacteriophages (2). Our knowledge about the RNA viruses in the marine virioplankton is much more limited, but the data available suggest that virtually all of them infect eukaryotic organisms, most likely protists (3). These RNA viruses were often assumed to be a minor component of the virioplankton, but recent data suggest that, at least at the one location sampled, they were as abundant in seawater as DNA viruses (4).
At present, our knowledge of the RNA viruses that infect the marine protistan plankton is limited to what we have learned from 13 isolates and the results of a few molecular surveys (3). The RNA viruses that have been isolated so far infect some of the major taxa of marine protists, including diatoms (5–9), dinoflagellates (10), raphidophytes (11), prasinophytes (12), and thraustochytrids (13). Phylogenies of viruses in the order Picornavirales based on alignments of the RNA-dependent RNA polymerase (RdRp) sequences are congruent with the established taxonomic assignments by the International Committee on Taxonomy of Viruses (ICTV) (14–16), and thus the RdRp is a useful molecular marker for the investigation of the diversity of viruses in the order Picornavirales (picornavirads). Cultivation-independent surveys targeting the RdRp of picornavirads in samples from temperate waters (17) and subtropical waters (18) have revealed a level of genetic diversity that is poorly represented by the limited number of existing cultures (19).
Metagenomic methods can provide a more comprehensive view of the genetic diversity of RNA viruses than single-gene surveys. This approach has been used to investigate RNA virus diversity in a variety of habitats, including reclaimed water (20), untreated wastewater (21, 22), hot springs (23), a freshwater lake (24), and various marine habitats (4, 25, 26). Application of this method to RNA viruses harvested from coastal waters of British Columbia suggested that most were predominantly positive-sense, single-stranded RNA (+ssRNA) viruses that are distantly related to established taxa (25). Very few dsRNA viral sequences were identified, and no sequences from RNA phage, negative-sense, single-stranded RNA (−ssRNA) viruses, or retroviruses were detected (25). These data offered a first glimpse into the genomic diversity of the natural RNA virioplankton in seawater but were limited to a single location in temperate coastal waters. To broaden our understanding of RNA viral ecology in the ocean, we estimated the abundance of RNA viruses relative to that of DNA viruses and the diversity of RNA viral communities harvested from coastal tropical waters. Because of their small genome sizes, RNA viruses cannot be accurately quantified in seawater even with RNA-specific stains (27, 28); therefore, we used an indirect approach in which the values corresponding to the relative masses of total viral RNA and DNA were divided by estimates of the mass of nucleic acid per RNA or DNA virion in the sample to determine relative abundances. Our data, from a tropical coastal site sampled on two occasions, indicated that the abundance of RNA viruses could at times exceed that of DNA viruses in seawater (4). An initial metagenomic analysis of RNA virus diversity in those samples suggested that, just as they did in one study in temperate waters (25), +ssRNA viruses in the order Picornavirales dominated in tropical coastal waters. The goal of our previous report was to estimate the relative abundances of RNA and DNA viruses. In this report, we expand our analysis of the two Kāne′ohe Bay viromes. Specifically, we present data on the buoyant density distributions of marine picornavirads, provide a more detailed analysis of the RNA virus metagenomes—including the reconstruction of six genomes—and provide estimates of the diversity of marine picornavirads using a number of different approaches.
RESULTS AND DISCUSSION
General description of the metagenomes.
A general characterization of the two metagenomes analyzed in this study can be found elsewhere (4), but the salient features are summarized here to provide a context for the new analyses that are the focus of this paper. Pyrosequencing of the two libraries prepared from samples collected in 2009 and 2010 from coastal O′ahu resulted in a combined total of 249,941 high-quality reads and approximately 89 Mbp of sequence. The majority of sequences in each library assembled into contigs (69% and 78% in the 2009 and 2010 libraries, respectively). Approximately 54% of the total sequences were most similar to those of RNA viruses, 4% appeared to derive from cells, and 42% could not be assigned. Of the sequences that were identified as viral, >97% were most similar to those of +ssRNA viruses (95% specifically to members of the order Picornavirales). The remaining 3% were most similar to those of dsRNA viruses, with the majority having similarity to Micromonas pusilla reovirus (MpRV), the sole member of the genus Mimoreovirus.
Buoyant densities of marine picornavirad-like viruses.
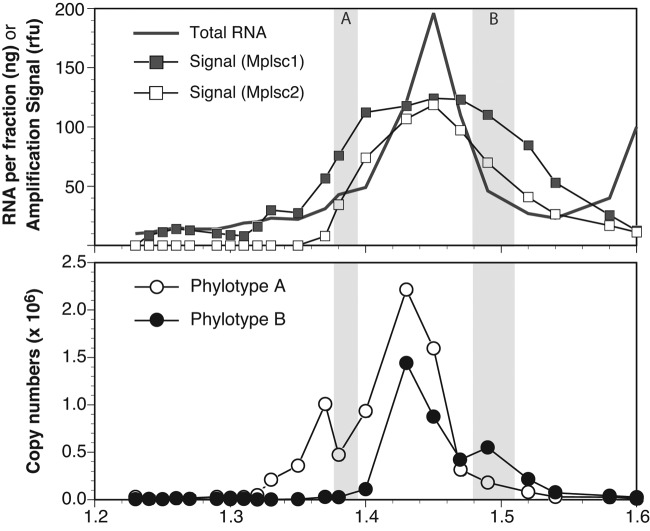
Buoyant density gradients are frequently used to purify viruses from natural assemblages for analysis (29–31), but there are few reports describing the density distribution of uncultivated assemblages of DNA-containing viruses (32–34) and none for the RNA-containing viruses. This information is useful for optimizing purification strategies (29). Although picornavirads dominated our metagenomic libraries, the libraries were prepared from pooled fractions representing a specific density range (1.38 to 1.53 g ml−1), which was chosen conservatively based only on the distribution of total RNA. To better understand the density distribution of marine picornavirads, we analyzed each fraction separately for the entire gradient (≤1.2 to ≥1.6 g ml−1), amplifying by PCR first with degenerate primers targeting two subclades of marine picornavirads and then with primers designed to specifically target the RdRp genes of a putative high-buoyant density phylotype and a putative low-buoyant density phylotype. Amplification with the degenerate primers resulted in variable amplicon yields that depended on which fraction of a CsCl buoyant density gradient was assayed. The patterns were similar for the two subclades assayed, with both showing a peak in the 1.45 g ml−1 fraction (Fig. 1). The distribution was somewhat broader for subclade 1, the primers for which were also found to capture a broader range of phylogenetic diversity (Culley and Steward [18]). Clone libraries prepared from subclade 1 RdRp amplicons derived from one of the lower-density fractions (1.38 g ml−1) and one of the higher-density fractions (1.49 g ml−1) on either side of the main amplification peak revealed that some sequences were present in both libraries but that others were detected only in one library or the other (see Fig. S1 in the supplemental material). Reverse transcription-quantitative PCR (RT-qPCR) using primers designed to target one of the sequences found only in the lower-density library (phylotype A) or one of the sequences found only in the higher-density library (phylotype B) revealed target distributions consistent with the clone library results (Fig. 1). Specifically, the concentration of phylotype A was much higher than that of phylotype B in the 1.38 g ml−1 fraction and the concentration of phylotype B was higher than that of phylotype A in the 1.49 g ml−1 fraction. In both cases, however, the distribution of target was bimodal, with a local maximum in or near the density fraction from which the sequence derived but an overall maximum occurring in the 1.43 g ml−1 fraction (Fig. 1).
