ABSTRACT
Thanks to high-throughput sequencing technologies, genome sequencing has become a common component in nearly all aspects of viral research; thus, we are experiencing an explosion in both the number of available genome sequences and the number of institutions producing such data. However, there are currently no common standards used to convey the quality, and therefore utility, of these various genome sequences. Here, we propose five “standard” categories that encompass all stages of viral genome finishing, and we define them using simple criteria that are agnostic to the technology used for sequencing. We also provide genome finishing recommendations for various downstream applications, keeping in mind the cost-benefit trade-offs associated with different levels of finishing. Our goal is to define a common vocabulary that will allow comparison of genome quality across different research groups, sequencing platforms, and assembly techniques.
EDITORIAL
Viruses represent the greatest source of biological diversity on Earth, and with the help of high-throughput (HT) sequencing technologies, great strides are being made toward the genomic characterization of this diversity (1–3). Genome sequences play a critical role in our understanding of viral evolution, disease epidemiology, surveillance, diagnosis, and countermeasure development and thus represent valuable resources which must be properly documented and curated to ensure future utility. Here, we outline a set of viral genome quality standards, similar in concept to those proposed for large DNA genomes (4) but focused on the particular challenges of and needs for research on small RNA/DNA viruses, including characterization of the genomic diversity inherent in all viral samples/populations. Our goal is to define a common vocabulary that will allow comparison of genome quality across different research groups, sequencing platforms, and assembly techniques.
Despite the small sizes of viral genomes, complications related to limited RNA quantities, host “contamination,” and secondary structure mean that it is often not time- or cost-effective to finish every genome, and given the intended use, finishing may be unnecessary (5). Therefore, we have used technology-agnostic criteria to define five standard categories designed to encompass the levels of completeness most often encountered in viral sequencing projects. Each viral family/species comes with its own challenges (e.g., secondary structure and GC content); therefore, we provide only loose guidance on the depth of sequence coverage likely required to obtain different levels of finishing. In reality, a similar amount of data will generate genomes with different levels of finishing for different viruses.
To alleviate any reliance on particular aspects of the different sequencing technologies, we have made two assumptions that should be valid in most viral sequencing projects. The first assumption is a basic understanding of the genomic structure of the virus being sequenced, including the expected size of the genome, the number of segments, and the number and distribution of major open reading frames (ORFs). Fortunately, genome structure is highly conserved within viral groups (6), and although new viruses are constantly being uncovered, the discovery of a novel family or even genus remains relatively uncommon (7). In the absence of such information, the defined standards can still be applied following further analysis to determine genome structure. The second assumption is that the genetic material of the virus being described can be accurately separated from the genomes of the host and/or other microbes, either physically or bioinformatically. Depending on the technology used, it is critical that the potential for cross-contamination of samples during the sample indexing/bar coding process and sequencing procedure be addressed with appropriate internal controls and procedural methods (8).
PROPOSED CATEGORIES FOR WHOLE-GENOME SEQUENCING OF VIRUSES
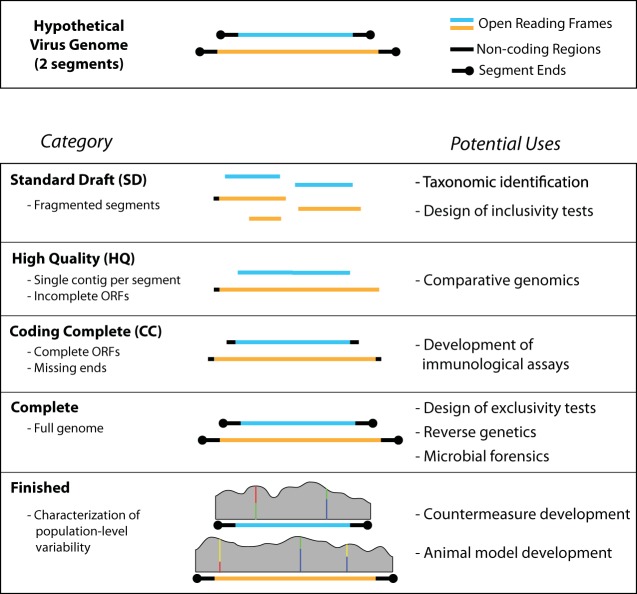
For a summary of the proposed categories for whole-genome sequencing of viruses, see Fig. 1 and Table 1.
