Abstract
Objective
Previous studies have replicated the association of variants within FTO (fat mass- and obesity-associated) intron 1 with obesity and adiposity quantitative traits in populations of European ancestry. Non-European populations, however, have not been so intensively studied. The goal of this investigation was to examine the association of FTO single-nucleotide polymorphisms (SNPs), prominent in the literature in a multiethnic sample of non-Hispanic White American (n = 458), Hispanic American (n = 373) and African American (n = 288) subjects from the Insulin Resistance Atherosclerosis Study (IRAS). This cohort provides the unique ability to evaluate how variation within FTO influences measures of adiposity and glucose homeostasis in three different ethnicities, which were ascertained and examined using a common protocol.
Design
A total of 26 FTO SNPs were genotyped, including those consistently associated in the literature (rs9939609, rs8050136, rs1121980, rs1421085, rs17817449 and rs3751812), and tested for association with adiposity and glucose homeostasis traits.
Results
For the adiposity phenotypes, these and other SNPs were associated with body mass index (BMI) in both non-Hispanic Whites (P-values ranging from 0.015 to 0.048) and Hispanic Americans (P-values ranging from 7.1 × 10−6 to 0.027). In Hispanic Americans, four other SNPs (rs8047395, rs10852521, rs8057044 and rs8044769) still showed evidence of association after multiple comparisons adjustment (P-values ranging from 5.0 × 10−5 to 5.2 × 10−4). The historically associated BMI SNPs were not associated in the African Americans, but rs1108102 was associated with BMI (P-value of 5.4 × 10−4) after accounting for multiple comparisons. For glucose homeostasis traits, associations were seen with acute insulin response in non-Hispanic Whites and African Americans. However, all associations with glucose homeostasis measures were no longer significant after adjusting for multiple comparisons.
Conclusion
These results replicate the association of FTO intron 1 variants with BMI in non-Hispanic Whites and Hispanic Americans but show little evidence of association in African Americans, suggesting that the effect of FTO variants on adiposity phenotypes shows genetic heterogeneity dependent on ethnicity.
Keywords: FTO, association, adiposity traits, multiethnic sample, glucose homeostasis traits
Introduction
A group of highly correlated single-nucleotide polymorphisms (SNPs) in the first intron of the fat mass- and obesity-associated (FTO) gene originally showed evidence of association with obesity and adiposity-related quantitative traits in three separate studies of European-derived populations.1–3 Multiple additional studies in Europeans and European-derived cohorts have replicated the association of several of the variants in this region.4–11 It has, however, been difficult to replicate association of these SNPs in other ethnicities, including Asians12–14 and African Americans.3,15,16 In addition, only three studies have examined the association of FTO variants with adiposity phenotypes in a standardized manner in several ethnicities: the family-based Insulin Resistance Atherosclerosis Family Study (IRASFS; Hispanic and African Americans),16 the family-based GenNet study (European, Hispanic and African Americans),3 and the Women’s Health Initiative-Observational Study (European, Hispanic, African and Asians/Pacific Islander American women).15 To date, no study has performed an in depth investigation of the association of FTO variants with both adiposity and glucose homeostasis measures in a multiethnic cohort examined using a common protocol. The goal of this study was to replicate the association of prominent FTO intron 1 SNPs with adiposity phenotypes and to test whether the SNPs are associated with measures of glucose homeostasis, that is, insulin sensitivity and β-cell function in the non-Hispanic White Americans, Hispanic Americans and African Americans of the Insulin Resistance Atherosclerosis Study (IRAS), a multiethnic study in which subjects were ascertained, recruited and examined using a common protocol.
Materials and methods
Recruitment
The study design, recruitment and phenotyping in IRAS have been previously described.17 Briefly, US individuals of non-Hispanic White, Hispanic and African American descent were recruited to reflect an equal representation of glucose tolerance status (normal, impaired glucose tolerance and diabetic), ethnicity, sex and age (40–49, 50–59 and 60–69 years). This report includes 458 non-Hispanic White Americans, 373 Hispanic Americans and 288 African Americans who completed a frequently sampled intravenous glucose tolerance test, anthropometric measures, and who provided consent for genetic studies.
Adiposity and glucose homeostasis phenotypes
Measures of adiposity were obtained using standard methods and included height, weight, waist and hip circumferences. Glucose effectiveness, insulin sensitivity, acute insulin response (AIR) and disposition index (DI) were obtained using the frequently sampled intravenous glucose tolerance test with minimal model analyses (MINMOD).18,19 AIR was measured 8 min following glucose infusion as the mean insulin increment in plasma insulin concentration above the basal concentration. DI was calculated from the product of insulin sensitivity and AIR. Fasting plasma glucose and insulin are also available.
Genotyping
Genomic DNA was purified using PUREGENE DNA isolation kits (Gentra Inc., Minneapolis, MN, USA). Total genomic DNA was quantified using a fluorometric assay provided by a Hoefer DyNA Quant 200 fluorometer (Hoefer Pharmacia Biotech Inc., San Francisco, CA, USA). The DNA samples were diluted to a final concentration of 5 ng μ1−1. FTO intron 1 variants in the region surrounding the SNPs associated with adiposity measures in the literature (60 kb region) were chosen using YRI HapMap (www.hapmap.org) tagSNPs with an r2 threshold of 0.8 and minor allele frequency > 5%. Next, these HapMap tagSNPs were incorporated in the CEU HapMap tagSNPs in order to cover variation in Caucasians. Genotypes for 26 SNPs were ascertained using the iPlex MassARRAY SNP genotyping system (Sequenom Inc., San Diego, CA, USA), which utilizes mass tagging to differentiate alleles.20
Statistical analysis
Maximum likelihood estimates of allele frequencies, departures from Hardy-Weinberg equilibrium proportions and linkage disequilibrium (LD) statistics were calculated separately for non-Hispanic White Americans (n = 458), Hispanic Americans (n = 373) and African Americans (n = 288). Power calculations were performed to approximate the ability of the IRAS non-Hispanic White, Hispanic and African American cohorts to detect statistically significant differences in phenotypic values when compared with a previous report by Scuteri et al.3 In order to accurately reflect the power estimates for the six SNPs prominently associated in the literature, a range of allele frequencies were used, and power was calculated using the program QUANTO.21
In order to account for admixture in the Hispanic and African Americans of IRAS, two sets of ancestry informative markers were genotyped. For the IRAS African Americans, ancestral allele estimates were obtained from 70 ancestry informative markers genotyped in 44 Yorubans and 39 European Americans. For the IRAS Hispanic Americans, 56 ancestry informative markers selected from the literature were genotyped and ancestral allele frequencies were obtained from the Yoruba, European American and Chinese HapMap populations. Ancestral proportions and estimates were computed using the expectation-maximization algorithm as implemented in FRAPPE22 and the Bayesian method implemented in the program ADMIXMAP.23
Variants were tested for association with each quantitative phenotype using a linear regression model implemented in SAS (version 9.1.3, SAS, Cary, NC, USA). Diabetics were removed from the analysis of glucose homeostasis traits. For each phenotype, the two degree of freedom test of genotypic association was performed. In addition, three individual contrasts defined by a priori genetic models were computed: additive (as additional copies of the minor allele are inherited, the phenotypic effect significantly increases), dominant (one copy of the minor allele produces a significant phenotypic effect) and recessive (two copies of the minor allele are necessary to create a significant phenotypic effect). Association tests for measures of adiposity and glucose homeostasis were adjusted for admixture, age, gender and additionally for body mass index (BMI). When necessary, quantitative traits were transformed to approximate the distributional assumptions of the test and to minimize heterogeneity of variance.
