Abstract
Bacteriophage endolysins have shown great efficacy in killing Gram-positive bacteria. PlyC, a group C streptococcal phage lysin, represents the most efficient lysin characterized to date, with a remarkably high specificity against different streptococcal species, including the important pathogen Streptococcus pyogenes. However, PlyC is a unique lysin, in terms of both its high activity and structure (two distinct subunits). We sought to discover and characterize a phage lysin active against S. pyogenes with an endolysin architecture distinct from that of PlyC to determine if it relies on the same mechanism of action as PlyC. In this study, we identified and characterized an endolysin, termed PlyPy (phage lysin from S. pyogenes), from a prophage infecting S. pyogenes. By in silico analysis, PlyPy was found to have a molecular mass of 27.8 kDa and a pI of 4.16. It was active against a majority of group A streptococci and displayed high levels of activity as well as binding specificity against group B and C streptococci, while it was less efficient against other streptococcal species. PlyPy showed the highest activity at neutral pH in the presence of calcium and NaCl. Surprisingly, its activity was not affected by the presence of the group A-specific carbohydrate, while the activity of PlyC was partly inhibited. Additionally, PlyPy was active in vivo and could rescue mice from systemic bacteremia. Finally, we developed a novel method to determine the peptidoglycan bond cleaved by lysins and concluded that PlyPy exhibits a rare d-alanyl-l-alanine endopeptidase activity. PlyPy thus represents the first lysin characterized from Streptococcus pyogenes and has a mechanism of action distinct from that of PlyC.
INTRODUCTION
While Streptococcus pyogenes (or group A streptococcus) is most commonly known as the causative agent of strep throat, it represents a major clinical issue causing severe invasive infections, such as streptococcal toxic shock-like syndrome and necrotizing fasciitis (1), as well as serious sequelae, such as rheumatic fever and rheumatic heart disease, resulting in over 500,000 deaths per year worldwide (2). Many of the virulence factors (and virulence attributes) of S. pyogenes are encoded by integrated prophages. Such prophages enable the bacteria to produce a number of pyrogenic exotoxins responsible for scarlet fever (3), exotoxin K and phospholipase A (4), as well as hyaluronidases, suggested to facilitate the degradation of the extracellular matrix (5). While the importance of these virulence factors for the pathogenicity of group A streptococcus-related diseases has been debated (6), most S. pyogenes strains contain between 4 and 6 integrated prophages, representing up to 12% of the total DNA content in the cell (7). Furthermore, the genetic differences among S. pyogenes strains in certain serotypes are almost exclusively due to prophages (8).
Despite this abundance of prophages in S. pyogenes, no one has, to our knowledge, studied the lysins encoded by these prophages or examined their ability to eradicate S. pyogenes. However, other streptococcal lysins displaying activity against S. pyogenes have been isolated and characterized, including those from group B streptococcus (GBS) (9, 10), Streptococcus uberis (Ply700) (11), group C streptococcal phage C1 (PlyC) (12), and Streptococcus pneumoniae Cpl-7 (13).
Phage-encoded lysins are transcribed late in the phage lytic cycle and accumulate in the cytoplasm until they are released by the presence of holins as a means to degrade the peptidoglycan layer of the host, thereby causing lysis of the bacterial cell and release of mature phages. Even though lysins harbor various enzymatic activities (amidases, muramidases, glucosaminidases, and endopeptidases), they all result in peptidoglycan degradation. Most Gram-positive bacterial lysins have a distinct two-domain structure with an N-terminal catalytic domain and a C-terminal binding domain, giving the lysin its specificity in most cases (14). Even though such architecture is common for most lysins, the group C streptococcal phage lysin, PlyC, is an exception. PlyC is not encoded on a single gene but is composed of two distinct gene products termed PlyCA and PlyCB separated by an intron-like sequence. These two products form a functional PlyC lysin by combining eight PlyCB (binding) subunits with one PlyCA (catalytic) subunit (12, 15). Clearly, PlyC represents a unique endolysin structure and is not representative of other lysins active against similar species. Earlier studies have shown, both in vitro and in vivo, that phage lysins have great potential as novel therapeutics, controlling several important Gram-positive bacterial pathogens, including Staphylococcus aureus (16, 17), Streptococcus pneumoniae (18), and Bacillus anthracis (19). In this study, we have identified the first phage lysin from a S. pyogenes lysogen, PlyPy (phage lysin from S. pyogenes), characterized its biochemical properties, identified its cleavage activity, and investigated its potential to specifically bind to and eradicate S. pyogenes both in vitro and in vivo.
MATERIALS AND METHODS
Bacterial strains.
All bacterial strains used for determining PlyPy specificity (see Table S1 in the supplemental material) were cultured in brain heart infusion (BHI) broth (Thermo Fischer Scientific, Waltham, MA) at 37°C. The strains are part of either the ATCC collection or The Rockefeller University collection.
Construction of the prophage lysin and corresponding binding domain.
Genomic DNA from S. pyogenes strain MGAS315 was purified using a Qiagen DNeasy blood and tissue kit (Qiagen, Germantown, MD), according to the manufacturer's protocol. The full-length lysin spyM3_1306 (GenBank accession no. AAM79913; gene identifier, 21905047) (PlyPy) was amplified by PCR using the primers 5′-TTAATAAAATTTACAATATGGATTGACATTGT-3′ and 5′-ACGACAGTTGGAGAGGAATAAAATATG-3′ (789-bp product), while the binding domain (PlyPyBD) was amplified using primers 5′-TCTAGAAGACCGCCTTATAGCGATA-3′ and 5′-CTGCAGTTAATAAAATTTACAATATGGA-3′ (327-bp product) and the Taq DNA polymerase from New England BioLabs. The PCR program was initiated by a denaturation step at 95°C for 5 min, followed by 35 cycles at 95°C for 30 s, 52°C for 30 s, and 72°C for 60 s and a final extension at 72°C for 10 min. The amplified DNA was analyzed on a 1% agarose gel and purified using a QIAquick gel purification kit (Qiagen). DNA fragments were ligated into a pBAD TOPO TA vector and transformed into TOP10 cells (Invitrogen, Carlsbad, CA).
Expression and purification of PlyPy.
Escherichia coli clones able to express active lysins were identified using an overlay of autoclaved bacteria, as described by Schmitz et al. (20), with modifications. Briefly, E. coli clones were grown overnight on LB plates supplemented with 0.2% arabinose and 50 μg/ml carbenicillin at 37°C. Lysin expression was further induced by exposing the E. coli clones to 20% (wt/vol) nebulized arabinose for 3 h at room temperature. The plates were then returned to 37°C, and the cells were incubated for an additional 3 h. Afterwards, the cells were permeabilized by chloroform treatment for 45 min, before a soft agar overlay of autoclaved MGAS315 cells was added. The cells were allowed to incubate overnight at 37°C, followed by inspection for clearing zones in the streptococcal soft agar, indicating the expression of active lysins.
Confirmed clones expressing active lysin were grown in LB supplemented with 50 μg/ml carbenicillin overnight at 37°C and 220 rpm. The culture was diluted 1:200 in fresh LB supplemented with carbenicillin and incubated for an additional for 3 h. Expression of the lysin was induced with 0.2% arabinose, and incubation continued overnight at 30°C. Cells were pelleted, resuspended in 20 mM Tris, pH 6.8, and homogenized using an Emulsiflex-C5 homogenizer (Avestin, Ottawa, Ontario, Canada). Cell debris was removed by centrifugation, and the sample was sterile filtered (pore size, 0.22 μm) to remove any remaining particles. To purify PlyPy, the lysin was precipitated with 30% ammonium sulfate, the supernatant was discarded, and the pellet was resuspended in 20 mM Tris, pH 6.8. To remove any potential inclusion bodies or precipitate, the sample was centrifuged and sterile filtered (pore size, 0.22 μm) before it was loaded on a Mono Q HiTrap FF column (GE Healthcare, Uppsala, Sweden) using an Äkta system (GE Healthcare), and bound lysin was eluted using a linear gradient of 0.0 to 1.0 M NaCl in 20 mM Tris, pH 6.8. Peaks were analyzed on SDS-polyacrylamide gels to determine the purity of the lysin. Fractions with high concentrations of lysin were dialyzed against 20 mM Tris, pH 6.8, overnight (molecular mass cutoff, 12 to 14 kDa; Spectrum Spectra/Por; Fischer Scientific) at 4°C.
Activity assays.
Overnight cultures of S. pyogenes strain MGAS315 were diluted 1:10 in prewarmed BHI broth and incubated until mid-log phase. Cells were then centrifuged (3,000 × g, 25 min, 14°C), washed 2 times in 20 mM Tris-HCl, pH 6.8, and finally resuspended in the same buffer to generate an optical density at 595 nm (OD595) of approximately 2.5.