FIG 1 .
Distribution of picornavirad-like viruses in a CsCl buoyant density gradient. (Upper panel) The total RNA from the 2009 sample measured in each fraction of a buoyant density gradient (redrawn using data from reference 4) is shown along with the average amplification signal from endpoint PCR with degenerate primers targeting marine picornavirad-like viruses. The amplification signal value (in arbitrary relative fluorescence units) was determined by image analysis of the amplicons on an agarose gel and is presented as the running average of the signal for two sets of primers broadly targeting marine picornavirad-like subclade 1 (Mplsc1) and Mplsc2. (Lower panel) The copy numbers of two specific RdRp phylotypes that were identified by cloning and sequencing of amplicons from two of the fractions as determined by RT-qPCR. The shaded regions in both panels indicate the fractions used to create clone libraries and are labeled according to the phylotype (A or B) that was found only in that library.
The reason for the bimodal peaks observed for both of the specific phylotypes assayed is unknown. The positions of the clearly separated minor peaks make sense, considering the criteria used to choose the targets, but the presence of major peaks for both targets in the same intermediate density fraction is curious. The pattern does not appear to be a result of nonspecific amplification, since melting curves of the amplicons indicated a single narrow peak at the same melting temperature for all fractions for a given primer set. Differences in total RNA levels among the fractions could have influenced the efficiency of the RT reactions (35), but the offset of the RT-qPCR peaks and the total RNA peak suggest this cannot be the sole explanation for the observed distributions. Two alternative explanations are that (i) identical target sequences are found in viruses that differ in their levels of buoyant density or (ii) many of the viruses of each phylotype were aggregated with each other, with other viruses, or with some other material that altered their equilibrium buoyant density, a phenomenon that has been observed previously (34).
Regardless of the details of the phylotype-specific distributions, the amplification data suggest that the buoyant density distribution of picornavirads was primarily in the density range from 1.35 to 1.5 g ml−1. The distribution was similar to that of total RNA, the most notable exception being the second peak in total RNA at high densities (ca. 1.6 g ml−1), which is not accompanied by an increasing signal for picornavirads (Fig. 1). This suggests that the RNA in the denser fractions is qualitatively different from that in the primary RNA peak and may not be of viral origin.
RdRp viral diversity in the metagenome.
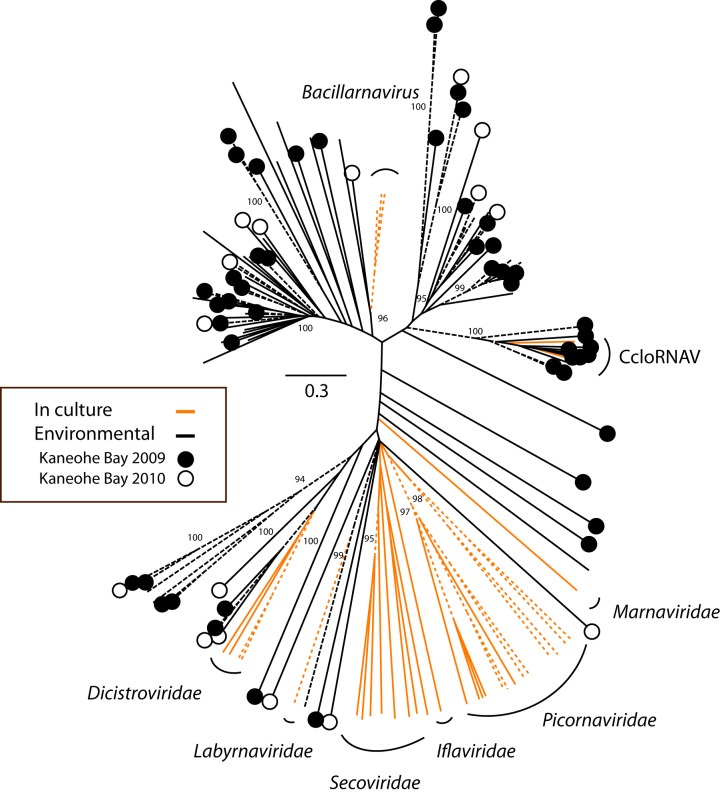
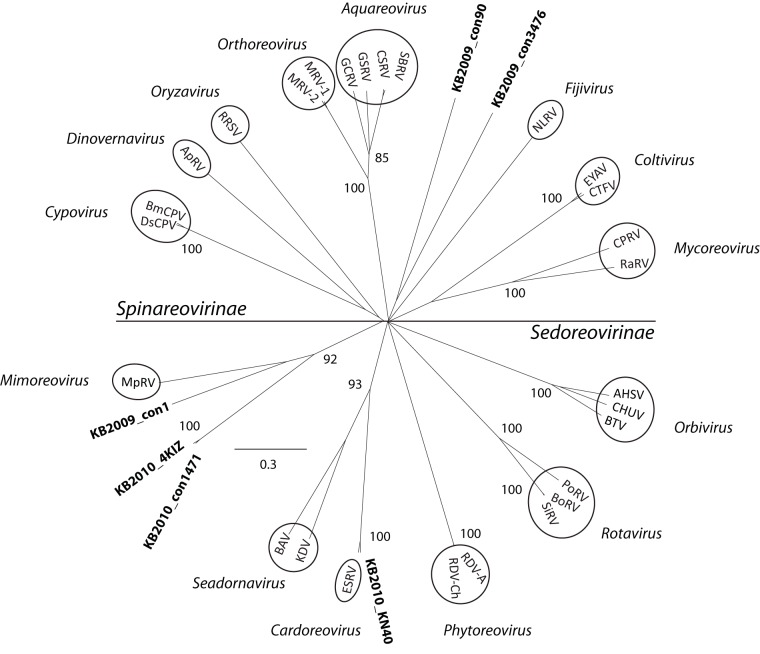
A search for all likely RdRp sequences (i.e., those that contained, at a minimum, the same two of seven conserved motifs) returned 531 (517 picornavirad-like and 14 reovirid-like) sequences in the 2009 library and 300 (292 picornavirad-like and 8 reovirid-like) in the 2010 library. A subset of these sequences that were longer and contained four of the seven conserved RdRp motifs were analyzed phylogenetically. These longer sequences (51 picornavirad-like and 3 reovirid-like sequences in 2009; 21 picornavirad-like and 3 reovirid-like sequences in 2010) formed large clusters with other environmental RdRp sequences, although some of these clusters were not well supported (maximum-likelihood support values < 80) and most were related only distantly to RdRp sequences from cultivated representatives (Fig. 2). One well-supported cluster (with a maximum-likelihood support value of 100) grouped 9 environmental sequences with two viruses (CloRNAV1 [Cylindrotheca closterium RNA virus 01] and CcloRNAV2) that infect a species of centric diatom. Of the six longer reovirid-like RdRp sequences analyzed, three formed a well-supported cluster (bootstrap value of 92) with MpRV, the only classified reovirid known to infect a marine protist (Fig. 3). Two other sequences formed a cluster distantly related to other genera within the family, and one sequence clustered closely (maximum-likelihood support value = 100) with ESRV (Eriocheir sinensis reovirus), a pathogen of the Chinese mitten crab.
FIG 2 .
