Abstract
Incorporation of modified nucleotides into in vitro RNA or DNA selections offer many potential advantages, such as the increased stability of selected nucleic acids against nuclease degradation, improved affinities, expanded chemical functionality, and increased library diversity. This unit provides useful information and protocols for in vitro selection using modified nucleotides. It includes a discussion of when to use modified nucleotides; protocols for evaluating and optimizing transcription reactions, as well as confirming the incorporation of the modified nucleotides; protocols for evaluating modified nucleotide transcripts as template in reverse transcription reactions; protocols for the evaluation of the fidelity of modified nucleotides in the replication and the regeneration of the pool; and a protocol to compare modified nucleotide pools and selection conditions.
In vitro selection is the process by which a pool of nucleic acids is enriched via iterative selection and amplification for those species that are capable of performing a particular task. Nucleic acids have been selected that bind to particular targets (aptamers), catalyze reactions (ribozymes or deoxyribozymes), or act as molecular switches (aptazymes). Similarly, nucleic acids have been found in nature that control gene expression upon binding an analyte (riboswitches).
Instructions for carrying out in vitro selection experiments have been detailed elsewhere in this chapter (i.e. UNITS 9.3, 9.4, and 9.5). This unit augments these units by describing how modified nucleotides can potentially be incorporated into a selection. It is strongly recommended that the researcher be conversant with a “normal” in vitro selection experiment prior to attempting selections with modified nucleotides. A normal in vitro selection experiment is already fraught with problems and pitfalls, and the addition of modified nucleotides adds an extra level of difficulty. For simplicity, this unit focuses on in vitro selection using RNA pools; however, similar procedures can be used for DNA pools.
CAUTION: When working with radioactivity, take appropriate precautions to avoid contamination of the experimenter and the surroundings. Carry out the experiment and dispose of wastes in an appropriately designated area, following the guidelines provided by the local radiation safety officer.
NOTE: Experiments involving RNA require careful precautions to prevent contamination and degradation by RNases (see APPENDIX 2A). The water used to make all solutions and buffers should be RNase free or treated with diethylpyrocarbonate (DEPC; APPENDIX 2A). Use sterile, disposable plasticware where possible.
STRATEGIC PLANNING
Advantages of Using Modified Nucleotides in In Vitro Selections
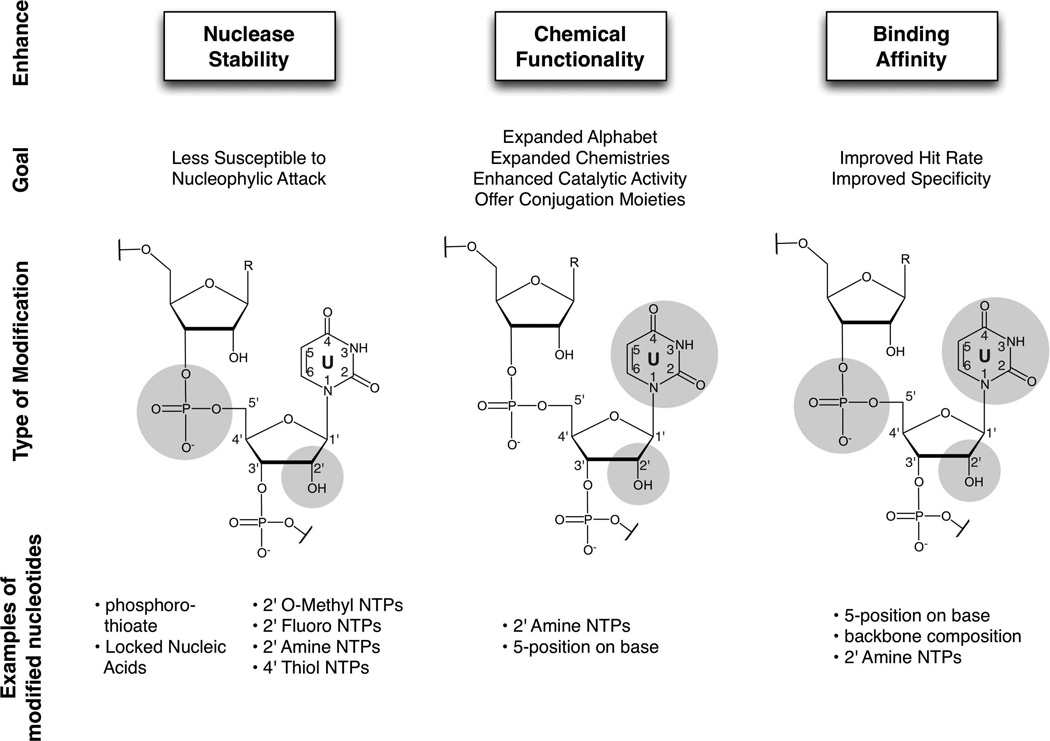
As discussed in other units (UNITS 9.3, 9.4 and 9.5), the desired outcome of a selection experiment should be determined in advance and the selection strategy should be designed accordingly. The decision to include modified nucleotides in a selection experiment and the choice of which nucleotide modifications to use should be based on how the nucleotides might benefit the selection (Figure 9.6.1). There are various advantages to using modified nucleotides, such as the increased stability of the selected nucleic acid against nuclease degradation, expanded chemical functionality, and / or improved aptamer binding affinity (recently reviewed in Bell and Micklefield, 2009; Mayer, 2009; Keefe and Cload, 2008; and Wilson and Keefe, 2006).
Figure 9.6.1.
Overview of the properties, advantages, and examples of modified nucleotides used in in vitro selections. For specific examples, refer to Table 9.6.1.
When nucleic acids are utilized as therapeutics or diagnostics, nuclease degradation is a common problem. Whereas unmodified nucleic acids are extremely susceptible to nuclease degradation, modification of the phosphate backbone or ribose moiety can render nucleic acids much more impervious to cleavage. For example, the incorporation of pyrimidine nucleosides modified at the 2′-position on the ribose moiety with amino or fluoro functional groups has been shown to drastically increase the stabilities of transcribed RNA molecules, in large measure because most ribonucleases polarize the 2’-hydroxyl group to attack the phosphodiester linkage. Numerous examples of nuclease-resistant aptamers and ribozymes have been published (cited in Table 9.6.1), including fully 2’-modified aptamers selected against vascular endothelial growth factor (Burmeister et al., 2005) and tissue factor pathway inhibitor (Waters et al., 2011).
Table 9.6.1.
Modified Nucleotides and Examples of Successful In Vitro Aptamer or Ribozyme/Deoxyribozyme Selections, if available.
| MODIFIED NUCLEOTIDE | POLYMERASE | TARGET/FUNCTION (CITATION)α | SPECIAL NOTES |
|---|---|---|---|
| Modified Sugars | |||
| 2’-Amino Pyrimidines* | Y639F T7 RNA Polymerase* |
VEGF/VPF (Green et al., 1995), Human neutrophil elastase (Lin et al., 1994), bFGF (Jellinek et al., 1995), hKGF (Pagratis et al., 1997), IFN-γ (Kubik et al., 1997) Ribozyme – trans cleavage of RNA (Beaudry et al., 2000) |
Several 2'-amino pyrimidine modified RNA aptamers have encountered synthesis difficulties and therefore have been abandoned as therapeutic candidates (reviewed in Keefe and Cload, 2008). |
| 2’-Fluoro Pyrimidines* | Y639F T7 RNA Polymerase* | VEGF 165 (Ruckman et al., 1998 and Chakravarthy et al., 2006), hKGF (Pagratis et al., 1997), IFN-γ (Kubik et al., 1997) | VEGF 165 aptamer is the first human aptamer therapeutic (Macugen ®). The aptamer was post modified after selection to include 2' O-Methyl nucleotides in a process called "back filling". |
| 2’-O-Methyl Nucleotides* | Y693F/H784A (Brieba and Sousa, 2000), "RGVG" (Chelliserrykattil and Ellington, 2004), "2P16" (Siegmund et al., 2012), or R425C T7 RNA Polymerase (Ibach et al., 2013) | VEGF 165 (Burmeister et al., 2005) | This nucleotide is less expensive to synthesize than 2'-amino or 2'-fluoro nucleotides. Also, 2'-O-methyl is a common post-transcriptional modification; for example, there are over 100 2'-O-methyl nucleotides are in each ribosome (Maden, 1990). Thus, the fact that these nucleotides are naturally occurring may make FDA approval more easily attainable. All 23 nucleotides in anti-VEGF aptamer are 2'-O-methyl nucleotides (Burmeister et al., 2005). |
| 2' Compositions, Mixed | Y639F/H784A/K378R T7 RNA Polymerase | Thrombin and IL-23 (Burmeister et al., 2006) | dRmY (dA/dG/mU/mC) was the chosen composition for in vitro selection, although others were considered. |
| TFPI (Waters et al., 2011) | dCmD (mA/mG/mU/dC) was the published composition for in vitro selection, although fRmY (fA/fG/mU/mC) was used in a patented variant against the same target. | ||
| 2’-O,4’-C-Methylene-Bridged ATP, GTP, TTP (Locked Nucleic Acids, LNAs) and 5-Methyl-CTP* | KOD Dash DNA Polymerase* and KOD mutant (KOD 2) DNA Polymerase (Kuwahara et al., 2000) (19:1 ratio of polymerases) (Kasahara et al., 2013) | Thrombin (Kasahara et al., 2013) | KOD Dash DNA polymerase is a mixture of a KOD polymerase and an archaeal DNA polymerase with proofreading activity (available from Toyobo Co. Ltd. (Osaka, Japan)). In the selection, 2’-O,4’-C-methylene bridged (B/L) TTP was incorporated enzymatically, whereas the B/L ATP, B/L GTP, and 5-methyl-CTP were present only in the pool (forward) primer region (Kasahara et al., 2013). |
| 4'-Thio Pyrimidines* | T7 RNA Polymerase* | Thrombin (Kato et al., 2005) | The thio-modified aptamer is 50 times more stable in the presence of RNase A and has an increase in thrombin inhibition compared to the corresponding unmodified RNA aptamer (Kato et al., 2005). |
| Modified Bases | |||
| 5-(3-Aminoally) Deoxycytidine* | Vent (exo -) DNA Polymerase* | RNase DNAzyme (Hollenstein et al., 2013) | This modified DNAzyme contains 3 modified nucleotides (5-guanidinoallyl-dUTP, 5-aminoallyl-dCTP, and 5-imidazolyl-dATP) and cleaves all-RNA targets independent of M2+ (Hollenstein et al., 2012). |
| 5-(3-Aminoallyl) Deoxyuridine* | Sequenase 2.0 DNA Polymerase* | RNase DNAzyme, sequence directed (Perrin et al., 2001) | This RNase DNAzyme contains both the 8-[2-(4-Imidazolyl)ethylamino] deoxyadenosine and the 5-(3-aminoallyl) deoxyuridine modified nucleotides and self-cleaved the internal rC. This is the first example of a metal-independent DNAzyme (Perrin et al., 2001). |
| 5-N-(6-Aminohexyl)Carbamoylmethyl Deoxyuridine | KOD DNA Polymerase* | Thalidomide (Shoji et al., 2007) | This aptamer is highly specific for the R-enantiomer of thalidomide (Shoji et al., 2007). |
| 5-(3-Aminopropynyl) Deoxyuridine | Vent DNA Polymerase* | ATP, ADP, and AMP (Battersby et al., 1999) | This is the first example of the incorporation of a positively charged functional group (Battersby et al., 1999). |
| 5-Benzylaminocarbonyl Deoxyuridine (BndU) (Vaught et al., 2010) | KOD DNA Polymerase* | Plasminogen Activator Inhibitor-1 (PAI-1), as well as multiple other targets (Gold et al., 2010) | Modified nucleotides were used to select against thirteen human proteins ("difficult targets") for which unmodified RNA / DNA in vitro selection did not yield an aptamer. The extent to which a given nucleotide modification was beneficial for a selection was highly dependent on the protein target (Gold et al., 2010). |
| 5-Boronic acid-modified Thymidine | Taq DNA Polymerase* | Fibrinogen (Li et al., 2008) | |
| 5-Bromo Deoxyuridine* | Taq DNA Polymerase* (Brody et al. 1999 and Golden et al. 2000) or E. coli DNA Polymerase I* (Smith et al. 2003) | bFGF (Brody et al. 1999 and Golden et al. 2000) and GP120MN (Smith et al. 2033) | Photocrosslinking aptamers were generated with this modified nucleotide. |
| 5-Carboxamide-modified Deoxyuridine (Vaught et al., 2010) | Deep Vent and KOD XL DNA Polymerases* | Tumor necrosis factor receptor super family member 9 (TNFRSF9) (Vaught et al., 2010) | PCR amplification could not be performed using any of the carboxide derivatives of dUTP, only primer extensions. Therefore, an intermediate PCR step using unmodified nucleotides was required to exponentially amplify, and then subsequent primer extension reactions reincorporated the modified nucleotides. Additionally, amide linkages increase the possibility of hydrogen bonding with the target (Vaught et al., 2010). |
| 5-Guanidinoallyl Deoxyuridine* | Vent (exo -) DNA Polymerase* | RNase DNAzyme (Hollenstein et al., 2013) | See "SPECIAL NOTES" for 4-(3-Aminoallyl) Deoxycytidine. |
| 5-Isobutylaminocarbonyl Deoxyuridine (iBudU) (Vaught et al., 2010) | KOD DNA Polymerase* | Human mobility group -1 (HMG-1), as well as multiple other targets (Gold et al., 2010) | See "SPECIAL NOTES" for 5-Benzylaminocarbonyl Deoxyuridine. |
| 5-Imidazolyl Deoxyadenosine* | Vent (exo -) DNA Polymerase* | RNase DNAzyme (Hollenstein et al., 2013) | See "SPECIAL NOTES" for 4-(3-Aminoallyl) Deoxycytidine. |
| 5-Imidizole Uridine | T7 RNA Polymerase* | Ribozyme with amide synthase activity (Wiegand et al., 1997); Ribozyme with urea synthase activity (Nieuwlandt et al., 2003) | A side-by-side comparison of a 5-imidazol uridine RNA pool versus an unmodified RNA pool used in a selection for urea bond catalysts resulted with significant catalytic activity of the modified pool after nine rounds of selection, while no significant increase in catalytic activity was observed over background with the unmodified RNA selection after fourteens rounds of selection (Nieuwlandt et al., 2003) |
| 5-Imidizole-Uridine analog (unnamed) | Thermostable DNA Polymerases* (possibly Taq, Vent, Pfu, and rTh DNA polymerases) | RNase DNAzyme, sequence directed (Santoro et al., 2000) | |
| 5-Naphtylmethylaminocarbonyl Deoxyuridine (NapdU) (Vaught et al., 2010) | KOD DNA Polymerase* | Human protein targets (Gold et al., 2010) | See "SPECIAL NOTES" for 5-Benzylaminocarbonyl Deoxyuridine. |
| 5-(2-Naphtylmethylaminocarbonyl) Deoxyuridine (2NapdU) (Vaught et al., 2010 and Ochsner et al., 2013) | KOD DNA Polymerase* | C. difficile toxins (Ochsner et al, 2013) | |
| 5-(1-Pentynyl) Deoxyuridine | Vent DNA Polymerase* | Thrombin (Latham et al., 1994) | |
| 5-Phenylethyl Deoxyuridine (PEdU) (Vaught et al., 2010 and Ochsner et al. 2013) | KOD DNA Polymerase* | C. difficile toxins (Ochsner et al, 2013) | |
| 5-Pyridylmethylcarboxamid Uridine | Not noted in paper | Ribozyme with Diels-Alderase activity (Tarasow et al., 1997) | |
| 5-Tyrosyl Deoxyuridine (TyrdU) (Vaught et al., 2010 and Ochsner et al., 2013) | KOD DNA Polymerase* | C. difficile toxins (Ochsner et al, 2013) | |
| 5-Tryptaminocarbonyl Deoxyuridine (TrpdU) (Vaught et al., 2010) | KOD DNA Polymerase* | Fractalkine(CX3CL-1), as well as multiple other targets (Gold et al., 2010) | See "SPECIAL NOTES" for 5-Benzylaminocarbonyl Deoxyuridine. |
| 6-Aminohexyl Adenosine | T7 RNA Polymerase* | Ribozyme with ligase activity (Teramoto et al., 2000) | The ribozyme catalyzed the ligation to its 5' end (Teramoto et al., 2000). |
| 7-(2-Thienyl)Imidazo[4,5-b] Pyridine* | AccuPrime Pfx DNA Polymerase* | VEGF 165 and IFN-γ (Kimoto et al., 2013) | Modified nucleotide exclusively pairs with diol-modified 2-nitro-4-propynlpyrrole, in essence creating a third base pair. The modified aptamers have >100-fold binding affinity over the unmodified aptamers (Kimoto et al., 2013). |
| 8-[2-(4-Imidazolyl)Ethylamino] Deoxyadenosine | Sequenase 2.0 DNA Polymerase* | RNase DNAzyme, sequence directed (Perrin et al., 2001) | See "SPECIAL NOTES" for 5-(3-Aminoallyl) Deoxyuridine. |
| Modified Phosphate Backbone | |||
| Boranophosphate linkages | T7 RNA Polymerase* | ATP (Lato et al., 2002) | |
| Phosphorothioate Linked DNA* (S-linked dNTPs) | Taq DNA Polymerase* |
NF-kB RelA (p65) (King et al., 2002) p50 (King et al., 2002) NF-IL6 (King et al., 1998) |
These modifications are nuclease resistant and efficiently internalized by cells (King et al., 2002). |
| Phosphorothioate Linked RNA* (S-linked NTPs) | T7 RNA Polymerase* | bFGF (Jhaveri et al., 1998) | |
Denotes the modified nucleotide or polymerase is commercially available
Abbreviations: bFGF, basic fibroblast growth factor; hKGF, human keratinocyte growth factor; IFN-γ, interferon γ; NF-IL6, nuclear factor for human interleukin 6; VEGF, vascular endothelial growth factor; VPF, vascular permeability factor.
