Abstract
We report a large-scale reduced expression of genes in pathways related to cell-type specific immunity functions that emerges from microarray analysis 48 h after ex vivo γ-ray irradiation (0, 0.5, 2, 5, 8 Gy) of human peripheral blood from five donors. This response is similar to that seen in patients at 24 h after the start of total-body irradiation and strengthens the rationale for the ex vivo model as an adjunct to human in vivo studies. The most marked response was in genes associated with natural killer (NK) cell immune functions, reflecting a relative loss of NK cells from the population. T- and B-cell mediated immunity genes were also significantly represented in the radiation response. Combined with our previous studies, a single gene expression signature was able to predict radiation dose range with 97% accuracy at times from 6–48 h after exposure. Gene expression signatures that may report on the loss or functional deactivation of blood cell subpopulations after radiation exposure may be particularly useful both for triage biodosimetry and for monitoring the effect of radiation mitigating treatments.
INTRODUCTION
In the light of growing concerns of potential terrorist attacks using radiological and nuclear materials, ionizing radiation represents a potential hazard to both public health and national security (1, 2). In the event of a nuclear detonation or even a “dirty bomb,” exposure doses would need to be determined for many thousands of individuals as quickly as possible to provide appropriate medical attention. The current gold-standard for radiation biodosimetry, the dicentric assay, is impractical for mass triage, as it requires several days to complete the assay (3, 4) and automation of the method has not been promising. The development of appropriate biodosimetry methods has, therefore, been identified as one of the highest priorities for nuclear threat countermeasures by the Homeland Security Council (5).
Gene expression profiling with human peripheral blood has been suggested as a viable alternative approach that can predict absorbed radiation dose (6–8). In addition, gene expression signatures can be adapted to a fully integrated biochip to provide rapid high throughput screening (9). Peripheral blood cells provide a good target for radiation biodosimetry, as they are relatively easily biopsied, sensitive to early radiation injury and gene expression changes can persist several days after exposure (6, 10). Emerging work also suggests that expression signatures can likely be selected that are little influenced by age, gender or smoking (11) further supporting their potential for biodosimetry.
Ex vivo studies provide a flexible platform for gene discovery for dosimetric assessment beyond the limited human in vivo exposure samples available from cancer patients undergoing radiotherapy. We previously demonstrated that a gene expression signature derived from donor samples irradiated ex vivo could predict with high accuracy the dose to samples from a heterogeneous population of patients undergoing total-body irradiation (TBI) (12). This supported the use of the ex vivo platform to develop radiation dosimetric signatures that are relevant for in vivo exposures.
Ideally, treatment of exposed individuals should be initiated within two to three days after radiation exposure, meaning dose estimates will be needed within this time-frame. As an event requiring large-scale radiological casualty screening is likely also to produce large-scale panic and infrastructure disruption, screening is unlikely to be completed within the first 24 h. In such a situation, a radiation signature useful across a relatively broad time range would be useful for radiological triage. Studies have indicated that gene expression changes persist at least several days after exposure (6, 13), although the dose-predictive capabilities of gene expression at times past 24 h have not been tested in whole blood through a dose range relevant for triage.
In this study we have investigated the ability of gene expression to predict radiation dose to human peripheral blood exposed ex vivo at up to 48 h after exposure. We describe a 72-gene classifier that can predict radiation exposure ranges at times from 6–48 h with 97% accuracy. Furthermore, we found that genes in pathways related to cell-type specific immunity functions, especially natural killer (NK) cell functions, were broadly under expressed at 48 h after exposure, but not at earlier times. The response of immune function genes is similar to that seen in patients 24 h after the start of a course of TBI. A relative depletion of NK cells in the blood cell population was also observed at 48 h, consistent with major observed gene expression changes. Therefore, we find that gene expression signatures may be useful to predict both radiation dose and relative abundance or function of specific blood cell types following radiation exposure. Such signatures may be useful not only for biodosimetric triage, but may also assist in monitoring the progress of treatment and recovery.
METHODS AND MATERIALS
Blood Irradiation and Culture
After obtaining informed consent, blood from healthy volunteers (2 male, 3 female) was drawn into 0.105 M sodium citrate vacutainer tubes (Becton Dickinson Co., Franklin Lakes, NJ). Aliquots of whole blood were exposed to 0, 0.5, 2, 5 or 8 Gy γ rays at the Center for Radiological Research using a Gammacell-40 137Cs irradiator (AECL, Ontario, Canada) at a dose rate of 0.82 Gy per min and then diluted 1:1 with RPMI 1640 medium (Mediatech Inc., Herndon, VA) supplemented with 10% heat-inactivated fetal bovine serum (Hy-Clone, Logan, UT) and incubated for 48 h at 37°C in a humidified incubator with 5% CO2. Except for the different incubation times and use of different donors, all conditions were the same as in experiments reported earlier (8). All experiments involving human subjects were approved by the Columbia University Medical Center Institutional Review Board IRB #3 and were conducted according to the principles expressed in the Declaration of Helsinki.
Purification of RNA
RNA was prepared using differential red and white cell lysis with the PerfectPure Blood RNA Purification kit (5 Prime, Inc., Gaithersburg, MD) following the manufacturer’s recommendations. This protocol differentially lyses red and white blood cells in whole blood and incorporates on-column DNA digestion. To maximize inter-comparability with our previous studies (8), globin mRNA levels were further reduced using GLOBINclear™ (Ambion Inc., Austin, TX). A NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Waltham, MA) was used to quantify RNA and quality was monitored with the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). All RNA samples had RNA integrity numbers (14) greater than 8.
Microarray Hybridization and Data Extraction
Cyanine-3 (Cy3) labeled cRNA was prepared from 0.5 µg RNA using the One-Color Quick Amp kit (Agilent) according to the manufacturer’s instructions, followed by RNAeasy column purification (Qiagen, Valencia, CA). Yield and Dye incorporation were monitored with the NanoDrop ND-1000 Spectrophotometer. 1.65 µg of cRNA with incorporation of >10 pmol Cy3 per µg cRNA was fragmented, hybridized to Agilent Whole Human Genome Oligo Microarrays (G4112A) for 17 h and then washed following the manufacturer’s recommendations. Immediately after washing, the microarrays were scanned with the Agilent DNA Microarray Scanner (G2404B), and the images were analyzed using Feature Extraction Software 9.1 (Agilent) with default parameters for background correction and flagging of nonuniform features.
Analyses in BRB ArrayTools
Background corrected hybridization intensities were imported into BRB-ArrayTools Version 3.8.0 beta (15) log2-transformed and median normalized. Data from prior experiments (8) was also imported to facilitate comparisons with results at earlier times postirradiation. Nonuniform outliers or features not significantly above background intensity in 25% or more of the samples, and features not changing at least 1.5-fold in at least 20% of the samples were filtered out. This yielded 15,152 features that were used in subsequent analyses. The microarray data are available through the GEO database using accession number GSE44201.
F test based class comparisons were conducted using BRB-ArrayTools to identify genes that were differentially expressed between the five radiation doses. Genes with P values less than 0.001 were considered statistically significant. The false discovery rate (FDR) was also estimated for each gene using the method of Benjamini and Hochberg (16), to control for false positives. Data from 48 h was analyzed separately, and data from all time points (including data from different independent donors assayed at 6 and 24 h after radiation exposure that was obtained previously) was also analyzed together without consideration of the time variable.