Mpl-region RdRp phylotypes. An unrooted maximum-likelihood (ML) tree based on an alignment of the translated RdRp region targeted by the degenerate Mpl primer sets described by Culley and Steward (18) is shown. Included in this analysis are environmental sequences (black branches) that are comprised of sequences from this study, as well as from three previous studies published by Culley and Steward (17, 18, 25), and homologous sequences from representative viruses from the established taxa (orange branches) in the order Picornavirales. The specific sequences in the alignment are listed in Table S1 in the supplemental material. The scale bar is the equivalent of 0.3 substitutions per site. The statistical support values shown are percentages calculated by the aLRT method. Dashed branches represent clades with support values greater than 80. CcloRNAV, Cylindrotheca closterium RNA virus 01 and 02.
FIG 3 .
Reovirid RdRp tree. An unrooted ML tree based on an alignment of sequences containing regions 1 to 4 conserved in the RdRp of reovirids (48) from this study and sequences from representative viruses from the established genera in the family Reoviridae is shown. All sequences above and below the horizontal line are in the subfamilies Spinareovirinae and Sedoreovirinae, respectively. The specific sequences in the alignment and the full names of the virus species represented by abbreviations in the figure are listed in Table S1 in the supplemental material. The scale bar is the equivalent of 0.3 substitutions per site. The statistical support values shown are percentages greater than 80 as calculated by the aLRT method.
Extrapolation from the frequency distribution of unique RdRp amino acid sequences using the mean Chao 1 estimator suggested that there were on the order of 600 to 1,000 phylotypes (95% confidence interval [CI] from 500 to 1,500) if extrapolating from the shorter sequences (2009 and 2010 samples) and around 400 sequences (95% CI from 200 to 1,000) if extrapolating from the longer sequences (2009 sample only). To put the sequence diversity into a taxonomic context, we clustered the sequences to a conservatively defined species level (≥68% amino acid [aa] identity) (18), which resulted in a minimum of 39 and 21 different clusters in the 2009 and 2010 libraries, respectively, with five of those appearing in both libraries. Extrapolation using the Chao 1 estimator for the 2009 sample resulted in an estimated species-level richness of 300 (95% CI from 100 to 900). This analysis could not be done for the 2010 sample, or for any of the reovirus-like sequences, because all of the sequences retrieved were unique in those instances.
None of the RdRp nucleotide sequences that we obtained from Kāne′ohe Bay were identical to sequences previously derived from waters of coastal British Columbia. Nor were there identical nucleotide sequences shared between the 2009 and 2010 samples. However, three of the nucleotide sequences from the 2009 sample in this study were identical to sequences from a prior sampling of Kāne′ohe Bay in 2006. After translation, five 2009 RdRp phylotypes were identical to phylotypes from the 2010 sample.
The RdRp amino acid sequences in our libraries spanned the distance represented in previous targeted gene surveys (17, 18) and include some new, deeply branching groups. Most sequences did not cluster near sequences from the few cultivated representatives. Since viral RdRp sequences tend to cluster based on host phylogeny (18), the phylogenetic distances among the RdRp sequences suggests that there are a great many protists from diverse clades in seawater that are being lysed by RNA viruses at any given time.
Assembly and analysis of picornavirad-like genomes.
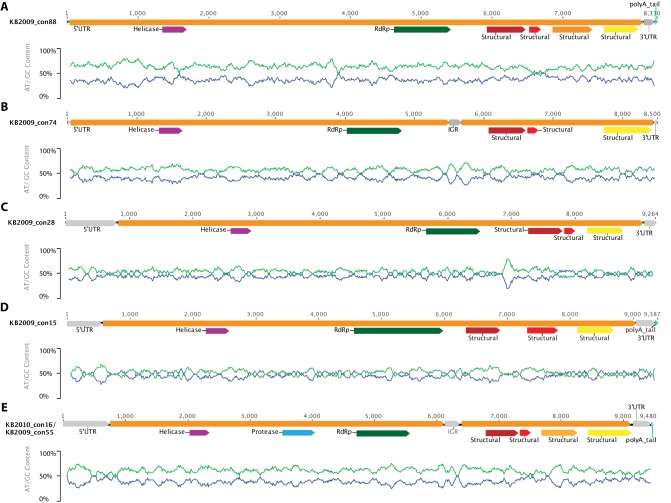
Six complete or near-complete genomes were assembled from the metagenomic libraries (Fig. 4). These contigs (KB2009_con55, KB2009__con15, KB2009__con28, KB2009__con74, and KB2009__con88 and KB2010_con16) ranged in size from 8,330 to 9,465 bp (mean, 9,008 bp) and had GC contents ranging from 36.4% to 46.8% (mean, 41.5%). Primer pairs designed to uniquely amplify a region within the RdRp gene of each assembled genome resulted in amplicons of the expected size and predicted sequences when the original RNA extract was used as the template (data not shown). Each genome contained either one or two large open reading frames (ORFs) that encoded polyproteins ranging in size from 906 to 2,827 amino acids (Fig. 4 and Table 1). These domains were similar to conserved domains of the nonstructural and structural proteins of known +ssRNA viruses in the order Picornavirales.
FIG 4 .
Maps of assembled genomes. Genome maps for the 6 complete, or near-complete, genomes assembled from this study (produced in Geneious version 6.1.7) are shown. The unit of measure for the horizontal scale bar is nucleotides. The 5′ untranslated region (UTR), intergenic region (IGR), and 3′ UTR are represented by gray arrows, and the open reading frames and a polyadenylated [poly(A)] tail are represented by orange and turquoise bars, respectively. The identified motifs (see Materials and Methods) are shown with arrows of different colors as follows: helicase, purple; protease, light blue; RNA-dependent RNA polymerase (RdRp), green; structural, wine, red, yellow, and light orange. Under each genome schematic is a graph of GC content along the genome (sliding window frame = 93) where % GC is shown with a blue line and % AT with a light-green one. KB2010_con16 and KB2009_con55 are represented by the same map (E), because they have identical genome organizations (see Results and Discussion). A protease is presumed to be present between the helicase and the RdRP in all genomes, but, in some cases (A to D), there was insufficient similarity to a known protease in this region to map its location.
TABLE 1 .
Summary information on assembled genomesa
| Genome name | Sample | Genome size (bp) | % GC | Avg coverage (no. of reads) | No. of ORFs | ORF 1 size (bp) | ORF 1 size (aa) | ORF 2 size (bp) | ORF 2 size (aa) | 5′ UTR size (bp) | IGR size (bp) | 3′ UTR size (bp) | % UTR | Poly(A) tail | % of totalb |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KB2010_con16 | 2010 | 9,465 | 38.9 | 529 | 2 | 5,388 | 1,796 | 2,718 | 906 | 773 | 296 | 289 | 14.3 | Y | 13 |
| KB2009_con55 | 2009 | 9,104 | 38.7 | 115 | 2 | 5,388 | 1,796 | 2,718 | 906 | 773 | 296 | 289 | 14.9 | Y | 2 |
| KB2009_con15 | 2009 | 9,387 | 46.8 | 169 | 1 | 8,481 | 2,827 | NA | NA | 585 | NA | 298 | 9.4 | Y | 3 |
| KB2009_con28 | 2009 | 9,264 | 45.3 | 259 | 1 | 8,268 | 2,756 | NA | NA | 904 | NA | 161 | 11.5 | ND | 5 |
| KB2009_con74 | 2009 | 8,500 | 42.6 | 131 | 2 | 5,415 | 1,805 | 2,718 | 906 | 61 | 259 | 47 | 4.3 | ND | 2 |
| KB2009_con88 | 2009 | 8,330 | 36.4 | 81 | 1 | 8,127 | 2,709 | NA | NA | 28 | NA | 138 | 2.0 | Y | 1 |
Estimated minimum genome size, % GC content, average coverage, the sizes of open reading frames (ORFs) and untranslated regions (UTRs), and whether a poly(A) tail was evident are presented for each of the six genomes assembled from the metagenomic libraries. IGR, intergenic region; NA, not available; ND, not detected; Y, yes.