Similarly, substitutions on the nucleic acid backbone, such as replacing the phosphate with a phosphorothioate (mono- or di-; Zon and Geiser, 1991) or linking the 2’- and 4’-positions of the ribose (reviewed in Veedu and Wengle, 2010), have also been shown to increase oligonucleotide stability in the presence of nucleases. An additional benefit is that phosphorothioate nucleotides have been shown to be incorporated into an elongating transcript by T7 RNA polymerase with little or no increase in KM (Griffiths et al., 1987). While DNA is not as vulnerable to hydrolysis as RNA, it is nonetheless susceptible to cleavage by a variety of deoxyribonucleases and phosphodiesterases. The stability of DNA can also be increased by the incorporation of phosphorothioate nucleotides, and these can be readily incorporated by Taq DNA polymerase (Nakamaye et al., 1988) and used for selection (King et al., 1998).
In addition to enhancing nuclease resistance, modified nucleotides potentially expand the chemical functionality of nucleic acids. Modified nucleotides have been included in selections for catalytic nucleic acids. The resultant catalysts have been shown to be highly dependent upon the modifications for activity (Tarasow et al., 1997; Wiegand et al., 1997; Beaudry et al., 2000; Santoro et al., 2000) and, in some examples, potentially improve the catalytic activity. For example, Nieuwlandt et al. (2003) selected a modified (5-imidazolyluridine) ribozyme that catalyzed the conjugation of the 3’-amino group of 3’-amino-3’-deoxycytidine to the N-terminus of a tripeptide substrate, forming a urea linkage. In the same work, a parallel unmodified selection was performed. Significant catalytic activity was observed from the modified pool after nine rounds of selection, but no significant increase in catalytic activity was seen in the unmodified RNA selection even after fourteen rounds of selection. In another example, Hollenstein et al. (2013) selected a metal-independent RNase active deoxyribozyme containing modified cytidine, uridine and adenine nucleosides (5-guanidinoallyl-dU, 5-aminoallyl-dC, and 8-2-(4-imidazolyl)ethylamino- dA) from a doped sequence pool that was in turn based on a previously identified unmodified deoxyribozyme. According to the first order rate constants, this modified deoxyribonuclease had improved reaction kinetics over similar, unmodified ribozymes (Geyer and Sen, 1997; Faulhammer and Famulok, 1997). The identification of magnesium (Mg2+)-independent catalysts may have been due to the inclusion of modified nucleotides or may have just been due to further selection; parallel experiments with an unmodified pool were not performed. In this regard, though, when a similar selection was performed using a deep random pool (i.e., not “doped”) as a starting point no RNase activity was found after nine rounds of selection.
In line with the hypothesis that modified nucleotides expand the functional repertoire of nucleic acids, there are multiple examples of modification dependent aptamers or deoxy/ribozymes (i.e., binding/activity is lost when modified nucleotides are replaced with unmodified nucleotides). For instance, the first RNA capable of catalyzing the formation of carbon-carbon bonds utilized 5-pyridylmethylcarboxamid-UTP in place of UTP (Tarasow et al., 1997). When the most active isolate from this selection was transcribed with unmodified UTP, it was inactive. Similar results were obtained for a ribozyme that catalyzed amide bond formation (Wiegand et al., 1997), whose activity was dependent on the incorporation of 5-imidazolyl -UTP. Additionally, two sequence-specific RNase deoxyribozymes were dependent on the incorporation of a 5-imidazolyl -dUTP (Santoro et al., 2000), and both 8-2-(4-imidazolyl)ethylamino-2′-dATP and 5-(3-aminoallyl)-2′-dUTP (Perrin et al., 2001). However, none of the sequences, motifs, or activities found in these selection experiments was directly compared with ribozymes that contained canonical nucleotides and that were sieved from the same pool using the same selection conditions.
Perhaps surprisingly, though, there are at least a few conflicting examples and counterexamples suggesting that modified nucleotides do not greatly contribute to binding or catalysis relative to unmodified nucleotides. Santoro and colleagues (Santoro and Joyce, 1997; Santoro et al., 2000) selected deoxyribozymes with RNA hydrolysis activity from different aliquots of the same, unamplified random sequence pool. According to the second order rate constants, the selection performed with unmodified nucleotides produced a much faster catalyst (Santoro and Joyce, 1997) than that identified through the modified selection (Santoro et al., 2000). Ultimately, it is unknown whether this indicates the superiority of unmodified nucleotides for this pool and this function, or whether the fraction of the original pool used for the selection of the unmodified catalyst just contained a “jackpot” sequence. In another contrasting example, whereas Tarasow et al. (1997) suggested that the inclusion of a modified nucleotide was the only reason they were able to select a Diels-Alder synthetase, Seelig et al. (1999) later selected ribozymes from an unmodified pool. The modified selection yielded a catalyst with a kcat/KM of ~4 M−1sec−1, while the unmodified selection yielded a catalyst with a kcat/KM of 167 M−1sec−1. However, these selections were performed by different research groups with different pools, and thus, again, are not directly comparable. For a final example, see Critical Parameters.
To the extent that a modified nucleotide will be included to hopefully enhance catalytic functionality, the choice of modification should complement the desired function. For example, an imidizole ring (with a pKa near neutrality; Battersby et al., 1999) may be beneficial in a pool to be screened for catalysis of an acid/base reaction, while the incorporation of a thiolated residue (Jhaveri et al., 1998) could allow nucleic acids to participate in disulfide bond formation or rearrangement, reactions normally denied to them. Additionally, 5-bromo and 5-iodo modifications of pyrimidine bases can serve as moieties for UV crosslinking to covalently linking a modified aptamer to its targeted protein. In those cases where the target protein is not already known, this can lead to biomarker identification (Mallikaratchy et al., 2007). Crosslinking can also be used to increase the stringency of washing and thereby decrease background (Brody et al., 1999).
While most of the modifications mentioned thus far retain native Watson-Crick pairing, this need not always be the case. Introducing modifications with novel base pairing may potentially provide additional chemical and functional properties, unrestricted by unmodified nucleotide base pairing. For example, Piccirilli et al. (1990) have shown that diaminopyrimidine and xanthosine can form a stable base pair, although the fidelity of the diaminopyrimidine during the polymerase chain reaction (PCR) was poor (Southworth et al., 1998). The Benner group developed an unnatural base pair (6-amino-5-nitro-3-(1’-β-d-2’-deoxyribofuranosyl)-2(1H)-pyridone and 2-amino-8-(1’-β-d-2’-deoxyribofuranosyl)-imidazo[1,2-α]-1,3,5-triazin-4(8H)-one) that is retained during PCR (Yang et al., 2011) and thus is suitable for in vitro selection. Further, Kimoto et al. (2013) have introduced the hydrophobic base pair 7-(2-thienyl)imidazole[4,5-b]pyridine and diol-modified 2-nitro-4-propynylpyrrole into selection experiments. Notably, these modified aptamers against human vascular endothelial cell growth factor-165 (VEGF-165) and interferon-γ (IFN-γ) have >100-fold improved dissociation constants over aptamers with unmodified nucleotides.
Modified nucleotide pools can also potentially increase the overall binding affinities of selected aptamers. For example, Vaught et al. (2010) selected a carboxamide-modified dUTP aptamer against tumor necrosis factor superfamily number 9 (TNFRSF9); previous unmodified DNA selections had failed. The increased affinity of this aptamer relative to similar RNA aptamers suggested that the modifications offered some binding advantage. Similar enhancements of modified aptamer affinities relative to unmodified aptamers have also been observed for the human α-thrombin aptamer (Kato et al., 2005), vascular endothelial growth factor (VEGF) aptamer, and interferon γ aptamer (Kimoto et al., 2013).
Finally, selections that targeted thirteen “difficult” human protein targets that had previously failed to yield aptamers with native DNA pools instead yielded high affinity aptamers when the selections were carried out with pools that contained a modification at the 5-position of uridine. Across nearly 1500 human proteins, pools containing 5-tryptaminocarbonyl-dU or 5-benzylaminocarbonyl-dU enhanced the success rate of selection to over 84% compared to selections containing the parental dU or dT nucleotide. Interestingly, the extent to which a given nucleotide modification was beneficial for a selection was highly dependent on the protein target (Gold et al., 2010). It should be noted however, that there were still some proteins in the repertoire that failed to produce any aptamers, suggesting that improvements in methods and novel modifications could still be beneficial.
Preparation and Use of Modified Nucleotides
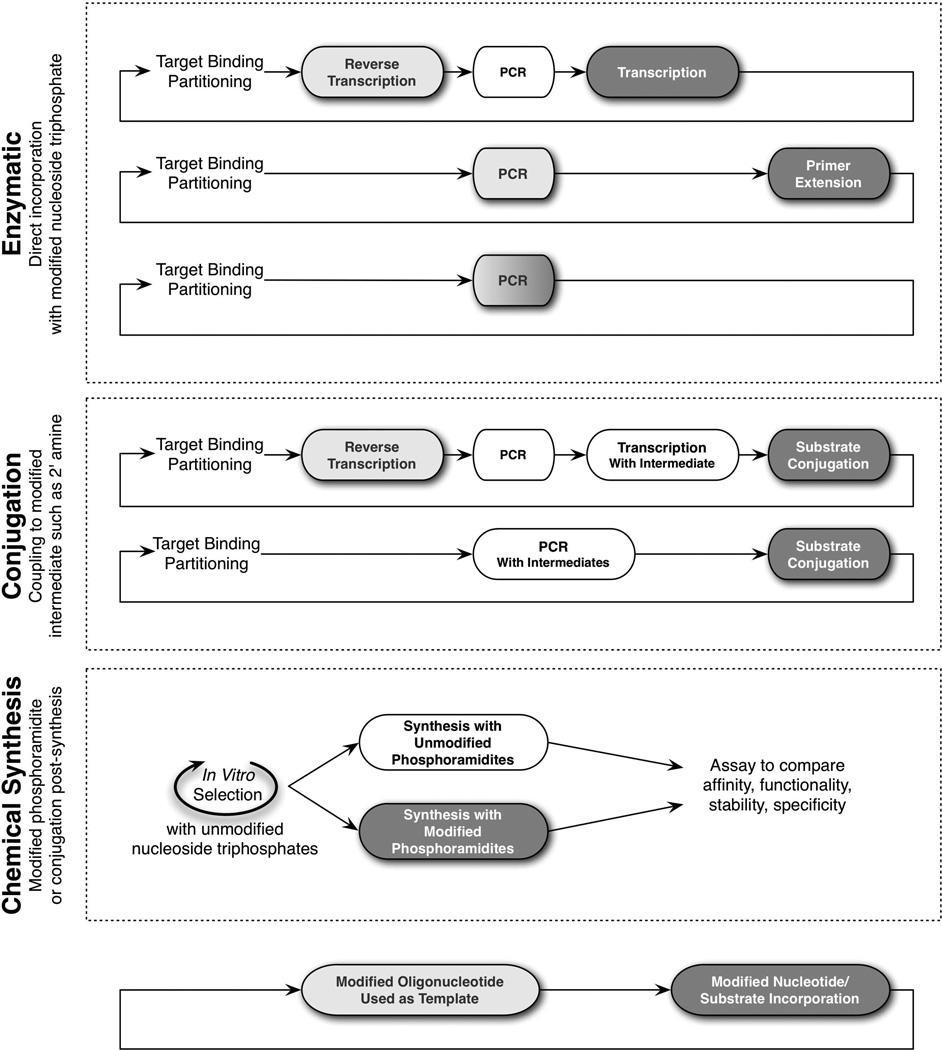
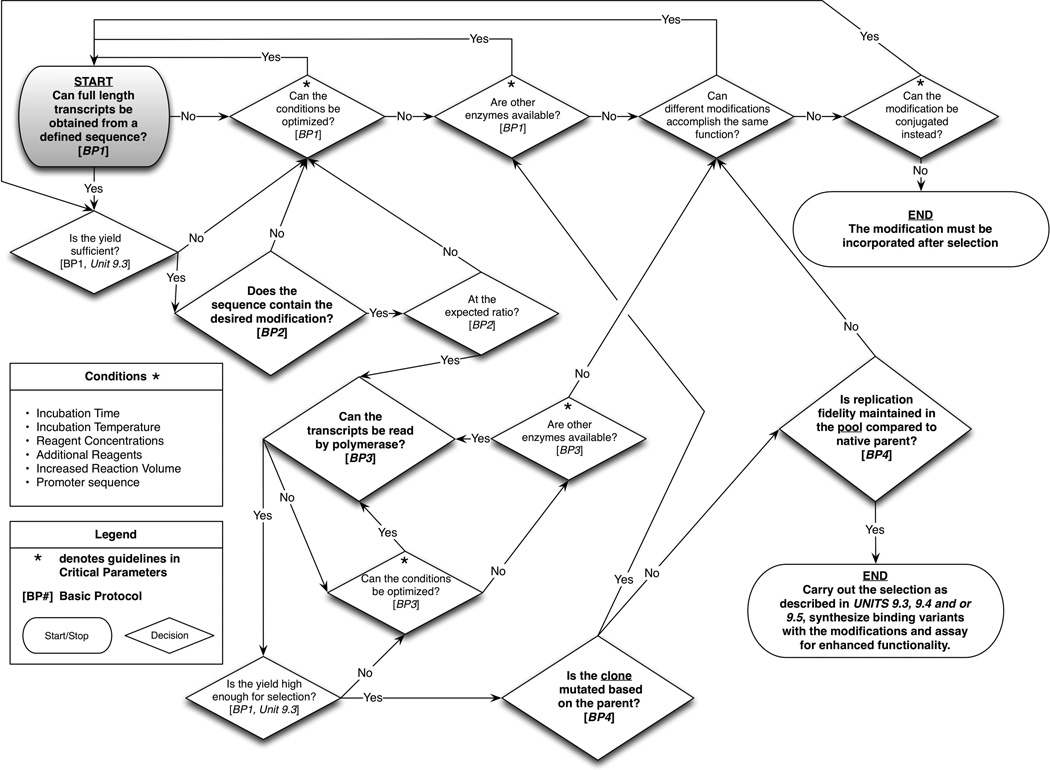
This unit presents procedures in two main sections: determination of the suitability of modified nucleotides for in vitro selection, followed by the determination of the modified pool most suitable for in vitro aptamer selection. Once the decision has been made that a modified nucleotide is necessary (see Figure 9.6.1), the main consideration is how to incorporate the given modification (Figure 9.6.2) and whether the given nucleoside triphosphate analog can be reproducibly and specifically introduced (Figure 9.6.3). For instance, can the modification be recognized and incorporated by a polymerase (i.e., “written” into the transcript)? If incorporated, can the modified nucleoside analog be specifically recognized by a polymerase and interpreted correctly without causing mutations (i.e., “read” from the transcript)? When potential enzymes have been identified, Figure 9.6.3 describes a validation schema that can be utilized to ensure both reproducibility and specificity of modified analog incorporation. It will also be critical to optimize reaction conditions to ensure sufficient yields for each round of the selection.