Class Prediction by the 3-Nearest Neighbor method was performed in BRB-ArrayTools using a vector of log-intensities for the genes in the multivariate predictor as the expression profile and Euclidean distance as the distance metric. The expression profile of each test sample is compared to all the other expression profiles and the three samples most similar to the expression profile of the test sample are determined. The majority class among the three closest samples is the class predicted for the test sample. Leave-one-out cross-validation was performed, in which one sample is omitted from the model, and its class is predicted based on proximity to the expression vectors of the other samples, with the process being repeated independently for each sample.
Multidimensional scaling (MDS) was performed in BRB-Array-Tools to create a low-dimensional graphical representation of the high-dimensional data from the identified gene expression signature. The Euclidian distance metric was used to compute a distance matrix and the principal components of the gene expression signature. The first three principal components were used as axes to generate a plot. A global test for clustering based on the first three principal components was used to test the null hypothesis that all the expression profiles were drawn from a single multivariate Gaussian distribution (one cluster). The distribution of nearest neighbor distances for the actual data were compared with Gaussian distributions of nearest neighbor distances generated by 10,000 random permutations of the data (17).
Gene Ontology Analysis
The significantly differentially expressed genes were imported into PANTHER (18) and the number of genes in each functional classification category was compared against the number of genes in that category in the NCBI human genome. The binomial test was used to determine statistical over-representation of PANTHER classification categories (19). Bonferroni corrected P values less than 0.05 were considered significant.
Quantitative Real-Time PCR
The High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA) was used to reverse transcribe 500 ng total RNA following the manufacturer’s instructions. Gene-specific primers and probes (Table 1) were designed with the aid of Applied Biosystems’ Primer Express® software and GeneScript Corporation’s online TaqMan Primer Design software, and sequences for ACTB, CDKN1A and PHPT1 were the same as used previously (8). Probes with 6-carboxyfluorescein (FAM) at the 5′ end and BHQ1 quencher at the 3′ end were synthesized by Operon Biotech, Inc. (Huntsville, AL). Standard curves were generated to optimize the amount of input cDNA for each gene (5 or 10 ng). Real-time PCR reactions were performed with the ABI 7300 Real Time PCR System using Universal PCR Master Mix from ABI following the manufacturer’s recommendations. All samples were run in duplicate and repeated a second time on a different day for each gene. Relative fold-inductions were calculated using the ΔΔCT method as previously described (20) with averaged relative levels of ACTB and GAPDH used for normalization.
TABLE 1.
Primers and Probes Used in qRT-PCR
| Gene | Primer–probe sequences | |
|---|---|---|
| DDB2 | Forward: | 5′-CAGGACACGGAAGTGAGAGA-3′ |
| Reverse: | 5′-CAAATCGCCACCTCTGCTTG-3′ | |
| Probe: | 5′-TCCAAGGCCTTGTCTGGCCC-3′ | |
| PCNA | Forward: | 5′-GCACTCAAGGACCTCATCAA-3′ |
| Reverse: | 5′-TCCATGCTCTGCAGGTTTAC-3′ | |
| Probe: | 5′-CCGCTGGAGCTAATATCCCAGCA-3′ | |
| CDKN1A | Forward: | 5′-CTGGAGACTCTCAGGGTCGAA-3′ |
| Reverse: | 5′-CGGCGTTTGGAGTGGTAGAA-3′ | |
| Probe: | 5′-TCATGCTGGTCTGCCGCCG-3′ | |
| PHPT1 | Forward: | 5′-CCACCAGAGTCAGGACAAG-3′ |
| Reverse: | 5′-GTGCTCAGTAGCCGTCGTTA-3′ | |
| Probe: | 5′-TATGGTCCTGCCCAGCACGC-3′ | |
| GNLY | Forward: | 5′-TCACCTTGTCCTGTGGAAGA-3′ |
| Reverse: | 5′-AGAGGTTGCTGAGGTTCCC-3′ | |
| Probe: | 5′-CACAGGCTCCTGTCCTCAGATCCC-3′ | |
| GZMA | Forward: | 5′-CTCGTGCAATGGAGATTCTG-3′ |
| Reverse: | 5′-AAGGCCAAAGGAAGTGACC-3′ | |
| Probe: | 5′-CACCCTCGCACAACAAAGGGC-3′ | |
| NKG7 | Forward: | 5′-CCCAGATCCAGACCTTCTTC-3′ |
| Reverse: | 5′-GGCACCTGTACAGAGCAAGA-3′ | |
| Probe: | 5′-CCCAGCCCAGGTAGAAGGACCA-3′ | |
| ACTB | Forward: | 5′-CACTCTTCCAGCCTTCCTTC-3′ |
| Reverse | 5′-GGATGTCCACGTCACACTTC-3′ | |
| Probe: | 5′-TGCCACAGGACTCCATGCCC-3′ | |
Flow Cytometric Determination of NK, T and B Cells in Irradiated Populations
Blood from healthy adult volunteers (different donors from those used for the gene expression work) was collected into sodium citrate vacutainer tubes (Becton Dickinson Co.), and aliquots of blood were irradiated and cultured as described above. After 48 h, antibodies were added directly to 50 µl aliquots of the diluted blood, followed by incubation for 30 min at room temperature. For NK cell determination, we used anti-human CD45/PerCP (BioLegend, San Diego, CA) and anti-human CD3-FITC/CD16+CD56-PE (BD Simultest, BD Biosciences, Franklin Lakes, NJ) antibodies, and for T and B cell determination, we used anti-human human CD45/PerCP (BioLegend) and anti-human CD3-FITC/CD19-PE (BD Simultest, BD Biosciences). The red blood cells were lysed by the addition of 450 µl of lysing solution (BD Biosciences) and the cell subsets were measured using an Accuri (BD Biosciences) flow cytometer. For NK cell identification we gated initially on the lymphocyte cell population (lymphocyte specific side scatter (SSC) and CD45+ cells) followed by gating on the CD3−, CD56+/CD16+ cell population. T and B cells were identified using the same initial gating followed by subsequent gating on CD3+ (T cells) and CD19+ (B cells). To obtain absolute cell counts, an AccuCount reference solution (Spherotech, Lake Forest, IL) with known number of particles per ml was used. The absolute blood cell counts were calculated from the volume of the sample that was measured by the Accuri flow cytometer and the bead counts for this volume.
RESULTS
Genes Distinguishing Between Radiation Doses
We measured global gene expression in human blood at 48 h after exposure to 0, 0.5, 2, 5 and 8 Gy γ rays using Agilent whole human genome microarrays and the Agilent one-color workflow. Five independent experiments were conducted with blood from different donors. Applying filters for minimum quality and changed expression gave a set of 15,152 features that were used for analysis.
The Class Comparison feature of BRB-ArrayTools identified 286 genes with significantly different (P < 0.001) expression across the five dose levels (see Supplementary Table S1; http://dx.doi.org/10.1667/RR13343.1.S1) 48 h after exposure to radiation. Of these genes, 281 had a false discovery rate (FDR) <5%. As we had previously found a substantial number of genes were differentially expressed at both 6 and 24 h postirradiation, we next performed a class comparison of the five doses pooling the data from the current study (48 h) with the data from 6 and 24 h obtained previously using blood from different donors (8). This analysis identified 269 differentially expressed genes (P < 0.001), 260 of which had a FDR <5% (see Supplementary Table S2; http://dx.doi.org/10.1667/RR13343.1.S2).