The activity of PlyPy was calculated on the basis of the reduction in the optical density (OD595) measured in a 96-well plate using a SpectraMax Plus reader (Molecular Devices, Sunnyvale, CA). To investigate any dependence of PlyPy on divalent cations, 100 μl of bacterial suspension was incubated with 100 μl of 20 mM Tris-HCl, pH 6.8, containing 5 μg PlyPy and various concentrations (0 to 100 mM) of CaCl2. The incubation proceeded for 5 to 20 min at 37°C.
To study the effect of pH on the activity of PlyPy, the same experimental settings described above were utilized with buffers containing 2 mM CaCl2 in 20 mM sodium acetate (pH 5.0, 5.5, and 6.0) or 20 mM Tris (pH 6.0, 6.5, 7.0, 7.5, 8.0, and 8.8). Other parameters tested using similar settings included NaCl concentration (0 to 1,000 mM) and PlyPy kinetics. All experiments were performed in triplicate, and results are shown as means ± standard deviations.
The activity of PlyPy was determined as described elsewhere (18). Briefly, 100 μl of lysin in different dilutions was incubated with 100 μl of bacteria suspended in 20 mM Tris-HCl, pH 6.8, 100 mM NaCl, 2 mM CaCl2 at 37°C for 15 min. The dilution corresponding to a 50% reduction of the starting OD595 was determined to correspond to 1 U.
PlyPy MIC.
The MIC of PlyPy was determined in a 96-well plate as described by Wiegand et al. (21). Briefly, an overnight culture of S. pyogenes strain MGAS315 was diluted to generate a final concentration of 5.0 × 105 CFU/ml in each well. Lysin activity buffer (2 mM CaCl2, 100 mM NaCl, 20 mM Tris-HCl, pH 6.8) was added together with PlyPy at concentrations ranging from 2 to 256 μg/ml. Incubation continued for 16 h, before an MIC value was read. The MIC was defined as the lowest concentration of PlyPy that inhibited any visible growth of the bacterium.
Specificity assay.
The susceptibility of different bacterial strains and species (see Table S1 in the supplemental material) to lysis by PlyPy was characterized as described above for the activity assay (15 min of incubation, 37°C, 1.5 U PlyPy).
PlyPy carbohydrate interaction.
To investigate whether the group A streptococcus-specific carbohydrate could interact with and affect the lysin, PlyPy and PlyC were preincubated with purified group A carbohydrate extracted from S. pyogenes (22) at a ratio of 4:1 (carbohydrate/lysin; wt/wt) for 15 min at 37°C. The lysins (1 U/ml) were then added to the washed bacteria, and the reduction in optical density was measured after 15 min at 37°C.
PlyPyBD fusion to a biotinylation site.
Primers 5′-GCGGAATTCATGAAACTGAAGGTAACAGTCAACG-3′ and 5′-GTGGGTACCGCGACCTTCGATGAGCTCGAG-3′ were used to amplify the biotin purification tag from the cloning vector PinPoint XA-1 (Promega, Madison, WI). The resulting PCR product was inserted into the EcoRI and KpnI sites of pBAD24 (23). PlyPyBD was cut out from the pBAD TOPO TA construct, using the restriction enzymes XbaI and PstI, and ligated in frame with the N-terminal biotinylation tag of the resulting plasmid. The construct was induced with arabinose using the above-mentioned method in the presence of 2 μM biotin. The cell pellet was resuspended in 20 mM Tris-HCl, pH 6.8, supplemented with 2 mM CaCl2 and homogenized. The lysate was centrifuged (10,000 × g, 45 min) and sterile filtered through a 0.22-μm-pore-size filter to remove any cell wall fragments. The crude lysate was then used for fluorescence microscopy studies.
Phage lysin binding assay using fluorescence microscopy.
For microscopy experiments, overnight cultures of bacterial strains were diluted 1:100 into fresh medium (BHI broth) and grown to mid-log phase, fixed with 2.6% paraformaldehyde in 30 mM phosphate buffer, pH 7.4, on ice for 1 h, and washed with 1× phosphate-buffered saline (PBS). Bacteria were attached to poly-l-lysine-coated coverslips and blocked with blocking buffer (PBS containing 2% bovine serum albumin and 1% gelatin). Clarified lysates of induced or noninduced E. coli bacteria containing the biotin-PlyPyBD expression vector were diluted 1:10 in blocking buffer and incubated with the cells for 1 h at room temperature in a moist chamber. The cells were washed with 1× PBS and incubated with a 1:100 dilution of streptavidin-fluorescein isothiocyanate (FITC) conjugate in blocking buffer for 1 h at room temperature. The cells were then washed with 1× PBS and mounted in 50% glycerol and 0.1% p-phenylenediamine in PBS, pH 8.0. Slides were imaged on a Delta-Vision image restoration microscope (Applied Precision/Olympus) equipped with a CoolSnap QE cooled charge-coupled-device camera (Photometrics). An Olympus ×100 (numerical aperture, 1.40) UPLS Apo oil-immersion objective was used in conjunction with a ×1.5 optovar. z-stacks were taken at 0.15-μm intervals. Images were deconvolved using SoftWoRx software (Applied Precision/DeltaVision).
Peptidoglycan purification and digestion with PlyPy.
S. pyogenes strain AR01, a ΔSrtA D471 strain (24), was cultured overnight in Todd-Hewitt broth supplemented with yeast extract, diluted 1:50 into fresh prewarmed medium, and cultured until it reached mid-log phase. The bacteria were spun down (4,000 × g, 30 min, 4°C), washed, and resuspended in PBS. Cells were then subjected to nitrous acid extraction to remove any attached carbohydrates (25). Briefly, a 0.125 volume of 4 M sodium nitrite and a 0.125 volume of glacial acetic acid were added to the cells. The reaction mixture was slowly mixed for 15 min, using a magnetic stirrer in a chemical hood, at which time the solution became yellow. The cells were collected by centrifugation, washed, and homogenized using an Emulsiflex-C5 homogenizer (Avestin). Cell debris was removed and discarded by slow centrifugation at 4,000 × g for 30 min at 4°C, while the cell wall fragments were collected by faster centrifugation (16,000 × g, 60 min, 4°C). Cell wall fragments were washed three times in double-distilled H2O, boiled in 4% SDS for 30 min, and washed in water four times before being resuspended in 20 mM Tris-HCl, pH 6.8 (1:2,000 of the original culture volume).
Purified peptidoglycan fragments were digested overnight with mutanolysin (5,000 U/ml, 37°C) and poured into an equilibrated wheat germ agglutinin (WGA) column (Vector Laboratories, Burlingame, CA). The column was then washed with 10 volumes of buffer (20 mM Tris-HCl, pH 6.8) and sealed until the column was reequilibrated with 20 mM Tris-HCl, 100 mM NaCl, and 2 mM CaCl2. PlyPy (50 μg) was then added, together with either regular water or H218O (Sigma, St. Louis, MO), and the column was incubated overnight at 37°C with a slow shaking. On the next day, the flowthrough was collected and the column was washed with 5 volumes of buffer. Bound peptidoglycan fragments were then eluted 5 times with 1 volume of 0.5 M N-acetyl-d-glucosamine (Sigma). The elution fractions were filtered through a Vivaspin 500 filter (molecular weight cutoff, 3,000; Sartorius Stedim Biotech, Goettingen, Germany).
MS analysis.
Samples were desalted and concentrated using a double Empore C18 membrane (3M, St. Paul, MN) inserted into a P200 pipette tip (26). The samples were then analyzed by nano-liquid chromatography (LC)-tandem mass spectrometry (MS) using an Q-Exactive mass spectrometer (Thermo, San Jose, CA) coupled to a Dionex NCP3200RS high-pressure liquid chromatography (HPLC) setup (Thermo, Sunnyvale, CA). The HPLC samples were loaded on a trap column prior to separation on a 75-μm by 12-cm column packed with 3-μm, reversed-phase C18 particles (Nikkyo Technos Co., Ltd., Japan). The analytical gradient was generated at 200 nl/min, increasing from 1% buffer B (0.1% formic acid in acetonitrile) and 99% buffer A (0.1% formic acid) to 45% buffer B and 35% buffer A in 40 min, followed by washing and conditioning. MS survey scans were scanned from m/z 300 to m/z 1,400 at a resolution of 70,000 (auto gain control [AGC], 1e6; maximum injection time [IT], 60 ms). Up to the 10 most abundant ions were subjected to tandem MS and measured at a resolution of 35,000 (AGC, 3e6; maximum IT, 120 ms) with m/z 100 as the lowest mass. For the tandem MS experiments, an isolation window of 3.5 Thomson was used.
In vivo murine model.