Figure 9.6.2.
Mechanisms to incorporate modified nucleotides into in vitro selections. Three mechanisms are routinely used to incorporate modified nucleotides into in vitro selections: “Enzymatic,” “Conjugation,” and “Chemical Synthesis.” These mechanisms and examples are discussed in detail in the Critical Parameter section. In this figure, the light grey bubble indicates that a modified nucleotide will serve as a template. The dark grey bubble indicates that the modified nucleotide will be the substrate for incorporation. If the modified nucleotide can serve both as a template and also be incorporated by the same enzyme, the bubble is shaded from light to dark grey.
Figure 9.6.3.
Evaluation and optimization schema to validate the successful incorporation of modified nucleotides into a pool for in vitro RNA selection. Start in the grey shaded box.
The protocols contained herein primarily focus on the enzymatic incorporation of modified nucleotides and the use of RNA pools, which require reverse transcriptases, DNA polymerases, and transcriptases to regenerate the pool. For selections requiring only DNA polymerases, the protocols can be taken as guidelines for designing similar workflows with methods (PCR, for instance) described elsewhere (UNIT 9.2 Support Protocol 3 and Basic Protocol 2). Methods for chemical synthesis and screening without an amplification step are not covered in this chapter.
The first set of procedures describe transcription reactions used to ensure that modified nucleotides can act as substrates to produce full-length transcripts (see Basic Protocol 1). Next, it is necessary to check the quality and prevalence of the modified nucleotide in the transcript (see Basic Protocol 2). Since selected transcripts must be amplified, it is necessary to verify that the modified transcript is a suitable template for reverse transcriptase (see Basic Protocol 3). If Basic Protocols 1, 2, and 3 give successful results, then it is likely that the random sequence pool can make it through the preparative steps leading to the selection. However, it is important to ensure the modification does not introduce unwanted mutations or base conversions into the transcript (see Basic Protocol 4). Finally, a mock selection (omitting the selective step) using a pool including the modifications should be performed with all the enzymatic steps in succession, and then the products should be sequenced to determine the fidelity of incorporation (see Basic Protocol 4 for a discussion). An overall outline of the process detailing key decisions is presented in Figure 9.6.3.
Once a given modified nucleotide(s) has been validated for use in selection, the starting pool or pool candidates can be constructed, and trial selections can be performed. In the second set of protocols, double-stranded DNA that codes for the modified pool (as generated in UNIT 9.2) is first transcribed, and the products are gel purified using the optimized set of conditions developed from Basic Protocols 1 and 3. Pool candidates possessing different modifications can be tested side-by-side to determine which is best for a given target (see Basic Protocol 5). The sequential step in the overall selection scheme, in vitro selections, is described in detail in UNITS 9.3, 9.4, and 9.5.
DETERMINATION OF THE SUITABILITY OF A MODIFIED NUCLEOTIDES FOR IN VITRO SELECTION
BASIC PROTOCOL 1
Evaluation and Optimization of Transcription with a Modified Nucleotide
In the case of RNA selections, the ability of a modified nucleotide to be incorporated into a transcript should be evaluated. It is best to perform a transcription reaction in which the modified nucleotide completely substitutes for its unmodified counterpart (e.g., 2’-deoxy-2′-fluorocytidine in place of cytidine) using a template of known sequence composition (initially). Once incorporation of the nucleotide has been validated, a small aliquot of pool template should be used as template to verify fidelity of incorporation thereby reserving most of the pool for the optimized large scale reaction and selections. A positive control reaction should always be performed in parallel on the same template with all unmodified nucleotides. Similarly, a negative control reaction, void of the modified nucleotide and unmodified counterpart, should always be performed on the same template. The comparative success of these reactions can be determined by including α-32P-labeled nucleotides in the reaction mix, separating transcripts by denaturing polyacrylamide gel electrophoresis, and visualizing the full-length transcripts with a phosphorimager or via autoradiography.
This protocol also includes optimizing the transcription reaction using different modified and unmodified nucleotide concentrations, buffer components, starting double stranded template concentration, and different combinations of modified nucleotides (see Critical Parameters for examples and references). If after optimization, yields (UNIT 9.3, Basic Protocol 1) are not high enough for the selection (see UNIT 9.3, Basic Protocol 2), then consider such options such as scaling up the reaction size, other modifications, or perhaps a single round of selection (or screening), followed by high throughput sequencing, and sequence analysis for aptamer candidates (see Figure 9.6.3 for a full range of options and validation schema).
Materials
3000 Ci/mmol α-32P-labeled ATP (e.g., Perkin Elmer)
- AmpliScribe T7 High Yield Transcription kit (Epicentre) for unmodified NTP incorporation, kit includes:
- T7 Enzyme solution,
- 100 mM ATP, CTP, GTP, and UTP solutions,
- 10× Reaction Mix, and
- 100 mM Dithiothreitol (DTT).
- DuraScribe T7 Transcription kit (Epicentre) for modified NTP incorporation, kit includes:
- DuraScribe T7 Enzyme solution,
- 50 mM ATP and GTP solutions
- 50 mM 2’-deoxy-2’-fluoro-CTP and 2’-deoxy-2’-fluoro-UTP solutions
- 10× Reaction Mix, and
- 100 mM Dithiothreitol (DTT).
100 mM modified nucleotide
500 ng DNA clone of known sequence composition (dsDNA containing a T7 promoter)
500 ng purified DNA pool or randomized clone per reaction (dsDNA containing a T7 promoter) (UNIT 9.2)
2× denaturing stop dye (see recipe)
10% (w/v) denaturing polyacrylamide gel (see recipe, APPENDIX 3B), 0.8 mm thick
Thermal cycler or 37 °C to 42 °C and 70 °C water baths or heating blocks
Gel blotting paper (Bio-Rad)
Plastic wrap
Gel dryer with vacuum (e.g., Bio-Rad)
Phosphorimager screen
Phosphorimager and image analysis software (e.g., GE Healthcare Life Sciences, ImageQuant)
Additional reagents and equipment for denaturing polyacrylamide gel electrophoresis (PAGE; APPENDIX 3B)
Perform transcription with modified and unmodified nucleotides
-
1Prepare 3 transcription reactions: (1) In the experimental reaction, the modified nucleotide is completely substituted for its unmodified counterpart (e.g., 2’-deoxy-2′-fluorocytidine in place of cytidine). (2) In the positive control reaction, all unmodified nucleotides are used. (3) In the negative control reaction, the modified nucleotide and unmodified counterpart is omitted. Assemble the transcription reactions according to the kit instructions (see also UNIT 9.3, Basic Protocol 1). Combine the following in three separate tubes (20 µL total volume):
- 2 µL 10× Transcription (TNX) Buffer (AmpliScribe kit)
- 2 µL 100 mM DTT (AmpliScribe or DuraScribe kit)
- 1.5 µL each NTP (AmpliScribe kit) or modified nucleotide
- 4 µL (~500 ng) DNA clone of known sequence composition
- 4 µL water
- 1.5 uLT7 RNA Polymerase (AmpliScribe or DuraScribe kit)
- 0.5 µL 3000 Ci/mmol α-32P-labeled ATP (this volume should be varied based on the age and subsequent activity of the radiolabeled ATP)
The AmpliScribe kit (Epicentre), containing T7 RNA polymerase, is a high-yield unmodified transcription kit. More affordable, conventional transcription reagents may be used in place of the AmpliScribe kit, however lower yields may result.The DuraScribe kit (Epicentre), containing a proprietary preparation of the Y639F variant of T7 RNA polymerase, is a transcription kit optimized for the incorporation of 2’-deoxy-2´-fluoro-CTP, 2’-deoxy-2´-fluoro-UTP, ATP, and GTP. Alternatively, other or additional polymerases may be tested for modified nucleotide incorporation (see Critical Parameters and multiple examples provided in Table 9.6.1).Many mutant T7 polymerases are not as active as their wild type counterparts, and therefore may require additional time, temperature, volume and buffer considerations that should be optimized as described below (see Critical Parameters). -
2Incubate at 37 °C to 42 °C.If α-32Phosphate substrates are not suitable or available (for instance if all unmodified nucleotides are replaced by a modification), the polyacrylamide gel can be stained with a dye such as SYBR gold, or the template can be end-labeled with γ-32P. However, an additional DNase step (such as with the use of Epicentre Baseline Zero DNase) will be required to digest template DNA, thus permitting just the detection of the RNA transcripts.If the template is the product of an extension or PCR reaction, care should be taken to remove the unincorporated dNTPs or assay extension efficiency in the absence of one nucleotide (preferably the modified nucleotide).
-
3At several time points (e.g., 0.25, 0.5, 1, and 2 h), gently mix and briefly centrifuge the sample to bring down the liquid. Transfer 1 µL of the reaction to separate tubes containing 4 µL nuclease free diH2O and 5 µL of 2× denaturing stop dye.Mixing and centrifuging the reaction ensures that each sample will contain a consistent portion of the reaction. The accumulation of condensation and the precipitation of pyrophosphate during transcription can sometimes lead to variations in reaction volumes or concentrations.
-
4Denature the time point samples by heating 3 min to 70 °C and separate transcripts from aborted transcripts and unincorporated nucleotides on a 0.8 mm thick, 10% denaturing polyacrylamide gel (APPENDIX 3B).The concentration of acrylamide can be varied to efficiently separate different length products, depending on their sizes (APPENDIX 3B).Pre-running the gel will heat it up and help to denature molecules that contain significant secondary structure.
Analyze transcription reactions
-
5Remove one of the two glass plates and place a sheet of gel blotting paper against the gel. Peel the gel off of the other glass plate (it will stick to the paper) and cover it with plastic wrap. Dry the gel under heat and vacuum using a commercial gel dryer.Drying the gel can be omitted, and a wet gel can be directly used for exposure. If this is done, leave the gel against one glass plate and carefully wrap with plastic wrap to avoid contaminating the phosphorimager screen and the exposure cassette with radiation.
-
6Expose the dried gel to a phosphorimager screen for 1 h.The exposure time may need to be increased or decreased, depending on the specific activity of the labeled nucleotide and the amount of transcript that is produced.Gels can also be exposed to X-ray film, although this makes quantitation somewhat more cumbersome. A standard laboratory densitometer can be used for quantitation.
-
7
Develop the phosphorimager screen and calculate the relative amounts of RNA transcripts produced using image analysis software such as ImageQuant.
-
8
Compare the intensities of product bands for the transcription reaction that includes the modified nucleotide to those for transcription with the normal nucleotide to determine the relative efficiency of incorporation for the modified nucleotide under the conditions tested.
-
9
Plot the amount of product that accumulates versus time to determine the optimal time for further transcriptions.
Optimize transcription reaction
-
10Following the validation schema outlined in Figure 9.6.3, repeat steps 1 to 7 using DNA pool or randomized clone (~500 ng) as template and different modified and unmodified nucleotides concentrations, buffer components, template concentrations, and modified nucleotides combinations (see Critical Parameters for examples and references). Take samples only at the optimal time determined in step 9.If pool template is used for optimization, it is advisable to amplify only a small portion of the pool that will actually be used in the selection for these test transcriptions. As this is a test reaction, do not commit the entire pool.Pre-running the gel will heat it up and help to denature molecules that contain significant secondary structure. Nonetheless, pools can sometimes appear “fuzzy” on a gel due to the presence of molecules that differ slightly in length or that have different secondary structures.Since it is likely that the modified nucleotide will not work as well as the normal nucleotide because it binds to the polymerase more poorly, increasing the modified nucleotide concentrations may be necessary to enhance transcription. When increasing the modified nucleotide concentration, care should be taken that the amount of modified nucleotide does not unnecessarily restrict the magnesium concentration in solution. If the total nucleotide concentration approaches that of magnesium, or the concentration of modified nucleotide exceeds the normal concentration used for unmodified (i.e., 2.5 mM), then additional magnesium should be added to the reaction. Magnesium can, however, decrease the fidelity of the polymerase and should be assessed (see Basic Protocol 4). Alternatively, increasing the polymerase concentration may increase the efficiency of the PCR reaction incorporating modified nucleotides (see, Kojima, et al. 2013).The initial sequence of the transcription product can also influence the incorporation of a modified nucleotide as can the promoter sequence when different polymerases are utilized (see Critical Parameters for examples and references).5’ End-labeling of the transcription products with γ-32P-ATP in a kinase reaction (see UNIT 9.3, Support Protocol 1) can also be utilized if internal labeling is undesirable.
-
11Plot the amount of product (band intensity) versus modified nucleoside triphosphate concentration or alternate reaction condition.If the modified nucleotide is expensive or difficult to produce, this step is important to determine the minimum quantity of modified to use that maximizes the yield of RNA. To conserve modified nucleotide, the lowest concentration of modified nucleotide should be chosen at which near-maximal levels of RNA are produced.
BASIC PROTOCOL 2
Confirmation of the Presence of Modified Nucleotides
The appearance of a full-length product in the transcription reaction (see Basic Protocol 1) is encouraging, but does not indicate the level at which a modified nucleotide has been incorporated into a pool. For example, full-length RNAs could be members of the original population that incorporated the modified nucleotide at only a few positions, could have been synthesized via contaminating unmodified nucleotides, could have been synthesized via the misincorporation of a unmodified nucleotide (e.g., via G:U mismatches), or could be the result of some combination of these possibilities. In order to determine with certainty the level of modified nucleotide incorporation, transcription products should be isolated, fully digested, and separated by high-performance liquid chromatograph (HPLC) to identify the component nucleosides.