Listed in this column are the percentages of total bp assigned to a genome calculated by multiplying the predicted size of the genome (in bp) by the average genome coverage value and dividing by the total no. of bp generated in the library (×100).
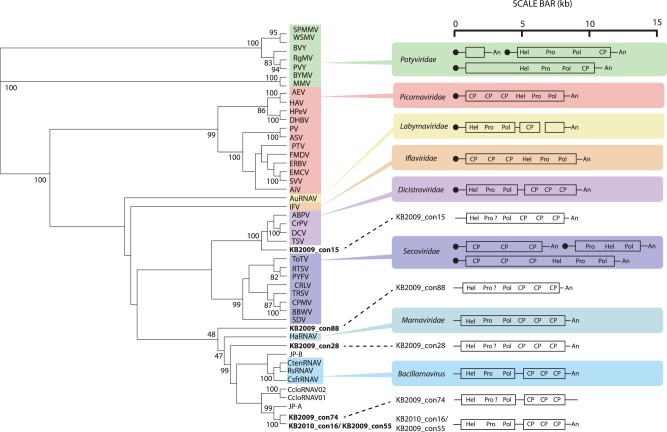
Phylogenetic analysis of the full-length RdRp genes from these assemblies and evidence from comparative genome organization analyses suggested that four of the assembled genomes (KB2009_con28, KB2009_con74, KB2009_con55, and KB2010_con16) were most similar to those of known diatom-infecting viruses, including the three classified members of the genus Bacillarnavirus (maximum-likelihood support value = 99) (Fig. 5). The RdRp of another near-complete genome (KB2009_con15) was most closely affiliated (maximum-likelihood support value = 100) with members of the family Dicistroviridae. One other genome (KB2009_con88) was more divergent and did not form any well-supported clades with any other viruses in the analysis.
FIG 5 .
Tree of the RdRp of assembled genomes. A rooted ML tree in which the tree branches have been transformed based on an alignment of the conserved regions of the RdRP (14) from the assembled genomes from this study, homologous sequences from representative viruses from the established taxa in the order Picornavirales (highlighted in color), CcloRNAV01 and CcloRNAV02, viruses that infect a pennate diatom, and JP-A and JP-B, putative viral genomes assembled from a marine metagenome, is shown. The specific sequences in the alignment and the full names of the virus species represented by abbreviations in the figure are listed in Table S1 in the supplemental material. The statistical support values shown are percentages greater than 80 as calculated by the aLRT method. The scale bar represents 0.4 substitutions per site. Adjacent to each picornavirad taxon and environmental sequence is a schematic, based on a study by Le Gall et al. (16), that shows the gene order and genome organization for each genome type. The symbols are as follows: a black circle = “VPg” = genome-linked protein, “Hel” = helicase, “Pro” = protease, “Pol” = polymerase, “CP” = capsid protein, a black line = untranslated region, and “An” = poly(A) tail. A question mark indicates that we detected no similarity to known protease sequences but that we presume that one is encoded.
Two of the assembled genomes (KB2009_con55 and KB2010_con16) were nearly identical (99.8%). The 2009 genome was 361 bp shorter than the 2010 genome, missing 231 bp on the 5′ end and 130 bp on the 3′ end, presumably a result of incomplete sequence coverage. The assembly of these genomes in which nearly all (19 of 22) differences were synonymous substitutions provides some confidence in the assemblies and suggests that the genomes derive from functionally equivalent strains. Whether these phylotypes coexist, or whether one replaced the other over time, we cannot discern from the present data. A high degree of genome sequence conservation over time (96% to 98% nucleic acid identity) was also observed among DNA-containing viruses over a 10-year period in coastal California (36). Although the collection dates for our samples were only 10 months apart, the data suggest that there could be similar genome stability among planktonic RNA viruses.
Each of the six assembled genomes contained a putative nonstructural gene with the highly conserved motifs of a nucleotide triphosphate (NTP)-binding domain and another gene with significant sequence similarity to the catalytic center of a family of +ssRNA virus RdRps. We located a region with sequence similarity to a family of viral 3C cysteine proteases in only two of the genomes (KB2009_con55 and KB2010_con16). The syntenous regions of the other genomes were of similar sizes but had no significant similarity to known proteases. Since this enzyme is critical for the reproduction of all known +ssRNA viruses, we believe that the syntenous regions in the other genomes also encode proteases that are highly divergent from known proteases. The structural genes (3 to 4 per genome) were homologous to the capsid-binding site of picornaviruses and to the VP4 and capsid proteins of dicistroviruses. In the untranslated regions (UTR), no similarities were found to any experimentally verified internal ribosomal entry site (IRES) structures. Poly(A) tails were present at the 3′ end of four of the six assembled genomes.
Of the nine genomes that have been assembled from marine RNA metagenomes—six from this study and three from a previous study (18)—all but one are related to the genomes of viruses in the order Picornavirales, which is consistent with the relatively high frequency of picornavirad-like RdRp sequences in the libraries. The assembled genomes from this study share several characteristics with picornavirads. These include a monopartite or bipartite genome, the helicase-protease-replicase nonstructural gene order, and a poly(A) tail (37). The nonstructural gene cassette is closest to the 5′ end and is followed by a gene block of structural genes. This configuration is similar to the gene order of viruses in the genus Bacillarnavirus and families Dicistroviridae and Marnaviridae, all taxa within the Picornavirales.
The number of sequences recruiting to various contigs provides some clues about the composition of the RNA viral community (Table 1). Based on the predicted size and the average coverage for the assembled genomes and the total number of nucleotides sequenced in the library, the recruitment data imply that the community structures of the RNA viruses in these samples differ. For example, on the basis of this type of analysis, we estimate that KB2010_con16 represents 13% of the total RNA viruses (Table 1). However, these estimates of dominance based on recruitment have considerable uncertainty because of the possibility of bias in the production of the metagenomic library as discussed below.
Comparison of RNA viral metagenomes.
In reciprocal BLAST analyses of the two libraries, 76% of the 2009 sequences had significant similarity (E value ≤ 10−5) to sequences from the 2010 library and 84% of the 2010 sequences had significant similarity to those from 2009. An intercomparison of these Kāne′ohe Bay metagenomes and two other aquatic metagenomes targeting RNA viruses, one from coastal British Columbia (25) and one from an artificial lake in Maryland (24), revealed that sequences most similar to +ssRNA viral genomes outnumbered those matching dsRNA viral genomes in all cases (Fig. 6). Furthermore, no ambisense, −ssRNA, or retroviral sequences were detected in any of the libraries.
FIG 6 .