The Rockefeller University's Institutional Animal Care and Use Committee approved all in vivo protocols. All experiments were conducted at The Rockefeller University's animal housing facility, an AAALAC-accredited research facility, with all efforts made to minimize suffering. The systemic infection model described by Gilmer et al. (27) and Daniel et al. (17) was used to test for the in vivo efficacy of PlyPy against S. pyogenes. Briefly, 4- to 5-week-old female FVB/NJ mice (weight range, 15 to 20 g) were obtained from The Jackson Laboratory (Bar Harbor, ME). After a period of acclimation, mice were injected intraperitoneally (i.p.) with 0.5 ml of mid-log-phase (OD600, 0.5) bacteria diluted with 5% hog gastric mucin (Sigma) in saline. Bacterial suspensions contained ∼1 × 107 CFU/ml of S. pyogenes MGAS5005. Actual bacterial inoculation titers were calculated by serial dilution and plating onto Columbia blood agar plates for each experiment.
Mice became bacteremic within 1 to 3 h and contained S. pyogenes within multiple organs, including spleen, liver, kidney, and heart/blood (unpublished observations) (27). At 3 h postinfection, the animals were randomly divided into 3 treatment groups and administered an i.p. injection of 0.5 ml containing either control buffer (20 mM Tris-HCl, pH 6.8, 150 mM NaCl, 2 mM CaCl2) or 0.25 mg or 0.50 mg of the PlyPy lysin in the control buffer.
The survival rate for each experimental group was monitored every 12 h for the first 24 h and then every 24 h up to 7 days postinfection. The results from 3 independent experiments were combined to evaluate a total of 18 mice for the buffer controls and 20 mice for each lysin treatment group. The data were statistically analyzed by the use of Kaplan-Meier survival curves, with standard errors, 95% confidence intervals, and significance (log-rank/Mantel-Cox test) calculated using the Prism computer program (GraphPad Software, La Jolla, CA).
RESULTS
Expression and purification of PlyPy.
Based on the high prevalence of prophages in S. pyogenes, we used a bioinformatics approach to identify putative endolysins from the S. pyogenes genome. We established three criteria for the identification of a group A streptococcal phage-encoded lysin: (i) the lysin should be a part of a phage (to exclude any autolysins), (ii) the lysin should have a common two-domain architecture, and (iii) the lysin should have low similarity to any other previously characterized lysins from other species. From these three criteria we chose the spyM3_1306 gene from S. pyogenes strain MGAS315, which we termed PlyPy (phage lysin from S. pyogenes) for further characterization. PlyPy has a typical two-domain structure, with an N-terminal catalytic CHAP domain and a C-terminal binding (SH3) domain (see Fig. S1 in the supplemental material).
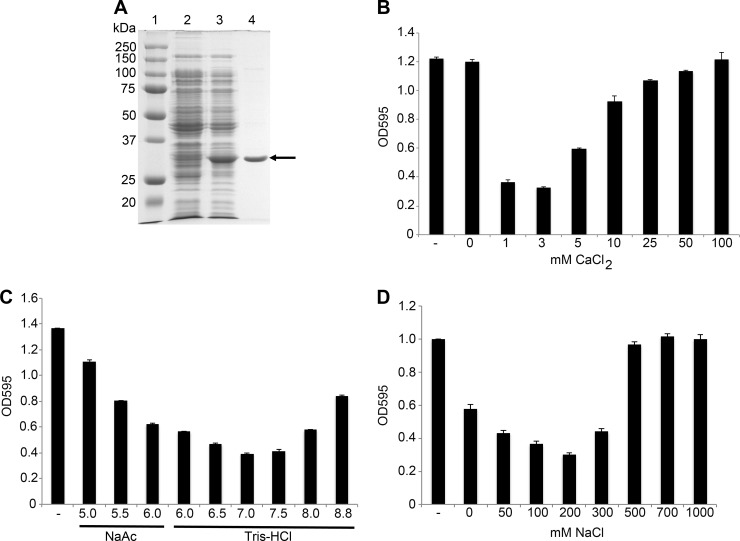
In our experience, the addition of protein purification tags to lysins often results in the formation of inclusion bodies and generally low yields. Thus, to reduce this complication, PlyPy was expressed as a native protein without any tags, using the arabinose-inducible pBAD24 vector. Due to the low theoretical pI of PlyPy (4.16), the protein precipitated at concentrations of ammonium sulfate much lower than those for the majority of contaminating E. coli proteins. This step, in combination with ion-exchange chromatography (on a Mono Q column), enabled us to purify PlyPy to >90% homogeneity, as determined by visual approximation (Fig. 1A). This procedure consistently resulted in approximately 25 mg lysin per liter of E. coli growth.
FIG 1.
Purification and characterization of PlyPy activity. (A) SDS-PAGE of the purification process for PlyPy. Lanes: 1, molecular size ladder; 2, crude extract from noninduced clone; 3, crude extract from induced clone; 4, final purified fraction of PlyPy (27.8 kDa; arrow). The effects of CaCl2 (B), pH (C), and NaCl (D) on the activity of PlyPy were all measured via determination of the reductions in the OD595 using 2 U/ml PlyPy at 37°C with incubation for 30 min. NaAc, sodium acetate.
Enzymatic characteristics of PlyPy.
The activity of several phage lysins, including the Streptococcus suis lysin PlySs2 (27) and the Streptococcus uberis lysin Ply700 (11), is increased by the presence of divalent cations at low concentrations. To investigate if the activity of PlyPy was affected by divalent cations as well, the lysin was incubated with different concentrations of CaCl2 (Fig. 1B). PlyPy showed a strong dependence on CaCl2, with no significant activity in the absence of CaCl2 or in the presence of MgCl2 (see Fig. S2 in the supplemental material). The optimal CaCl2 concentration was determined to be 3 mM, but CaCl2 concentrations ranging from 1 to 3 mM showed similar effects, while higher concentrations of the salt significantly decreased the activity of PlyPy (Fig. 1B).
In addition to divalent cations, several other parameters may influence the enzymatic activity of PlyPy. We investigated the pH, NaCl, dithiothreitol (DTT), and temperature sensitivity of the lysin. PlyPy had its highest activity at pH 7.0 (Fig. 1C) but also showed considerable activity in the pH range of 6.0 to 8.0. Furthermore, the addition of 50 to 300 mM NaCl increased the activity of PlyPy, while higher concentrations (>500 mM) completely inhibited its activity (Fig. 1D). The activity of PlyPy was not affected by 5 mM DTT (see Fig. S1A in the supplemental material). The optimal temperature for PlyPy activity was 37°C, but the enzymatic activity was high at temperatures of between 22 and 42°C (see Fig. S1B in the supplemental material). Incubation at temperatures over 60°C denatured PlyPy and thus inhibited its activity (see Fig. S1C in the supplemental material). Taken together, these findings suggest that the lysin should be active in an in vivo setting and thus have a potential therapeutic use. For further experiments, we used 20 mM Tris-HCl, pH 6.8, supplemented with 100 mM NaCl and 2 mM CaCl2 as our activity buffer. Using the above-mentioned conditions, PlyPy was determined to have a lytic activity corresponding to 0.5 U/μg, as defined in Materials and Methods (18), which is considered quite active compared to other reported lysins (28).
To further study the kinetics of the enzyme, we resuspended mid-log-phase S. pyogenes strain MGAS315 in activity buffer, added different concentrations of PlyPy (1.5 to 50 U/ml), and incubated the cells at 37°C while measuring the reduction of turbidity at an OD595. When using high concentrations of the lysin (25 to 50 U/ml), the OD595 dropped to a baseline of 0.25 within 3 min and remained at this OD595 (see Fig. S1A in the supplemental material), representing only bacterial cell debris (data not shown). At more dilute lysin concentrations, however, there was an initial lag phase before optical reduction. PlyPy was more active against exponentially growing bacteria than stationary-phase bacteria (see Fig. S1B in the supplemental material) and killed S. pyogenes in a concentration-dependent manner. Using S. pyogenes strain MGAS315, the MIC was determined to be 64 μg/ml (data not shown).
Specificity of PlyPy.