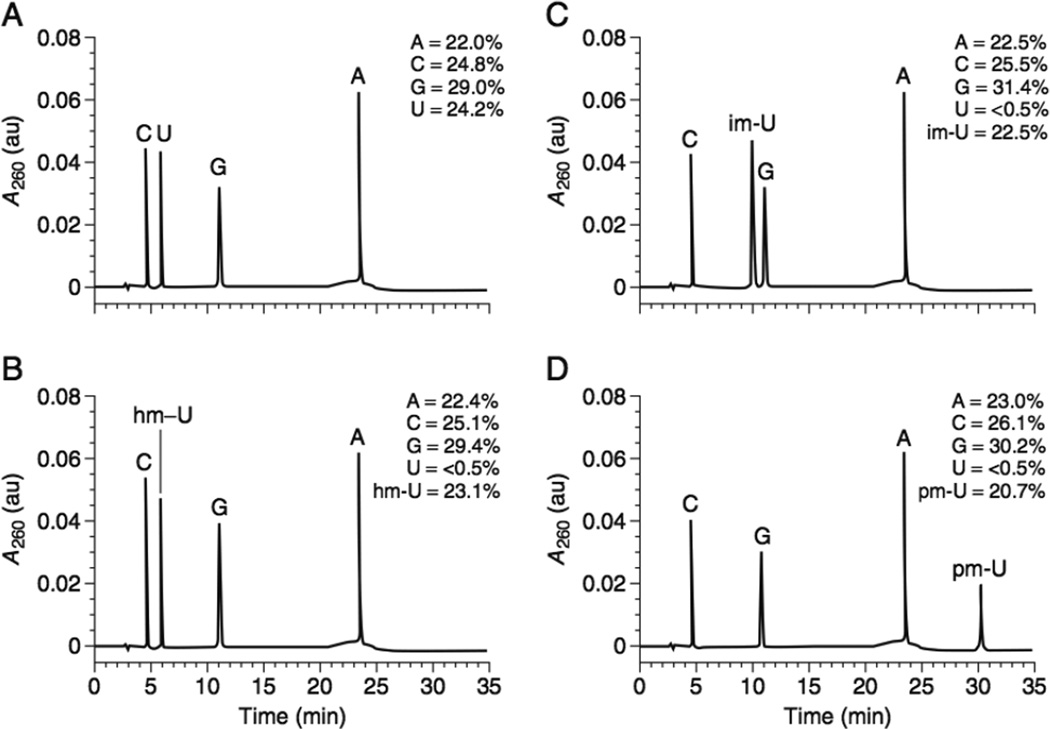
This protocol assumes that one has already gel-purified and calculated the yields of the full-length transcription products without radioisotopes (UNIT 9.3, Basic Protocol 1). The RNA will be digested to mononucleotides using nuclease P1 and then treated with alkaline phosphatase to generate nucleosides. Separation of individual nucleotides will be carried out by reversed-phase HPLC. Digestions will be compared to standards containing both modified and unmodified nucleotides. Sample HPLC data are given in Figure 9.6.4 (Robertson, 2001).
Figure 9.6.4.
HPLC elution profile of pool RNA that has been treated with nuclease P1 and alkaline phosphatase as described in Basic Protocol 2. (A) Unmodified pool, (B) pool containing 5-hydroxymethyluridine, (C) pool containing 5-imidazolemethyluridine, (D) pool containing 5-phenolmethyluridine (Robertson, 2001).
Materials
Transcribed RNA pools, modified and unmodified (Basic Protocol 1 without radiolabeled ATP)
Nuclease P1 digestion mix (see recipe)
Alkaline phosphatase reaction mix (see recipe)
HPLC mobile phase solution: 5% (v/v) methanol in 0.1 M sodium phosphate, pH 6.0 (APPENDIX 2A)
Thermal cycler or 37 °C and 50 °C water baths
HPLC with reversed-phase C18 column (5 µm, 250 × 4.5 mm; Waters, Inc.) and UV detector
- Combine 200 pmol of each RNA pool (modified and unmodified) in separate reactions with nuclease P1 digestion mix for a total reaction volume of 5 µL and incubate 1.5 h at 37 °C.This step will digest the nucleic acid into its component nucleotides. Nuclease P1 will digest both RNA and DNA. It is possible that nuclease P1 will not digest after a specific modified nucleotide. If incomplete digestion is seen in the HPLC results compared to an unmodified sample digestion, this in itself is convincing evidence of modified nucleotide incorporation.
Combine each entire nuclease P1 digestion reaction with alkaline phosphatase reaction mix in a final volume of 25 µL. Incubate 1.5 hr at 37 °C, followed by 1 h at 50 °C.
- Inject 2.5 µL of one reaction onto a reversed-phase C18 HPLC column and separate using a mobile phase flow rate of 1 mL/min and detection at 260 nm. Repeat with the other reaction.If the pool initially had an equimolar mix of nucleotides, then the digested RNA should contain ~25% of the modified nucleoside and no peak corresponding to the replaced nucleoside. Of course, the actual composition of pools can vary greatly, depending on the synthesis of the pool (e.g., see UNIT 9.2). Therefore, the composition of the original pool should first be determined either by sequencing individual clones (e.g., see CPMB, UNIT 7.4) or by first digesting and separating a transcription product made solely from unmodified nucleotides.Nucleotides are detected at 260 nm as this is close to the absorbance maxima for their aromatic bases. Nucleotides with heavily modified bases that disrupt their aromaticity may absorb at different maxima; however, A260 should still be used because the RNA will likely be mostly unmodified.The unmodified transcription reaction should be run separately through the same column using the same conditions to provide a series of standard retention times for comparison. This control can also be used to obtain the extinction coefficient of the modified nucleoside, if it is not otherwise known.
BASIC PROTOCOL 3
Evaluation of Modified RNA as a Template for Reverse Transcriptase
During each round of in vitro selection, the selected RNAs must be amplified. Therefore, it is necessary to determine if an RNA transcript containing modified nucleotides will serve as a suitable template for reverse transcription (RT), the first step of the amplification process. A control reaction can be performed on the same template, except using unmodified nucleotides. The relative success of the reverse transcription reactions can again be visualized by incorporation of α-32P-labeled nucleotide, gel electrophoresis, and analysis on a phosphorimager.
The success of the reverse transcription reaction will depend primarily on the reverse transcriptase that is used. While reverse transcriptases, in general, tend to be somewhat forgiving with respect to template chemistry (e.g., they can recognize both RNA and DNA as templates), different reverse transcriptases may have distinct capabilities with modified bases as substrates (for instance, higher reaction temperatures for structurally stable RNAs). If one reverse transcriptase does not prove efficient, then another one or a combination should be used. See Figure 9.6.3 for guidelines in optimizing this reaction.
Materials
Transcribed RNA pools, modified and unmodified (prepared as in Basic Protocol 1 but without radiolabeled ATP)
3000 Ci/mmol α-32P-labeled dATP (e.g., Perkin Elmer)
- ThermoScript reverse transcriptase kit (Invitrogen, Life Technologies), kit includes:
- 5× Buffer,
- 100 mM DTT,
- RNase OUT (ribonuclease inhibitor), and
- RT enzyme
100 µM reverse (3’-end) primer
10 mM dNTP mix (containing 10 mM each of dATP, dCTP, dGTP, and dTTP)
2× denaturing stop dye (see recipe)
10% (w/v) denaturing polyacrylamide gel (see recipe; APPENDIX 3B), 0.8 mm thick
Thermal cycler
Gel blotting paper (Bio-Rad)
Plastic wrap
Gel dryer with heat and vacuum (e.g., Bio-Rad)
Phosphorimager screen
Phosphorimager
Image analysis software (e.g., GE Healthcare Life Sciences ImageQuant)
Additional reagents and equipment for denaturing polyacrylamide gel electrophoresis (PAGE; APPENDIX 3B)
Optimize RT reaction
-
1Combine the following RT reactions in two separate tubes (13 µL total volume):
- 10.5 µL (10 pmol or ~0.5 µg) transcribed RNA pool (modified in one tube, unmodified in the other)
- 1.0 µL 100 µM reverse (3’-end) primer
- 0.5 µL α-32P-labeled dATP (this volume should be varied based on the age and subsequent activity of the radiolabeled dATP)
- 1.0 µL 10 mM dNTP mix
Do not use radiolabeled RNA as the input. It may be difficult to differentiate product from input if both are labeled. See UNIT 9.3, Basic Protocol 3, step 1 for suggested control reactions. -
2
Heat denature the RT reactions at 65 °C in a thermal cycler for 5 min and cool to room temperature over ~10 min.
-
3Add the following components to each reaction (20 µL total volume) and mix well:
- 4 µL 5× buffer (from ThermoScript reverse transcription kit)
- 1 µL 100 mM DTT (from ThermoScript reverse transcription kit)
- 1 µL RNaseOUT (40 units/µL, optional) (from ThermoScript reverse transcription kit)
- 1 µL ThermoScript reverse transcriptase (15 U/µL) (from ThermoScript reverse transcription kit)
If optimizing difficult reverse transcription reactions, multiple reverse transcriptases (or even combinations thereof) may be tested in parallel. In addition to ThermoScript (an AMV reverse transcriptase variant), AMV reverse transcriptase, and SuperScript II reverse transcriptase (Invitrogen) have been used in modified nucleotide reverse transcription reactions.RNA secondary structures may hinder the reverse transcription of some templates. Because of this, thermostable reverse transcriptases (such as ThermoScript) with reaction temperatures at the primer annealing temperature are recommended to minimize secondary structure formation. In extreme cases, a thermophilic DNA polymerase, Tth polymerase, has been shown to have significant reverse transcriptase activity, and can potentially be used to copy recalcitrant RNA molecules. -
4
Incubate the reaction at the primer annealing temperature (such as 50 °C; up to 70 °C) for 30–60 min. Heat inactivate the enzyme at 85 °C for 5 min.
-
5
Remove 2 µL and add it to 5 µL of 2× denaturing stop dye.
-
6Denature by heating 3 min to 70 °C and run on a 0.8 mm thick 10% denaturing polyacrylamide gel (APPENDIX 3B).A sample of radiolabeled RNA, as produced in the test transcriptions (Basic Protocol 1), can be used as a convenient size standard. RNA runs slightly slower on a gel than DNA of the same size.The concentration of acrylamide can be varied to efficiently separate different products (APPENDIX 3B).
Analyze RT reaction
-
7Remove one of the two glass plates. Place a sheet of gel blotting paper against the gel. Peel the gel off of the other glass plate (it will stick to the paper) and cover with plastic wrap. Dry the gel in a gel dryer under heat and vacuum.This latter step is not strictly necessary as a wet gel can be directly exposed to the phosphorimager plate. If this is done, carefully wrap the gel with plastic wrap to avoid contaminating the screen and exposure cassette with radiation.
-
8Expose the dried gel to a phosphorimager screen for 1 h.The exposure time may need to be increased or decreased, depending on the specific activity of the labeled nucleotide and the amount of transcript produced.Alternatively, gels can be exposed to X-ray film, and the film developed to visualize bands.
-
9Develop the phosphorimager screen and view the output with image analysis software such as ImageQuant.Full-length cDNA products are desired from both modified and unmodified RNA templates. However, it is possible that smaller, discrete bands (i.e., incomplete cDNA products) will be observed. These may be due to the failure of reverse transcriptase to read past RNA secondary structures, which could artificially decrease the pool diversity in early rounds of selection. In this case, multiple reverse transcriptases should be examined (see Step 3) to optimize the reaction for the modification.
BASIC PROTOCOL 4
Determination of the Fidelity of Replication
If Basic Protocols 1, 2, and 3 give successful results, then it is likely that the random sequence pool can make it through the preparative steps leading to the selection. However, it is possible that the incorporation of modified nucleotides may alter the dynamics of selection experiments in one of two ways: first, modified nucleotides may in and of themselves be extremely mutagenic; second, it may be difficult to either make full-length transcripts that contain modified nucleotides or to fully copy transcripts containing modified nucleotides. In the latter case, there will be an unseen selection against transcripts that contain modified nucleotides over the course of several cycles. This protocol determines the mutagenic or replicative potential of modified nucleotides in an in vitro selection.
To test the mutagenic potential, a single sequence or clone from the pool should be subjected to a “mock” round of selection (i.e., omitting the selective step). Refer to UNITS 9.3, 9.4, and/or 9.5 for in vitro selection protocols. Following the selection, ~30 clones should be sequenced (refer to CPMB UNIT 7.4 for sequencing protocol) and the number of mutations counted.
To test the replicative potential, compare the nucleotide ratios of the unselected pool to a pool participating in a “mock” round of selection (i.e., omitting the selective step). To accomplish this, initially the pool is subjected to a round of “mock” selection (see UNITS 9.3, 9.4, and/or 9.5 for selection protocols). Before and after the selection, ~30 clones should be sequenced (see CPMB UNIT 7.4) and the base composition (i.e., relative ratio of nucleotides) of the pool should be analyzed compared to the initial ssDNA pool and RNA pool in which unmodified nucleotides were incorporated. In recent years, it has become cost effective to perform high throughput sequencing on the samples and compare nucleotide ratios for many more sequences.
Through these protocols, it should be possible to isolate any bias in addition to the process responsible (see also UNIT 9.2, Support Protocol 2). If the nucleotide frequency of the original pool is skewed, the entire pool should be resynthesized as described in UNIT 9.2. If the nucleotide frequency of the pool incorporating modified nucleotides is highly skewed relative to the original unmodified pool, then one of the enzymatic steps should be investigated further. If the nucleotide composition is highly skewed after the “mock” selection relative to before selection, then selection against a given residue may prove to be a problem. It should be recalled that even small skewing can prove to be significant over many rounds of selection. For example, if a modified cytidine is present at 25% of the random sequence positions in a starting pool, but is present at only 22.5% of the positions following one round of selection, after 10 rounds of selection its frequency may fall to 0.25[(0.225/0.25)10] = 0.087, or <9% of the random sequence positions in a pool.
As mentioned previously, when examining pool bias, it may only be possible to detect statistically significant differences in nucleotide frequencies by comparing high throughput sequence data from sequences (perhaps hundreds of thousands). If such problems persist and inhibit the enrichment process, it may prove useful to perform a single round of selection in the presence and absence of target, sequence clones, and use computational analyses to identify enriched motifs specific to incubation with the target that may be suggestive of function and that could either identify potential aptamer candidates for assay or assist with the design of a new, better pool.
DETERMINATION OF MODIFIED POOL MOST SUITABLE FOR IN VITRO APTAMER SELECTION
BASIC PROTOCOL 5
Comparing Modified Nucleotide Pools and Selection Conditions
One of the key decisions to make when designing a selection experiment is to choose the modification (or set of modifications) to incorporate in the nucleic acid pool. Gold et al. (2010) nicely showed that certain modifications work better than others with a given protein. Choosing a modified pool with an initially higher affinity for the target can potentially increase the success of the selection. If there is a decision between equally beneficial modified nucleotides that have passed each of the previous criteria (Basic Protocols 1–4), parallel “mini” selections of two to three rounds of selection (see UNITS 9.3, 9.4, and/or 9.5) should be performed with the different modifications. Affinity assays can determine the pool with the lowest dissociation constant to take forward in the full selection. It should be noted that certain recalcitrant proteins may require more rounds of selection before a modified pool reveals preferential binding affinity. Alternatively, high throughput sequencing, followed by sequence analysis and binding assays has previously been used to compare selection conditions in early rounds when affinity assay detection limits prevent determination of differences.