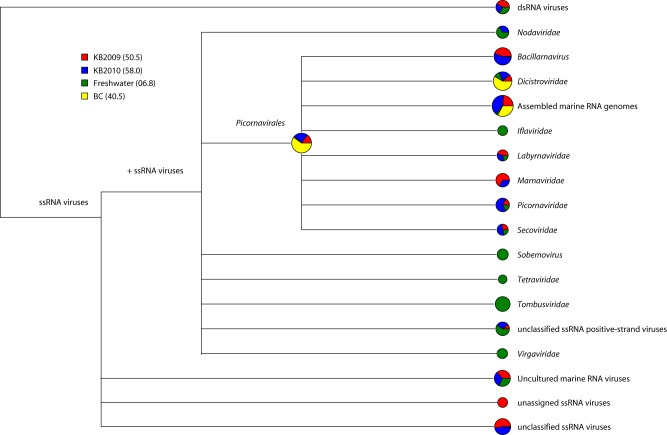
Comparison of KB viromes with those of coastal British Columbia waters (BC) and freshwater. Results of comparisons of the RNA viromes from this study with an RNA virome from the Strait of Georgia, British Columbia (17), and from a man-made freshwater lake (24) generated with MEGAN metagenomic analysis software (53) are shown. The comparison was normalized to accommodate the drastic differences in the numbers of sequences among the libraries. In this figure, the area of each pie is proportional to the number of sequences in it, and the area of each wedge in a pie is proportional to the representation of sequences from each sample in that pie. The percentages of total sequences classified as viral for each library are shown in parentheses in the key.
Most of the +ssRNA in the marine samples, but not those in the freshwater sample, were assigned to the order Picornavirales. Among the picornavirad-like sequences, those with similarity to sequences of dicistrovirids and JP-B (a putative viral genome of unknown affiliation assembled from a marine sample) were identified in all four of the metagenomes. Sequences related to the diatom-infecting bacillarnaviruses were common in the Kāne′ohe Bay libraries but were not detected in the coastal British Columbia or freshwater libraries. The relative high representation of bacillarnavirus-like sequences in the libraries from Kāne′ohe Bay is consistent with the importance of diatoms in this system, blooms of which often dominate the eukaryotic phytoplankton community (38).
Also notable was the detection, only in the freshwater library, of sequences most similar to those of viruses that infect land plants (e.g., tombusvirids, sobemovirids, and members of the Virgaviridae) or insects (iflavirids). The detection of sequences similar to those of plant and insect viruses in the lake, but not the sea, might be attributable in part to viruses entering this shallow retention basin in terrestrial runoff (24). However, many of the novel virus sequences recovered may derive from uncharacterized viruses that infect benthic or planktonic freshwater organisms.
The predominance of +ssRNA virus-like sequences in all of the libraries analyzed may reflect a higher relative abundance of these types of RNA viruses in aquatic environments, but we cannot yet rule out biases from the steps involved in library preparation. The amplification method we used in this study was found to introduce little intragenomic bias when used to sequence individual viruses having a variety of genome configurations (39), but there is no data on the relative efficiencies with which ssRNA versus dsRNA genomes are recovered in a mixture of the two. A hypothesis that has yet to be tested is that the apparent low representation of dsRNA viruses in all of the libraries reflects interference from the reannealing of complementary strands during reverse transcription. Even if this were found to be true, it does not explain the dearth of sequences similar to those of other viruses having single-stranded RNA genomes (negative sense, ambisense, retroviral). Ultimately, quantitative assays of the major viral groups identified by metagenomic analyses will be needed to confirm the apparent dominance of picornavirads among marine RNA viruses.
Conclusion.
The six genomes assembled as part of this study represent a significant increase in the number of marine picornavirad genome sequences available and will be useful for designing future experiments to test the ecological contributions of these viruses.
The apparent dominance of picornavirad-like sequences in our samples from coastal tropical waters is consistent with a previous metagenomic analysis of RNA viruses in coastal temperate waters (25), despite the use of very different methods in the two studies for each of three major steps: viral harvesting (ultrafiltration versus flocculation), amplification (sequence-independent single-primer amplification [SISPA] versus linker ligation), and sequencing (454 versus Sanger). This suggests that, as documented for metagenomes of DNA viruses (31, 40), the results are robust with respect to at least some methodological biases and that picornavirads may generally dominate the pool of RNA viruses in the ocean (but perhaps not in freshwater). However, quantitative analyses of additional marine habitats (e.g., polar, open ocean) and a thorough evaluation of potential biases during reverse transcription are needed to either bolster or banish this incipient paradigm.
MATERIALS AND METHODS
A description of the study site and methodological details concerning sample collection, processing, and construction of the metagenome was provided in a companion study (4). Summaries of those elements are provided here, as a convenience to the reader, along with more-detailed descriptions of the analyses specific to this paper.
Sample collection and processing.
Surface seawater (<0.5-m depth) was collected on 1 August 2009 (35 liters) and 3 June 2010 (40 liters) in polycarbonate carboys from a pier in Kāne′ohe Bay, Hawai’i (21°25′46.80″N, 157°47′31.51″W), a reef-protected, subtropical embayment located on the windward side of O′ahu. Samples were filtered (Sterivex; Millipore) (0.22 µm pore size) to remove cells and larger particles, and then viruses in the filtrate were concentrated by chemical flocculation and filtration (41) followed by centrifugal ultrafiltration (Amicon 15; Millipore) (30 kDa). Viruses from concentrated, virus-enriched samples were purified using sequential step and continuous CsCl buoyant density gradients (29). Fractions of approximately 0.5 ml were collected from the continuous gradient (22 to 23 fractions per gradient). After buffer exchange into TE (10 mM Tris, 1 mM EDTA, pH 8), nucleic acids were extracted from a portion of each density fraction (QIAamp MinElute Viral Spin kit; Qiagen). Each nucleic acid extract was treated with DNase to remove any copurified DNA, and the RNA concentration was determined by fluorometry (4).
Amplification, cloning, and sequencing of RdRp genes.
Reverse-transcription PCR (RT-PCR) with two sets of degenerate primers targeting marine picorna-like virus subclades 1 and 2 (Mplsc1 and Mplsc2; see Table S2 in the supplemental material) was performed with RNA template from each buoyant density fraction according to the protocol described by Culley and Steward (18). The resulting endpoint PCR products were separated on a 1% agarose gel and visualized on a digital gel documentation system. The intensity of PCR amplification in each fraction was measured using Molecular Imaging Software (Kodak). Amplified products from the 1.38 g ml−1 and the 1.49 g ml−1 buoyant density fractions were excised from the gel and purified separately (MinElute Gel Extraction kit; Qiagen). The ends of the purified products were repaired (PCRTerminator End Repair kit; Lucigen) and ligated into the pSMART-HCKan vector (Lucigen). Ligated vector was transformed into Ecloni 10G Supreme cells (Lucigen) via electroporation using the supplier’s recommended conditions. Clones were screened for insertions by PCR amplification, and products in the correct size range were purified and sequenced by Sanger sequencing with fluorescent dye terminators (Applied Biosystems).
Quantification of RdRp phylotypes in the buoyant density gradient.