Gene Ontology Analysis
We used PANTHER (19), which assigns genes to ontology groups using both gene families and protein sequence information, to look for enrichment of gene groups among the differentially expressed genes. The biological processes significantly affected at 48 h after radiation exposure were dominated by immune functions, in contrast to the earlier times, when processes such as cell proliferation and differentiation, apoptosis, and signal transduction dominated (Table 2). Only apoptosis and signal transduction were significantly over-represented at all the times studied.
TABLE 2.
Gene Ontology Classification of Dose Responsive Genes by PANTHER
| Bonferroni corrected P values | |||
|---|---|---|---|
| Biological process | 48 h | 24 ha | 6 ha |
| Natural killer cell mediated immunity | 4.12 × 10−20 | NSb | NS |
| Immunity and defense | 1.37 × 10−15 | 2.75 × 10−2 | NS |
| Signal transduction | 3.55 × 10−9 | 1.21 × 10−2 | 4.55 × 10−4 |
| Cytokine/chemokine mediated immunity | 1.99 × 10−3 | NS | NS |
| Cytokine and chemokine mediated signaling pathway | 2.47 × 10−3 | NS | NS |
| Granulocyte-mediated immunity | 3.01 × 10−3 | NS | NS |
| Cell surface receptor mediated signal transduction | 3.43 × 10−3 | NS | 6.81 × 10−3 |
| Ligand-mediated signaling | 3.46 × 10−3 | NS | NS |
| Apoptosis | 6.22 × 10−3 | 1.97 × 10−3 | 1.72 × 10−5 |
| T-cell mediated immunity | 7.08 × 10−3 | NS | NS |
| B-cell-and antibody-mediated immunity | 2.89 × 10−2 | 7.51 × 10−3 | NS |
| Cell cycle | NS | NS | 7.27 × 10−3 |
| Induction of apoptosis | NS | 9.03 × 10−3 | 7.48 × 10−3 |
| NF-κB cascade | NS | NS | 1.83 × 10−2 |
| Cell proliferation and differentiation | NS | 2.23 × 10−3 | 2.29 × 10−2 |
| Protein phosphorylation | NS | NS | 2.75 × 10−2 |
| DNA repair | NS | 1.41 × 10−2 | NS |
| DNA metabolism | NS | 1.69 × 10−2 | NS |
Analysis of data from S. Paul (2008).
Not significant (P > 0.05).
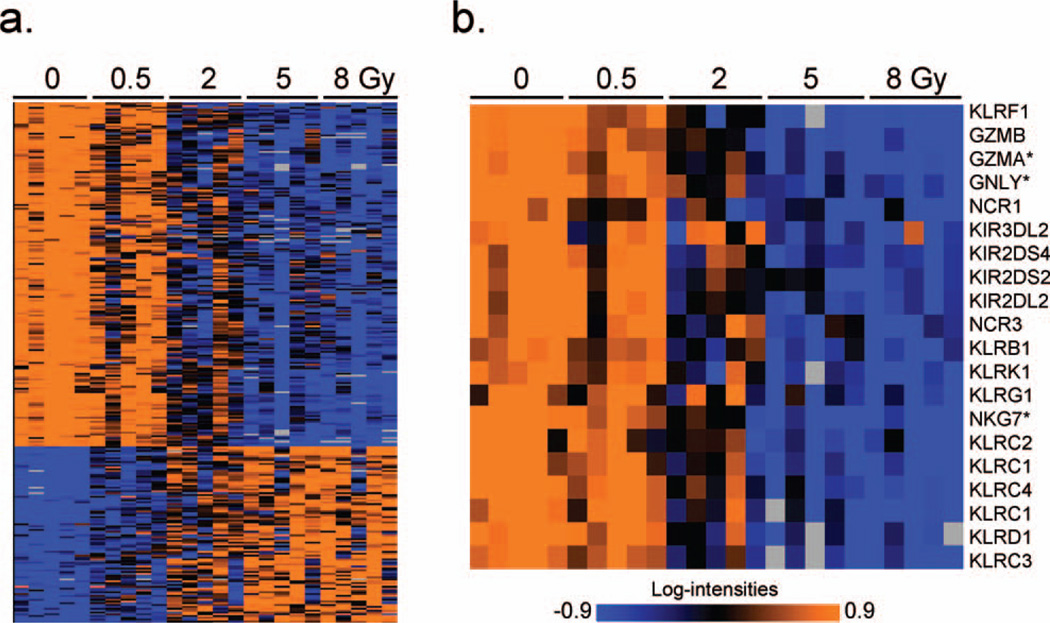
The top biological process among genes differentially expressed 48 h after irradiation was NK cell mediated immunity (P = 4.1 × 10−20), which was not significant at any of the earlier times. Examination of the genes with this annotation revealed that they all showed reduced expression after irradiation, being among the most strongly under-expressed genes found, especially at the higher doses (see Supplementary Table S1; http://dx.doi.org/10.1667/RR13343.1.S1).
Dose Responses of Individual Genes
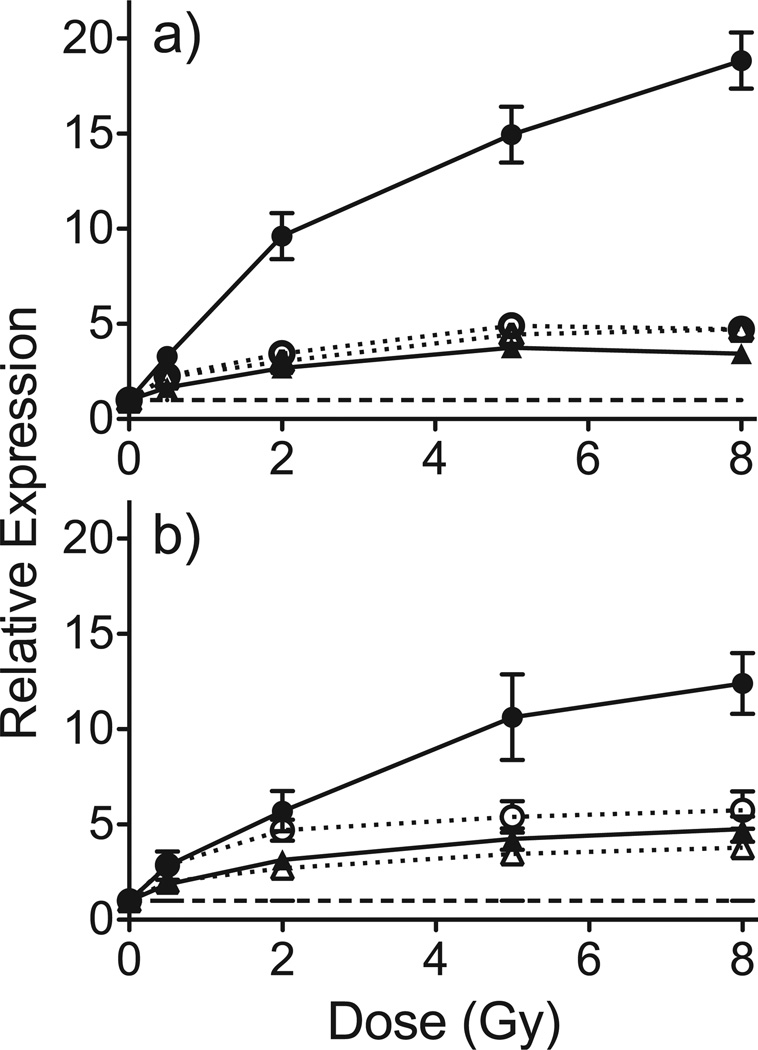
We used quantitative real-time PCR (qRT-PCR) to measure the radiation dose-response of CDKN1A, DDB2, PCNA and PHPT1 as representative genes significantly differentially expressed 48 h after radiation exposure that also responded in our earlier study at 6 and 24 h post-exposure (8). qRT-PCR was performed on all samples and plotted as average response ratios normalized to controls (Fig. 1a). All genes tested responded with the same general pattern as that seen on the microarrays (Fig. 1b). CDKN1A, DDB2, PCNA and PHPT1 increased with increasing dose, but with a decreased slope above 2 Gy, consistent with prior observations (8, 21).