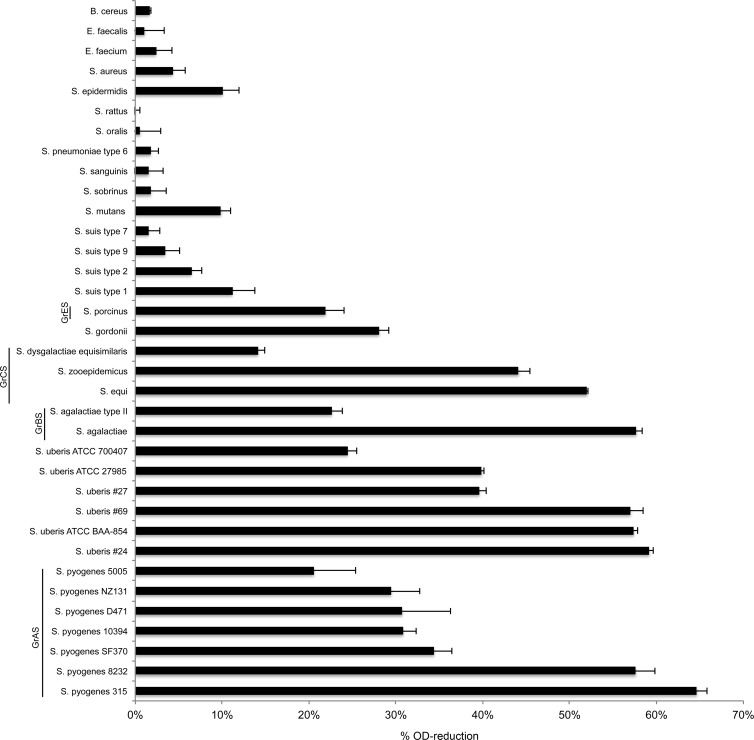
Phage lysins generally exhibit high specificity (28), displaying elevated activity against a few closely related species, though lysins with broader activity do exist (29). To study the specificity of PlyPy, bacterial isolates of different species (see Table S1 in the supplemental material) were cultured until mid-exponential phase, washed, and resuspended in activity buffer. PlyPy was added (1.5 U/ml), and the mixture was incubated for 15 min before the reduction in the optical density was measured. The lysin was found to be highly active against most S. pyogenes isolates used in the study, though strain-specific differences were observed (Fig. 2). In addition to S. pyogenes, PlyPy was able to reduce the optical density of several group B and C streptococci, as well as that of S. uberis. The lysin showed moderate activity against the oral commensal organism Streptococcus gordonii and the group E streptococcus Streptococcus porcinus, but it showed little or no activity against strains of Enterococcus, Staphylococcus, and Bacillus. Interestingly, several other species of streptococci, including Streptococcus rattus, Streptococcus oralis, S. pneumoniae, Streptococcus sanguinis, Streptococcus sobrinus, and S. suis, appeared to be resistant to the enzyme.
FIG 2.
Specific activity of PlyPy. PlyPy (1.5 U/ml) was incubated with various bacterial species and isolates, and relative OD595 reductions after 15 min at 37°C are reported. The main streptococcal groups (group A streptococci [GrAS], group B streptococci [GrBS], group C streptococci [GrCS], and group E streptococci [GrES]) are represented, as are species from the viridans streptococcal mutans group (S. mutans, S. sobrinus, S. rattus) and mitis group (S. oralis, S. sanguinis, S. gordonii).
Binding affinity of PlyPy.
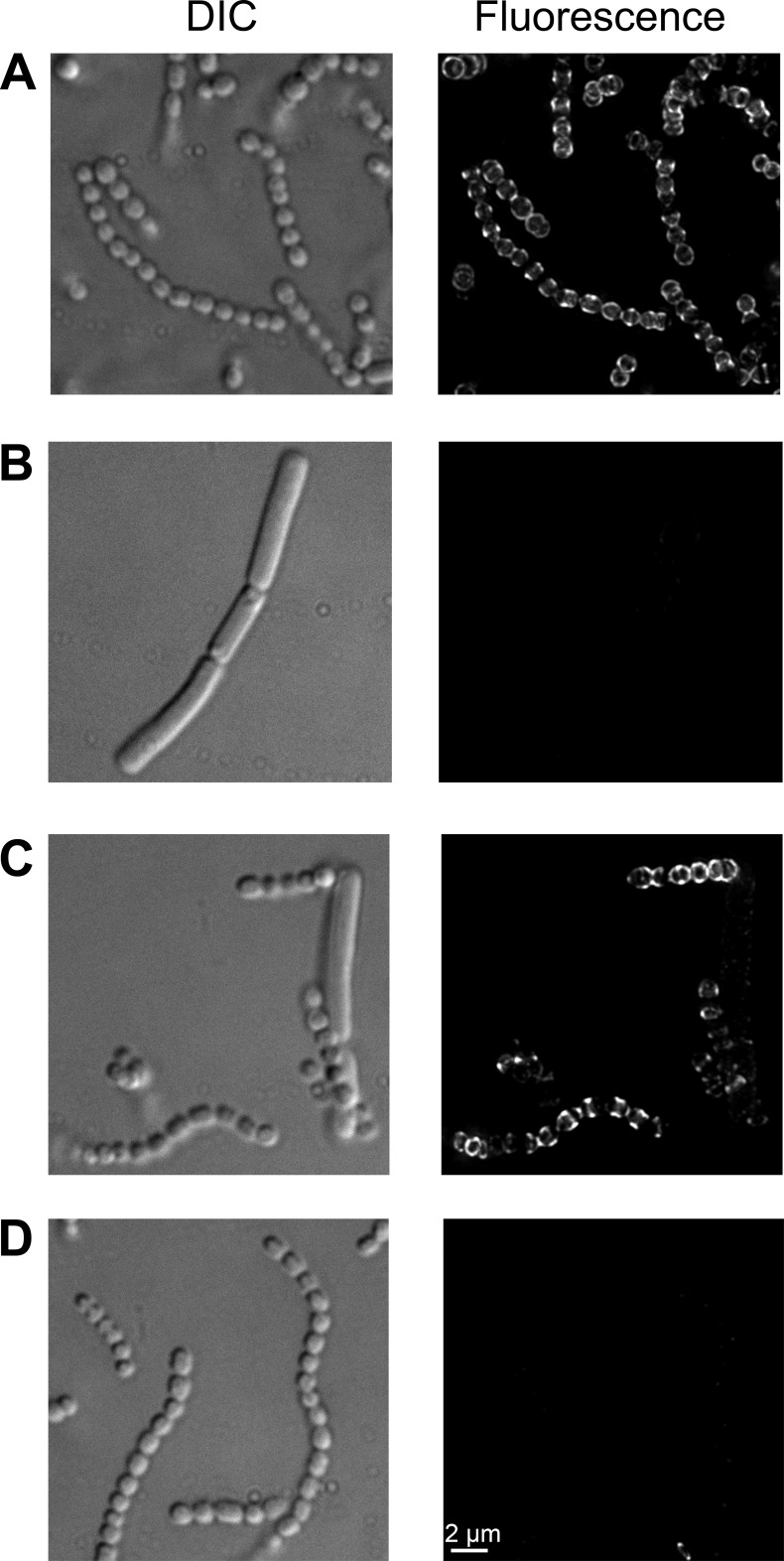
While we observed PlyPy activity against several streptococci, including S. pyogenes, it was apparent that some streptococci, as well as other bacterial species, were not affected by the lysin. As most lysins display a two-domain structure with a catalytic N-terminal domain and a C-terminal binding domain (14), we wanted to determine if the inactivity of PlyPy against certain bacterial strains was due to the inability of the binding domain to bind to cell wall receptors of these organisms. To test this possibility, we expressed the binding domain of PlyPy fused to an N-terminal biotinylated tag (biotin-PlyPyBD). Cells of S. pyogenes and Bacillus cereus were fixed, mounted on microscopy slides, and reacted with the biotin-PlyPyBD construct. Binding of the biotinylated construct to bacteria was visualized using a streptavidin-FITC conjugate. PlyPyBD displayed binding activity, as it bound to the cell walls of S. pyogenes (Fig. 3A), but it could not bind to B. cereus (Fig. 3B). Furthermore, in a mixed population of S. pyogenes and B. cereus cells, PlyPyBD bound only S. pyogenes, stressing its high specificity (Fig. 3C). To rule out the possibility that a nonspecific biotinylated E. coli protein could in part be responsible for the observed signal, we analyzed the induced lysate by Western blotting. Only a single biotinylated band with a molecular mass corresponding to that of biotin-PlyPyBD was detected (data not shown). Furthermore, the noninduced lysate did not generate any fluorescent signal on S. pyogenes cells (Fig. 3D).
FIG 3.
Characterization of the PlyPy binding domain. The binding domain of PlyPy was expressed as a fusion protein with a biotinylation tag. S. pyogenes (A), B. cereus (B), and a mixture of S. pyogenes and B. cereus (C) were incubated with biotin-PlyPyBD and subsequently with a streptavidin-FITC conjugate. Slides were visualized by deconvolution immunofluorescence microscopy and differential interference contrast (DIC) microscopy. The binding of the noninduced crude lysate of biotin-PlyPyBD to S. pyogenes (D) served as a background control.