Reverse transcription quantitative PCR (RT-qPCR) was used to determine the abundances of two RNA virus phylotypes identified in the 2009 sample. Primers were designed to target regions of the RdRp unique to each phylotype (see Table S2 in the supplemental material). Reactions were performed with RNA template from each of the RNA viral buoyant density fractions from the 2009 sample. Reaction mixtures for cDNA synthesis (Superscript III; Invitrogen Corporation) consisted of 5 µl of the extracted, DNase-treated RNA template, a 0.2 mM concentration of each deoxynucleoside triphosphate, and a 0.5 µM concentration of reverse primer. Samples were denatured at 65°C for 5 min and cooled on ice and then supplemented with 1× First-Strand Buffer (Invitrogen Corporation), 5 mM dithiothreitol, 40 U RNase (RNaseOUT; Invitrogen Corporation), and 200 U reverse transcriptase (Superscript III; Invitrogen Corporation) to obtain a final reaction mixture volume of 20 µl. The reaction mixtures were brought to 55°C for 60 min and to 70°C for 15 min as a final termination step and were then supplemented with 1 µl RNase H (Invitrogen Corporation) and incubated for 20 min at 37°C. The qPCR amplification was performed on a 7300 real-time PCR system (Applied Biosystems) with Power SYBR green PCR Mastermix (Applied Biosystems). The reaction mixtures contained 12.5 µl SYBR green PCR master mix (Applied Biosystems), a 0.2 µM concentration of each primer, and 2 µl of sample cDNA template, with a final volume of 25 µl. For each primer set, reactions were replicated two times, and each set of reaction mixtures contained duplicate samples, standards, and negative controls. Standards consisted of 10-fold serial dilutions (3 × 101 to 3 × 109 molecules per reaction) of target molecule that had been cloned, amplified using appropriate primers, purified by agarose gel electrophoresis, and extracted with a MinElute Gel Purification kit (Qiagen). Quantities of DNA were determined fluorometrically with a Quant-iT DNA Assay kit (Invitrogen Corporation). The thermal cycling protocol consisted of a denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at the primer-specific temperature (see Table S2) for 30 s, extension at 72°C for 35 s, and a final extension at 72°C for 14 min and 25 s. The specificity of each primer set was verified by an analysis of the DNA melting curve of the amplification products over the course of the reaction as well as independent experiments in which the amplification efficiency of the primers was determined in samples spiked with various amounts of nontarget template.
Metagenome construction.
For each sampling date, RNA purified from density fractions in the range from 1.38 to 1.49 g ml−1 was pooled and subjected to a random-priming-mediated sequence-independent single-primer amplification (RP-SISPA) reaction (39, 42). After size selection (500 to 1,000 bp) on an agarose gel, RP-SISPA products were sequenced with a GS FLX Titanium platform (Roche Diagnostics Corporation). Sequences were run through a quality-control pipeline (43) to remove short and low-quality reads and presumed artificial replicates, and the ends were trimmed to remove any remaining primer sequences. Rarefaction curves for each RNA viral metagenome were generated in METAVIR (44) using equal samplings of the unassembled, quality-controlled sequences and a clustering percentage of 75.
Identification and analysis of RdRp genes in the metagenome.
A hidden Markov model was built and used to identify RdRp-like sequences in each of the metagenomes using HMMER v 3.0 (45). Markov models were first produced based on amino acid alignments of conserved RdRp regions (46) of representative picornavirad-like viruses and conserved RdRp regions (47) of representative reovirids (see Table S1 in the supplemental material). Sequences fitting the criteria of the models were retrieved from the libraries translated into all six frames. RdRp sequences that matched a known RdRp gene by BLAST with an E value ≤ 10−3 were considered to be significant. We used this approach to identify (i) sequences that contained the smallest, still-identifiable region of the RdRp (conserved domains 6 to 7) (14), (ii) picornavirad-like sequences in the libraries that contained the RdRp regions targeted by Mpl primers, which includes conserved domains 4 to 7 (14), and (iii) sequences containing regions 1 to 4 conserved in the RdRp of reovirids (48). Translated picornavirad-like RdRP sequences were clustered at the putative species level based on a conservative phylogenetic distance criterion as previously described (18). In essence, the greatest distance between any two officially classified strains of picornavirads belonging to the same species (Human Rhinovirus 2 and Human Rhinovirus 89) was taken as the species threshold. Any sequences whose distance from one another was lesser than the distance represented by that threshold were considered members of the same species and those whose distance from one another was greater than the distance represented by that threshold were considered to be different species. The Chao 1 estimator (49) of total phylotype or species richness was calculated with EstimateS (50). In light of the large uncertainty in the estimator, values were rounded to the nearest hundred.
Maximum-likelihood trees were constructed with PhyML (51) from protein sequences aligned with MAFFT (52) using the auto function.
Sequence assembly and library comparison.
Sequences were assembled using CLC Genomics Workbench version 5.0 and the following parameters: global alignment, a minimum contig length of 200, mismatch, insertion, and deletion costs set to 3, length fraction set to 0.5, similarity threshold set to 0.8, automatic word value set to 20, and bubble size set to 50. Assembly statistics were presented elsewhere (4). Contigs and singletons from the assembled libraries were classified with MEGAN (53) based on blastx (54) searches of the NCBI nonredundant nucleotide database where hits with an E value ≤ 10−5 were considered significant. We used this conservative, but frequently used, cutoff value to further reduce the likelihood of the misclassification of a sequence. All of the individual reads comprising a given contig inherited the taxonomic assignment given to the contig.
We compared the KB libraries by BLAST analysis (blastx; E value, ≤10−5 cutoff) where the unassembled reads from one library were used to query the second library and vice versa.
Analysis of assembled genomes.
Open reading frames were identified with the heuristic approach for gene prediction described by Besemer and Borodovsky (55). Searches of the Conserved Domain Database (CDD) NCBI database (56) were conducted with the translated ORFs from each genome (Table 1). Searches of a database of experimentally verified internal ribosomal entry site (IRES) structures (57) were conducted with the untranslated regions (UTR) of the six genomes.
Accession numbers.
Metagenomic data referred to in this paper are available at the CAMERA website (http://camera.calit2.net/) under accession number CAM_PROJ_BROADPHAGE and sample names CAM_SMPL_000815 (1 August 2009) and CAM_SMPL_000824 (3 June 2010). The accession numbers of the MPL phylotypes described in this research are listed in Table S1 in the supplemental material.
SUPPLEMENTAL MATERIAL
Tree of Mpl phylotypes from 1.38 and 1.49 g/ml buoyant density fractions. A tree of 80 translated RdRp sequences amplified with Mplsc1 and Mplsc2 primers from the 1.38 and 1.49 g/ml fractions from the 2009 sample is shown. Accession numbers for the sequences in this analysis can be found in Table S1 in the supplemental material. The scale bar represents 0.2 substitutions per site. Download
Rarefaction curves. Rarefaction curves generated in METAVIR (44) for the 2009 and 2010 metagenomic libraries. In this analysis, the total number of clusters generated (threshold = 75%) from each library are plotted as a function of the total number of sequences. Download
Description of viruses used in phylogenetic analyses. The families that are highlighted with color comprise the order Picornavirales. Accession numbers for the sequences shown in Fig. S1 in the supplemental material are listed.
Primers used in the study. The Mplsc set consists of degenerate primers (18) used to amplify picornavirad-like viruses from the KB2009 and KB2010 samples. FR primers (39) were used in the SISPA reaction to produce viromes from viral RNA template, and the Sc1 primer sets were used in qPCR reactions to determine the abundance of specific phylotypes in different density fractions.
ACKNOWLEDGMENTS
We thank Gordon O. Walker for assistance with sampling and Shulei Sun and the Broad Institute for assistance with the processing and sequencing of the viromes.
This research was funded in part by the Gordon and Betty Moore Foundation through grant GBMF1799 to the Broad Institute. Samples G4008 (KB2009) and G8294 (KB2010) were sequenced at the Broad Institute. Bioinformatics analyses were supported in part through the use of the COBRE/INBRE Bioinformatics core, supported jointly by NIH Grants from the National Institute of General Medical Sciences (P20GM103516 and P20GM103466). This work was supported by NSF grants to G.F.S. and A.I.C. (OCE 08-26650) and the Center for Microbial Oceanography Research and Education (EF 04-24599).