A three-stage approach utilizing principle component analyses was used to adjust for multiple comparisons. This allowed for the estimation of the number of independent dimensions for each hypothesis while accounting for LD and trait correlations. The number of estimated independent dimensions for each hypothesis was used to determine a more precise hypothesis-specific Bonferroni adjustment.24 The first stage identified SNPs with historical association in FTO intron 1. These SNPs were not adjusted because of the strong a priori hypothesis that FTO variants are associated with BMI1,3,5,7,8,25 The second step computed a principal component analysis on the other SNPs in the region to determine how many independent dimensions exist (that is, accounting for LD) and applied a Bonferroni adjustment based on the number of independent dimensions for the trait BMI. The third step was designed to estimate the number of independent dimensions in the other phenotypes (that is, WAIST, WHR, SAT, VAT, VSR, fasting plasma glucose, fasting plasma insulin, insulin sensitivity, AIR and DI). This was then used to adjust: (1) the SNPs with historic association with BMI and (2) all remaining SNPs.
For SNPs prominent in the literature (for example, rs1421085, rs1121980, rs17817449, rs8050136, rs3751812 and rs9939609), a SNP-by-ethnicity interaction analysis for the association with BMI was computed. Specifically, a linear regression model was computed to test the SNP-by-ethnicity interaction, adjusting for admixture, age, gender, SNP and ethnicity. A global test of the SNP-by-ethnicity interaction (all ethnicities jointly) and the pairwise SNP-by-ethnicity pair interactions were computed (non-Hispanic Whites versus Hispanic Americans, non-Hispanic Whites versus African Americans and Hispanic Americans versus African Americans). Interaction plots for each of the BMI-associated SNPs were generated using the least square means for BMI as implemented in SAS.
Results
A total of 26 SNPs within FTO intron 1, the region focused on previous studies, were genotyped and tested for association with adiposity and glucose homeostasis traits. Demographic statistics for the IRAS cohorts are summarized in Table 1. Briefly, the non-Hispanic Whites, Hispanics and African Americans were similar in age and had comparable BMI, WAIST and WHR values. However, the non-Hispanic Whites were more insulin sensitive (insulin sensitivity = 2.37 ± 2.05) than both the Hispanic (1.92 ± 1.97) and African Americans (1.91 ± 1.88).
Table 1.
IRAS demographic summary of non-Hispanic White, Hispanic and African American participants
|
Non-Hispanic Whites
|
Hispanics
|
African Americans
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| N | Mean ± s.d. | Median | N | Mean ± s.d. | Median | N | Mean ± s.d. | Median | |
| Subjects | 458 | 373 | 289 | ||||||
| Demographics | |||||||||
| Age (years) | 458 | 56.2 ±8.2 | 57 | 373 | 55.1 ± 8.6 | 55 | 289 | 55.4 ± 8.3 | 56 |
| Female gender (%) | 238 | 52.0% | 218 | 58.5% | 170 | 58.8% | |||
| Type 2 Diabetes (%) | 98 | 21.4% | 84 | 22.5% | 95 | 32.9% | |||
| Obesity | |||||||||
| BMI (kg m−2) | 28.7 ± 5.7 | 27.6 | 28.9 ± 5.3 | 28.1 | 30.5 ± 5.9 | 29.3 | |||
| WAIST (cm) | 92.8 ± 13.5 | 92.4 | 92.1 ± 12.7 | 92.7 | 93.6 ± 13.1 | 92.4 | |||
| WHR | 0.872 ± 0.086 | 0.878 | 0.886 ± 0.089 | 0.885 | 0.856 ± 0.081 | 0.861 | |||
| Glucose homeostasis | |||||||||
| Fasting glucose (mg per 100 ml) | 99.1 ± 11.4 | 98 | 97.7 ± 10.7 | 96 | 101.9 ± 11.6 | 101 | |||
| Fasting insulin (μU ml−1) | 14.2 ± 11.9 | 11.5 | 17.1 ± 12.4 | 15 | 16.6 ± 14.8 | 14 | |||
| SG | 0.019 ± 0.008 | 0.019 | 0.02 ± 0.008 | 0.019 | 0.019 ± 0.009 | 0.018 | |||
| SI (× 10−5min−1 (pmoll−1)−1) | 2.37 ± 2.05 | 1.8 | 1.92 ± 1.97 | 1.38 | 1.91 ± 1.88 | 1.39 | |||
| AIR (pmoll−1) | 514 ± 395 | 415 | 732 ± 618 | 533 | 673 ± 510 | 553 | |||
| DI (SI × AIR; × 10−5min−1) | 992 ± 918 | 761 | 1140 ± 1172 | 761 | 1168 ± 1387 | 701 | |||
Abbreviations: AIR, acute insulin response; BMI, body mass index; DI, disposition index; IRAS, Insulin Resistance Atherosclerosis Study; SG, glucose effectiveness; SI, insulin sensitivity; WAIST, waist circumference; WHR, waist-to-hip ratio.
Power analysis was performed using the effect sizes found from Scuteri et al., with European Americans having β-values of 0.12 and Hispanic Americans having β-values of 0.15.3 These ethnic-specific effect sizes for prominent SNPs associated in the literature were used to estimate the power of the non-Hispanic White and Hispanic American IRAS cohorts using a range of allele frequencies. The allele frequency for the six prominent SNPs was about 40% in the non-Hispanic Whites and ranged between 25 and 30% in Hispanic Americans. Non-Hispanic Whites had 43% power to detect 0.12 s.d.s for variants with an allele frequency of 40%. Hispanic Americans had 43 and 47% power to detect 0.15 s.d.s for variants with an allele frequency of 25 and 30%, respectively. Power estimates for the African Americans were calculated from known European effect sizes in the literature.3 The six historically associated BMI SNPs had allele frequencies ranging between 8 and 30% in African Americans. To detect 0.12 s.d.s for variants with an allele frequency ranging from 8% to 42%, African Americans had 12–30% power.