FIG 1 .
Graphical representation of viral genome standards. Bullets on the left represent primary distinctions between categories. Bullets on the right indicate potential downstream applications of genomes in each category.
TABLE 1 .
Overview of viral genome standards
| Feature | Standard drafta | High qualitya | Coding completea | Complete | Finished |
|---|---|---|---|---|---|
| No. of contigs | >1 for some segments | 1 per segment | 1 per segment | 1 per segment | 1 per segment |
| Open reading frames | Incomplete | Incomplete | Complete | Complete | Complete |
| Estimated % of genome coveredb | ≥50% | ~80-90% | ~90-99% | 100% | 100% |
| Population-level characterization | Optional | Optional | Optional | Optional | Required |
| Contaminant analysis | Optional | Optional | Optional | Optional | Optional |
It is suggested that all bases included in any incomplete genome meet a minimum quality standard, with ≥5 reads supporting the consensus base call with individual base qualities of ≥20 on the Phred scale.
Percentages of genome covered are not meant to serve as criteria for categorizing a genome; they are simply estimates of expected levels of coverage.
Standard draft (SD).
The “standard draft” category is for whole shotgun genome assemblies with coverage that is low and/or uneven enough to prevent the assembly of a single contig for ≥1 genome segments. Genomes in this category are likely to result from samples with low viral titers, such as clinical and environmental samples, or to be those containing regions that are difficult to sequence across (e.g., intergenic hairpin regions) (9). To distinguish standard drafts from targeted amplification of partial viral sequences, standard drafts should contain at least 1 contig for each genomic segment and should be prepared in a manner that allows the possibility of sequencing the vast majority of a virus’s genome. To avoid the inclusion of small pieces of genomes as “drafts,” there needs to be some type of minimum cutoff for breadth of coverage. Therefore, we suggest that at least a majority (≥50%) of the genome be present for a set of sequences to be considered a draft genome.
High quality (HQ).
Genomes should be considered high quality if no gaps remain (i.e., a single contig per genome/segment), even if one or more ORFs remain incomplete due to missing sequence at the ends of segments. An HQ genome can often be achieved with modest levels of HT sequencing coverage (~15 to 30×) or through Sanger-mediated gap resolution of an SD.
Coding complete (CC).
The “coding complete” category indicates that in addition to the lack of gaps, all ORFs are complete. This level of completion is typically possible with high levels of HT sequencing coverage (>100×) or may require the use of conserved PCR primers targeting the ends of the segments.
Complete.
A genome is complete when the genome sequence has been fully resolved, including all non-protein-coding sequences at the ends of the segment(s). This is typically achieved through rapid amplification of cDNA ends (RACE) or similar procedures.
Finished.
This final category represents a special instance in which, in addition to having a completed consensus genome sequence, there has been a population-level characterization of genomic diversity. Typically this requires ~400 to 1,000× coverage (see below). This provides the most complete picture of a viral population; however, this designation will apply only for a single stock. Additional characterizations will be necessary for future passages.
ADDITIONAL HIGH-THROUGHPUT SEQUENCE-BASED GENOME CHARACTERIZATIONS
Population-level characterization.
HT sequencing technologies provide powerful platforms for investigating the genetic diversity within viral populations, which is integral to our understanding of viral evolution and pathogenesis (10, 11). Population-level characterization requires very high levels of HT sequencing coverage (12, 13); however, the exact level will depend on the background error profiles of the sequencing technology and the desired level of sensitivity. As an example, Wang et al. (12) determined that for pyrosequencing data, ~400× coverage is necessary to identify minor variants present at 1% frequency with 99.999% confidence, and ~1,000× coverage is needed for variants with a frequency of 0.5%. Targeted amplification of the viral genome is often necessary to achieve these coverage requirements. Due to the modest sequence lengths of most HT technologies, the state of the art for population-level analysis has been the characterization of unphased polymorphisms. However, single-molecule technologies, with maximum read lengths of >20 kb, are opening the door for complete genome haplotype phasing (14).
Identification of contaminants or adventitious agents.