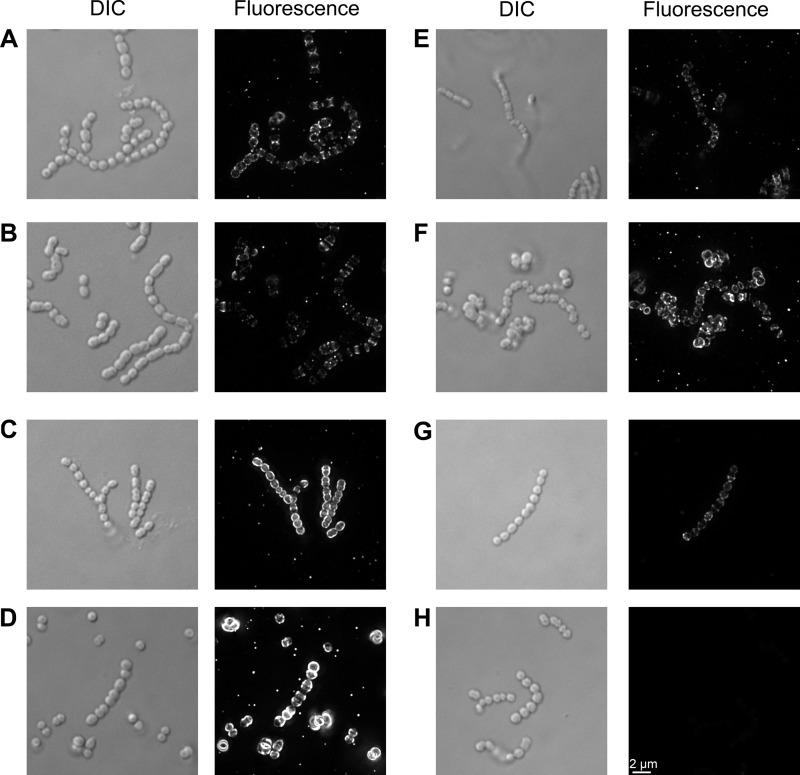
To further expand on the binding of PlyPyBD to different species, we included six more streptococcal species for which the lysin had various levels of activity (Fig. 4A to H). PlyPyBD bound to all tested species with various intensities. It showed a low degree of binding (compared to that to S. pyogenes) to S. suis (Fig. 4B), S. rattus (Fig. 4E), and Streptococcus equi (Fig. 4G), while it had a moderate degree of binding to S. uberis (Fig. 4F). Both Streptococcus agalactiae (Fig. 4C) and S. porcinus (Fig. 4D) strongly bound PlyPyBD.
FIG 4.
PlyPy binds to several streptococcal species. Biotin-PlyPyBD was incubated with S. pyogenes (A), S. suis (B), S. agalactiae (GBS) (C), S. porcinus (group E streptococcus) (D), S. rattus (E), S. uberis (F), and S. equi (group G streptococcus) (G) and subsequently with a streptavidin-FITC conjugate. Slides were visualized by deconvolution immunofluorescence microscopy and differential interference contrast (DIC) microscopy. The binding of the noninduced crude lysate of biotin-PlyPyBD to S. pyogenes (H) served as a background control.
Binding receptor.
Additional lysins with activity against S. pyogenes, including the highly active PlyC lysin, which has been thoroughly studied and whose crystal structure was recently solved (15), have been characterized. It has been reported that a portion of the group-specific carbohydrate (polyrhamnose) for group A streptococci functions as part of the receptor for PlyC, providing its specificity (30). Thus, we wanted to investigate if PlyPy also relies on such structures. To test this, we took OD595 measurements of S. pyogenes cells after the addition of PlyC or PlyPy with or without the presence of group A-specific carbohydrate. We found that addition of PlyC lysin to S. pyogenes cells rapidly decreased the OD595 by 50% (Fig. 5). However, the addition of the group A-specific carbohydrate to the mixture inhibited the activity of the PlyC lysin but did not do so completely. In addition, the group A carbohydrate did not affect PlyPy activity, indicating that it interacts with another, hitherto unknown conserved cell wall structure of the bacterium. Furthermore, the absence of M protein on the surface of S. pyogenes did not influence the activity of PlyPy (see Fig. S3 in the supplemental material).
FIG 5.
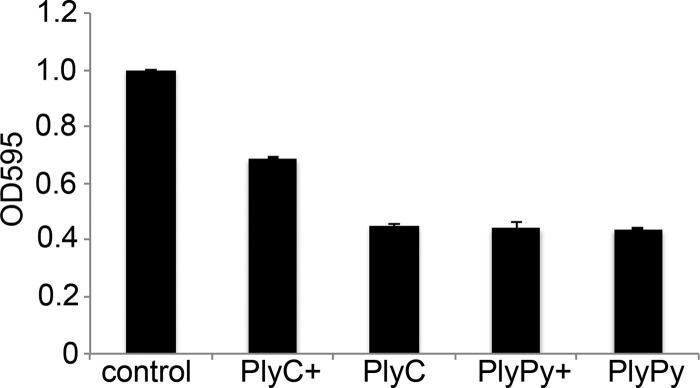
PlyC and PlyPy interaction with group A streptococcus-specific carbohydrates. PlyC and PlyPy (1 U/ml) were preincubated with purified group A streptococcus-specific carbohydrates, before adding the mixture to S. pyogenes cells. The reduction in the OD595 was measured after 15 min at 37°C. +, the sample contained a mixture of the lysin and the group A streptococcus carbohydrate at a ratio of 1:4 (wt/wt).
Cleavage activity of PlyPy.
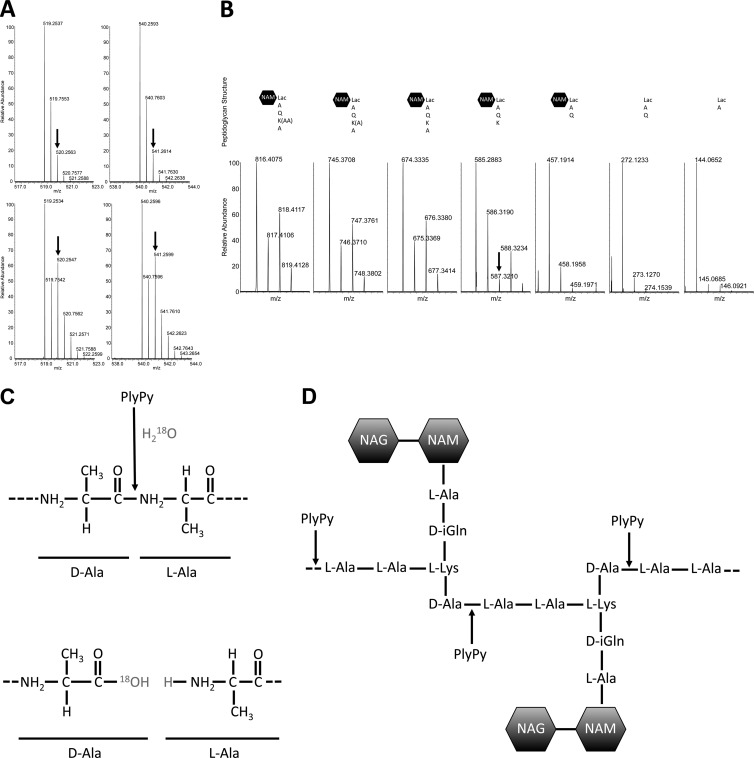
Peptidoglycan is a very repetitious structure, specifically in the case of streptococcal and staphylococcal peptidoglycans, with their alanine and glycine cross bridges, respectively. For this reason, to correctly identify the enzymatic activity of lysins active against these species, it might prove insufficient or even misleading if tandem MS is not used to decipher specific digestion fragments, particularly if the lysin has endopeptidase activity (31). Until recently, this issue posed difficulties in the studies of lysins, until Gargis et al. published a novel method using 4-sulfophenyl isothiocyanate to pinpoint the site of digestion for zoocin A (31). Though this method was effective in addressing the cleavage activity of the lysin, it is quite time-consuming and technically challenging. We therefore developed a novel method to determine the cleavage activity of lysins based on lectin binding proteins and the incorporation of H218O, with the latter being a common technique used for protein sequencing in proteomics (32). Briefly, we digested the purified peptidoglycan with mutanolysin (an N-acetylmuramidase) and applied the digest to a wheat germ agglutinin column to specifically bind molecules with the free GlcNAc termini created by the cleavage. The column was then washed to remove any unbound products (e.g., mutanolysin and uncleaved peptidoglycan). The bound peptidoglycan was then incubated with PlyPy in the presence of ordinary water or H218O, enabling the incorporation of 18O at the site of hydrolysis, assuming that H2O takes part in the enzymatic reaction. Cleavage products were then eluted with free GlcNAc, and the released peptidoglycan fragments were analyzed using liquid chromatography and mass spectrometry. By comparing the results of the LC-MS experiments with the digested and undigested (control) samples, we identified two doubly charged ions that were unique for the PlyPy-treated peptidoglycan, namely, m/z 519.2537 and m/z 540.2593 (see Fig. S4 in the supplemental material). When we repeated the PlyPy digestion experiment but this time replacing normal water with H218O, the isotopic pattern of these two ions changed to a pattern indicating the incorporation of 18O for these two ions (the third isotope peak increased in abundance) (Fig. 6A). Tandem MS experiments suggested that these two PlyPy-generated cleavage products represent the same parental structure but that they differ by one acetyl group. To identify the particular amino acid or glycan where the 18O was incorporated, we manually interpreted the tandem MS spectrum acquired for doubly charged ions at m/z 519.2537 and plotted and traced the isotopic pattern (Fig. 6B; see Fig. S5 in the supplemental material). Studying the tandem MS-generated fragment ions, we observed that the 18O-induced isotopic pattern disappeared by the removal of the d-Ala (Fig. 6B; see Table S2 in the supplemental material). Thus, in the presence of H218O, PlyPy incorporates 18O at the carboxy terminus of d-Ala when the peptide bond between d-Ala and l-Ala is cleaved (Fig. 6C and D) and thus represents a d-alanyl-l-alanine endopeptidase.