Non-Hispanic White Americans
The non-Hispanic White association results under the best-fitting additive model for the FTO SNPs with adiposity and glucose homeostasis phenotypes are summarized in Table 2a. All of the SNPs historically associated with obesity and measures of adiposity (that is, rs8050136, rs9939609, rs1121980, rs1421085, rs17817449 and rs3751812) were located within the same LD block at the 3′ end of the FTO intron 1 (see Supplementary Figure 1). These SNPs, denoted by b, as well as other variants within this LD block, were associated with BMI (P-values ranging from 0.015 to 0.048) and AIR (P-values ranging from 0.012 to 0.048). The minor allele for those SNPs prominent in the literature was consistently associated with an increased BMI and AIR under the additive model (Supplementary Table 1). For example, individuals with one copy of the minor allele for the most significantly associated BMI SNP (rs1421085) had a 1.12kg m−2 increase in BMI, whereas adding an additional allele increased BMI by another 0.87kg m−2. Individuals with one copy of the minor allele for the most significantly associated AIR SNP (rs1121980) had a 48 pmoll−1 increase in AIR, whereas the addition of another minor allele increased AIR by another 136 pmoll−1. None of the non-a priori SNPs were statistically significant at the non-Hispanic White Bonferroni threshold of 0.001 for the non-a priori SNPs and phenotypes.
Table 2.
Association P-values of FTO SNPs with adiposity and glucose homeostasis phenotypes in the IRAS (a) non-Hispanic White Americans, (b) Hispanic Americans and (c) African Americans under the additive model
| SNP | Allelesa | MAF | Adjusted for age and gender BMI |
Adjusted for age, gender and BMI
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| WAIST | WHR | GFAST | SG | FINS | SI | AIR | DI | ||||
| (a) | |||||||||||
| rs7186637 | C/T | 0.19 | 0.41 | 0.66 | 0.85 | 0.97 | 0.83 | 0.16 | 0.29 | 0.42 | 0.78 |
| rs11643744 | A/G | 0.34 | 0.026 | 0.24 | 0.44 | 0.32 | 0.65 | 0.51 | 0.70 | 0.80 | 0.64 |
| rs9940700 | G/C | 0.19 | 0.28 | 0.69 | 0.86 | 0.80 | 0.73 | 0.19 | 0.45 | 0.47 | 0.66 |
| rs13334933 | A/G | 0.18 | 0.098 | 0.98 | 0.73 | 0.45 | 0.90 | 0.20 | 0.33 | 0.37 | 0.56 |
| rs16952517 | G/A | 0.10 | 0.16 | 0.10 | 0.27 | 0.38 | 0.01 | 0.89 | 0.22 | 0.34 | 0.055 |
| rs6499642 | C/T | 0.04 | 0.67 | 0.17 | 0.67 | 0.18 | 0.20 | 0.50 | 0.48 | 0.65 | 0.80 |
| rs4784323 | G/A | 0.35 | 0.91 | 0.23 | 0.24 | 0.81 | 0.39 | 0.44 | 0.39 | 0.26 | 0.82 |
| rs8047395 | G/A | 0.498 | 0.051 | 0.33 | 0.27 | 0.054 | 0.95 | 0.69 | 0.96 | 0.048 | 0.23 |
| rs1421085b | T/C | 0.42 | 0.015 | 0.15 | 0.25 | 0.0999 | 0.38 | 0.70 | 0.64 | 0.018 | 0.071 |
| rs10852521 | C/T | 0.496 | 0.048 | 0.44 | 0.56 | 0.026 | 0.92 | 0.63 | 0.79 | 0.012 | 0.0998 |
| rs12447107 | G/C | 0.02 | 0.38 | 0.13 | 0.50 | 0.87 | 0.025 | 0.81 | 0.10 | 0.42 | 0.46 |
| rs11075986 | C/G | 0.07 | 0.76 | 0.75 | 0.41 | 0.26 | 0.82 | 0.88 | 0.57 | 0.92 | 0.76 |
| rs2058908 | C/T | 0.29 | 0.21 | 0.098 | 0.093 | 0.79 | 0.84 | 0.94 | 1.00 | 0.77 | 0.99 |
| rs1121980b | G/A | 0.44 | 0.035 | 0.29 | 0.36 | 0.10 | 0.78 | 0.74 | 0.97 | 0.012 | 0.11 |
| rs8057044 | G/A | 0.49 | 0.044 | 0.44 | 0.75 | 0.071 | 0.79 | 0.37 | 0.71 | 0.058 | 0.14 |
| rs17817449b | T/G | 0.42 | 0.027 | 0.19 | 0.45 | 0.16 | 0.43 | 0.68 | 0.53 | 0.043 | 0.061 |
| rs8063946 | C/T | 0.05 | 0.26 | 0.64 | 0.44 | 0.58 | 0.72 | 0.14 | 0.092 | 0.064 | 0.86 |
| rs8050136b | C/A | 0.42 | 0.028 | 0.19 | 0.44 | 0.17 | 0.44 | 0.67 | 0.53 | 0.047 | 0.065 |
| rs3751812b | G/T | 0.41 | 0.021 | 0.17 | 0.40 | 0.16 | 0.47 | 0.71 | 0.58 | 0.046 | 0.074 |
| rs9939609b | T/A | 0.42 | 0.021 | 0.18 | 0.47 | 0.18 | 0.52 | 0.75 | 0.53 | 0.043 | 0.057 |
| rs7199182 | A/G | 0.003 | 0.50 | 0.83 | 0.73 | 0.67 | 0.64 | 0.96 | 0.54 | 0.82 | 0.49 |
| rs13337696 | C/T | 0.02 | 0.32 | 0.069 | 0.21 | 0.70 | 0.28 | 0.82 | 0.18 | 0.30 | 0.72 |
| rs7204609 | T/C | 0.02 | 0.25 | 0.79 | 0.23 | 0.69 | 0.22 | 0.18 | 0.49 | 0.26 | 0.14 |
| rs8044769 | C/T | 0.47 | 0.074 | 0.29 | 0.71 | 0.16 | 0.84 | 0.85 | 0.68 | 0.045 | 0.30 |
| (b) | |||||||||||
| rs7186637 | C/T | 0.161 | 0.023 | 0.50 | 0.86 | 0.55 | 0.93 | 0.66 | 0.81 | 0.74 | 0.82 |
| rs16952508 | A/G | 0.001 | 0.53 | 0.31 | 0.28 | 0.17 | 0.51 | 0.65 | 0.87 | 0.14 | 0.19 |
| rs1108102 | T/A | 0.003 | 0.29 | 0.40 | 0.083 | 0.25 | 0.82 | 0.34 | 0.53 | 0.36 | 0.20 |
| rs11643744 | A/G | 0.408 | 0.73 | 0.89 | 0.85 | 0.85 | 0.49 | 0.94 | 0.68 | 0.62 | 0.33 |
| rs9940700 | G/C | 0.164 | 0.063 | 0.59 | 0.71 | 0.43 | 0.66 | 0.78 | 0.74 | 0.90 | 0.88 |
| rs13334933 | A/G | 0.162 | 0.027 | 0.43 | 0.89 | 0.48 | 0.95 | 0.73 | 0.88 | 0.95 | 0.92 |
| rs16952517 | G/A | 0.104 | 0.70 | 0.34 | 0.44 | 0.58 | 0.094 | 0.51 | 0.41 | 0.90 | 0.20 |