After isolation, viruses are often maintained as stocks, which are propagated within host cells in tissue culture and thus amplified and preserved for future use. Despite careful laboratory practices, it is possible for these stocks to become contaminated with additional microbes. Contaminating microbes are often detrimental to subsequent applications such as vaccine development or the testing of therapeutics, making it imperative to monitor the purity of viral stocks. HT sequencing provides a powerful method for not only detecting the presence of contaminants within a sample but also for identification and characterization of any contaminants. The level of sequencing required for contamination analysis is dependent on the desired sensitivity, with more sequencing required to ensure detection of contaminants present at very low levels. For most approaches, HQ-level sequencing should be sufficient. Depending on the intended applications, analysis may need to be repeated after further passaging to ensure that no additional contaminants have been introduced.
RECOMMENDED STANDARDS FOR DOWNSTREAM APPLICATIONS
Description of novel viruses.
Despite the rapidly growing collection of viral sequences, the description of novel viruses is likely to remain an important aspect of viral genome sequencing (7, 15, 16). This is true in part because viruses evolve rapidly and are capable of recombining to form novel genotypes (17, 18). It is also true that most of the viruses that are currently circulating remain uncharacterized (15). Particularly lacking are representatives from groups that are not currently known to infect humans or organisms of economic importance. It would be imprudent, however, to continue to ignore these uncharacterized reservoirs of diversity, because it is difficult to predict the source of future emerging diseases (19–21). Additionally, with the current suite of primarily sequence similarity-based pathogen identification tools, the ability to detect novel pathogens is wholly dependent on high-quality reference databases (22). There is a trend toward requiring a complete genome sequence when a description of a novel virus is being published, and we agree that this is a good goal; however, the amount of time and resources required to complete the last 1 to 2% of a viral genome is often cost and time prohibitive for projects sequencing a large number of samples, and in most cases the very ends of the segments are not essential for proper identification and characterization. Therefore, for the majority of viral characterization projects, we recommend, at a minimum, a CC genome. This will ensure a complete description of the viral proteome and will allow accurate phylogenetic placement.
Molecular epidemiology.
One of the most common and important applications for viral genomes is in the study of viral epidemiology, which encompasses our understanding of the patterns, causes, and effects of disease. Early studies of molecular epidemiology targeted small pieces of viral genomes; however, this type of analysis is likely to miss important changes elsewhere in the genome. Therefore, there has been a strong focus in recent years toward the sequencing of “full” viral genomes. Institutes such as the Broad Institute and the J. Craig Venter Institute (JCVI) have been instrumental in breaking ground in the collection of large numbers of good-quality viral sequences. Their newly identified genomes typically fall within our CC category. This is likely to remain the gold standard for studies involving a large number of genome sequences, especially when some samples come from low-titer clinical samples, often necessitating amplicon-based sequencing methods. CC genomes allow for interrogation of changes throughout the coding portion of the viral genome and often include partial noncoding regions. In the absence of high-throughput RACE alternatives, the time and resources required to complete hundreds or thousands of genomes are likely to continue to outweigh the potential information gained from completing the terminal sequences.
Countermeasure development.
Advancements in our capabilities to sequence viral genomes are changing the way we counteract global pandemics and acts of bioterrorism. There are two important aspects of countermeasure development that can benefit strongly from the availability of genome sequences and HT sequencing data: the detection of the infectious agent and the treatment of the disease caused by the agent. Taxonomic classification and detection through DNA/RNA-based inclusivity assays (i.e., using techniques such as PCR to detect the presence of a pathogen) can be designed using fragmented and incomplete genomes (e.g., SD and HQ sequences). Fully resolved ORFs (CC) further enable the development of immunological assays, such as enzyme-linked immunosorbent assays (ELISA) and immunofluorescence assays (IFA), for protein-based detection, and obtaining a complete genome opens the door to a plethora of additional downstream applications, including the design of exclusivity tests, the establishment of reverse genetics systems, and the design of robust forensics protocols. However, for effective development and testing of animal models, therapeutics, vaccines, and prophylactics, it is necessary to obtain a complete picture of the variability present within both the challenge stock and postinfection populations, thereby necessitating finished genomes. In these medical applications, it is also important to demonstrate the absence of adventitious agents.