FIG. 1.
Relative expression of CDKN1A (●), DDB2 (○), PCNA (▲) and PHPT1 (△) at 48 h after ex vivo irradiation as determined by qRT-PCR (panel a) and microarray hybridization (panel b). Points denote mean response of 5 independent donors and error bars denote standard error of the mean (SEM). The dashed line indicates the expression level in unirradiated controls.
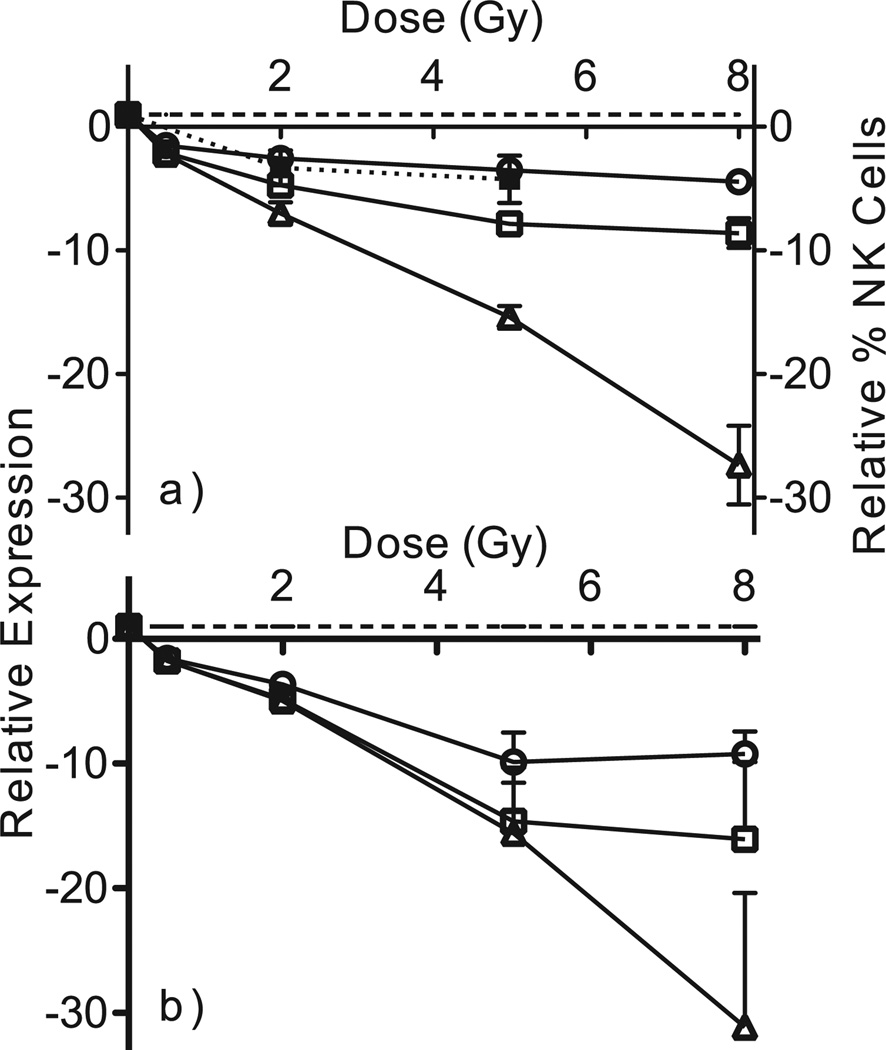
In contrast to the earlier times examined previously, at 48 h postirradiation almost two-thirds of differentially expressed genes were down-regulated (Fig. 2a). Prominent among the most significant differentially expressed genes were NK cell mediated cytotoxicity genes, which were strongly under-expressed compared with unirradiated controls at 48 h (Fig. 2b), but not at earlier times. To verify this finding, the dose-responses of three genes from this functional class (GZMA, NKG7 and GNLY, selected to span the range of fold changes seen by microarray) were measured by qRT-PCR (Fig. 3a) and found to show the same pattern as in the microarray data (Fig. 3b).
FIG. 2.
Panel a: Average linkage clustering of genes differentially expressed across doses at 48 h postirradiation. Annotation of all genes in clustered order is presented in Supplementary Table S1. High expression is depicted as orange and low expression is depicted as blue. Exposure dose (in Gy) is labeled across the top of the panel. Panel b: Heat map illustrating the expression pattern of 20 NK cell associated genes (identified by BRB annotation) significantly down-regulated at 48 h post exposure. Gene names are shown along the right edge of the figure and dose (in Gy) is labeled across the top of the panel. Genes used for qRT-PCR are marked with an asterisk (*).
FIG. 3.
Relative expression of three NK cell associated cytotoxic genes, GNLY (□), GZMA (△) and NKG7 (○), measured by qRT-PCR (panel a) and microarray hybridization (panel b) 48 h after radiation exposure. Each point represents the mean of relative induction above background in five independent donors. Error bars are standard errors of the mean. The dashed line shows the basal level of expression in unirradiated controls. The percentage of NK cells in the leukocyte population at 48 h relative to unirradiated controls (■) is also shown in panel a. This point represents the mean of three independent donors and error bars are standard error of the mean.