| rs6499642 | C/T | 0.028 | 0.62 | 0.62 | 0.19 | 0.54 | 0.22 | 0.56 | 0.15 | 0.70 | 0.26 |
| rs4784323 | G/A | 0.322 | 0.056 | 0.44 | 0.055 | 0.49 | 0.34 | 0.98 | 0.25 | 0.49 | 0.31 |
| rs8047395 | G/A | 0.427 | 4.7 × 10−4 | 0.42 | 0.27 | 0.60 | 0.028 | 0.59 | 0.37 | 0.020 | 0.027 |
| rs1421085b | T/C | 0.237 | 1.5 × 10−4 | 0.13 | 0.011 | 0.77 | 0.085 | 0.71 | 0.50 | 0.80 | 0.59 |
| rs10852521 | T/C | 0.404 | 5.2 × 10−4 | 0.61 | 0.25 | 0.91 | 0.21 | 0.85 | 0.72 | 0.075 | 0.18 |
| rs12447107 | G/C | 0.053 | 0.33 | 0.66 | 0.98 | 0.85 | 0.38 | 0.096 | 0.26 | 0.15 | 0.92 |
| rs11075986 | C/G | 0.113 | 0.27 | 0.33 | 0.12 | 0.93 | 0.85 | 0.62 | 0.71 | 0.22 | 0.26 |
| rs2058908 | C/T | 0.339 | 0.059 | 0.54 | 0.14 | 0.48 | 0.25 | 0.70 | 0.24 | 0.47 | 0.17 |
| rs1121980b | G/A | 0.303 | 3.6 × 10−3 | 0.13 | 0.012 | 0.65 | 0.074 | 0.90 | 0.64 | 0.20 | 0.24 |
| rs8057044 | G/A | 0.352 | 5.0 × 10−5 | 0.60 | 0.32 | 0.49 | 0.019 | 0.58 | 0.54 | 0.017 | 0.065 |
| rs17817449b | T/G | 0.255 | 6.0 × 10−5 | 0.11 | 0.031 | 0.58 | 0.031 | 0.57 | 0.37 | 0.51 | 0.27 |
| rs8063946 | C/T | 0.125 | 0.76 | 0.65 | 0.58 | 0.65 | 0.37 | 0.56 | 0.97 | 0.39 | 0.49 |
| rs8050136b | C/A | 0.255 | 6.9 × 10−5 | 0.10 | 0.026 | 0.50 | 0.022 | 0.56 | 0.37 | 0.43 | 0.23 |
| rs3751812b | G/T | 0.234 | 7.1 × 10−6 | 0.32 | 0.057 | 0.81 | 0.077 | 0.74 | 0.74 | 0.69 | 0.70 |
| rs9939609b | T/A | 0.257 | 1.8 × 10−5 | 0.19 | 0.052 | 0.55 | 0.022 | 0.63 | 0.54 | 0.45 | 0.34 |
| rs7199182 | A/G | 0.031 | 0.74 | 0.30 | 0.69 | 0.14 | 0.23 | 0.45 | 0.34 | 0.53 | 0.13 |
| rs13337696 | C/T | 0.043 | 0.87 | 0.31 | 0.86 | 0.46 | 0.31 | 0.28 | 0.27 | 0.31 | 0.86 |
| rs7204609 | T/C | 0.093 | 0.67 | 0.38 | 0.87 | 0.94 | 0.60 | 0.85 | 0.84 | 0.98 | 0.49 |
| rs8044769 | T/C | 0.431 | 2.5 × 10−4 | 0.80 | 0.40 | 0.77 | 0.07 | 0.74 | 0.89 | 0.033 | 0.15 |
| (c) | |||||||||||
| rs7186637 | C/T | 0.405 | 0.12 | 0.74 | 0.53 | 0.52 | 0.33 | 0.45 | 0.19 | 0.27 | 0.050 |
| rs16952508 | A/G | 0.042 | 0.013 | 0.64 | 0.62 | 0.91 | 0.12 | 0.75 | 0.61 | 0.51 | 0.70 |
| rs1108102 | T/A | 0.125 | 5.4 × 10−4 | 0.76 | 0.46 | 0.32 | 0.028 | 0.65 | 0.27 | 0.045 | 0.048 |
| rs11643744 | A/G | 0.119 | 0.37 | 0.33 | 0.18 | 0.21 | 0.94 | 4.6 × 10−3 | 0.21 | 0.92 | 0.20 |
| rs9940700 | C/G | 0.401 | 0.32 | 0.94 | 0.51 | 0.083 | 0.99 | 0.21 | 0.46 | 0.70 | 0.25 |
| rs13334933 | G/A | 0.397 | 0.59 | 0.77 | 0.71 | 0.11 | 0.93 | 0.20 | 0.56 | 0.80 | 0.33 |
| rs16952517 | G/A | 0.194 | 0.84 | 0.18 | 0.41 | 0.96 | 0.43 | 0.27 | 0.61 | 0.39 | 0.63 |
| rs6499642 | C/T | 0.285 | 0.18 | 0.96 | 0.83 | 0.16 | 0.92 | 0.54 | 0.98 | 0.57 | 0.81 |
| rs4784323 | G/A | 0.13 | 0.46 | 0.62 | 0.98 | 0.54 | 0.13 | 0.39 | 0.64 | 0.32 | 0.28 |
| rs8047395 | A/G | 0.34 | 0.011 | 0.58 | 0.56 | 0.87 | 0.81 | 0.25 | 0.43 | 0.71 | 0.39 |
| rs1421085b | T/C | 0.08 | 0.24 | 0.34 | 0.46 | 0.39 | 0.028 | 0.91 | 0.61 | 0.24 | 0.64 |
| rs10852521 | C/T | 0.316 | 4.4 × 10−3 | 0.65 | 0.32 | 0.36 | 0.90 | 0.51 | 0.44 | 0.73 | 0.16 |
| rs12447107 | G/C | 0.27 | 0.54 | 0.61 | 0.27 | 0.90 | 0.73 | 0.33 | 0.65 | 0.59 | 0.64 |
| rs11075986 | C/G | 0.279 | 0.11 | 0.33 | 0.37 | 0.61 | 0.90 | 0.88 | 0.36 | 0.21 | 0.19 |
| rs2058908 | C/T | 0.065 | 0.15 | 0.10 | 0.36 | 0.94 | 0.32 | 0.19 | 0.60 | 7.0 × 10−3 | 0.21 |
| rs16952524 | C/A | 0.051 | 0.41 | 0.74 | 0.29 | 0.034 | 0.64 | 0.71 | 0.86 | 0.042 | 0.30 |
| rs1121980b | G/A | 0.424 | 0.43 | 0.38 | 0.42 | 0.45 | 0.47 | 0.37 | 0.90 | 0.042 | 0.50 |
| rs8057044 | A/G | 0.314 | 0.039 | 0.48 | 0.45 | 0.49 | 0.87 | 0.57 | 0.54 | 0.97 | 0.35 |
| rs17817449b | T/G | 0.33 | 0.55 | 0.090 | 0.094 | 0.15 | 0.37 | 0.74 | 0.90 | 0.12 | 0.70 |
| rs8063946 | C/T | 0.349 | 0.45 | 0.44 | 0.83 | 0.12 | 0.48 | 0.58 | 0.76 | 0.074 | 0.20 |
| rs8050136b | C/A | 0.391 | 0.20 | 0.18 | 0.32 | 0.12 | 0.24 | 0.78 | 0.45 | 0.072 | 0.93 |
| rs3751812b | G/T | 0.083 | 0.22 | 0.33 | 0.42 | 0.29 | 0.084 | 0.94 | 0.64 | 0.29 | 0.64 |
| rs9939609b | T/A | 0.418 | 0.71 | 0.25 | 0.48 | 0.18 | 0.056 | 0.41 | 0.40 | 0.033 | 0.97 |
| rs7199182 | A/G | 0.26 | 0.85 | 0.25 | 0.30 | 0.33 | 0.71 | 0.80 | 0.33 | 0.28 | 0.73 |
| rs13337696 | C/T | 0.054 | 0.43 | 0.37 | 0.67 | 0.74 | 0.44 | 0.40 | 0.25 | 0.45 | 0.14 |
| rs7204609 | T/C | 0.344 | 0.89 | 0.57 | 0.53 | 0.34 | 0.85 | 0.17 | 0.44 | 0.58 | 0.80 |
| rs8044769 | C/T | 0.27 | 2.4 × 10−3 | 0.71 | 0.50 | 0.66 | 0.78 | 0.54 | 0.98 | 0.80 | 0.73 |
Abbreviations: AIR, acute insulin response; BMI, body mass index; DI, disposition index; FINS, fasting insulin; GFAST, fasting glucose; IRAS, Insulin Resistance Atherosclerosis Study; MAF, minor allele frequency; SG glucose effectiveness; SI, insulin sensitivity; SNP, single-nucleotide polymorphism; WAIST, waist circumference; WHR, waist-to-hip ratio.