REPOSITORIES OF GENOMIC INFORMATION AND DATA CURATION
In addition to standardizing the vocabulary of viral genome assemblies, it is also critical for researchers to routinely provide raw sequencing reads. Without these, it is impossible for others to independently verify the quality of an assembly. Data repositories such as GenBank already provide a platform for depositing HT sequencing reads, but this is not a requirement for the submission of a genome, nor is this option typically utilized. Wider analysis of data will ultimately result in higher-quality assemblies. It is worth considering broader implementation of a wiki-like, crowd-sourcing strategy to genome assembly, similar to the annotation strategies that have been adopted for specific genomes of high interest (23, 24). This approach would allow multiple parties to work on genome assembly and annotation at the same time and would provide instant updates for the entire community to evaluate and utilize in their own research.
Our primary goal here is to initiate a conversation. The rate at which viral genomes are being sequenced is only going to increase in the coming years, and without some standardization, it will be impossible for these valuable resources to be utilized to their full potential. We present these categories as a starting point, with the goal of adjusting and refining them over time as our capabilities and needs continue to change.
ACKNOWLEDGMENTS
We acknowledge the valuable discussions held during the meetings of the Filovirus Animal Nonclinical Group (FANG) Well Characterized Challenge Material Working Group. We also thank Vivien Dugan, Susan Garges, and Lucy Ward (FANG members, NIAID-DMID), Jared Evans (JHU), and JSTO (CBA/CBM) for their useful comments, which helped to improve the manuscript.
This work was funded, in part, by Defense Threat Reduction Agency Project no. 1881290. J.H.K. performed this work as an employee of Tunnell Government Services, Inc., a subcontractor to Battelle Memorial Institute under its prime contract with NIAID, under contract no. HHSN272200700016I.
The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position, expressed or implied, of the Department of the Navy, JPEO-CBD-MCS-CRP, the U.S. Army, the FDA, the U.S. Department of Defense, the U.S. Department of Health and Human Services, the U.S. government, or the institutions and companies with which the authors are affiliated.
The views expressed in this article do not nec essarily reflect the views of the journal or of ASM.
Footnotes
Citation Ladner JT, Beitzel B, Chain PSG, Davenport MG, Donaldson EF, Frieman M, Kugelman JR, Kuhn JH, O’Rear J, Sabeti PC, Wentworth DE, Wiley MR, Yu G-Y, the Threat Characterization Consortium, Sozhamannan S, Bradburne C, Palacios G. 2014. Standards for sequencing viral genomes in the era of high-throughput sequencing. mBio 5(3):e01360-14. doi:10.1128/mBio.01360-14.
REFERENCES
- 1. Suttle C. 2005. Crystal ball. The viriosphere: the greatest biological diversity on Earth and driver of global processes. Environ. Microbiol. 7:481–482. 10.1111/j.1462-2920.2005.803_11.x [DOI] [PubMed] [Google Scholar]
- 2. Culley AI, Lang AS, Suttle CA. 2006. Metagenomic analysis of coastal RNA virus communities. Science 312:1795–1798. 10.1126/science.1127404 [DOI] [PubMed] [Google Scholar]
- 3. Daszak P, Lipkin WI. 2011. The search for meaning in virus discovery. Curr. Opin. Virol. 1:620–623. 10.1016/j.coviro.2011.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chain PS, Grafham DV, Fulton RS, Fitzgerald MG, Hostetler J, Muzny D, Ali J, Birren B, Bruce DC, Buhay C, Cole JR, Ding Y, Dugan S, Field D, Garrity GM, Gibbs R, Graves T, Han CS, Harrison SH, Highlander S, Hugenholtz P, Khouri HM, Kodira CD, Kolker E, Kyrpides NC, Lang D, Lapidus A, Malfatti SA, Markowitz V, Metha T, Nelson KE, Parkhill J, Pitluck S, Qin X, Read TD, Schmutz J, Sozhamannan S, Sterk P, Strausberg RL, Sutton G, Thomson NR, Tiedje JM, Weinstock G, Wollam A, Genomic Standards Consortium Human Microbiome Project Jumpstart Consortium, Detter JC. 2009. Genomics. Genome project standards in a new era of sequencing. Science 326:236–237. 10.1126/science.1180614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marston DA, McElhinney LM, Ellis RJ, Horton DL, Wise EL, Leech SL, David D, de Lamballerie X, Fooks AR. 2013. Next generation sequencing of viral RNA genomes. BMC Genomics 14:444. 