Flow Cytometric Determination of NK Cells after Irradiation
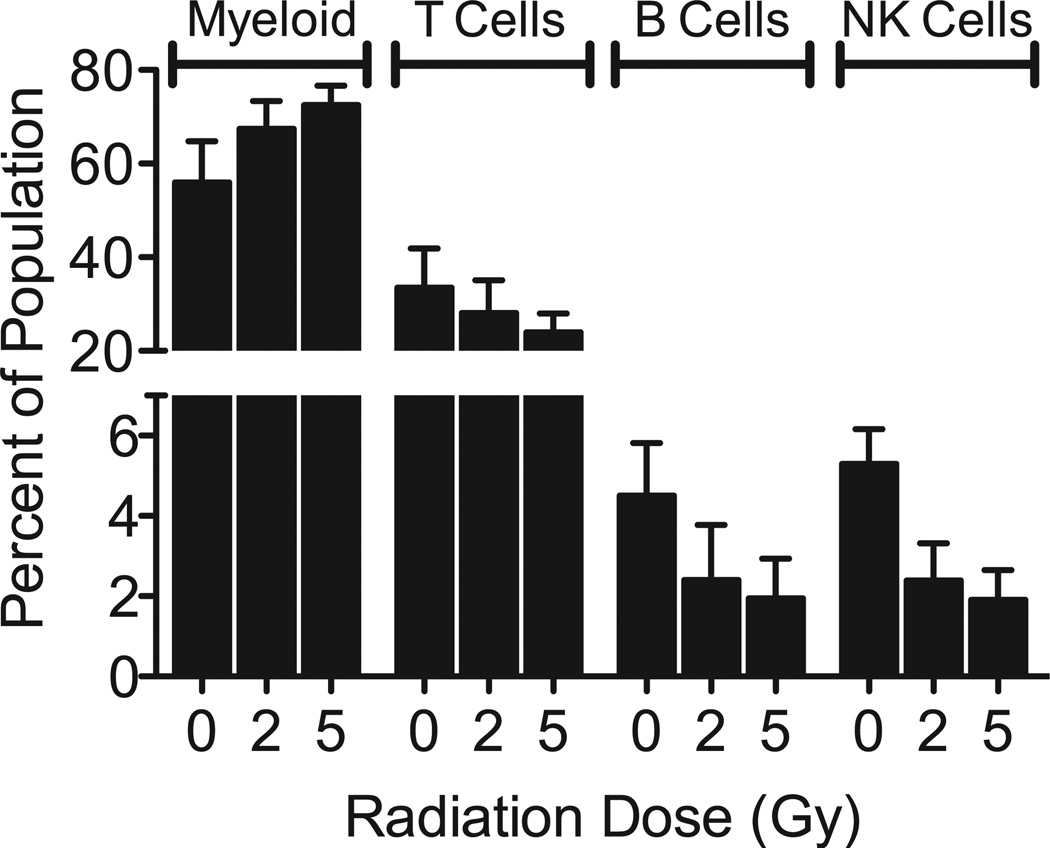
As NK cell mediated immunity and immune functions appeared so prominently among the genes under-expressed 48 h after irradiation, in contrast to earlier times (8), we questioned if this could be explained by the disappearance of NK cells from the ex vivo culture population. Reports on radiation sensitivity of NK cells have been contradictory (22–26), and our ex vivo culture conditions might affect radiation sensitivity, so we used flow cytometry to measure the numbers of different blood cell subtypes (T-cells, B-cells and NK cells) in our ex vivo culture model at 48 h after exposure to 0, 2, 5 or 8 Gy γ rays. The samples exposed to 8 Gy did not yield sufficient cell counts in the flow cytometry assay to provide reliable data. The results from the 0, 2 and 5 Gy samples are presented as the percentage of lymphocyte subpopulations identified in the culture at the time when RNA was harvested (Fig. 4), along with the remaining cells in the CD45+ population, representing mainly cells of myeloid lineage. Among lymphocytes, T-cells appeared to be the most radioresistant, whereas B-cell and NK cells were more radiosensitive. Overall the relative radiosensitivity presented as follows: NK cells > B-cells > T-cells > myeloid cells.
FIG. 4.
Flow cytometric analysis of peripheral blood subsets in the ex vivo cultures 48 h after irradiation. Each bar represents the mean percentage (n = 3 individual donors) of T-cell (CD3+, CD56−, CD16−), B-cell (CD3−, CD19+) and NK cell (CD3−, CD56+, CD16+/−) among CD45+ cells in the ex vivo population. The remaining CD45+ leukocytes that were not positive for any of the lymphocyte markers are indicated as “myeloid”. Error bars are standard error of the mean.
Prediction of Radiation Dose to Individual Samples
We used BRB-ArrayTools to build and test classifiers to predict the exposure dose to individual blood samples using gene expression 48 h after exposure. Using a 3-nearest neighbors (3NN) algorithm we were able to predict the dose to 72% of the 48 h samples correctly. As in our previous study of earlier times postirradiation, the majority of misclassification occurred between samples treated with 5 and 8 Gy. Combining these two doses into a single “high dose” category improved correct classification of the 48 h samples to 100%.
As we had previously developed a single classifier that performed well on samples from both 6 and 24 h after irradiation (8), we next tested the ability of that signature to predict dose across all times tested (6, 24 and 48 h). It predicted all 5 doses with 70% accuracy and combining the two highest doses as a single group increased accuracy considerably, to 95%.
To test if inclusion of the 48 h data would produce a dose classifier more robust to time since irradiation, we next built new classifiers incorporating the gene expression results from all three times to determine if this could improve prediction accuracy. The algorithm selected 72 features (see Supplementary Table S3; http://dx.doi.org/10.1667/RR13343.1.S3) and the resulting classifiers yielded a modest improvement in accuracy over the 6–24 h classifier, with 76% correct prediction of all five doses. Grouping the two highest doses again improved prediction accuracy, in this case to 97% correct classification of the 4 dose ranges across all times from 6–48 h. The sensitivity and specificity of all classifiers is summarized in Supplementary Table 3.
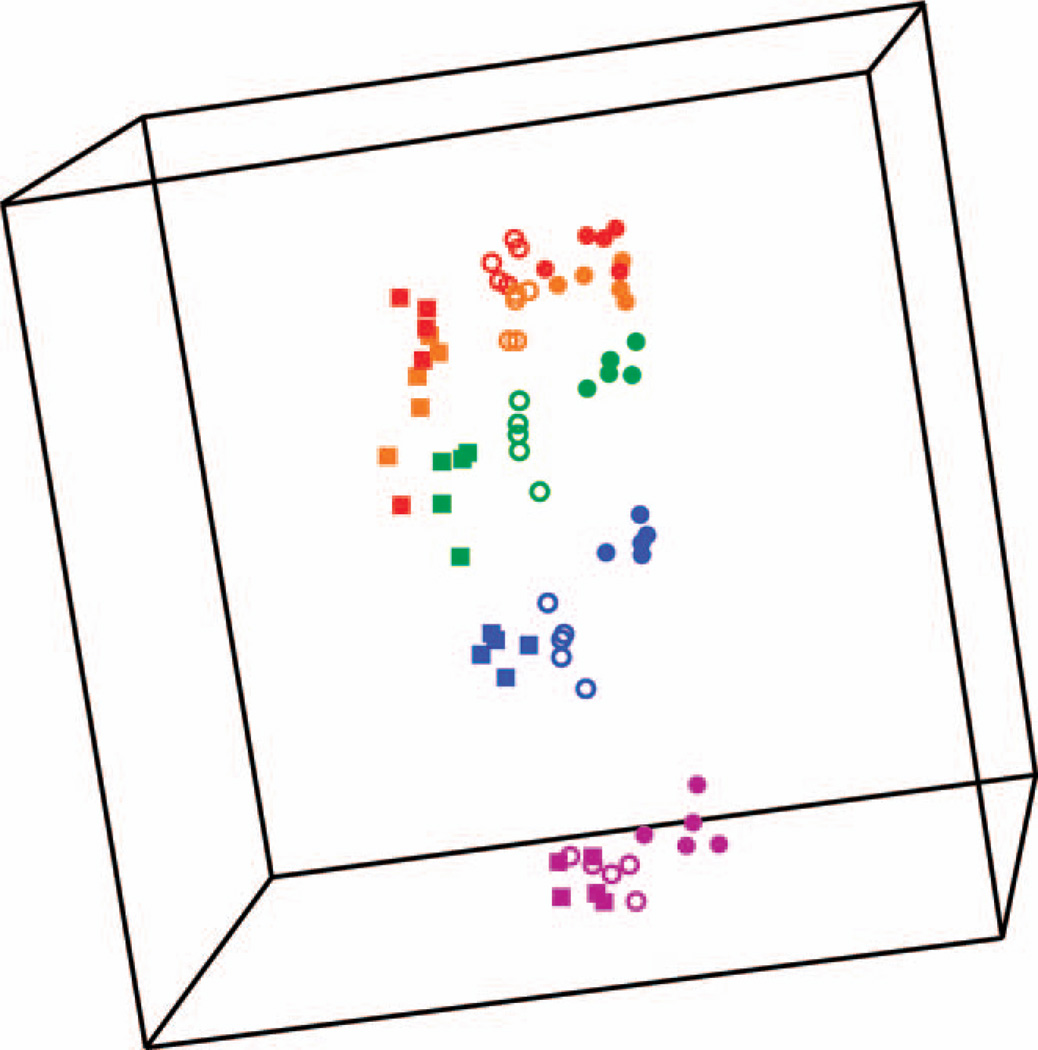
Separation by dose of samples taken 6, 24 and 48 h after exposure (Fig. 5) is illustrated using Multidimensional scaling (MDS) with the genes in the newly-derived 6–48 h classifier (see Supplementary Table S3; http://dx.doi.org/10.1667/RR13343.1.S3). Multidimensional scaling is a visualization tool for graphically representing high-dimensional data in two or three dimensions, while preserving all pair-wise similarities between samples. The first three principal components of the gene expression data are used as axes and a plot is created where each point represents an individual sample, and the distance between any two points reflects the overall similarity of the expression levels of all features in the signature. Clustering by dose was significant [P < 0.0001; global test for clustering (17)], indicating a low probability that all samples came from a single homogeneous cluster.