Major/minor allele.
SNPs prominent in the literature. Numbers in bold indicate P-value ≤0.05 and numbers in italics indicate P-value ≤0.1.
Hispanic Americans
The Hispanic American association analysis results for FTO variants and adiposity and glucose homeostasis phenotypes under the additive model are depicted in Table 2b. All of the historically associated SNPs clustered in the second LD block near the 3′ end of FTO intron 1, except for rs9939609 (see Supplementary Figure 2). Variants within this LD block were statistically significantly associated with BMI (P-values ranging from 7.1 × 10−6 to 3.6 × 10−3) and WHR (P-values ranging from 0.011 to 0.031). The minor allele for those SNPs prominent in the literature (SNPs denoted with asterisks) was associated with an increased BMI under the additive model (Supplementary Table 1). Individuals with one copy of the minor allele for the most highly associated SNP (rs3751812) had a 1.4kg m−2 increase in BMI, and the addition of another allele caused an additional 3.8kg m−2 increase. Four SNPs (rs8047395, rs10852521, rs8057044 and rs8044769) remained statistically significant (P-values ranging from 5.0 × 10−5 to 5.2 × 10−4) at the Hispanic American Bonferroni threshold of 9.1 × 10−4 for the non-a priori SNPs and phenotypes.
The historically associated BMI SNPs were not associated with measures of glucose homeostasis; however, other SNPs (rs8047395, rs10852521, rs8057044 and rs8044769) within this LD block were trending or marginally associated with AIR (P-values ranging from 0.017 to 0.075) and DI (P-values of 0.027 and 0.065; Table 2b). These SNPs were no longer associated after accounting for multiple comparisons.
African Americans
The African American association analysis results (Table 2c) of FTO variants with adiposity and glucose homeostasis phenotypes under the additive model were not consistent with the results found for the non-Hispanic Whites and Hispanic Americans. The African Americans also had a different LD pattern, with prominent SNPs in the literature, except rs1421085, clustered in the fourth and fifth LD blocks (see Supplementary Figure 3). None of the SNPs prominent in the literature were associated with BML. However, there were four other moderately correlated SNPs (r2 between 0.3 and 0.6) within the third and fourth LD blocks that were associated with BMI (rs8047395, rs10852521, rs8057044 and rs8044769; with P-values ranging from 2.4 × 10−3 to 0.039). In addition, two SNPs (rs16952508 and rs1108102) at the 5′ end of intron 1 were associated with BMI (P-values of 0.013 and 5.4 × 10−4, respectively). These two SNPs were not correlated with the historically associated BMI SNPs. Several SNPs were associated or trending towards association with AIR (P-values ranging from 7.0 × 10−3 to 0.074) in the LD blocks three and four, including three of the SNPs prominent in the literature (Table 2c). The effects of SNPs on measures of BMI and AIR are illustrated in Supplementary Table 1. Except for the association of rs1108102 with BMI noted above, none of the non-a priori SNPs reached the African American Bonferroni threshold of statistical significance of 6.0 × 10−4 for either adiposity or glucose homeostasis measures.
An SNP-by-ethnicity interaction analysis was performed to address the lack of association of historically associated FTO intron 1 variants with BMI in the African American cohort (Table 3). The results of this analysis indicated that there is statistically significant evidence that the phenotypic effect of the SNP on BMI differs by ethnicity. This was seen for all of the SNPs with historic BMI association (P-values ranging from 5.2 × 10−4 to 0.035) when comparing all ethnicities. The effect of ethnicity on BMI in African Americans was more statistically significant when compared with Hispanic Americans (P-values ranging from 1.0 × 10−4 to 0.012) than when compared with non-Hispanic Whites (P-values ranging from 0.015 to 0.084). The difference between non-Hispanic Whites and Hispanic Americans was not significant for all SNPs, except for trending towards association with rs3751812 (P-value of 0.075). Plotting the BMI genotypic means for each ethnicity clearly illustrates the differences between the cohorts (Figure 1). African Americans had a clear shift in the mean of BMI by genotype when compared with non-Hispanic Whites and Hispanic Americans.
Table 3.
FTO SNP-by-ethnicity interaction P-values under the additive model comparing the IRAS cohort for SNPs historically associated with BMI
|
Adjusted for age, gender and admixturea
|
||||
|---|---|---|---|---|
| SNP × ethnicity P-value for BMI | ||||
| SNP | All ethnicities | NHW vs HA | NHW vs AA | HA vs AA |
| rs1421085 | 2.7 × 10−3 | 0.23 | 0.015 | 6.9 × 10−4 |
| rs1121980 | 0.035 | 0.46 | 0.051 | 0.012 |
| rs17817449 | 8.1 × 10−3 | 0.13 | 0.065 | 1.9 × 10−3 |
| rs8050136 | 1.5 × 10−3 | 0.14 | 0.018 | 3.4 × 10−4 |
| rs3751812 | 5.2 × 10−4 | 0.075 | 0.016 | 1.0 × 10−4 |
| rs9939609 | 7.8 × 10−3 | 0.10 | 0.084 | 1.8 × 10−3 |
Abbreviations: AA, African Americans; BMI, body mass index; HA, Hispanic Americans; IRAS, Insulin Resistance Atherosclerosis Study; NHW, non-Hispanic White Americans; SNP, single-nucleotide polymorphism.