FIG 6.
PlyPy has d-Ala-l-Ala endopeptidase activity. Purified peptidoglycan from S. pyogenes MGAS315 was incubated with PlyPy in the presence of ordinary water (A, top) or H218O (A, bottom). Two peptides (m/z 519.2537 and 540.2593; z = 2) having the distinct H218O isotopic pattern, characterized by an increased abundance of the third isotope peak (arrow), were identified. (B) Tandem MS of the m/z 519.2537 ion (the m/z 540.2593 ion is identical but has the addition of an acetyl group) revealed that the isotopic pattern disappeared once the carboxy-d-Ala was removed (the third isotope peak is no longer more intense than the second peak. (C and D) PlyPy, in the presence of H218O, inserts 18O in the carboxy-terminal part of d-Ala when digesting the d-Ala–l-Ala bond (C), enabling the determination of the activity of PlyPy as a d-Ala-l-Ala endopeptidase (D). NAG, N-acetylglucosamine; NAM, N-acetylmuramic acid.
Protection from systemic S. pyogenes infection.
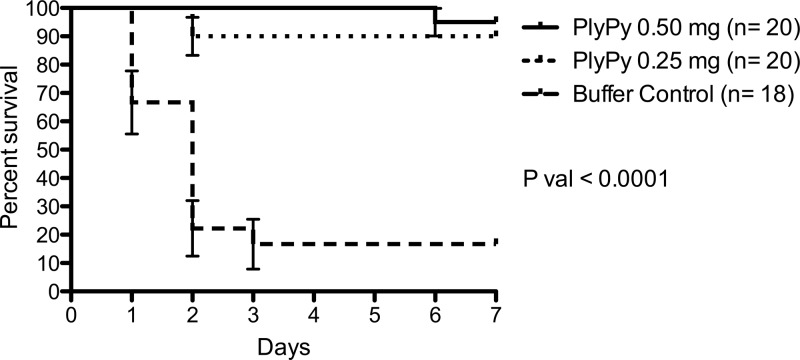
In order to assess whether PlyPy treatment could prevent death resulting from systemic S. pyogenes infections, 4- to 5-week-old FVB/NJ mice were intraperitoneally injected with ∼5 × 106 CFU of strain MGAS5005 in 5% mucin. Treatment occurred at 3 h postinfection (a time when the animals were bacteremic; data not shown) by i.p. injection of either control buffer or PlyPy (0.25 mg or 0.5 mg). Mice were then monitored for survival over 7 days. The results from three independent experiments were combined (PlyPy treatments, n = 20; buffer treatment, n = 18), and the mouse survival data were plotted by use of a Kaplan-Meier survival curve (Fig. 7). Within 72 h of S. pyogenes infection, 83% (n = 15) of the control mice died of streptococcal bacteremia, while only 10% (n = 2) of the 0.25 mg PlyPy-treated mice died during this period. Overall, just 3/18 (17%) of the buffer-treated control mice survived infection with S. pyogenes. Conversely, mice could be significantly protected from death (log-rank test, P < 0.0001) by treatment with PlyPy at both 0.25 mg (90%; 18/20) and 0.50 mg (95%; 19/20) over the 7 days of the experiment (Fig. 7).
FIG 7.
PlyPy is able to rescue mice with S. pyogenes bacteremia. FVB/NJ mice were intraperitoneally injected with ∼5 × 106 CFU of S. pyogenes strain MGAS5005 in 5% mucin. Three hours after infection, mice were treated with i.p. injection of control buffer or 0.5 mg or 0.25 mg of PlyPy, and survival was monitored over 7 days. The results from three independent experiments were combined.
DISCUSSION
The potential use of bacteriophages as treatments against bacterial infections has long been recognized since their initial discovery (33). However, with the introduction of antibiotics, research in this field drastically decreased, only to find new momentum as antibiotic resistance emerged as a new threat. However, rather than using whole bacteriophages as therapeutics, the focus has now shifted toward the sole use of the lytic enzymes of phages, or phage endolysins (34).
Using a bioinformatics approach, we identified one lysin encoded by an S. pyogenes prophage, termed PlyPy. This lysin has an unusually low pI of 4.16, due to a high abundance of aspartic acids (>9%) (Table 1), giving it a highly negative charge at neutral pH. While a low pI is not uncommon for lysins active against streptococcal species (10, 11, 18), the pI of PlyPy is in the lower part of this low range of pIs. The aspartic acids are spread throughout the protein and are not preferentially clustered in either the N-terminal active domain (which has CHAP domain characteristics) or in the C-terminal binding domain (which is a typical SH3 domain) (see Fig. S1 in the supplemental material).
TABLE 1.
Characterized lysins and their pIs
| Lysin | Origin | pI | Reference or source |
|---|---|---|---|
| Cpl-7 | S. pneumoniae ΦCP-7 | 3.99 | 43 |
| PlyPy | S. pyogenes | 4.16 | This article |
| Cpl-1 | S. pneumoniae ΦCpl-1 | 4.52 | 44 |
| Ply700 | S. uberis | 4.54 | 11 |
| GBS B30 lysin | S. agalactiae (GBS) ΦB30 | 4.84 | 10 |
| PlyGBS | S. agalactiae (GBS) ΦNCTC11261 | 4.91 | 45 |
| PlyCA | S. pyogenes ΦC1 | 4.98 | 46 |
| Pal | S. pneumoniae ΦDp-1 | 5.01 | 18 |
| LambdaSa1 | S. agalactiae LambdaSa1 | 5.24 | 9 |
| PlyG | B. anthracis Φγ | 7.12 | 19 |
| LambdaSa2 | S. agalactiae LambdaSa2 | 7.65 | 9 |
| LySMP | S. uberis ΦSMP | 7.88 | 47 |
| PlyPH | B. anthracis | 8.13 | 48 |
| Ply3626 | Clostridium perfringens Φ3626 | 8.36 | 49 |
| Phi11 | S. aureus Φ11 | 9.43 | 50 |
| MV-L | S. aureus ΦMR11 | 9.51 | 51 |
| LysH5 | S. aureus ΦH5 | 9.52 | 52 |
| LysWMY | Staphylococcus warneri M ΦWMY | 9.55 | 53 |
| Lys16 | S. aureus ΦP68 | 9.58 | 54 |
| PlySs2 | S. suis | 9.65 | 27 |
| CD27 | Clostridium difficile ΦCD27 | 9.69 | 55 |
| LysGH15 | S. aureus ΦGH15 | 10.15 | 56 |
| Ply500 | Listeria monocytogenes ΦA500 | 10.15 | 57 |
| LysK | S. aureus ΦK | 10.20 | 58 |
| PlyV12 | E. faecalis Φ1 | 10.26 | 29 |
| Ply511 | L. monocytogenes ΦA511 | 10.34 | 59 |
| Ply118 | L. monocytogenes ΦA118 | 10.39 | 59 |
The low pI of the lysin may influence its activity, as has been suggested for some bacillus lysins (35), as well as for the streptococcal lysin Cpl-7 (13). Low et al. manipulated the Bacillus subtilis lysin XlyA to specifically study the effect of a catalytic domain's net charge (35). By removing the cell wall binding domain of XlyA, the lysin lost all of its activity, possibly due to the negative net charge (−3) of the catalytic domain. This negative charge could interfere with attachment of the lysin to the negatively charged peptidoglycan, inhibiting its activity. In accordance with this, mutagenesis of five selected amino acids not directly involved in the catalytic cleft altered the net charge of the XlyA catalytic domain from −3 to +3, and in doing so, the catalytic subunit of the lysin regained almost all of its activity compared to that of the full-length lysin (35). The authors further suggested that by changing the net charge, the specificity of the lysin might also be altered. Additional experiments to change the net charge of PlyPy are under way to ascertain how charge effects may influence lysin activity.