FIG. 5.
Signature predicting dose at all times is visualized by multidimensional scaling. Axes represent the first three principal components of gene expression. Each point represents the relative expression of all 72 features in the signature (Supplementary Table S3) for an individual sample. The distance between any two points reflects their overall similarity of expression across all genes. The points are colored according to dose: 0 Gy (purple), 0.5 Gy (blue), 2 Gy (green), 5 Gy (orange) and 8 Gy (red). The shape of the symbols indicates the time after irradiation: 48 h (square), 24 h (open circle) and 6 h (closed circle).
DISCUSSION
In the present work, we have demonstrated that a gene expression profile derived from ex vivo irradiated blood samples at 6 and 24 h after radiation exposure can predict the dose to samples from different donors measured at 48 h after exposure with only a slight degradation in performance. This was somewhat surprising, considering the substantial shifts in significantly over-represented gene ontology (GO) classifications seen among the differentially expressed genes at these times. Use of gene expression data from all three times to build a more representative expression classifier restored the correct prediction rate to within 1–2% of that obtained previously for 6–24 h samples alone.
This work further supports the development of a core gene expression signature that can be widely applicable across various doses, times and radiation modalities. We have previously shown that our initial 6–24 h signature (8) was capable of accurate dose prediction outside the dose range used to develop it (11) and in patients undergoing total body irradiation (12). Our studies continue to support the feasibility of developing response signatures that are robust against the interindividual variations that have been reported in the peripheral blood cells of unirradiated volunteers (27) and in irradiated lymphoblastoid cells (28). Further optimization is likely to allow reduction in the number of genes needed to provide robust dosimetric estimates that are valid across a range of times. While is not yet clear how much beyond the first 48 h after exposure the initial signature may be useful, it is highly encouraging to find that it is unlikely to be necessary to develop separate signatures for very small increments of time after a large-scale radiological event. Even as many of the early responding genes return towards baseline expression levels, new responses develop, some being maintained for weeks or months after exposure (29, 30). As research in this area goes forward, especially with studies incorporating in vivo models and more protracted times, additional biodosimetric signatures and their optimally informative times are being developed. Such signatures could be readily incorporated into developing platforms for rapid high-throughput or field analysis (9, 31) to provide a practical approach for radiation biodosimetry.
Although the ex vivo system used here serves as a useful adjunct to available cancer patient models, simply holding blood in culture results in changes in gene expression (32), and also affects expression of genes in our dosimetric signature as reflected in the separation of samples by time [Fig. 5 and (8)]. This does not greatly impact discrimination between doses, however.
It is interesting that the GO categories that were most significant in our recent in vivo study (12) bore a striking similarity to those identified in the present study at 48 h ex vivo. Notably, these included many immune and cytokine functions, as well as T-cell, B-cell and NK-cell mediated immunity, which were not seen at earlier times ex vivo. It may be that alterations in the programming of these functions occur more rapidly in vivo, but are eventually recapitulated ex vivo as well.
In our GO summary of ex vivo responses (Table 2), signal transduction and apoptosis were significantly over-represented at all times. These are among the top biological processes commonly seen in response to radiation. In addition to our earlier ex vivo time points, significant enrichment of these GO categories has also been found in total body irradiation patients (12), mice irradiated in vivo (30, 33), and cell culture models (34, 35).
To gain some insight into the specific signal transduction pathways implicated in our 48 h response, we imported the genes from this functional annotation category into Ingenuity Pathway Analysis (IPA) (Ingenuity® Systems, www.ingenuity.com) and examined their network interactions. We found that the major signaling networks represented among the responding genes in this GO category were the MAPK pathways ERK1/2 and Jnk, the Akt pathway, and the NFκB pathway (see Supplementary Fig S1; http://dx.doi.org/10.1667/RR13343.1.S4). All of these pathways have previously been shown to be activated by ionizing radiation (36–38). Genes in the signal transduction category coded for proteins with activity in the plasma membrane, including many transmembrane receptors. This category also included genes with products active in the nucleus, the cytoplasm, and the extracellular space (see Supplementary Fig S1; http://dx.doi.org/10.1667/RR13343.1.S4).
At earlier times, we found the ex vivo response was dominated by genes associated with cell cycle, induction of apoptosis and the p53 pathway. TP53-regulated genes, such as CDKN1A, DDB2 and PCNA (Fig. 1), and functions regulated by p53 were represented among the significant in vivo and 48 h responses, but the p53 pathway was not a significantly enriched GO category at this time after radiation exposure.
The top biological process enriched among genes differentially expressed 48 h after irradiation was NK cell-mediated immunity, which was not significant at the earlier time points ex vivo (8). Although not among the GO categories reported in a recent study focusing on lower dose responses in lymphocytes separated from human peripheral blood (39), NK cell-mediated immunity was also the top GO category in that study when we performed GO analysis on the genes they reported to be altered 48 h after 2 and 4 Gy exposures. The NK immunity genes were strongly under-expressed in a dose-dependent manner, perhaps reflecting a relative loss of this cell sub-population in the irradiated 48 h samples, or a functional shift in cellular programming. Flow cytometric analysis indicated that the NK cell population was the most radiosensitive in our model, relative to B-cells, and the more abundant myeloid and T-cell populations (Fig. 4).