Admixture adjustment uses separate ancestry estimates for Hispanics and African Americans as a covariate in the analysis. Numbers in bold indicate P-value ≤0.05 and numbers in italics indicate P-value ≤0.1.
Figure 1.
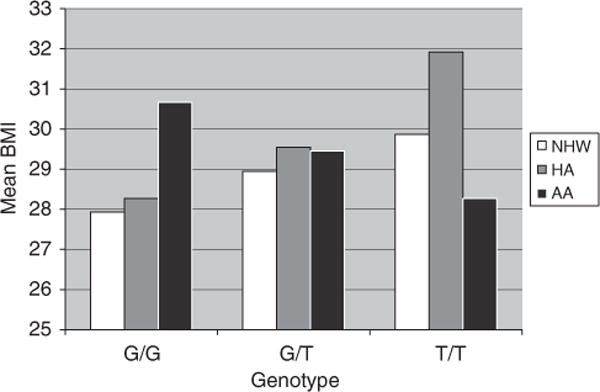
Genotypic means for the SNP rs3751812 for BMI in the Non-Hispanic White Americans, Hispanic Americans and African Americans of IRAS. Mean BMI for each rs3751812 genotype for the IRAS cohorts, where G is the major allele and T is the minor allele. AA, African Americans; BMI, body mass index; HA, Hispanic Americans; IRAS, Insulin Resistance Atherosclerosis Study; NHW, non-Hispanic White Americans.
Discussion
In the IRAS cohorts, 26 SNPs were analyzed for association with measures of adiposity and glucose homeostasis. These SNPs included variants prominent in the literature, rs9939609,1 rs8050136,4,6 and rs1121980, rs1421085, rs17817449 and rs3751812.2
Non-Hispanic White Americans
Non-Hispanic Whites illustrated nominal association with all of the historically associated FTO intron 1 SNPs with BMI (P-values ranging from 0.015 to 0.048). However, the other SNPs in this region did not pass the modified Bonferroni correction for multiple comparisons. Other studies have consistently replicated the association of one or more of these SNPs in European-derived populations1,2,5,7–10 and European Americans.3,11 The minor allele frequencies for these SNPs in the non-Hispanic Whites (41–44%) were comparable with previous reports (39–49%).1–4,6 In addition, the magnitude and direction of the effects for BMI (0.14–0.16) in the IRAS non-Hispanic Whites were comparable with those found in the literature (0.08–0.19).1–3
FTO intron 1 variants also showed evidence of association with AIR (P-values ranging from 0.012 to 0.048), although these SNPs were no longer significant after accounting for multiple comparisons. Several reports in Europeans have shown modest association with AIR. A cohort of Swedish obese children showed trending towards association with AIR calculated using the minimal model approach.26 Two other studies showed borderline significant associations with oral glucose tolerance test (OGTT)-derived measures for AIR (BIGTT-AIR).5,27 However, these studies differ from the present study because the glucose homeostasis measures were not adjusted for BMI.
Hispanic Americans
The IRAS Hispanic Americans showed stronger evidence of association between FTO intron 1 SNPs and BMI (P-values ranging from 7.1 × 10−6 to 3.6 × 10−3) than two previously studied Hispanic populations.3,15 These results are consistent with our previously described results from a Hispanic sample in the IRASFS.16 In addition, four moderately to highly correlated SNPs (r2 ranging from 0.68 to 0.9) were still significantly associated with BMI after adjustment for multiple comparisons. These SNPs were moderately correlated (r2 from 0.37 to 0.74) with the six historically associated SNPs in non-Hispanic White populations. This suggests that these SNPs are most likely in LD with the causal variant, which is also correlated with the historically associated BMI SNPs. Minor allele frequencies in the Hispanic Americans of IRAS (23–30%) were comparable with frequencies seen in other Hispanic populations (20–30%),3,15,16 but slightly lower than those observed in European-ancestry populations (39–49%).1–4,6 The magnitude and direction of the effects of the historically associated SNPs on BMI (0.21–0.35) were higher than those reported in the literature for Hispanic and Europeans (0.08–0.19).1–3
For the glucose homeostasis measures in the IRAS Hispanic Americans, there were several SNPs trending or associated with AIR (P-values ranging from 0.017 to 0.075) and DI (P-values of 0.027 and 0.065). However, after multiple comparison adjustment, these associations were no longer significant. The only other studies of FTO and glucose homeostasis in Hispanics have been with our IRASFS. One study in the IRASFS Hispanic Americans showed no evidence of association of rs8050136 with glucose homeostasis measures.28 The other more in-depth study of FTO intron 1 variants illustrated SNPs associated with AIR and DI in the same region as rs8050136, but there was little overlap between this study and our IRAS investigation.16 Only one SNP (rs10852521) showed consistent results, with trending towards association with AIR in both the IRAS Hispanics (P-value of 0.075) and the IRASFS Hispanic Americans (P-value of 0.056).16
African Americans
We failed to replicate the historic association of FTO intron 1 SNPs in the IRAS African Americans. However, there were five variants (rs16952508, rs1108102, rs8047395, rs10852521, rs8057044 and rs8044769) in the FTO intron 1 region that were associated with BMI (P-values ranging from 5.4 × l0−4 to 0.039). Few previous studies have investigated the role of FTO SNPs in African Americans. Our group has found nominal associations with BMI for the variants rs8050136 and rs9939609.16 Two studies have reported evidence of association with BMI in African American children, one with rs3751812,29 and the other with rs8057044.30 Another study in adults has also shown association of rs3751812 and an additional SNP, rs9941349.31 However, three other studies were unable to replicate association with one or more of the historically associated SNPs in African Americans3,15 and Africans.32
In this study, the five SNPs that were associated with BMI were not highly correlated (r2 ranging from 0.04 to 0.34) with those variants historically associated in the literature. Our recent study in the IRASFS African Americans illustrated that three of these five SNPs (rs8047395, rs10852521 and rs8057044) were associated with BMI (P-values ranging from 0.024 to 0.045) under the dominant model, with borderline significant association with BMI (P-values ranging from 0.053 to 0.10) under the two degree of freedom model, but were not associated under the additive model.16 None of these SNPs were associated after adjustment for multiple comparisons. There was only one SNP that passed the multiple comparisons threshold, rs1108102. This SNP had very low correlation with the historically associated SNPs (r2 ranging from 0 to 0.06). However, this SNP was not associated in our study in IRASFS African Americans.16 Further replication in other African American cohorts is needed to determine whether this SNP may influence BMI independently of the other historically associated SNPs.