After performing two to three rounds of selection (see UNITS 9.3, 9.4, and/or 9.5) against a target using different pool candidates, their dissociation constants should be estimated using the filter-based protocol provided below (see also UNIT 9.3, Support Protocol 2 for filter-based assaying and UNIT 9.5, Support Protocol 1 for bead-based assaying).
Materials
32P end-labeled nucleic acid pools with the specific modifications (modified pool in one tube, unmodified in another; refer to UNIT 9.5, Support Protocol 1 for the radiolabeling protocol)
Binding Buffer - The binding buffer should promote binding and be specific to the aptamer application. Common examples are phosphate-buffered saline (PBS) and PCR buffer (e.g., 10 mM Tris•Cl, pH 8.4, 50 mM KCl, and 1.5 mM MgCl2). For more discussion see UNIT 9.5.
Target molecule (range of concentrations from 1×10−7 M to 1×10−12 M)
Thermal cycler or 37 °C and 65 °C water baths or heating blocks
Minifold I Dot Blot System, 96-well plate vacuum manifold (e.g., Whatman)
Vacuum pump or water aspirator
Nylon membrane (e.g., Hybond N+ Nylon Sheet)
0.45 µm nitrocellulose transfer and immobilization membrane (e.g., Protran BA-83, Whatman)
Methanol
0.5 M Potassium hydroxide (KOH)
Phosphorimager screen
Phosphorimager and image analysis software (e.g., GE Healthcare Life Sciences ImageQuant)
Incubate the pool with target
-
1Heat denature the pool at 65 °C for 3 min and then cool to room temperature for at least 10 min. This should let the RNA assume a more stable structure.Prepare ~10 RNA aliquots for each pool candidate to be tested. A low RNA concentration of ~1×10−11 M should be used for each reaction/aliquot.
-
2Add the protein target in binding buffer to the RNA pool to generate “protein-pool binding reactions.”A range of about ten different concentrations (1×10−7 M to 1×10−12 M) of protein should be used. Prepare a “no protein control” to determine the pool’s background binding to filters, beads, or other matrices. If background binding is sufficiently high, (>10%) then tRNA or other nonspecific additives should be included.
-
3Incubate the “protein-pool binding reactions” at 37 °C for 30–45 min.Alternatively, different incubation temperatures and times may be used depending on the downstream applications for the selected aptamers.
Perform an affinity assay
-
4Assemble the dot blot system, with the nitrocellulose sheet (top) and nylon membrane (below the nitrocellulose membrane) sandwiched between the dot blot plates (Figure 9.6.5). Moisten the nylon membrane and the nitrocellulose sheet with binding buffer. Be sure the sheets are layered such that no air becomes trapped between the sheets before final assembly of the system. Connect the assembled system to a vacuum pump or water aspirator.It may be necessary to soak the filters prior to assembly to enhance protein capture (nitrocellulose) and unbound oligonucleotide (nylon). Typically, the nitrocellulose sheet is soaked in 0.5M KOH for 5 min, whereas the nylon membrane is soaked in 100% methanol for 5 min. Both filters should then be rinsed in diH2O for3 min and soaked in binding buffer for at least 5 min. The buffer should not contain bovine serum albumin (BSA) or other nonspecific blocking proteins that may saturate the nitrocellulose filter.To check for system leaks and verify proper assembly, prewash a few wells with binding buffer. The buffer should flow freely through the well at a rate of roughly 100 µL every 3 sec. Bubbles can clog the well preventing filtration and may be unclogged by simply pipetting the applied solution up and down.
-
5Apply each “protein-pool binding reaction” to a well and wash with 3 to 10 volumes of binding buffer. When all the reactions have passed through the filters, dry the filters in an 80 °C oven on a glass plate for 5 min.Upon dispensing the “protein-pool binding reaction” into the well, wait for the solution to completely filter through the membranes before administering the washes.When dispensing solution, pipet slowly to minimize the formation of bubbles, which may clog the filter. Additionally, keep the pipet tip close to the membrane without touching the membrane, as contact may damage the surface.
-
6Wrap the filters in plastic wrap and place in a developing cassette along with a blanked phosphorimager screen for 15–30 min.Depending on the age of the radioactive nucleotide, a longer exposure time might be necessary.
-
7Measure the radioactivity using a phosphorimager and calculate the dissociation constants of the different pools (see UNIT 9.3 for calculations).Calculate and compare the dissociation constants for each of the modified and unmodified pools tested. In general, the pool (modified or unmodified) with the lowest dissociation constant may be the best candidate for continuing a selection.Since the candidate pools are the result of several rounds of selection, additional rounds may continue from this point using the best candidate pool. Alternatively, the selection can be reinitiated from a naïve pool using either more or less stringent selection conditions, which may include negative selections, which may further refine aptamer selection. Refer to UNITS 9.3, 9.4, and/or 9.5 for in vitro selection protocols.
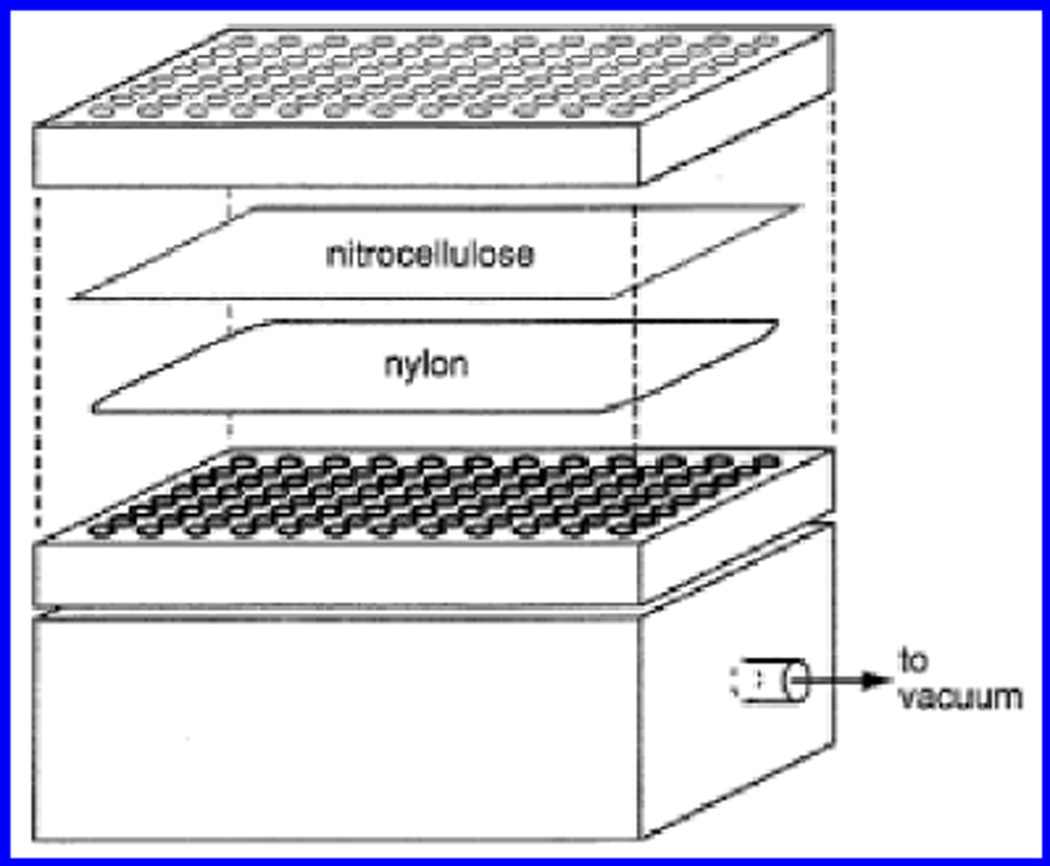
Figure 9.6.5.
Dot-blot assembly for filter based binding assays. See UNIT 9.3.2.
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocol steps. For common stock solutions, see APPENDIX 2A; for suppliers, see SUPPLIERS APPENDIX.
Alkaline phosphatase reaction mix
50 mM Tris·Cl, pH 8.5 (APPENDIX 2A)
0.1 mM EDTA
2 U alkaline phosphatase per 25 µL reaction
Store buffer without enzyme indefinitely at –20 °C. Add enzyme fresh.
Denaturing polyacrylamide gel, 10% (w/v)
1×TBE electrophoresis buffer (APPENDIX 2A) containing:
10% (w/v) acrylamide
0.5% (w/v) bisacrylamide
7 M urea
Prepare fresh
See APPENDIX 3B for full details on pouring and running gels.
Denaturing stop dye, 2×
Deionized formamide containing:
0.1% (w/v) bromophenol blue
50 mM EDTA
Store indefinitely at –20 °C. Daily-use aliquot may be stored up to 1 month at room temperature.
Nondenaturing dye, 6×
60% (v/v) glycerol
0.6% (w/v) bromophenol blue
10× ethidium bromide
Store indefinitely at –20 °C. Daily-use aliquot may be stored up to 1 month at room temperature.
Nuclease P1 digestion mix
15 mM sodium acetate, pH 5.2 (APPENDIX 2A)
0.1 mM zinc sulfate
1 U P1 nuclease per 5 µL reaction
Store buffer indefinitely at –20 °C. Add enzyme fresh.
COMMENTARY
Background Information
Nucleic acid selection experiments have generated a wide variety of binding species (aptamers) and catalysts (ribozymes and deoxyribozymes). However, nucleic acids are, by and large, not as functional as proteins: most aptamers bind their ligands less well than comparable antibodies, whereas ribozymes are generally slower catalysts than protein enzymes. It is possible that these limitations are a function of the relative infancy of aptamer and catalyst selections relative to a better-established field like protein engineering; however, it is also possible that functional nucleic acids are inherently limited by the diversity of the genetic alphabet. The binding interactions and chemical reactions performed by nucleic acid biopolymers may be constrained by the functional groups they contain. The ability to expand the functional groups available to a DNA or RNA polymer through the incorporation of modified nucleotides could potentially open up binding interactions and reaction chemistries that were previously inaccessible.
Due to advances in chemical synthesis technologies, decreases in the overall cost of starting materials, and the availability of mutant polymerases, modified nucleotides can now be easily incorporated within in vitro selections. Whereas once limited to the five canonical nucleotides, numerous aptamers, ribozymes, and deoxyribozymes with a variety of functions and applications have been selected that utilize a plethora of modified nucleotides (see Table 9.6.1). The modifications regularly used in in vitro selections can be coarsely divided into three main groups: ribose modifications, phosphate backbone modifications, and base modifications (see Figure 9.6.1). In particular, 2’-ribose modifications (such as 2’-amino or 2’-fluoro) and phosphate backbone modifications offer increased stabilities, in large measure because most ribonucleases polarize the 2’-hydroxyl group to attack the phosphodiester linkage. Base modifications (in particular at the 5-position of pyrimidines nucleosides, such as uridine) can provide additional chemical functionalities that are accessible in the major groove of helices.
Critical Parameters
How to choose a modified nucleotide
Modified versus unmodified nucleotide
Initially, the most important decision is what modification(s) to introduce. While multiple pools containing different modified nucleotides can be used, depending on availability and incorporation efficiency, the potential utility of not using modifications at all should be kept in mind. It is entirely possible that while modified nucleotides will increase the probability of finding functional species, it is also possible that simpler selections with unmodified pools will also find such species. For example, it has been shown that a pool containing ≥1015 unique sequences may contain extremely rare binding or catalytic species (e.g., Bartel and Szostak, 1993).
Expanding the variety of modified nucleotides
Whereas there are numerous modified nucleotides to choose from that have been successfully used in in vitro selections (see Table 9.6.1), additional modified nucleotides have been proposed, but no prominent cases of their use can be found. Some examples of these modified nucleotides include 5-aminoallyl-2'-fluoro-dUTP, 5-aminoallyl-UTP, and N(6) (6-aminohexyl) carbamoylmethyl-ATP (Schoetzau et al., 2003), as well as 5-iodo-dUTP (Brody et al., 1999), 2’-azido-dUTP (Padilla and Sousa, 2002), 2’-Se-methyl nucleotides (Chelliserrykattil and Ellington, 2004, and Siegmund et al., 2012), 6-amino-5-nitro-3-(1’ -β-d-2’ -deoxyribofuranosyl)-2(1H)-pyridone, and 2-amino-8-(1’-β-d-2’-deoxyribofuranosyl)-imidazo[1,2-a]-1,3,5-triazin-4(8H)-one) (Yang et al., 2011). Additionally, 7-deaza-dATP, which can be readily incorporated using Taq DNA polymerase, can be conjugated at the carbon 7-position with aminopropyl, aminopropynyl, and Z-aminopropenyl side chains (Gourlain et al., 2001). Numerous conjugates of 5-(3-aminopropenyl)-2’deoxyuridine suitable for selection have been identified (Sakthivel and Barbas 1998) along with deoxynucleoside triphosphate analogues of all four unmodified nucleotides bearing basic, acidic, or lipophilic groups (Jäger et al. 2005).
Understanding modified nucleotide chemistry
The incorporation of additional functional groups into the context of an RNA backbone is expected to increase the diversity of its available interactions, both with itself and with its desired substrates. Thiol groups, for example, can participate directly in catalysis as nucleophiles. Additionally, disulfide bonds could be formed intramolecularly between thiols. This may add to the structural diversity of nucleic acids, normally limited to hydrogen bonding, salt bridges with metals, and stacking interactions. Charged groups, such as a lysine-like side chain, could potentially add to the structural repertoire of nucleic acids by allowing the formation of electrostatic interactions and salt bridges. The inclusion of chemical moieties with a pKa closer to neutrality, such as an imidazole group, is also expected to benefit nucleic acid chemistry. For instance, a primary amine on 2’-amino nucleotides, with a pKa of 6.2 and increased nucleophilicity compared to unmodified nucleotides, may react with an aldehyde near physiological conditions to generate 2’-imino conjugated oligonucleotides (Bugaut et al., 2004, Bugaut et al., 2005). However, unmodified nucleotide bases contain no functional groups with unperturbed pKa values between 3.5 and 9.2 inherently limiting the proton “push-pull” chemistry found in so many protein enzymes. The introduction of these and other functional groups can also potentially increase the abilities of nucleic acids to bind metals.
While attempts to associate nucleotide functionality with binding or catalytic properties are at best still guesswork, it is nonetheless true that failing to appreciate the chemical properties of modified nucleotides can potentially adversely affect how a selection experiment proceeds. For example, the inclusion of thiolated nucleotides can potentially lead to the unwanted formation of disulfide-bonded products if care is not taken to include a reducing agent in enzyme and/or selection reactions. For example, deoxyribozyme ligases that form an unnatural internucleotide linkage between a 5′-iodinated pool and an oligonucleotide substrate with a 3′-phosphorothioate have been selected from random sequence pools (Levy and Ellington, 2001; Levy and Ellington 2002). Unless reducing agents are kept in the selection reaction, the oligonucleotide substrates will dimerize, reducing their effective concentration. Similarly, the inclusion of modified nucleotides that have altered pKas could very easily change the pH of a concentrated stock solution, and care should be taken to make sure that all such stocks are at the desired pH and appropriately buffered. The accidental alteration of pH in enzyme or selection reactions can, of course, lead to decreases in product yield. Finally, the inclusion of modified nucleotides that have new metal binding or chelating properties may alter the available metal concentrations in an enzyme or selection reaction. In particular, the chelation of magnesium can lead to large changes in the efficiency of product formation by many different polymerases.