It is noteworthy that PlyPy shows little similarity to any other characterized streptococcal lysin, except the high-pI lysin PlySs2 (9.65), to which it shows 31.5% overall amino acid similarity and 56.7% amino acid similarity within the cell wall binding domain. Despite this similarity, it is indeed interesting to note that their respective pIs (4.16 and 9.65) and, thus, net charges (−15 and +6) are remarkably different. However, the two lysins do show different specificities, with PlyPy being active mainly against streptococci (Fig. 2), while PlySs2 shows a wide range of activity, including activity against S. pyogenes, S. aureus, S. suis, and even certain strains of Listeria (27). Whether this difference in specificity is mediated through the different binding domain sequences or at least partly through net charges remains to be determined.
Despite the low pI of PlyPy, the enzyme is most active at about neutral pH (Fig. 1C). However, the activity is heavily influenced by the presence of salts and ions (Fig. 1D and B, respectively). The necessity of CaCl2 for lysin activity has been demonstrated previously and most recently for another streptococcal lysin, Ply700 (11). However, Ply700 tolerated much higher concentrations of CaCl2, with the optimal CaCl2 concentration for Ply700 being 10 mM (versus 3 mM for PlyPy), yet it still retained activity even at 100 mM CaCl2. The activity of PlyPy is also influenced by the presence of NaCl, though it shows partial activity even in the absence of salts (Fig. 1D). Furthermore, as is commonly observed for lysins, stationary-phase bacteria were less susceptible to PlyPy than exponential-phase bacteria (see Fig. S1B in the supplemental material).
While the ion concentration can affect lysin activity, so does the concentration of the lysin itself. By lowering the initial dose of PlyPy, we were able to detect a phase before an OD drop (see Fig. S1A in the supplemental material). Similar kinetics has been seen for other lysins (data not shown), but thus far, no conclusive mechanism has been attributed to this phenomenon. While the data may suggest the necessity for PlyPy to form a dimer or multimer to gain activity, this appears unlikely, as elution of PlyPy from size exclusion columns occurred at a volume corresponding to its monomeric size only (data not shown). However, we cannot rule out the possibility that a critical interaction may occur on the cell surface before catalytic activity can occur. Further experiments are needed to elucidate the precise interaction between PlyPy and the bacterial cell wall and to determine whether the kinetic pattern can be attributed to the catalytic and/or binding domain.
Intriguingly, when analyzing the binding potential of PlyPyBD, it was able to bind to all streptococcal species tested, though at different intensities, while it was not able to interact with a Bacillus species (Fig. 3 and 4), indicating that the interaction partner of the lysin is a conserved structure on most, if not all, streptococcal species. However, binding activity did not necessarily correlate with lytic activity, and vice versa. For example, both S. agalactiae and S. uberis had strong binding to PlyPy and were sensitive to the lytic activity of PlyPy (Fig. 4 and 2, respectively). S. porcinus, on the other hand, had very strong binding to PlyPy (Fig. 4D), despite being only moderately sensitive to the lysin (Fig. 2), suggesting that even though the receptor is present, the peptidoglycan might be masked by other cell wall structures, as has been suggested for other peptidoglycan-hydrolyzing agents (36), or have an alternative peptidoglycan structure that the lysin cannot hydrolyze. Both S. suis and S. rattus had a rather low degree of binding to PlyPyBD, which is to be expected, since they are typically not sensitive to the lytic action of PlyPy. However, the opposite could be seen for S. equi, which had only a low degree of binding to PlyPyBD but was highly sensitive to the lytic action of the lysin. Further investigations in this area will, it is hoped, shed some light on what binding receptor the lysin is targeting.
The use of bacteriophage lysins as therapeutics is suggested to have several advantages over the use of conventional antibiotics, most notably, the low possibility that bacteria will develop resistance against the lysin (27, 28). This has been attributed to the cell wall binding domain of the lysin, which targets essential conserved cell wall structures (28). Indeed, the binding domain of lysins active against S. pneumoniae interacts with the critical cell wall structure choline (37), while the group C streptococcal lysin PlyC interacts with the polyrhamnose backbone of the streptococcus-specific carbohydrate antigen (38). Since PlyC and PlyPy have similar specificities mainly targeting the same streptococci (i.e., group A, C, and E streptococci, as well as S. uberis and S. equi), we hypothesized that they might bind to the common cell wall structure in these organisms (i.e., the polyrhamnose backbone of the surface carbohydrate). However, we could demonstrate that the presence of soluble group A-specific carbohydrate competitively lowered the activity of PlyC (Fig. 5), as previously shown (38), but had no effect on PlyPy activity (Fig. 5), suggesting that PlyPy is interacting with another, hitherto unknown structure commonly found in the cell wall of these other organisms.
To further study the enzymatic characteristics of the enzyme, we wished to determine the cleavage site of the lysin, i.e., where it cuts in the peptidoglycan. Several studies have described this for other lysins and mainly used either Edman sequencing or mass spectrometry (9, 39). However, on the basis of the cleavage site of the lysins, these approaches can have varying success rates. Due to the highly repetitious nature of the peptidoglycan, several distinct fragments could yield the same molecular weight, making it difficult to unequivocally identify the cleavage bond. By digesting the S. pyogenes peptidoglycan with PlyPy, we identified a muropeptide (m/z 519.25, z = 2) possibly representing five distinct muropeptides and, thus, five distinct enzymatic activities (31). Rather than using the technically difficult and time-consuming method of adding 4-sulfophenyl isothiocyanate to the muropeptide, as suggested by Gargis et al., to identify the activity (31), we digested the peptidoglycan with PlyPy in the presence of normal H2O or H218O. When combined with LC-tandem MS, this allowed us to use changes in isotopic patterns to (i) identify targets of interest and (ii) pinpoint the cleavage site. In doing so, we could demonstrate that PlyPy represents a d-alanyl-l-alanine endopeptidase. This particular enzymatic activity has been demonstrated only once before in phage lysins (a GBS lysin) (10) and once for bacterium-produced enzymes (zoocin A) (31). The activity determined is in accordance with all of our other findings and explains, among other things, why PlyPy is not active against Streptococcus suis, which has a peptidoglycan that has a direct cross-link to the ε-amino group of the lysine in the stem peptide instead of an l-Ala cross bridge. In fact, all of the bacterial species lysed by PlyPy have peptidoglycans with d-Ala–l-Ala bonds, while the majority of species not being lysed do not have this particular cross bridge but, rather, have direct cross-linking or alternative peptide cross bridges (Table 2). Despite this good correlation, there are exceptions. For example, both S. rattus and Streptococcus mutans have two or three l-Ala in the cross bridge (l-Ala2-3) but are nevertheless nonsensitive to PlyPy. A similar phenomenon was also reported earlier by Pritchard et al., when studying the GBS lysin (10). The GBS lysin contains two lytic domains, a lysozyme and an endopeptidase, with the latter having a d-alanyl-l-alanine endopeptidase activity identical to that of PlyPy. The GBS lysin showed a species specificity similar to that of PlyPy, but it also failed to lyse S. rattus and S. mutans. The authors speculated that this might be due to the presence of high levels of cell wall teichoic acids in these particular bacterial species, blocking the access of the lysin (10). The mechanism behind the insensitivity of Enterococcus faecalis, which also has an l-Ala2-3 (40), to PlyPy is currently unknown. Nevertheless, our method represents a novel and efficient way to accurately define the cleavage activity of lysins.
TABLE 2.
Reported cross bridges in examined bacterial species
| Species | Cross bridge | PlyPy sensitive | Reference |
|---|---|---|---|
| B. cereus | Direct | No | 40 |
| E. faecalis | l-Ala2-3 | No | 40 |
| Enterococcus faecium | d-Asp | No | 40 |
| S. aureus | Gly5 | No | 60 |
| Staphylococcus epidermidis | Gly5, variable (Ser) | No | 60 |
| S. rattus | l-Ala2-3 | No | 36 |
| S. oralis | Direct | No | 36 |
| S. pneumoniae | Direct, variable | No | 61 |
| S. sanguinis | l-Ala2-3, direct | No | 40 |
| S. sobrinus | l-Thr–l-Ala | No | 62 |
| S. mutans | l-Ala2-3 | No | 36 |
| S. suis | Direct | No | 63 |
| S. porcinus | l-Ala2-4 | Yes | 64 |
| S. gordonii | l-Ala2-3 | Yes | 36 |
| Streptococcus dysgalactiae subsp. equisimilaris | l-Ala2-3 | Yes | 40 |
| Streptococcus zooepidemicus | l-Ala2-3 | Yes | 40 |
| S. equi | l-Ala2-3 | Yes | 40 |
| S. agalactiae | l-Ala–l-Ala (Ser) | Yes | 40 |
| S. uberis | l-Ala3-4 | Yes | 40 |
| S. pyogenes | l-Ala2-3 | Yes | 40 |
Importantly, PlyPy not only can kill S. pyogenes in vitro, but also it is efficient in vivo at low concentrations (Fig. 7). The S. pyogenes strain MGAS5005 is known for its invasive characteristics and virulence in both mice and humans (41, 42). Although PlyPy shows moderate in vitro activity against this strain (Fig. 2) compared to its activity against other S. pyogenes strains, it significantly protected infected mice from death in a bacteremia model (Fig. 7). Therefore, PlyPy represents a novel S. pyogenes lysin with high therapeutic potential.