The lack of association of SNPs prominent in the literature within the IRAS African Americans could be because of differences in the genetic structure between ethnicities or a smaller sample size (n = 288). Although there was limited power (12–30%) to detect association between FTO intron 1 SNPs in the African Americans, a SNP-by-ethnicity interaction also illustrates that there is statistical evidence that the historically associated SNPs’ effect on BMI differs depending on the ethnicity. This indicates that African Americans may truly have a different genetic architecture when compared with other ethnicities, which would explain the lack association in the IRAS African Americans as well as the lack of consistency of association studies in the literature for SNPs historically associated with BMI.3,29
The value of comparative studies: multiethnic samples
This study illustrates the potential power of genetic analysis in multiethnic cohorts. The different evolutionary histories of these populations have resulted in differences in genetic structure between ethnically diverse populations. Thorough analysis of other ethnicities can also identify ethnic-specific patterns of association.33,34 The IRAS, the focus of work here, is especially appropriate as ascertainment and recruitment were performed using a common research design. Studies in African Americans and Hispanic Americans are especially relevant as they have the highest rates of obesity35 and other diseases, such as type 2 diabetes.36 African Americans also have smaller LD blocks when compared with other ethnicities,37 resulting in smaller areas to look for functional or causal variants and making them an ideal population to narrow in on regions associated with disease. Examining multiethnic cohorts allows for direct comparison of LD patterns and allele frequencies of SNPs within candidate genes, and can possibly result in the identification of other causal variants.38 We have demonstrated this approach with comparative LD mapping in the TCF7L2 gene and diabetes.39 In addition, ‘comparative genomics’ approaches can provide insight into the origins of disease risk in populations. For example, we have shown that polymorphisms that are compellingly associated with type 2 diabetes in European-derived populations show, at best, little evidence for association in African Americans.40 These results, however, are balanced by the observation that multiple risk SNPs in Europeans approach being monomorphic, or are monomorphic, for the risk allele in African populations. Thus, variation in one ethnicity likely provides information about the origins of risk in a second population of a very different structure.
The lack of association of FTO intron 1 SNPs with BMI in the African Americans in this study may be due, in part, to differences in body composition between ethnicities. BMI, although the most popular trait for association with obesity, is highly correlated with both fat mass and lean mass. Studies have shown that at similar BMI values, African Americans have higher fat-free mass (bone mineral density and skeletal muscle mass) than non-Hispanic Whites.41 In addition at similar BMI levels, African Americans carry less visceral fat when compared with non-Hispanic Whites and Hispanic Americans.42 These differences suggest that ethnic-specific cutoffs are necessary for current obesity measures in relation to disease.43 This study also illustrates the importance of being mindful of ethnic differences in phenotypes when testing for genetic association.
Conclusion
This study investigated the association of 26 FTO intron 1 variants with measures of adiposity and glucose homeostasis in a multiethnic cohort. Standard ascertainment and examination of non-Hispanic White, Hispanic and African American subjects allowed for a unique opportunity to compare the association results between ethnicities and reduced potential bias.
For glucose homeostasis traits, non-Hispanic Whites showed nominal evidence of association of AIR with SNPs prominent in the literature. African American showed less consistent association with AIR, with only three of the six SNPs illustrating marginal evidence of association. Hispanic Americans showed no association with glucose homeostasis traits for these SNPs. Accounting for multiple comparisons removed the evidence of association with glucose homeostasis. Further studies are necessary to determine whether FTO intron 1 SNPs have an effect on glucose homeostasis traits independent of BML.
Consistent with previous studies that included non-Hispanic Whites, Hispanics and African Americans, we replicated the association of FTO variants with BMI in the non-Hispanic White and Hispanic Americans, but not in African Americans. In addition, in Hispanic Americans, we identified four variants that were still significant after accounting for multiple comparisons. These results add to the growing evidence that common variants in intron 1 of FTO predispose non-Hispanic Whites and Hispanic Americans to obesity. This study also suggests that the genetic architecture of obesity related to FTO is different in African Americans.
Supplementary Material
Acknowledgments
This research was supported in part by NIH grants HL060894, HL060931, HL060944, HL061019 and HL061210. We acknowledge the support of the Wake Forest University Health Sciences Center for Public Health Genomics.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on International Journal of Obesity website (http://www.nature.com/ijo)
References
- 1.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 3.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreasen CH, Stender-Petersen KL, Mogensen MS, Torekov SS, Wegner L, Andersen G, et al. Low physical activity accentuates the effect of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes. 2008;57:95–101. doi: 10.2337/db07-0910. [DOI] [PubMed] [Google Scholar]
- 6.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pascoe L, Tura A, Patel SK, Ibrahim IM, Ferrannini E, Zeggini E, et al. Common variants of the novel type 2 diabetes genes CDKAL1 and HHEX/IDE are associated with decreased pancreatic beta-cell function. Diabetes. 2007;56:3101–3104. doi: 10.2337/db07-0634. [DOI] [PubMed] [Google Scholar]
- 8.Peeters A, Beckers S, Verrijken A, Roevens P, Peeters P, Van Gaal L, et al. Variants in the FTO gene are associated with common obesity in the Belgian population. Mol Genet Metab. 2008;93:481–484. doi: 10.1016/j.ymgme.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Haupt A, Thamer C, Machann J, Kirchhoff K, Stefan N, Tschritter O, et al. Impact of variation in the FTO gene on whole body fat distribution, ectopic fat, and weight loss. Obesity (Silver Spring) 2008;16:1969–1972. doi: 10.1038/oby.2008.283. [DOI] [PubMed] [Google Scholar]
- 10.Do R, Bailey SD, Desbiens K, Belisle A, Montpetit A, Bouchard C, et al. Genetic variants of FTO influence adiposity, insulin sensitivity, leptin levels, and resting metabolic rate in the Quebec Family Study. Diabetes. 2008;57:1147–1150. doi: 10.2337/db07-1267. [DOI] [PubMed] [Google Scholar]
- 11.Hunt SC, Stone S, Xin Y, Scherer CA, Magness CL, Iadonato SP, et al. Association of the FTO gene with BMI. Obesity (Silver Spring) 2008;16:902–904. doi: 10.1038/oby.2007.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Wu Y, Loos RJ, Hu FB, Liu Y, Wang J, et al. Variants in the fat mass- and obesity-associated (FTO) gene are not associated with obesity in a Chinese Han population. Diabetes. 2008;57:264–268. doi: 10.2337/db07-1130. [DOI] [PubMed] [Google Scholar]