Modified nucleotides and mutations
Although a selection starts with a large number of sequences, this number is usually a small fraction of the total number of sequences possible for the length of the random region. Additionally, with each round, the number of sequences within the pool is diminished. As such, it may be useful to explore the sequence space around the selected winners in order to discover functional variants. To some extent, this occurs in any selection due to the inherent mutations that arise in the amplification process; however, this background mutation rate is small, and one may wish to increase the frequency of mutations. For these purposes, the mutagenic potential of a nucleotide analog that serves as a nonspecific template can be used to increase the diversity of a pool (see Basic Protocol 4 for assessment of mutagenic potential). Similar to mutagenic PCR (UNIT 9.4), these techniques can be used at the outset of a selection to mutate an existing ribozyme or aptamer for either optimization or reselection for altered specificity. For example, Kore et al. (2000) have used modified nucleotides to create a degenerate pool based on the hammerhead ribozyme, from which they selected a variant that cleaves at an alternate sequence. Additionally, a mutagenic step can be incorporated between rounds in a selection to more efficiently explore sequence space. However, it should be noted that in the latter case, one does not want the mutation rate to be so high that a few of each winning sequence from one round do not survive unchanged into the next round.
One benefit of using modified nucleotides rather than standard mutagenic PCR is that it is easy to control the amount of mutation introduced into the sample simply by adjusting the relative rate of modified to unmodified nucleotide in the amplification process. As such, a higher rate of mutation can be achieved than with mutagenic PCR, which would be particularly useful for diversifying an initial pool. Zaccolo et al. (1996) describe such a system using both a purine and pyrimidine analog in a PCR and have calculated the frequencies of each base transition. It should be mentioned that when using mutagenic modified nucleotides, one would not want them to be included in the active pool, as their decreased fidelity would make it less likely that they would be in the same position during the next round. For example, if a DNA selection were being performed, a second amplification of the pool would be required using only unmodified nucleotides.
When to Incorporate Modified Nucleotides for In Vitro Selection
Pre- vs. post in vitro selection modifications
Overall, the most important considerations in deciding whether to use a modified nucleotide in a selection experiment are purely technical ones, i.e., can the modification be incorporated and will it inhibit amplification of the nucleic acid pool? A number of modified nucleotides have been incorporated with varying degrees of success into selection experiments (Table 9.6.1), while other modified nucleotides cannot yet be enzymatically incorporated. As previously described, there are three main ways to incorporate modified nucleotides into the selection (summarized in Figure 9.6.2) including enzymatic processing, conjugation, and chemical synthesis, each with their own benefits and disadvantages.
When modifications are enzymatically incorporated into the naïve pool (i.e., “pre-selection”), nucleoside triphosphates (NTPs) are the pool “building blocks,” conferring unmodified or modified moieties. An advantage of this “pre-selection” incorporation is that the modified library yields oligonucleotides that exhibit better nuclease resistance from that start. Specifically, 2’-modifications (i.e., nuclease resistance modifications) are commonplace in selections, as it is often more difficult to modify an aptamer after the selection with nuclease stable nucleotides than to perform the selection with a pool containing these nucleotides. However, once aptamers are selected, downstream applications typically require large-scale syntheses, thus requiring phosphoramidites (chemically equivalent to the original NTPs) for oligonucleotide solid phase synthesis, (see UNITS 3 and 4). Currently, the commercially available modified NTPs are not equally available as modified phosphoramidites.
In addition to solid phase synthesis, modifications may be incorporated “post-selection” using conjugation techniques. While Bugaut et al. (2006) (described below in Incorporation after amplification (conjugation or extension)) used the conjugation methodology to generate a naïve pool, the method could similarly be applied “post-selection.” As with all “post-selection” modifications and site-specific modified nucleotide incorporations, however, a detailed understanding, at the nucleotide position, of the chemical variant’s impact on binding, functionality, catalysis, stability, specificity, and other properties is necessary.
Furthermore, when enzymatic incorporation is not possible, chemically synthesized libraries containing modified nucleotides can instead be subjected to successive, often less stringent, screens for function without amplification. Successful motifs can either be used directly, or a new library can be synthesized and screened based on the perceived change in information content. One advantage of this approach is that chemically synthesized pools permit addition of multiple chemical modifications in parallel. As an example, He et al. (2012) screened for so-called X-aptamers that contained modified nucleotides and a phosphorodithioate oligonucleotide backbone. However, rather than being replicated, X-aptamers were synthesized by a split-bead method that led to one aptamer variant clonally represented in many copies per bead. The beads were screened for ligand-binding properties, and the sequences from successful beads were cleaved and sequenced. Using this method, a drug conjugate that bound CD44 was found in a single round of screening. It is also worth noting that the elimination of PCR amplification removes the biases that may be related to amplification, such as the disfavored amplification of structurally stable species.
Alternatively, Spiegelmers, or aptamers comprised of l-ribose instead of the enzymatically incorporated d-ribose, are generated using a novel approach to modified nucleotide incorporation (Wlotzka et al., 2002, reviewed in Eulberg and Klussman, 2003 and in Wilson and Keefe, 2006). Specifically, Spiegelmer precursors are identified through in vitro selections against a biological protein’s enantiomer (synthesized using d-amino acids), thus generating traditional aptamers (although against non-biological targets). Remarkably, the mirror-image aptamer (i.e., Spiegelmer) recognizes the native biological protein
Efficiency of enzymatic incorporation
In general, modified nucleotides may incorporate less efficiently than unmodified nucleotides, depending on the polymerase, modified nucleotide, as well as other reaction conditions. For example, in the authors’ experience, the incorporation of 5-Br-dUTP with Taq DNA polymerase decreased the replicability of sequences and created an inherent and unintended selection in favor of those members of a population that had fewer modified nucleotides. Lutz et al. (1998), using xanthosine in the template, showed that diamino pyrimidine nucleoside triphosphate was incorporated by the 9°N-7 thermostable DNA polymerase (Southworth et al., 1998) in a sequence-specific fashion but with only 45% incorporation of the correct nucleotide analog. Additionally, Vaught et al. (2010) compared primer extension yields of dUTP analogues modified at the 5-position and found that Vent DNA polymerase provided yields similar to or better than TTP for 4 of the 6 modified nucleotides tested, while KOD XL DNA polymerase provided yields similar to TTP for only 1 of the 6 modified nucleotides tested.
Incorporation after amplification (conjugation or extension)
However, amplification complications using modified nucleotides in aptamer selections have not stunted recent aptamer advances. Instead, Vaught et al. (2010) developed a new DNA aptamer selection scheme to accommodate the poor enzymatic incorporation of modified nucleotides. The novel scheme involves an intermediate PCR step that relied on unmodified nucleotides to exponentially amplify a selected pool. A subsequent primer extension reaction reincorporates the modified nucleotides to generate an enriched, modified pool (see Figure 9.6.2). Somalogic has used this strategy (Ochsner et al., 2013) to successfully generate aptamers against >1,000 human proteins (Mehan et al., 2013).
Alternatively, Bugaut et al. (2006) offered a “conjugation” approach to incorporate modified nucleotides post-amplification into an in vitro selection (see Figure 9.6.2). At each round, following selection, amplification, and the regeneration of the 2’-amino-2’-deoxyUTP pool, a reductive amination reaction conjugated the aldehyde-functionalized ligands to the 2’-amino groups. This unique approach to modified nucleotide incorporation was successfully used to generate novel modified aptamers against HIV-1 trans-activation response (TAR).
Making the most of modified nucleotide incorporation: Further Optimizations
As has been apparent throughout, there are few hard and fast rules regarding what will and will not work when selecting functional nucleic acids. Because of this, researchers need to be somewhat versatile in their approach to selection experiments involving modified nucleotides. If a given approach does not work, this does not mean that the selection experiment inherently has no chance of working, but instead indicates that it may be necessary to alter one or more parameters. Simply put – optimization of just three variables is all that may be needed to improve the incorporation of modified nucleotides: pool, polymerase, and reaction buffer.
Pool considerations
A good portion of this protocol has been devoted to testing reactions and optimizing conditions. Although this process will likely take several weeks, it is time wisely spent. The first round of selection is by far the most important, as this is the round when selection conditions will query the greatest number of possible answers. Concomitantly, creating the largest possible number of unique sequences (1013 to 1015) for the first round of selection is a critical task (UNIT 9.2, Design, Synthesis, and Amplification of DNA Pools for In Vitro Selection). However, because creating a large library requires a large amount of effort and large volumes of relatively expensive reagents, initial optimization and practice should always be performed on a small scale.
Similarly, it is expected that only a small number of the input molecules will survive any given round; therefore, it is essential that these successful sequences be efficiently carried to the next round. Inefficient reverse transcription or amplification reactions may lead to a selection for molecules that can be efficiently replicated (amplification artifacts) rather than to molecules that are highly functional, therefore the optimization of these reactions and the entire process as a whole is necessary (see Basic Protocols 1 and 3).
Additionally, the length and composition of a random sequence pool can vary according to the type of selection that is carried out (also see UNIT 9.2) and some attention should be paid to the choice of constant sequence regions. While the largest possible pool is preferred to begin the selection, shorter pools are easier and less costly to generate (see UNIT 9.2). Similarly, aptamers and deoxy/ribozymes tend to be synthetically produced for testing and downstream applications; thus, considering solid phase phosphoramidite synthesis limitations is important. For instance, whereas DNA pool lengths can be well over 100 nt, RNA pools should be kept below 80 nt when full length modified variants are being considered for synthesis. In addition, current phosphoramidite reagents costs should be taken into account. For instance, 2’-deoxy-2’-fluoro purine nucleoside phosphoramidites are considerably more expensive than their respective pyrimidines or 2’-O-methyl containing phosphoramidites.
In consideration of composition, RNA polymerases tend to have difficulty incorporating modified nucleotides prior to positions 8 to 10 following the transcription start site (Milligan and Uhlenbeck, 1989; Kujau et al., 1997), most likely because the attempted incorporation of modified nucleotides leads to increased abortive initiation. Thus, it is wise to design a pool that lacks or limits the incorporation of the modified base within this region. Some modified T7 polymerases can overcome this limitation through incorporating a purine leader sequence of 10 to 15 nucleotides after the T7 promoter. Inasmuch as the first 18 or more nucleotides of a transcribed pool typically remain constant to allow for second-strand DNA synthesis, this limitation should not affect the overall diversity of the pool.
Finally, the sequence of the pool itself can have a surprisingly large effect on selection experiments. Robertson and Ellington (1999) originally selected a relatively fast ribozyme ligase from an RNA pool that contained 90 random sequence positions. A variant of this pool was generated in which only four positions in the constant region were altered, substituting a GAAA tetraloop in place of a UUCG tetraloop. Selection experiments for ribozyme ligases were initiated from amplified aliquots of this pool; one aliquot was used for the selection of ribozymes containing canonical nucleotides, other aliquots of identical complexity were used for the selection of ribozymes containing one of three different modified nucleotides (Robertson, 2001). After six rounds of selection and amplification, all of the pools had collapsed to contain a relatively few winning sequences. However, the rates of all of these winning sequences were remarkably slow (500-fold slower) compared to the ribozyme ligase originally selected from the slightly different pool. There was no discernible difference between the ribozymes that contained canonical or modified nucleotides; they were all very slow. Robertson (2001) hypothesized that the pools had somehow become artificially narrowed during the course of the selection, and therefore repeated the selection experiments with new aliquots of the original, amplified pool. Again, only very slow ribozymes were obtained.
Choosing the right enzyme for modified nucleotide incorporation
Different polymerases clearly have different potentialities for the incorporation of modified nucleotides (see Table 9.6.1). For example, the Benner group has tested several thermostable DNA polymerases for their ability to incorporate a variety of modified nucleotides (Lutz et al., 1998, 1999). Likewise, Vaught et al. (2010), using modified DNA templates, examined the extension efficiency of D. Vent DNA polymerase and KOD XL DNA polymerase to incorporate modified nucleotides (5-position of uridine). They found that Vent DNA polymerase provided yields similar to or better than TTP for 4 of the 6 modified nucleotides tested, while KOD XL DNA polymerase provided yields similar to TTP for only 1 of the 6 modified nucleotides tested
Similar variations in RNA polymerases to incorporate modified nucleotides, especially 2’-modifications, have been observed. While 2’-modified ribonucleotides can be incorporated into RNA by the wild-type T7 RNA polymerase, the kcat/KM values for incorporation of 2′-modified ribonucleotides are substantially higher than for unmodified nucleotides, with a preference order of 2′-OH > 2′-NH2 > 2′-F > 2′-H (Huang et al., 1997). In order to improve incorporation, some groups have engineered or evolved polymerases that can incorporate modified nucleotides. For example, a single mutation in T7 RNA polymerase (Y639F) reduces discrimination at the 2′-position and allows more efficient incorporation of deoxyribonucleotides into RNA (Kostyuk et al., 1995; Sousa and Padilla, 1995). Additionally, the Y639F T7 RNA polymerase, which is commercially available in DuraScribe kits (Epicentre, Madison, WI) allows the incorporation of NTPs with fluoro, amino, and thiol groups at their 2′-positions (see Basic Protocol 1). The Y639F mutant of T7 RNA polymerase has been shown to have up to 24-fold greater specificity for incorporation of 2′-modified NTPs, with a preference order of 2′-OH > 2′-F > 2′-H > 2′-NH2 (Huang et al., 1997). It is, however, inefficient at incorporating bulkier 2’-groups such as 2’-OMe, that when incorporated, greatly enhance nuclease stability of transcripts.
Histidine 784 (H784) plays an important role in ribose discrimination by T7 RNA polymerase, as it is thought to cause premature termination following the incorporation of a 2’-modified ribonucleotides (Padilla and Sousa, 2002). An H784A mutation relaxes this discrimination and, when coupled with the Y639F mutation, allows for bulkier groups to be incorporated such as 2’-O-methyl (2’-OMe) and 2’-azido (Padilla and Sousa, 2002). Further, the Y639F H784A double mutant has reduced preference for NTPs over dNTPs (Brieba and Sousa, 2000) and 2’-F NTPs (author’s observation).
A directed evolution approach was taken, in which R425, G542, Y639, and H784 were randomized and the T7 RNA polymerase was selected for normal in vivo activity (Chelliserrykattil and Ellington, 2004). Screening the resulting population yielded a particular mutant known as “RGVG” (R425, G542, Y639V, H784G plus additional E593G V685A mutations that arose during the selection), which demonstrated the ability to incorporate 2’-OMe-modified ribonucleotides. RGVG was later shown to incorporate 2’-SeMe modified ribonucleotides (Siegmund et al., 2011). RGVG was further improved by error-prone PCR and high-throughput screening. A particular mutant “2P16” (which contained seven additional mutations beyond the RGVG background) shows comparable 2’-OMe- and enhanced 2’-SeMe-modified ribonucleotide incorporation as compared to RGVG (Siegmund et al., 2012). A similar approach was used to evolve the R425C mutant T7 RNA polymerase, which is capable of synthesizing RNA entirely from 2’-OMe-modified NTPs (Ibach et al., 2013).