Footnotes
Citation Culley AI, Mueller JA, Belcaid M, Wood-Charlson EM, Poisson G, Steward GF. 2014. The characterization of RNA viruses in tropical seawater using targeted PCR and metagenomics. mBio 5(3):e01210-14. doi:10.1128/mBio.01210-14.
REFERENCES
- 1. Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5:801–812. 10.1038/nrmicro1750 [DOI] [PubMed] [Google Scholar]
- 2. Edwards RA, Rohwer F. 2005. Viral metagenomics. Nat. Rev. Microbiol. 3:504–510. 10.1038/nrmicro1163 [DOI] [PubMed] [Google Scholar]
- 3. Lang AS, Rise ML, Culley AI, Steward GF. 2009. RNA viruses in the sea. FEMS Microbiol. Rev. 33:295–323. 10.1111/j.1574-6976.2008.00132.x [DOI] [PubMed] [Google Scholar]
- 4. Steward GF, Culley AI, Mueller JA, Wood-Charlson EM, Belcaid M, Poisson G. 2013. Are we missing half of the viruses in the ocean? ISME J 7:672–679. 10.1038/ismej.2012.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nagasaki K, Tomaru Y, Katanozaka N, Shirai Y, Nishida K, Itakura S, Yamaguchi M. 2004. Isolation and characterization of a novel single-stranded RNA virus infecting the bloom-forming diatom Rhizosolenia setigera. Appl. Environ. Microbiol. 70:704–711. 10.1128/AEM.70.2.704-711.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shirai Y, Tomaru Y, Takao Y, Suzuki H, Nagumo T, Nagasaki K. 2008. Isolation and characterization of a single-stranded RNA virus infecting the marine planktonic diatom Chaetoceros tenuissimus Meunier. Appl. Environ. Microbiol. 74:4022–4027. 10.1128/AEM.00509-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tomaru Y, Takao Y, Suzuki H, Nagumo T, Nagasaki K. 2009. Isolation and characterization of a single-stranded RNA virus infecting the bloom-forming diatom Chaetoceros socialis. Appl. Environ. Microbiol. 75:2375–2381. 10.1128/AEM.02580-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tomaru Y, Toyoda K, Kimura K, Hata N, Yoshida M, Nagasaki K. 2012. First evidence for the existence of pennate diatom viruses. ISME J 6:1445–1448. 10.1038/ismej.2011.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tomaru Y, Toyoda K, Kimura K, Takao Y, Sakurada K, Nakayama N, Nagasaki K. 2013. Isolation and characterization of a single-stranded RNA virus that infects the marine planktonic diatom Chaetoceros sp. (SS08-C03). Phycol. Res. 61:27–36. 10.1111/j.1440-1835.2012.00670.x [DOI] [Google Scholar]
- 10. Tomaru Y, Katanozaka N, Nishida K, Shirai Y, Tarutani K, Yamaguchi M, Nagasaki K. 2004. Isolation and characterization of two distinct types of HcRNAV, a single-stranded RNA virus infecting the bivalve-killing microalga Heterocapsa circularisquama. Aquat. Microb. Ecol. 34:207–218 http://www.int-res.com/abstracts/ame/v34/n3/p207-218/ [Google Scholar]
- 11. Tai V, Lawrence JE, Lang AS, Chan AM, Culley AI, Suttle CA. 2003. Characterization of HaRNAV, a single-stranded RNA virus causing lysis of Heterosigma akashiwo (Raphidophyceae). J. Phycol. 39:343–352. 10.1046/j.1529-8817.2003.01162.x [DOI] [Google Scholar]
- 12. Brussaard CPD, Noordeloos AAM, Sandaa R-A, Heldal M, Bratbak G. 2004. Discovery of a dsRNA virus infecting the marine photosynthetic protist Micromonas pusilla. Virology 319:280–291. 10.1016/j.virol.2003.10.033 [DOI] [PubMed] [Google Scholar]
- 13. Takao Y, Nagasaki K, Mise K, Okuno T, Honda D. 2005. Isolation and characterization of a novel single-stranded RNA virus infectious to a marine fungoid protist, Schizochytrium sp. (Thraustochytriaceae, Labyrinthulea). Appl. Environ. Microbiol. 71:4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koonin EV, Dolja VV. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375–430. 10.3109/10409239309078440 [DOI] [PubMed] [Google Scholar]
- 15. Zanotto PMD, Gibbs MJ, Gould EA, Holmes EC. 1996. A reevaluation of the higher taxonomy of viruses based on RNA polymerases. J. Virol. 70:6083–6096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Le Gall O, Christian P, Fauquet CM, King AM, Knowles NJ, Nakashima N, Stanway G, Gorbalenya AE. 2008. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T=3 virion architecture. Arch. Virol. 153:715–727. 10.1007/s00705-008-0041-x [DOI] [PubMed] [Google Scholar]
- 17. Culley AI, Lang AS, Suttle CA. 2003. High diversity of unknown picorna-like viruses in the sea. Nature 424:1054–1057. 10.1038/nature01886 [DOI] [PubMed] [Google Scholar]
- 18. Culley AI, Steward GF. 2007. New genera of RNA viruses in subtropical seawater, inferred from polymerase gene sequences. Appl. Environ. Microbiol. 73:5937–5944. 10.1128/AEM.01065-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Steward GF, Culley AI, Wood-Charlson EM. 2013. Marine viruses, p 127–144 In Levin SA, Encyclopedia of biodiversity, vol 5 Elsevier, London, United Kingdom [Google Scholar]
- 20. Rosario K, Nilsson C, Lim YW, Ruan Y, Breitbart M. 2009. Metagenomic analysis of viruses in reclaimed water. Environ. Microbiol. 11:2806–2820. 10.1111/j.1462-2920.2009.01964.x [DOI] [PubMed] [Google Scholar]
- 21. Cantalupo PG, Calgua B, Zhao G, Hundesa A, Wier AD, Katz JP, Grabe M, Hendrix RW, Girones R, Wang D, Pipas JM. 2011. Raw sewage harbors diverse viral populations. mBio 2:e00180-11. 10.1128/mBio.00180-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ng TFF, Marine R, Wang C, Simmonds P, Kapusinszky B, Bodhidatta L, Oderinde BS, Wommack KE, Delwart E. 2012. High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J. Virol. 86:12161–12175. 10.1128/JVI.00869-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bolduc B, Shaughnessy DP, Wolf YI, Koonin EV, Roberto FF, Young M. 2012. Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated Yellowstone Hot Springs. J. Virol. 86:5562–5573. 10.1128/JVI.07196-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Djikeng A, Kuzmickas R, Anderson NG, Spiro DJ. 2009. Metagenomic analysis of RNA viruses in a fresh water lake. PLoS One 4:e7264. 10.1371/journal.pone.0007264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Culley AI, Lang AS, Suttle CA. 2006. Metagenomic analysis of coastal RNA virus communities. Science 312:1795–1798. 10.1126/science.1127404 [DOI] [PubMed] [Google Scholar]
- 26. Andrews-Pfannkoch C, Fadrosh DW, Thorpe J, Williamson SJ. 2010. Hydroxyapatite-mediated separation of double-stranded DNA, single-stranded DNA, and RNA genomes from natural viral assemblages. Appl. Environ. Microbiol. 76:5039–5045. 10.1128/AEM.00204-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brussaard CPD, Marie D, Bratbak G. 2000. Flow cytometric detection of viruses. J. Virol. Methods 85:175–182 [DOI] [PubMed] [Google Scholar]
- 28. Tomaru Y, Nagasaki K. 2007. Flow cytometric detection and enumeration of DNA and RNA viruses infecting marine eukaryotic microalgae. J. Oceanogr. 63:215–221. 10.1007/s10872-007-0023-8 [DOI] [Google Scholar]
- 29. Lawrence JE, Steward GF. 2010. Purification of viruses by centrifugation, p 166–181 In Wilhelm SW, Weinbauer MG, Suttle CA. (ed), Manual of aquatic viral ecology. American Society of Limnology and Oceanography, Waco, TX [Google Scholar]