- 13.Horikoshi M, Hara K, Ito C, Shojima N, Nagai R, Ueki K, et al. Variations in the HHEX gene are associated with increased risk of type 2 diabetes in the Japanese population. Diabetologia. 2007;50:2461–2466. doi: 10.1007/s00125-007-0827-5. [DOI] [PubMed] [Google Scholar]
- 14.Al-Attar SA, Pollex RL, Ban MR, Young TK, Bjerregaard P, Anand SS, et al. Association between the FTO rs9939609 polymorphism and the metabolic syndrome in a non-Caucasian multi-ethnic sample. Cardiovasc Diabetol. 2008;7:5. doi: 10.1186/1475-2840-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song Y, You NC, Hsu YH, Howard BV, Langer RD, Manson JE, et al. FTO polymorphisms are associated with obesity but not diabetes risk in postmenopausal women. Obesity (Silver Spring) 2008;16:2472–2480. doi: 10.1038/oby.2008.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wing MR, Ziegler J, Langefeld CD, Ng MC, Haffner SM, Norris JM, et al. Analysis of FTO gene variants with measures of obesity and glucose homeostasis in the IRAS Family Study. Hum Genet. 2009;125:615–626. doi: 10.1007/s00439-009-0656-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagenknecht LE, Mayer EJ, Rewers M, Haffner S, Selby J, Borok GM, et al. The insulin resistance atherosclerosis study (IRAS) objectives, design, and recruitment results. Ann Epidemiol. 1995;5:464–472. doi: 10.1016/1047-2797(95)00062-3. [DOI] [PubMed] [Google Scholar]
- 18.Pacini G, Bergman RN. MINMOD: a computer program to calculate insulin sensitivity and pancreatic responsivity from the frequently sampled intravenous glucose tolerance test. Comput Methods Programs Biomed. 1986;23:113–122. doi: 10.1016/0169-2607(86)90106-9. [DOI] [PubMed] [Google Scholar]
- 19.Bergman RN, Finegood DT, Ader M. Assessment of insulin sensitivity in vivo. Endocr Rev. 1985;6:45–86. doi: 10.1210/edrv-6-1-45. [DOI] [PubMed] [Google Scholar]
- 20.Buetow KH, Edmonson M, MacDonald R, Clifford R, Yip P, Kelley J, et al. High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc Natl Acad Sci USA. 2001;98:581–584. doi: 10.1073/pnas.021506298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gauderman WJ. Sample size requirements for matched case-control studies of gene-environment interaction. Stat Med. 2002;21:35–50. doi: 10.1002/sim.973. [DOI] [PubMed] [Google Scholar]
- 22.Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 23.McKeigue PM, Carpenter JR, Parra EJ, Shriver MD. Estimation of admixture and detection of linkage in admixed populations by a Bayesian approach: application to African-American populations. Ann Hum Genet. 2000;64:171–186. doi: 10.1017/S0003480000008022. [DOI] [PubMed] [Google Scholar]
- 24.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32:361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 25.Hinney A, Nguyen TT, Scherag A, Friedel S, Bronner G, Muller TD, et al. Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PLoS ONE. 2007;2:e1361. doi: 10.1371/journal.pone.0001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobsson JA, Danielsson P, Svensson V, Klovins J, Gyllensten U, Marcus C, et al. Major gender difference in association of FTO gene variant among severely obese children with obesity and obesity related phenotypes. Biochem Biophys Res Commun. 2008;368:476–482. doi: 10.1016/j.bbrc.2008.01.087. [DOI] [PubMed] [Google Scholar]
- 27.Kring SI, Holst C, Zimmermann E, Jess T, Berentzen T, Toubro S, et al. FTO gene associated fatness in relation to body fat distribution and metabolic traits throughout a broad range of fatness. PLoS ONE. 2008;3:e2958. doi: 10.1371/journal.pone.0002958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmer ND, Goodarzi MO, Langefeld CD, Ziegler J, Norris JM, Haffner SM, et al. Quantitative trait analysis of type 2 diabetes susceptibility loci identified from whole genome association studies in the Insulin Resistance Atherosclerosis Family Study. Diabetes. 2008;57:1093–1100. doi: 10.2337/db07-1169. [DOI] [PubMed] [Google Scholar]
- 29.Grant SF, Li M, Bradfield JP, Kim CE, Annaiah K, Santa E, et al. Association analysis of the FTO gene with obesity in children of Caucasian and African ancestry reveals a common tagging SNP. PLoS ONE. 2008;3:e1746. doi: 10.1371/journal.pone.0001746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bollepalli S, Dolan LM, Deka R, Martin LJ. Association of FTO gene variants with adiposity in African-American adolescents. Obesity (Silver Spring) 2010;18:1959–1963. doi: 10.1038/oby.2010.82. [DOI] [PubMed] [Google Scholar]
- 31.Hassanein MT, Lyon HN, Nguyen TT, Akylbekova EL, Waters K, Lettre G, et al. Fine mapping of the association with obesity at the FTO locus in African-derived populations. Hum Mol Genet. 2010;19:2907–2916. doi: 10.1093/hmg/ddq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hennig BJ, Fulford AJ, Sirugo G, Rayco-Solon P, Hattersley AT, Frayling TM, et al. FTO gene variation and measures of body mass in an African population. BMC Med Genet. 2009;10:21. doi: 10.1186/1471-2350-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 34.Elbein SC. Evaluation of polymorphisms known to contribute to risk for diabetes in African and African-American populations. Curr Opin Clin Nutr Metab Care. 2007;10:415–419. doi: 10.1097/MCO.0b013e3281e2c99a. [DOI] [PubMed] [Google Scholar]
- 35.Flegal KM, Carroll MD, Ogden CL, Curtin LR. Prevalence and trends in obesity among US adults, 1999–2008. JAMA. 2010;303:235–241. doi: 10.1001/jama.2009.2014. [DOI] [PubMed] [Google Scholar]
- 36.Cowie CC, Rust KF, Byrd-Holt DD, Eberhardt MS, Flegal KM, Engelgau MM, et al. Prevalence of diabetes and impaired fasting glucose in adults in the US population: National Health And Nutrition Examination Survey 1999–2002. Diabetes Care. 2006;29:1263–1268. doi: 10.2337/dc06-0062. [DOI] [PubMed] [Google Scholar]
- 37.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 38.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hester JM, Palmer ND, Ng MCY, Adeyemo A, Rotimi C, Freedman BI, et al. Comprehensive genetic evaluation of variation at the TCF7L2 locus in African Americans. Diabetes. 2009;58(suppl 1) [Google Scholar]
- 40.Lewis JP, Palmer ND, Hicks PJ, Sale MM, Langefeld CD, Freedman BI, et al. Association analysis in African Americans of European-derived type 2 diabetes single nucleotide polymorphisms from whole-genome association studies. Diabetes. 2008;57:2220–2225. doi: 10.2337/db07-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner DR, Heyward VH. Measures of body composition in blacks and whites: a comparative review. Am J Clin Nutr. 2000;71:1392–1402. doi: 10.1093/ajcn/71.6.1392. [DOI] [PubMed] [Google Scholar]
- 42.Carroll JF, Chiapa AL, Rodriquez M, Phelps DR, Cardarelli KM, Vishwanatha JK, et al. Visceral fat, waist circumference, and BMI: impact of race/ethnicity. Obesity (Silver Spring) 2008;16:600–607. doi: 10.1038/oby.2007.92. [DOI] [PubMed] [Google Scholar]
- 43.Wells JC, Cole TJ, Bruner D, Treleaven P. Body shape in American and British adults: between-country and inter-ethnic comparisons. Int J Obes (Lond) 2008;32:152–159. doi: 10.1038/sj.ijo.0803685. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