Buffer optimization
The buffer conditions for incorporation can be adjusted and optimized for modified nucleotide incorporation. For example, Padilla and Sousa (1999) have systematically investigated several buffer conditions that aid the incorporation of nucleotides modified specifically at the 2’-position. These authors found better incorporation upon supplementing a transcription reaction with 0.5 mM MnCl2, 1 U/µL pyrophosphatase, and either 8 mM spermidine for plasmid templates or 8 mM spermine for short DNA templates. Similarly, Burmeister et al. (2006) optimized their 2’-deoxy purine and 2’-O-methyl pyrimidine (dRmY) transcription reactions with 2 mM spermine, 9.6 mM MgCl2, and 2.9 mM MnCl2. However, Minakawa et al. (2008), in an attempt to optimize the transcription efficiency of their fully modified 4’-thio RNA, failed to improve the efficiency through MnCl2 addition, instead of MgCl2, or with an increased spermidine concentration. However, increasing the ratio of modified purines to unmodified purines resulted in more modified nucleotides being incorporated. While this did cause a slight decrease in transcription efficiency it resulted in the fully modified transcripts needed for selection.
Archiving reactions
In vitro selection experiments are particularly grueling because the ultimate outcome, the successful isolation of binding species or catalysts, may not be known for days, weeks, or even months. This is especially true when working with modified nucleotides, because of the strong possibility that one or more amplification reactions or selection steps will not work at some point during the course of the selection. Therefore, it is desirable to keep an archived copy of each round. After the initial round, there should be multiple copies of each winning sequence present in the selected population. Thus, it is not necessary to use all of the RNA, cDNA, or double-stranded DNA template in a given selection or amplification step. Rather, some portion of the pool should be saved at any convenient point (e.g., as a double-stranded PCR product). In the event an unsuccessful round of selection should occur, it will not be necessary to repeat the entire experiment. Rather, selection can continue from the last successful round, with a minimal loss of time and effort.
A second added benefit of archiving each round is that “branched” selections can be considered. Such “branched” selections may begin with the same starting material (i.e., selected pool), but then parallel selections are performed under different stringencies, selection conditions, or using mutagenic conditions. Sampling a wider range of selection conditions potentially increases the chances of teasing out a high affinity binding variant, especially once the pool exhibits some binding potential.
Necessity of modified nucleotide (i.e,. did the modified nucleotide improve the functional oligonucleotide?)
To understand the contribution of a modified nucleotide in an in vitro selection, parallel modified nucleotide and unmodified nucleotide selections must be performed. An example of this is in seen in the Somalogic work comparing modified (i.e., 5-position of uridine) and unmodified nucleotide selections against thirteen difficult targets (Gold et al., 2010). The results indicated that only the selections with modified nucleotide generated aptamers; however, the extent of benefit of the modified nucleotide was highly dependent on the modified nucleotide and target. Alternative to selections in parallel with different modifications, the selections can be performed in parallel with one modification and different targets, then the targets that prove difficult can be reselected with a different modification. Similar modification contributions may be estimated upon substituting in unmodified nucleotides in place of modified nucleotides. Multiple examples of this are provided above in Strategic Planning: Advantages of Using Modified Nucleotides in In Vitro Selections. However, nucleotide substitutions may interfere with the oligonucleotide fold and presentation to the target, thus obfuscating a direct comparison of the modified with the unmodified species. Similarly, others have compared unmodified selections with modified selections performed using different pools, conditions, etc. (see examples in Strategic Planning: Advantages of Using Modified Nucleotides in In Vitro Selections). However, altering multiple variables obscures the cause of the results, whether it is the superiority of the nucleotide, pool, and/or selection conditions.
Anticipated Results
As with any selection experiments (UNITS 9.3, 9.4, and 9.5), it is virtually impossible to anticipate the outcome of any given experiment. This is especially true when considering the incorporation of modified nucleotides, considering the variety of modified nucleotides available and resulting combinatorial permutations. However, if the protocols for the production of RNA and DNA pools that have been outlined are followed, it can be anticipated that it should be possible to generate and purify upwards of at least 1014 different nucleic acid sequences (~10 µg) that contain a particular modified nucleotide. For successful selection experiments, the population should be sieved by a factor of 100 to 1000 each round. That is, <1% of the total population will be recovered following a round of selection. Thus, most successful selections will show significant functional improvement or be completed within 5 to 10 rounds. However, in some selections, it may take very successful functional sequences a longer period of time to overcome a great sea of mediocre functional sequences. For example, it took 18 rounds of selection to isolate cGMP-dependent aptazymes from a random sequence population (Koizumi et al., 1999). Following selection, the population should contain a relatively small number (1 to 10) of sequence classes. A sequence class is a set of related sequences that contain a common motif. To date, the sequences of aptamers or ribozymes selected from pools containing modified nucleotides do not correspond to the sequences of aptamers or ribozymes selected from pools that contain canonical nucleotides. Thus, any selection that includes modified nucleotides is likely to produce new sequence motifs.
Time Considerations
The preparative portions of each round of selection can be expected to take several days. A transcription could take several hours to overnight, followed by ~2 h gel purification and an overnight elution from the excised gel pieces. The amount of time spent on the selective steps will vary greatly with each individual selection. However, in initial rounds, the reaction time alone is typically several hours to an entire day. The selective segregation of winning sequences from losing sequences will vary from a few to several hours, depending on the method used. Reverse transcription followed by PCR, isolation, and quantitation of new template DNA will take a few more hours, depending mainly on the number of cycles required for amplification. The largest variable, the number of rounds of selection required, will vary greatly depending on the activity sought, from only a few to ≥12 rounds. Cloning and assaying of individual sequences will likely also take several days. Thus, even if no problems are encountered during a selection, it is likely to take ≥1 month to complete.
Acknowledgments
Gwendolyn Stovall was primarily supported by the Freshman Research Initiative at the University of Texas at Austin and partially supported by both the National Science Foundation (Cooperative Agreement No. DBI-0939454) and the National Institute of Health (5R03HD068691). These methods were refined by undergraduate students (Robert Bedenbaugh and Shruti Singh) from the Freshman Research Initiative based on generous funding from the National Science Foundation (CHE 0629136). Additionally, Robert Bedenbaugh was partially funded by the National Institute of Health (5R03HD068691) and Shruti Singh was partially funded by the National Science Foundation (Cooperative Agreement No. DBI-0939454). Adam Meyer’s work was supported by the National Security Science and Engineering Faculty Fellowship (FA9550-10-1-0169) and the Welch Foundation (F-1654). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation, The University of Texas at Austin, or Altermune Technologies, LLC. This work was a revision of the original protocol written by Scott M. Knudsen and Michael P. Robertson.
Literature Cited
- Bartel DP, Szostak JW. Isolation of new ribozymes from a large pool of random sequences. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- Battersby TR, Ang DN, Burgstaller P, Jurczyk SC, Bowser MT, Buchanan DD, Kennedy RT, Benner SA. Quantitative analysis of receptors for adenosine nucleotides obtained via in vitro selection from a library incorporating a cationic nucleotide analog. J. Am. Chem. Soc. 1999;121:9781–9789. doi: 10.1021/ja9816436. [DOI] [PubMed] [Google Scholar]
- Beaudry A, DeFoe J, Zinnen S, Burgin A, Beigelman L. In vitro selection of a novel nuclease-resistant RNA phosphodiesterase. Chem. Biol. 2000;7:323–334. doi: 10.1016/s1074-5521(00)00110-1. [DOI] [PubMed] [Google Scholar]
- Bell NM, Micklefield J. Chemical modification of oligonucleotides for therapeutic, bioanalytical and other applications. Chembiochem. 2009;10:2691–2703. doi: 10.1002/cbic.200900341. [DOI] [PubMed] [Google Scholar]
- Brieba LG, Sousa R. Roles of histidine 784 and tyrosine 639 in ribose discrimination by T7 RNA polymerase. Biochemistry. 2000;39:919–923. doi: 10.1021/bi992324+. [DOI] [PubMed] [Google Scholar]
- Brody EN, Willis NC, Smith JD, Jayasena S, Zichi D, Gold L. The use of aptamers in large arrays for molecular diagnostics. Molecular Diagnostics. 1999;4:381–388. doi: 10.1016/s1084-8592(99)80014-9. [DOI] [PubMed] [Google Scholar]
- Bugaut A, Toulmé JJ, Rayner B. Use of dynamic combinatorial chemistry for the identification of covalently appended residues that stabilize oligonucleotide complexes. Angew. Chem. Int. Ed. Engl. 2004;43:3144–3147. doi: 10.1002/anie.200454041. [DOI] [PubMed] [Google Scholar]
- Bugaut A, Bathany K, Schmitter JM, Rayner B. Target-induced selection of ligands from a dynamic combinatorial library of mono- and bi-conjugated oligonucleotides. Tetrahedron Lett. 2005;46:687–690. [Google Scholar]
- Bugaut A, Toulmé JJ, Rayner B. SELEX and dynamic combinatorial chemistry interplay for the selection of conjugated RNA aptamers. Org. Biomol. Chem. 2006;4:4082–4088. doi: 10.1039/b610890c. [DOI] [PubMed] [Google Scholar]
- Burmeister PE, Lewis SD, Silva RF, Preiss JR, Horwitz LR, Pendergrast PS, McCauley TG, Kurz JC, Epstein DM, Wilson C, Keefe AD. Direct in vitro selection of a 2’-o-methyl aptamer to VEGF. Chem. Biol. 2005;12:25–33. doi: 10.1016/j.chembiol.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Burmeister PE, Wang C, Killough JR, Lewis SD, Horwitz LR, Ferguson A, Thompson KM, Pendergrast PS, McCauley TG, Kurz M, Diener J, Cload ST, Wilson C, Keefe AD. 2‘-Deoxy purine, 2’-O-methyl pyrimidine (dRmY) aptamers as candidate therapeutics. Oligonucleotides. 2006;16:337–351. doi: 10.1089/oli.2006.16.337. [DOI] [PubMed] [Google Scholar]
- Chakravarthy U, Adamis AP, Cunningham ET, Goldbaum M, Guyer DR, Katz B, Patel M. Year 2 efficacy results of 2 randomized controlled clinical trials of pegaptanib for neovascular age-related macular degeneration. Ophthalmology. 2006;113:1–25. doi: 10.1016/j.ophtha.2006.02.064. [DOI] [PubMed] [Google Scholar]
- Chelliserrykattil J, Ellington AD. Evolution of a T7 RNA polymerase variant that transcribes 2'-O-methyl RNA. Nat. Biotechnol. 2004;22:1155–1160. doi: 10.1038/nbt1001. [DOI] [PubMed] [Google Scholar]
- Eulberg D, Klussman S. Spiegelmers: biostable aptamers. Chembiochem. 2003;4:979–983. doi: 10.1002/cbic.200300663. [DOI] [PubMed] [Google Scholar]
- Faulhammer D, Famulok M. Characterization and divalent metal-ion dependence of in vitro selected deoxyribozymes which cleave DNA/RNA chimeric oligonucleotides. J. Mol. Biol. 1997;269:188–202. doi: 10.1006/jmbi.1997.1036. [DOI] [PubMed] [Google Scholar]
- Geyer CR, Sen D. Evidence for the metal-cofactor independence of an RNA phosphodiester-cleaving DNA enzyme. Chem. Biol. 1997;4:579–593. doi: 10.1016/s1074-5521(97)90244-1. [DOI] [PubMed] [Google Scholar]
- Gold L, Ayer D, Bertino J, Bock C, Bock A, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden MC, Collins BD, Willis MC, Koch TH. Diagnostic potential of photo-SELEX-evolved ssDNA aptamers. J. Biotechnol. 2000;81:167–178. doi: 10.1016/s0168-1656(00)00290-x. [DOI] [PubMed] [Google Scholar]
- Gourlain T, Sidorov A, Mignet N, Thorpe SJ, Lee SE, Grasby JA, Williams DM. Enhancing the catalytic repertoire of nucleic acids. II. Simultaneous incorporation of amino and imidazolyl functionalities by two modified triphosphates during PCR. Nucl. Acids Res. 2001;29:1898–1905. doi: 10.1093/nar/29.9.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green LS, Jellinek D, Bell C, Beebe LA, Feistner BD, Gill SC, Jucker FM, Janjic N. Nuclease-resistant nucleic acid ligands to vascular permeability factor/vascular endothelial growth factor. Chem. Biol. 1995;2:683–695. doi: 10.1016/1074-5521(95)90032-2. [DOI] [PubMed] [Google Scholar]
- Griffiths AD, Potter BV, Eperon IC. Stereospecificity of nucleases towards phosphorothioate-substituted RNA: Stereochemistry of transcription by T7 RNA polymerase. Nucl. Acids Res. 1987;15:4145–4162. doi: 10.1093/nar/15.10.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Elizondo-Riojas MA, Li X, Lokesh GLR, Somasunderam A, Thiviyanathan V, Volk DE, Durland RH, Englehardt J, Cavasotto CN, Gorenstein DG. X-Aptamers: a bead based selection method for random incorporation of druglike moieties onto next-generation aptamers for enhanced binding. Biochemistry. 2012;51:8321–8323. doi: 10.1021/bi300471d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenstein M, Hipolito CJ, Lam CH, Perrin DM. Toward the combinatorial selection of chemically modified DNAzyme RNase A mimics active against all-RNA substrates. ACS Combinatorial Science. 2013;15:174–182. doi: 10.1021/co3001378. [DOI] [PubMed] [Google Scholar]
- Huang Y, Eckstein F, Padilla R, Sousa R. Mechanism of ribose 2'-group discrimination by an RNA polymerase. Biochemistry. 1997;36:8231–8242. doi: 10.1021/bi962674l. [DOI] [PubMed] [Google Scholar]
- Ibach J, Dietrich L, Koopmans KR, Nöbel N, Skoupi M, Brakmann S. Identification of a T7 RNA polymerase variant that permits the enzymatic synthesis of fully 2'-O-methyl-modified RNA. J. Biotechnol. 2013;167:287–295. doi: 10.1016/j.jbiotec.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Jäger S, Rasched G, Kornreich-Leshem H, Engeser M, Thum O, Famulok M. A versatile toolbox for variable DNA functionalization at high density. J. Am. Chem. Soc. 2005;127:15071–15082. doi: 10.1021/ja051725b. [DOI] [PubMed] [Google Scholar]