Comparison of the relative decline in the NK-cell population (solid symbols and dotted line in Fig. 3a) with expression of NK cytotoxicity genes revealed that NK cell abundance declined to a similar extent as expression of NKG7, but to a lesser extent and at a slower rate than expression of GNLY and GZMA. This suggests that while shifts in cell subpopulations may play a role in shaping the gene expression profiles seen at 48 h after irradiation, regulation of expression of genes involved in specialized functions may also play a role. Many genes with immune-related functions were expressed at decreased levels by 48 h after irradiation in this study, including those associated with T- and B-cells, suggesting the possibility of a broad shut-down of cell-type specific immunity functions in damaged or dying cells prior to their actual removal from the population. Further studies will be required to fully elucidate the mechanisms behind the broad decrease in expression of specific immune function genes observed 48 h after radiation exposure.
There have been conflicting reports in the literature regarding the radiation sensitivity of NK cells. An in vivo study with TBI patients (40) suggested that NK cells were relatively resistant to ionizing radiation compared with other lymphocyte subsets. Mori and Desaintes (24) similarly reported that NK cells were relatively radioresistant among lymphocyte subpopulations 8 h after a 1 Gy dose in vitro. However, Seki et al. (23) reported that NK cells were the most radiosensitive lymphocyte subpopulation 24 h after irradiation with a 15 Gy dose in vitro. These contradictory findings might have been due to the use of different treatment and assay parameters, particularly in light of the finding that low doses of radiation enhanced the cytotoxic activity of NK cells, while higher doses caused a loss of activity (41), with the latter effect being especially pronounced 48 h after exposure (22). These findings support the potential contribution of both cell killing and reprogramming of cytotoxic functions to the gene expression patterns seen at 48 h after radiation exposure. More detailed analyses within NK cell subpopulations as well as comparison of in vitro and in vivo effects will be required for a clearer understanding.
In conclusion, we have shown that the same gene expression signature can predict radiation dose range with high accuracy at times from 6–48 h after exposure (see Supplementary Table S3; http://dx.doi.org/10.1667/RR13343.1.S3). Moreover, we report a large-scale decreased expression of genes in pathways related to cell-type specific immunity functions that occurs at 48 h post-exposure in the ex vivo model, but is not apparent at earlier times. This response is similar to that manifesting at earlier times in a human TBI model, and strengthens the rationale for the ex vivo model as an adjunct to human in vivo studies. Signatures that may report on the loss or functional activity of blood cell subpopulations after radiation exposure may be particularly useful not only for biodosimetry, but also for monitoring the progress of mitigating treatments. However, broader testing, including larger populations and potential confounding factors such as co-morbidities and the potential impact of medications or lifestyle factors, will be needed to confirm the utility of such gene expression signatures for radiological triage.
Supplementary Material
ACKNOWLEDGMENTS
Analyses were performed using BRB-ArrayTools developed by Dr. Richard Simon and Amy Peng Lam. This work was supported by the Center for High-Throughput Minimally-Invasive Radiation Biodosimetry, National Institute of Allergy and Infectious Diseases grant No. U19AI067773.
Footnotes
SUPPLEMENTARY INFORMATION
Supplementary Table S1. Genes differentially expressed across doses at 48 h post-exposure. (http://dx.doi.org/10.1667/RR13343.1.S1)
Supplementary Table S2. Genes differentially expressed across doses pooling data from 6, 24 and 48 h post-exposure. (http://dx.doi.org/10.1667/RR13343.1.S2)
Supplementary Table S3. Classifier summary and genes. (http://dx.doi.org/10.1667/RR13343.1.S3)
Supplementary Fig. S1. IPA network of signal transduction genes. (http://dx.doi.org/10.1667/RR13343.1.S4)
REFERENCES
- 1.Mettler FAJ, Voelz GL. Major radiation exposure–what to expect and how to respond. N Engl J Med. 2002;346:1554–1561. doi: 10.1056/NEJMra000365. [DOI] [PubMed] [Google Scholar]
- 2.Waselenko JK, Macvittie TJ, Blakely WF, Pesik N, Wiley AL, Dickerson WE, et al. Medical management of the acute radiation syndrome: Recommendations of the strategic national stockpile radiation working group. Ann Intern Med. 2004;140:1037–1051. doi: 10.7326/0003-4819-140-12-200406150-00015. [DOI] [PubMed] [Google Scholar]
- 3.Flegal FN, Devantier Y, Mcnamee JP, Wilkins RC. Quickscan dicentric chromosome analysis for radiation biodosimetry. Health Phys. 2010;98:276–281. doi: 10.1097/HP.0b013e3181aba9c7. [DOI] [PubMed] [Google Scholar]
- 4.Vaurijoux A, Gruel G, Pouzoulet F, Gregoire E, Martin C, Roch-Lefevre S, et al. Strategy for population triage based on dicentric analysis. Radiat Res. 2009;171:541–548. doi: 10.1667/RR1664.1. [DOI] [PubMed] [Google Scholar]
- 5.Pellmar TC, Rockwell S. Priority list of research areas for radiological nuclear threat countermeasures. Radiat Res. 2005;163:115–123. doi: 10.1667/rr3283. [DOI] [PubMed] [Google Scholar]
- 6.Amundson SA, Shahab S, Bittner M, Meltzer P, Trent J, Fornace AJ., Jr Identification of potential mrna markers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiation Res. 2000;154:342–346. doi: 10.1667/0033-7587(2000)154[0342:iopmbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 7.Dressman HK, Muramoto GG, Chao NJ, Meadows S, Marshall D, Ginsburg GS, et al. Gene expression signatures that predict radiation exposure in mice and humans. PLoS Med. 2007;4:e106. doi: 10.1371/journal.pmed.0040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paul S, Amundson SA. Development of gene expression signatures for practical radiation biodosimetry. Int J Radiat Oncol Biol Phys. 2008;71:1236–1244. doi: 10.1016/j.ijrobp.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brengues M, Paap B, Bittner M, Amundson S, Seligmann B, Korn R, et al. Biodosimetry on small blood volume using gene expression assay. Health Phys. 2010;98:179–185. doi: 10.1097/01.HP.0000346706.44253.5c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turtoi A, Brown I, Oskamp D, Schneeweiss FH. Early gene expression in human lymphocytes after gamma-irradiation-a genetic pattern with potential for biodosimetry. Int J Radiat Biol. 2008;84:375–387. doi: 10.1080/09553000802029886. [DOI] [PubMed] [Google Scholar]