10.1186/1471-2164-14-444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed). 2012. Virus taxonomy. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 7. Woolhouse M, Scott F, Hudson Z, Howey R, Chase-Topping M. 2012. Human viruses: discovery and emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367:2864–2871. 10.1098/rstb.2011.0354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kircher M, Sawyer S, Meyer M. 2012. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40:e3. 10.1093/nar/gks632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sánchez AB, de la Torre JC. 2006. Rescue of the prototypic arenavirus LCMV entirely from plasmid. Virology 350:370-380. 10.1016/j.virol.2006.01.012 [DOI] [PubMed] [Google Scholar]
- 10. Domingo E, Martin V, Perales C, Grande-Perez A, Garcia-Arriaza J, Arias A. 2006. Viruses as quasispecies: biological implications. Curr. Top. Microbiol. Immunol. 299:51–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439:344–348. 10.1038/nature04388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. 2007. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome Res. 17:1195–1201. 10.1101/gr.6468307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Macalalad AR, Zody MC, Charlebois P, Lennon NJ, Newman RM, Malboeuf CM, Ryan EM, Boutwell CL, Power KA, Brackney DE, Pesko KN, Levin JZ, Ebel GD, Allen TM, Birren BW, Henn MR. 2012. Highly sensitive and specific detection of rare variants in mixed viral populations from massively parallel sequence data. PLoS Comput. Biol. 8:e1002417. 10.1371/journal.pcbi.1002417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roberts RJ, Carneiro MO, Schatz MC. 2013. The advantages of SMRT sequencing. Genome Biol. 14:405. 10.1186/gb-2013-14-6-405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anthony SJ, Epstein JH, Murray KA, Navarrete-Macias I, Zambrana-Torrelio CM, Solovyov A, Ojeda-Flores R, Arrigo NC, Islam A, Ali Khan S, Hosseini P, Bogich TL, Olival KJ, Sanchez-Leon MD, Karesh WB, Goldstein T, Luby SP, Morse SS, Mazet JA, Daszak P, Lipkin WI. 2013. A strategy to estimate unknown viral diversity in mammals. mBio 4:e00598-13. 10.1128/mBio.00598-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lipkin WI. 2013. The changing face of pathogen discovery and surveillance. Nat. Rev. Microbiol. 11:133–141. 10.1038/nrmicro2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nelson MI, Holmes EC. 2007. The evolution of epidemic influenza. Nat. Rev. Genet. 8:196–205. 10.1038/nrg2053 [DOI] [PubMed] [Google Scholar]
- 18. Palacios G, Tesh R, Travassos da Rosa A, Savji N, Sze W, Jain K, Serge R, Guzman H, Guevara C, Nunes MR, Nunes-Neto JP, Kochel T, Hutchison S, Vasconcelos PF, Lipkin WI. 2011. Characterization of the candiru antigenic complex (Bunyaviridae: phlebovirus), a highly diverse and reassorting group of viruses affecting humans in tropical America. J. Virol. 85:3811–3820. 10.1128/JVI.02275-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, Guan YJ, Butt KM, Wong KL, Chan KW, Lim W, Shortridge KF, Yuen KY, Peiris JS, Poon LL. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276–278. 10.1126/science.1087139 [DOI] [PubMed] [Google Scholar]
- 20. Chan JF, Lau SK, Woo PC. 2013. The emerging novel Middle East respiratory syndrome coronavirus: the “knowns” and “unknowns.” J. Formos. Med. Assoc. 112:372–381. 10.1016/j.jfma.2013.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang C, Wang J, Su W, Gao S, Luo J, Zhang M, Xie L, Liu S, Liu X, Chen Y, Jia Y, Zhang H, Ding H, He H. 2014. Relationship between domestic and wild birds in live poultry market and a novel human H7N9 virus in China. J. Infect. Dis. 209:34–37. 10.1093/infdis/jit478 [DOI] [PubMed] [Google Scholar]
- 22. Fancello L, Raoult D, Desnues C. 2012. Computational tools for viral metagenomics and their application in clinical research. Virology 434:162–174. 10.1016/j.virol.2012.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee E, Helt GA, Reese JT, Munoz-Torres MC, Childers CP, Buels RM, Stein L, Holmes IH, Elsik CG, Lewis SE. 2013. Web Apollo: a web-based genomic annotation editing platform. Genome Biol. 14:R93. 10.1186/gb-2013-14-8-r93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. 2011. Pseudomonas genome database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39:D596–D600. 10.1093/nar/gkq869 [DOI] [PMC free article] [PubMed] [Google Scholar]