- Jellinek D, Green LS, Bell C, Lynott CK, Gill N, Vargeese C, Kirschenheuter G, McGee DP, Abesinghe P, Pieken WA, et al. Potent 2′-amino-2′-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry. 1995;34:11363–11372. doi: 10.1021/bi00036a009. [DOI] [PubMed] [Google Scholar]
- Jhaveri S, Olwin B, Ellington AD. In vitro selection of phosphorothiolated aptamers. Bioorg. Med. Chem. Lett. 1998;8:2285–2290. doi: 10.1016/s0960-894x(98)00414-4. [DOI] [PubMed] [Google Scholar]
- Kasahara Y, Irisawa Y, Ozaki H, Obika H, Kuwahara M. 2’,4’-BNA/LNA aptamers: CS-SELEX using a DNA-based library of full-length 2’-O,4’-C-methylene-bridged/linked bicyclic ribonucleotides. Bioorg. Med. Chem. Lett. 2013;23:1288–1292. doi: 10.1016/j.bmcl.2012.12.093. [DOI] [PubMed] [Google Scholar]
- Kato Y, Minakawa N, Komatsu Y, Kamiya H, Ogawa M, Harashima H, Matsuda A. New NTP analogs: the synthesis of 4’-thioUTP and 4’-thioCTP and their utility for SELEX. Nucl. Acids Res. 2005;33:2942–2951. doi: 10.1093/nar/gki578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefe AD, Cload ST. SELEX with modified nucleotides. Curr. Opin. Chem. Biol. 2008;12:448–456. doi: 10.1016/j.cbpa.2008.06.028. [DOI] [PubMed] [Google Scholar]
- Kimoto M, Yamashige R, Matsunaga K, Yokoyama S, Hirao I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotech. 2013;31:453–457. doi: 10.1038/nbt.2556. [DOI] [PubMed] [Google Scholar]
- King DJ, Ventura DA, Brasier AR, Gorenstein DG. Novel combinatorial selection of phosphorothioate oligonucleotide aptamers. Biochemistry. 1998;37:16489–16493. doi: 10.1021/bi981780f. [DOI] [PubMed] [Google Scholar]
- King DJ, Bassett SE, Li X, Fennewald SA, Herzog NA, Luxong BA, Shope R, Gorenstein DG. Combinatorial selection and binding phosphorothioate aptamers targeting human NF-kappa B RelA (p65) and p50. Biochemistry. 2002;41:9696–9706. doi: 10.1021/bi020220k. [DOI] [PubMed] [Google Scholar]
- Kojima T, Furukawa K, Maruyama H, Inoue N, Tarashima N, Matsuda A, Minakawa N. PCR amplification of 4’-thioDNA using 2’-deoxy-4’-thionucleoside 5’-triphosphates. ACS Synth Biol. 2013;2:529–536. doi: 10.1021/sb400074w. [DOI] [PubMed] [Google Scholar]
- Koizumi M, Soukup GA, Kerr JN, Breaker RR. Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nat. Struct. Biol. 1999;6:1062–1071. doi: 10.1038/14947. [DOI] [PubMed] [Google Scholar]
- Kore AR, Vaish NK, Morris JA, Eckstein F. In vitro evolution of the hammerhead ribozyme to a purine-specific ribozyme using mutagenic PCR with two nucleotide analogues. J. Mol. Biol. 2000;301:1113–1121. doi: 10.1006/jmbi.2000.4020. [DOI] [PubMed] [Google Scholar]
- Kostyuk DA, Dragan SM, Lyakhov DL, Rechinsky VO, Tunitskaya VL, Chernov BK, Kochetkov SN. Mutants of T7 RNA polymerase that are able to synthesize both RNA and DNA. FEBS Lett. 1995;369:165–168. doi: 10.1016/0014-5793(95)00732-o. [DOI] [PubMed] [Google Scholar]
- Kubik MF, Bell C, Fitzwater T, Watson SR, Tasset DM. Isolation and characterization of 2′-fluoro-,2′-amino-, and 2′-fluoro-/amino-modified RNA ligands to human IFN-gamma that inhibit receptor binding. J. Immunol. 1997;159:259–267. [PubMed] [Google Scholar]
- Kujau MJ, Siebert A, Wolfl S. Design of leader sequences that improve the efficiency of the enzymatic synthesis of 2′-amino-pyrimidine RNA for in vitro selection. J. Biochem. Biophys. Methods. 1997;35:141–151. doi: 10.1016/s0165-022x(97)00039-0. [DOI] [PubMed] [Google Scholar]
- Kuwahara M, Takano Y, Kasahara Y, Nara H, Ozaki H, Sawai H, Sugiyama A, Obika S. Study on suitability of KOD DNA polymerase for enzymatic production of artificial nucleic acids using base/sugar modified nucleoside triphosphates. Molecules. 2010;15:8229–8240. doi: 10.3390/molecules15118229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham JA, Johnson R, Toole JJ. The application of a modified nucleotide in aptamer selection: Novel thrombin aptamers containing 5-(1-pentynyl)-2′-deoxyuridine. Nucl. Acids Res. 1994;22:2817–2822. doi: 10.1093/nar/22.14.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lato SM, Ozerova NDS, He K, Sergueeva Z, Shaw BR, Burke DH. Boron-containing aptamers to ATP. Nucl. Acids Res. 2002;30:1401–1407. doi: 10.1093/nar/30.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Ellington AD. Section of deoxyribozyme ligases that catalyze the formation of an unnatural internucleotide linkage. Bioorg. Med. Chem. 2001;9:2581–2587. doi: 10.1016/s0968-0896(01)00033-5. [DOI] [PubMed] [Google Scholar]
- Levy M, Ellington AD. In vitro selection of a deoxyribozyme that can utilize multiple substrates. J. Mol. Evol. 2002;54:180–190. doi: 10.1007/s00239-001-0066-1. [DOI] [PubMed] [Google Scholar]
- Li M, Lin N, Huang Z, Du L, Altier C, Fang H, Wang B. Selecting aptamers for a glycoprotein through the incorporation of boronic acid moiety. J. Am. Chem. Soc. 2008;130:12636–12638. doi: 10.1021/ja801510d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Qiu Q, Gill SC, Jayasena SD. Modified RNA sequence pools for in vitro selection. Nucl. Acids Res. 1994;22:5229–5234. doi: 10.1093/nar/22.24.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz MJ, Horlacher J, Benner SA. Recognition of a non-standard base pair by thermostable DNA polymerases. Bioorg. Med. Chem. Lett. 1998;8:1149–1152. doi: 10.1016/s0960-894x(98)00177-2. [DOI] [PubMed] [Google Scholar]
- Lutz S, Burgstaller P, Benner SA. An in vitro screening technique for DNA polymerases that can incorporate modified nucleotides. Pseudo-thymidine as a substrate for thermostable polymerases. Nucl. Acids Res. 1999;27:2792–2798. doi: 10.1093/nar/27.13.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden BEH. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- Marshall KA, Ellington AD. Molecular parasites that evolve longer genomes. J. Mol. Evol. 1999;49:656–663. doi: 10.1007/pl00006586. [DOI] [PubMed] [Google Scholar]
- Mayer G. The chemical biology of aptamers. Angew. Chem. Int. Ed. Engl. 2009;48:2672–2689. doi: 10.1002/anie.200804643. [DOI] [PubMed] [Google Scholar]
- Mehan MR, Ostroff R, Wilcox SK, Steele F, Schneider D, Jarvis TC, Baird GS, Gold L, Janjic N. Highly multiplexed proteomic platform for biomarker discovery, diagnostics, and therapeutics. Advances in Experimental Medicine and Biology. 734:283–300. doi: 10.1007/978-1-4614-4118-2_20. [DOI] [PubMed] [Google Scholar]
- Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Mallikaratchy P, Tang Z, Kwame S, Meng L, Shangguan D, Tan W. Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt’s lymphoma cells. Mol. Cell. Proteomics. 2007;6:2230–2238. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- Minakawa N, Sanji M, Kato Y, Matsuda A. Investigations toward the selection of fully-modified 4’-thioRNA aptamers: Optimization of in vitro transcription steps in the presence of 4’-thioNTPs. Bioorganic and Medicinal Chemistry. 2008;16:9450–9456. doi: 10.1016/j.bmc.2008.09.048. [DOI] [PubMed] [Google Scholar]
- Nakamaye KL, Gish G, Eckstein F, Vosberg HP. Direct sequencing of polymerase chain reaction amplified DNA fragments through the incorporation of deoxynucleoside alpha-thiotriphosphates. Nucl. Acids Res. 1988;16:9947–9959. doi: 10.1093/nar/16.21.9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwlandt D, West M, Cheng X, Kirshenheuter G, Eaton BE. The first example of an RNA urea synthetase: selection through the enzyme active site of human neutrophile elastase. Chem. Bio. Chem. 2003;4:649–662. doi: 10.1002/cbic.200300610. [DOI] [PubMed] [Google Scholar]
- Ochsner UA, Katilius E, Janjic N. Detection of Clostridium difficile toxins A, B and binary toxin with slow off-rate modified aptamer. Diagn. Microbiol. Infect. Dis. 2013;76:278–285. doi: 10.1016/j.diagmicrobio.2013.03.029. [DOI] [PubMed] [Google Scholar]
- Padilla R, Sousa R. Efficient synthesis of nucleic acids heavily modified with non-canonical ribose 2′-groups using a mutantT7 RNA polymerase (RNAP) Nucl. Acids Res. 1999;27:1561–1563. doi: 10.1093/nar/27.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla R, Sousa R. A Y639F/H784A T7 RNA polymerase double mutant displays superior properties for synthesizing RNAs with non-canonical NTPs. Nucl. Acids Res. 2002;30:e138. doi: 10.1093/nar/gnf138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagratis NC, Bell C, Chang YF, Jennings S, Fitzwater T, Jellinek D, Dang C. Potent 2′-amino- and 2′-fluoro-2′-deoxyribonucleotide RNA inhibitors of keratinocyte growth factor. Nat. Biotechnol. 1997;15:68–73. doi: 10.1038/nbt0197-68. [DOI] [PubMed] [Google Scholar]
- Perrin DM, Garestier T, Helene C. Bridging the gap between proteins and nucleic acids: A metal-independent RNAseA mimic with two protein-like functionalities. J. Am. Chem. Soc. 2001;123:1556–1563. doi: 10.1021/ja003290s. [DOI] [PubMed] [Google Scholar]
- Piccirilli JA, Krauch T, Moroney SE, Benner SA. Enzymatic incorporation of a new base pair into DNA and RNA extends the genetic alphabet. Nature. 1990;343:33–37. doi: 10.1038/343033a0. [DOI] [PubMed] [Google Scholar]
- Pieken WA, Olsen DB, Benseler F, Aurup H, Eckstein F. Kinetic characterization of ribonuclease-resistant 2′-modified hammerhead ribozymes. Science. 1991;253:314–317. doi: 10.1126/science.1857967. [DOI] [PubMed] [Google Scholar]
- Robertson MP. Ph.D. dissertation. Austin: University of Texas; 2001. Engineered regulation of an RNA ligase ribozyme. [Google Scholar]
- Robertson MP, Ellington AD. In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat. Biotechnol. 1999;17:62–66. doi: 10.1038/5236. [DOI] [PubMed] [Google Scholar]
- Robertson MP, Hesselberth JR, Ellington AD. Optimization and optimality of a short ribozyme ligase that joins non-Watson-Crick base pairings. RNA. 2001;7:513–523. doi: 10.1017/s1355838201002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD, Claesson-Welsh L, Janjic N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998;273:20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- Sakthivel K, Barbas CF. Expanding the potential of DNA for binding and catalysis: highly functionalized dUTP derivatives that are substrates for thermostable DNA polymerases. Angew. Chem., Int. Ed. 2011;37:2872–2875. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2872::AID-ANIE2872>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SW, Joyce GF, Sakthivel K, Gramatikova S, Barbas CF., III RNA cleavage by a DNA enzyme with extended chemical functionality. J. Am. Chem. Soc. 2000;122:2433–2439. doi: 10.1021/ja993688s. [DOI] [PubMed] [Google Scholar]
- Schoetzau T, Langner J, Moyround E, Roehl I, Vonhoff S, Klussmann S. Aminomodified nucleobases: functionalized nucleoside triphosphates applicable for SELEX. Bioconjugate Chemistry. 2003;14:919–926. doi: 10.1021/bc0256547. [DOI] [PubMed] [Google Scholar]
- Seelig B, Jaschke A. A small catalytic RNA motif with Diels-Alderase activity. Chem. Biol. 1999;6:167–176. doi: 10.1016/S1074-5521(99)89008-5. [DOI] [PubMed] [Google Scholar]
- Siegmund V, Santner T, Micura R, Marx A. Enzymatic synthesis of 2’-methylseleno-modified RNA. Chemical Science. 2011;2:2224–2231. [Google Scholar]
- Siegmund V, Santner T, Micura R, Marx A. Screening mutant libraries of T7 RNA polymerase for candidates with increased acceptance of 2’-modified nucleotides. Chemical Communications. 2012;48:9870–9872. doi: 10.1039/c2cc35028a. [DOI] [PubMed] [Google Scholar]
- Shoji A, Kuwahara M, Ozaki H, Sawai H. Modified DNA aptamer that binds the (R)-isomer of thalidomide derivative with high enantioselectivity. J. Am. Chem. Soc. 2007;129:1456–1464. doi: 10.1021/ja067098n. [DOI] [PubMed] [Google Scholar]
- Smith D, Collins BD, Heil J, Koch TH. Sensitivity and specificity of photoaptamer probes. Mol Cell Proteomics. 2003;2:11–18. doi: 10.1074/mcp.m200059-mcp200. [DOI] [PubMed] [Google Scholar]
- Sousa R, Padilla R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 1995;14:4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southworth MW, Kong H, Kucera RB, Ware J, Jannasch HW, Perler FB. Cloning of thermostable DNA polymerases from hyperthermophilic marine Archaea with emphasis on Thermococcus sp. 9°N-7 and mutations affecting 3′-5′ exonuclease activity. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5281–5285. doi: 10.1073/pnas.93.11.5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasow TM, Tarasow SL, Eaton BE. RNA-catalysed carbon-carbon bond formation. Nature. 1997;389:54–57. doi: 10.1038/37950. [DOI] [PubMed] [Google Scholar]
- Teramoto N, Imanishi Y, Ito Y. In vitro selection of a ligase ribozyme carrying alkylamino groups in the side chains. Bioconjug. Chem. 2000;11:744–748. doi: 10.1021/bc990146r. [DOI] [PubMed] [Google Scholar]
- Vaught JD, Bock C, Carter J, Fitzwater T, Otis M, Schneider D, Rolando J, Waugh S, Wilcox SK, Eaton BE. Expanding the chemistry of DNA for in vitro selection. J Am Chem Soc. 2010;132:4141–4151. doi: 10.1021/ja908035g. [DOI] [PubMed] [Google Scholar]
- Veedu RN, Wengel J. Locked nucleic acids: promising nucleic acid analogs for therapeutic applications. Chem. Biodivers. 2010;7:536–542. doi: 10.1002/cbdv.200900343. [DOI] [PubMed] [Google Scholar]
- Waters EK, Genga RM, Schwartz MC, Nelson JA, Schaub RG, Olson KA, Kurz JC, McGinness KE. Aptamer ARC19499 mediates a procoagulant hemostatic effect by inhibiting tissue factor pathway inhibitor. Blood. 2011;117:5277–5278. doi: 10.1182/blood-2010-10-311936. [DOI] [PubMed] [Google Scholar]
- Wilson C, Keefe AD. Building oligonucleotide therapeutics using non-natural chemistries. Curr. Opin. Chem. Biol. 2006;10:607–614. doi: 10.1016/j.cbpa.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Wiegand TW, Janssen RC, Eaton BE. Selection of RNA amide synthases. Chem. Biol. 1997;4:675–683. doi: 10.1016/s1074-5521(97)90223-4. [DOI] [PubMed] [Google Scholar]
- Wlotzka B, Leva S, Eschgfäller B, Burmeister J, Kleinjung F, Kaduk C, Muhn P, Hess-Stumpp H, Klussmann S. In vivo properties of an anti-GnRH Spiegelmer: an example of an oligonucleotide-based therapeutic substance class. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8898–8902. doi: 10.1073/pnas.132067399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Chen F, Alvarado JB, Benner SA. Amplification, mutation, and sequencing of a six-letter synthetic genetic system. J. Am. Chem. Soc. 2011;133:15105–15112. doi: 10.1021/ja204910n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccolo M, Williams DM, Brown DM, Gherardi E. An approach to random mutagenesis of DNA using mixtures of triphosphate derivatives of nucleoside analogues. J. Mol. Biol. 1996;255:589–603. doi: 10.1006/jmbi.1996.0049. [DOI] [PubMed] [Google Scholar]
- Zon G, Geiser TG. Phosphorothioate oligonucleotides: Chemistry, purification, analysis, scale-up and future directions. Anticancer Drug Des. 1991;6:539–568. [PubMed] [Google Scholar]