- 11.Paul S, Amundson SA. Gene expression signatures of radiation exposure in peripheral white blood cells of smokers and non-smokers. Int J Radiat Biol. 2011;87:791–801. doi: 10.3109/09553002.2011.568574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paul S, Barker CA, Turner HC, Mclane A, Wolden SL, Amundson SA. Prediction of in vivo radiation dose status in radiotherapy patients using ex vivo and in vivo gene expression signatures. Radiat Res. 2011;175:257–265. doi: 10.1667/RR2420.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fält S, Holmberg K, Lambert B, Wennborg A. Long-term global gene expression patterns in irradiated human lymphocytes. Carcinogenesis. 2003;24:1837–1845. doi: 10.1093/carcin/bgg134. [DOI] [PubMed] [Google Scholar]
- 14.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, et al. The rin: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon R, Lam A, Li M-C, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using brb-array tools. Cancer Informatics. 2007;2:11–17. [PMC free article] [PubMed] [Google Scholar]
- 16.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 17.Mcshane LM, Radmacher MD, Freidlin B, Yu R, Li MC, Simon R. Methods for assessing reproducibility of clustering patterns observed in analyses of microarray data. Bioinformatics. 2002;18:1462–1469. doi: 10.1093/bioinformatics/18.11.1462. [DOI] [PubMed] [Google Scholar]
- 18.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. Panther: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, et al. Applications for protein sequence-function evolution data: Mrna/protein expression analysis and coding snp scoring tools. Nucleic Acids Res. 2006;34:W645–W650. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amundson SA, Grace MB, Mcleland CB, Epperly MW, Yeager A, Zhan Q, et al. Human in vivo radiation-induced biomarkers: Gene expression changes in radiotherapy patients. Cancer Res. 2004;64:6368–6371. doi: 10.1158/0008-5472.CAN-04-1883. [DOI] [PubMed] [Google Scholar]
- 21.Amundson SA, Do KT, Fornace AJ., Jr Induction of stress genes by low doses of gamma rays. Radiat Res. 1999;152:225–231. [PubMed] [Google Scholar]
- 22.Rana R, Vitale M, Mazzotti G, Manzoli L, Papa S. Radiosensitivity of human natural killer cells: Binding and cytotoxic activities of natural killer cell subsets. Radiat Res. 1990;124:96–102. [PubMed] [Google Scholar]
- 23.Seki H, Iwai K, Kanegane H, Konno A, Ohta K, Ohta K, et al. Differential protective action of cytokines on radiation-induced apoptosis of peripheral lymphocyte subpopulations. Cell Immunol. 1995;163:30–36. doi: 10.1006/cimm.1995.1095. [DOI] [PubMed] [Google Scholar]
- 24.Mori M, Benotmane MA, Vanhove D, Van Hummelen P, Hooghe-Peters EL, Desaintes C. Effect of ionizing radiation on gene expression in cd4+ t lymphocytes and in jurkat cells: Unraveling novel pathways in radiation response. Cell Mol Life Sci. 2004;61:1955–1964. doi: 10.1007/s00018-004-4147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mori M, Benotmane MA, Tirone I, Hooghe-Peters EL, Desaintes C. Transcriptional response to ionizing radiation in lymphocyte subsets. Cell Mol Life Sci. 2005;62:1489–1501. doi: 10.1007/s00018-005-5086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vokurková D, Vávrová J, Šinkora J, Stoklasová A, Bláha V, Řezáčová M. Radiosensitivity of cd3-cd8+cd56+ nk cells. Radiation Measurements. 2010;45:1020–1023. [Google Scholar]
- 27.Eady JJ, Wortley GM, Wormstone YM, Hughes JC, Astley SB, Foxall RJ, et al. Variation in gene expression profiles of peripheral blood mononuclear cells from healthy volunteers. Physiol Genomics. 2005;22:402–411. doi: 10.1152/physiolgenomics.00080.2005. [DOI] [PubMed] [Google Scholar]
- 28.Jen KY, Cheung VG. Transcriptional response of lymphoblastoid cells to ionizing radiation. Genome Res. 2003;13:2092–2100. doi: 10.1101/gr.1240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rube CE, Uthe D, Wilfert F, Ludwig D, Yang K, Konig J, et al. The bronchiolar epithelium as a prominent source of pro-inflammatory cytokines after lung irradiation. Int J Radiat Oncol Biol Phys. 2005;61:1482–1492. doi: 10.1016/j.ijrobp.2004.12.072. [DOI] [PubMed] [Google Scholar]
- 30.Datta K, Hyduke DR, Suman S, Moon BH, Johnson MD, Fornace AJJ. Exposure to ionizing radiation induced persistent gene expression changes in mouse mammary gland. Radiat Oncol. 2012;7:205. doi: 10.1186/1748-717X-7-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang TC, Paul S, Gong P, Levicky R, Kymissis J, Amundson SA, et al. Gene expression analysis with an integrated cmos microarray by time-resolved fluorescence detection. Biosens Bioelectron. 2011;26:2660–2665. doi: 10.1016/j.bios.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Moser K, Ortmann WA, et al. Expression levels for many genes in human peripheral blood cells are highly sensitive to ex vivo incubation. Genes Immun. 2004;5:347–353. doi: 10.1038/sj.gene.6364098. [DOI] [PubMed] [Google Scholar]
- 33.Rudqvist N, Parris TZ, Schuler E, Helou K, Forssell-Aronsson E. Transcriptional response of balb/c mouse thyroids following in vivo astatine-211 exposure reveals distinct gene expression profiles. EJNMMI Res. 2012;2:32. doi: 10.1186/2191-219X-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sokolov M, Panyutin IG, Neumann R. Genome-wide gene expression changes in normal human fibroblasts in response to low-let gamma-radiation and high-let-like 125iudr exposures. Radiat Prot Dosimetry. 2006;122:195–201. doi: 10.1093/rpd/ncl423. [DOI] [PubMed] [Google Scholar]
- 35.Ghandhi SA, Yaghoubian B, Amundson SA. Global gene expression analyses of bystander and alpha particle irradiated normal human lung fibroblasts: Synchronous and differential responses. BMC Med Genomics. 2008;1:63. doi: 10.1186/1755-8794-1-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dent P, Yacoub A, Contessa J, Caron R, Amorino G, Valerie K, et al. Stress and radiation-induced activation of multiple intracellular signaling pathways. Radiat Res. 2003;159:283–300. doi: 10.1667/0033-7587(2003)159[0283:sariao]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 37.Valerie K, Yacoub A, Hagan MP, Curiel DT, Fisher PB, Grant S, et al. Radiation-induced cell signaling: Inside-out and outside-in. Mol Cancer Ther. 2007;6:789–801. doi: 10.1158/1535-7163.MCT-06-0596. [DOI] [PubMed] [Google Scholar]
- 38.Lu TP, Lai LC, Lin BI, Chen LH, Hsiao TH, Liber HL, et al. Distinct signaling pathways after higher or lower doses of radiation in three closely related human lymphoblast cell lines. Int J Radiat Oncol Biol Phys. 2010;76:212–219. doi: 10.1016/j.ijrobp.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knops K, Boldt S, Wolkenhauer O, Kriehuber R. Gene expression in low- and high-dose-irradiated human peripheral blood lymphocytes: Possible applications for biodosimetry. Radiat Res. 2012;178:304–312. doi: 10.1667/rr2913.1. [DOI] [PubMed] [Google Scholar]
- 40.Clave E, Socie G, Cosset JM, Chaillet MP, Tartour E, Girinsky T, et al. Multicolor flow cytometry analysis of blood cell subsets in patients given total body irradiation before bone marrow transplantation. Int J Radiat Oncol Biol Phys. 1995;33:881–886. doi: 10.1016/0360-3016(95)00213-6. [DOI] [PubMed] [Google Scholar]
- 41.Brovall C, Schacter B. Radiation sensitivity of human natural killer cell activity: Control by x-linked genes. J Immunol. 1981;126:2236–2239. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.