- 30. Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. 2009. Laboratory procedures to generate viral metagenomes. Nat. Protoc. 4:470–483. 10.1038/nprot.2009.10 [DOI] [PubMed] [Google Scholar]
- 31. Hurwitz BL, Deng L, Poulos BT, Sullivan MB. 2013. Evaluation of methods to concentrate and purify ocean virus communities through comparative, replicated metagenomics. Environ. Microbiol. 15:1428–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Steward GF, Montiel JL, Azam F. 2000. Genome size distributions indicate variability and similarities among marine viral assemblages from diverse environments. Limnol. Oceanogr. 45:1697–1706. 10.4319/lo.2000.45.8.1697 [DOI] [Google Scholar]
- 33. Steward GF, Preston CM. 2011. Analysis of a viral metagenomic library from 200 m depth in Monterey Bay, California constructed by direct shotgun cloning. Virol. J. 8:287. 10.1186/1743-422X-8-287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brum JR, Culley AI, Steward GF. 2013. Assembly of a marine viral metagenome after physical fractionation. PLoS One 8:e60604. 10.1371/journal.pone.0060604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Levesque-Sergerie JP, Duquette M, Thibault C, Delbecchi L, Bissonnette N. 2007. Detection limits of several commercial reverse transcriptase enzymes: impact on the low- and high-abundance transcript levels assessed by quantitative RT-PCR. BMC Mol. Biol. 8:93. 10.1186/1471-2199-8-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Angly F, Youle M, Nosrat B, Srinagesh S, Rodriguez-Brito B, McNairnie P, Deyanat-Yazdi G, Breitbart M, Rohwer F. 2009. Genomic analysis of multiple Roseophage SIO1 strains. Environ. Microbiol. 11:2863–2873. 10.1111/j.1462-2920.2009.02021.x [DOI] [PubMed] [Google Scholar]
- 37. Sanfaçon H, Gorbalenya AE, Knowles NJ, Chen YP. 2011. Picornavirales, p 835–839 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic, London, United Kingdom [Google Scholar]
- 38. Hoover RS, Hoover D, Miller M, Landry MR, DeCarlo EH, Mackenzie FT. 2006. Zooplankton response to storm runoff in a tropical estuary: bottom-up and top-down controls. Mar. Ecol. Prog. Ser. 318:187–201. 10.3354/meps318187 [DOI] [Google Scholar]
- 39. Djikeng A, Halpin R, Kuzmickas R, Depasse J, Feldblyum J, Sengamalay N, Afonso C, Zhang X, Anderson NG, Ghedin E, Spiro DJ. 2008. Viral genome sequencing by random priming methods. BMC Genomics 9:5. 10.1186/1471-2164-9-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Solonenko SA, Ignacio-Espinoza JC, Alberti A, Cruaud C, Hallam S, Konstantinidis K, Tyson G, Wincker P, Sullivan MB. 2013. Sequencing platform and library preparation choices impact viral metagenomes. BMC Genomics 14:320. 10.1186/1471-2164-14-320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. John SG, Mendez CB, Deng L, Poulos B, Kauffman AK, Kern S, Brum J, Polz MF, Boyle EA, Sullivan MB. 2011. A simple and efficient method for concentration of ocean viruses by chemical flocculation. Environ. Microbiol. Rep. 3:195–202. 10.1111/j.1758-2229.2010.00208.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Culley AI, Suttle CA, Steward GF. 2010. Characterization of the diversity of marine RNA viruses, p 193–201 In Wilhelm SW, Weinbauer MG, Suttle CA. (ed), Manual of aquatic viral ecology. ASLO, Waco, TX [Google Scholar]
- 43. Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. 10.1186/1471-2105-9-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roux S, Faubladier M, Mahul A, Paulhe N, Bernard A, Debroas D, Enault F. 2011. Metavir: a web server dedicated to virome analysis. Bioinformatics 27:3074–3075. 10.1093/bioinformatics/btr519 [DOI] [PubMed] [Google Scholar]
- 45. Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Koonin EV, Wolf YI, Nagasaki K, Dolja VV. 2008. The big bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 6:925–939. 10.1038/nrmicro2030 [DOI] [PubMed] [Google Scholar]
- 47. Belhouchet M, Mohd Jaafar F, Tesh R, Grimes J, Maan S, Mertens PP, Attoui H. 2010. Complete sequence of Great Island virus and comparison with the T2 and outer-capsid proteins of Kemerovo, Lipovnik and Tribec viruses (genus Orbivirus, family Reoviridae). J. Gen. Virol. 91:2985–2993. 10.1099/vir.0.024760-0 [DOI] [PubMed] [Google Scholar]
- 48. Rao S, Carner GR, Scott SW, Omura T, Hagiwara K. 2003. Comparison of the amino acid sequences of RNA-dependent RNA polymerases of cypoviruses in the family Reoviridae. Arch. Virol. 148:209–219. 10.1007/s00705-002-0923-2 [DOI] [PubMed] [Google Scholar]
- 49. Chao A. 1987. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 43:783–791 [PubMed] [Google Scholar]
- 50. Colwell RK. 2013, posting date EstimateS: statistical estimation of species richness and shared species from samples, version 9. User’s guide and application. http://purl.oclc.org/estimates.
- 51. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 52. Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518. 10.1093/nar/gki198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. 2011. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 21:1552–1560. 10.1101/gr.120618.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 55. Besemer J, Borodovsky M. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27:3911–3920. 10.1093/nar/27.19.3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mokrejs M, Masek T, Vopálensky V, Hlubucek P, Delbos P, Pospísek M. 2010. IRESite—a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic Acids Res. 38:D131–D136. 10.1093/nar/gkq224 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tree of Mpl phylotypes from 1.38 and 1.49 g/ml buoyant density fractions. A tree of 80 translated RdRp sequences amplified with Mplsc1 and Mplsc2 primers from the 1.38 and 1.49 g/ml fractions from the 2009 sample is shown. Accession numbers for the sequences in this analysis can be found in Table S1 in the supplemental material. The scale bar represents 0.2 substitutions per site. Download
Rarefaction curves. Rarefaction curves generated in METAVIR (44) for the 2009 and 2010 metagenomic libraries. In this analysis, the total number of clusters generated (threshold = 75%) from each library are plotted as a function of the total number of sequences. Download
Description of viruses used in phylogenetic analyses. The families that are highlighted with color comprise the order Picornavirales. Accession numbers for the sequences shown in Fig. S1 in the supplemental material are listed.
Primers used in the study. The Mplsc set consists of degenerate primers (18) used to amplify picornavirad-like viruses from the KB2009 and KB2010 samples. FR primers (39) were used in the SISPA reaction to produce viromes from viral RNA template, and the Sc1 primer sets were used in qPCR reactions to determine the abundance of specific phylotypes in different density fractions.