While several other lysins with activity against S. pyogenes have been characterized, PlyPy is unique in various aspects. Not only does it have an unusually low pI (even in comparison to the pIs of other streptococcal lysins), but it also displays an unusual dependence on a low concentration of CaCl2. Furthermore, in contrast to PlyC, PlyPy does not seem to interact with the polyrhamnose backbone of the streptococcal carbohydrate antigen, suggesting that these two lysins may interact with different conserved structures in the cell wall. This suggests that these lysins could be used individually or in combination to increase their beneficial antimicrobial effect and stresses the importance of studying several lysins to gain a broader knowledge of how these important molecules interact with the bacterial cell wall.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by USPHS grant AI11822 and by a grant from Contrafect Inc. to V.A.F. and by a grant from the Tegger Foundation to R.L.
We are grateful to Rachel Shively for generously supplying us with the group A-specific carbohydrate and to Douglas Deutsch for critically reading the manuscript.
Footnotes
Published ahead of print 17 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00115-14.
REFERENCES
- 1.Cole JN, Barnett TC, Nizet V, Walker MJ. 2011. Molecular insight into invasive group A streptococcal disease. Nat. Rev. Microbiol. 9:724–736. 10.1038/nrmicro2648 [DOI] [PubMed] [Google Scholar]
- 2.Carapetis JR, McDonald M, Wilson NJ. 2005. Acute rheumatic fever. Lancet 366:155–168. 10.1016/S0140-6736(05)66874-2 [DOI] [PubMed] [Google Scholar]
- 3.Johnson LP, L'Italien JJ, Schlievert PM. 1986. Streptococcal pyrogenic exotoxin type A (scarlet fever toxin) is related to Staphylococcus aureus enterotoxin B. Mol. Gen. Genet. 203:354–356. 10.1007/BF00333979 [DOI] [PubMed] [Google Scholar]
- 4.Banks DJ, Lei B, Musser JM. 2003. Prophage induction and expression of prophage-encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. Infect. Immun. 71:7079–7086. 10.1128/IAI.71.12.7079-7086.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hynes WL, Ferretti JJ. 1989. Sequence analysis and expression in Escherichia coli of the hyaluronidase gene of Streptococcus pyogenes bacteriophage H4489A. Infect. Immun. 57:533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vlaminckx BJ, Schuren FH, Montijn RC, Caspers MP, Beitsma MM, Wannet WJ, Schouls LM, Verhoef J, Jansen WT. 2007. Dynamics in prophage content of invasive and noninvasive M1 and M28 Streptococcus pyogenes isolates in The Netherlands from 1959 to 1996. Infect. Immun. 75:3673–3679. 10.1128/IAI.01695-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238–276. 10.1128/MMBR.67.2.238-276.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smoot JC, Barbian KD, Van Gompel JJ, Smoot LM, Chaussee MS, Sylva GL, Sturdevant DE, Ricklefs SM, Porcella SF, Parkins LD, Beres SB, Campbell DS, Smith TM, Zhang Q, Kapur V, Daly JA, Veasy LG, Musser JM. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. U. S. A. 99:4668–4673. 10.1073/pnas.062526099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pritchard DG, Dong S, Kirk MC, Cartee RT, Baker JR. 2007. LambdaSa1 and LambdaSa2 prophage lysins of Streptococcus agalactiae. Appl. Environ. Microbiol. 73:7150–7154. 10.1128/AEM.01783-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pritchard DG, Dong S, Baker JR, Engler JA. 2004. The bifunctional peptidoglycan lysin of Streptococcus agalactiae bacteriophage B30. Microbiology 150:2079–2087. 10.1099/mic.0.27063-0 [DOI] [PubMed] [Google Scholar]
- 11.Celia LK, Nelson D, Kerr DE. 2008. Characterization of a bacteriophage lysin (Ply700) from Streptococcus uberis. Vet. Microbiol. 130:107–117. 10.1016/j.vetmic.2007.12.004 [DOI] [PubMed] [Google Scholar]
- 12.Nelson D, Schuch R, Chahales P, Zhu S, Fischetti VA. 2006. PlyC: a multimeric bacteriophage lysin. Proc. Natl. Acad. Sci. U. S. A. 103:10765–10770. 10.1073/pnas.0604521103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diez-Martinez R, de Paz H, Bustamante N, Garcia E, Menendez M, Garcia P. 2013. Improving the lethal effect of Cpl-7, a pneumococcal phage lysozyme with broad bactericidal activity, by inverting the net charge of its cell wall-binding module. Antimicrob. Agents Chemother. 57:5355–5365. 10.1128/AAC.01372-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischetti VA. 2005. Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol. 13:491–496. 10.1016/j.tim.2005.08.007 [DOI] [PubMed] [Google Scholar]
- 15.McGowan S, Buckle AM, Mitchell MS, Hoopes JT, Gallagher DT, Heselpoth RD, Shen Y, Reboul CF, Law RH, Fischetti VA, Whisstock JC, Nelson DC. 2012. X-ray crystal structure of the streptococcal specific phage lysin PlyC. Proc. Natl. Acad. Sci. U. S. A. 109:12752–12757. 10.1073/pnas.1208424109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pastagia M, Euler C, Chahales P, Fuentes-Duculan J, Krueger JG, Fischetti VA. 2011. A novel chimeric lysin shows superiority to mupirocin for skin decolonization of methicillin-resistant and -sensitive Staphylococcus aureus strains. Antimicrob. Agents Chemother. 55:738–744. 10.1128/AAC.00890-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniel A, Euler C, Collin M, Chahales P, Gorelick KJ, Fischetti VA. 2010. Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 54:1603–1612. 10.1128/AAC.01625-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loeffler JM, Nelson D, Fischetti VA. 2001. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 294:2170–2172. 10.1126/science.1066869 [DOI] [PubMed] [Google Scholar]
- 19.Schuch R, Nelson D, Fischetti VA. 2002. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 418:884–889. 10.1038/nature01026 [DOI] [PubMed] [Google Scholar]
- 20.Schmitz JE, Schuch R, Fischetti VA. 2010. Identifying active phage lysins through functional viral metagenomics. Appl. Environ. Microbiol. 76:7181–7187. 10.1128/AEM.00732-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3:163–175. 10.1038/nprot.2007.521 [DOI] [PubMed] [Google Scholar]
- 22.Swanson J, Hsu KC, Gotschlich EC. 1969. Electron microscopic studies on streptococci. I. M antigen. J. Exp. Med. 130:1063–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raz A, Fischetti VA. 2008. Sortase A localizes to distinct foci on the Streptococcus pyogenes membrane. Proc. Natl. Acad. Sci. U. S. A. 105:18549–18554. 10.1073/pnas.0808301105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pancholi V, Fischetti VA. 1988. Isolation and characterization of the cell-associated region of group A streptococcal M6 protein. J. Bacteriol. 170:2618–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2:1896–1906. 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- 27.Gilmer DB, Schmitz JE, Euler CW, Fischetti VA. 2013. Novel bacteriophage lysin with broad lytic activity protects against mixed infection by Streptococcus pyogenes and methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 57:2743–2750. 10.1128/AAC.02526-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischetti VA. 2008. Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 11:393–400. 10.1016/j.mib.2008.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoong P, Schuch R, Nelson D, Fischetti VA. 2004. Identification of a broadly active phage lytic enzyme with lethal activity against antibiotic-resistant Enterococcus faecalis and Enterococcus faecium. J. Bacteriol. 186:4808–4812. 10.1128/JB.186.14.4808-4812.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischetti VA. 2006. Using phage lytic enzymes to control pathogenic bacteria. BMC Oral Health 6(Suppl 1):S16. 10.1186/1472-6831-6-S1-S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gargis SR, Heath HE, Heath LS, Leblanc PA, Simmonds RS, Abbott BD, Timkovich R, Sloan GL. 2009. Use of 4-sulfophenyl isothiocyanate labeling and mass spectrometry to determine the site of action of the streptococcolytic peptidoglycan hydrolase zoocin A. Appl. Environ. Microbiol. 75:72–77. 10.1128/AEM.01647-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schnölzer M, Jedrzejewski P, Lehmann WD. 1996. Protease-catalyzed incorporation of 18O into peptide fragments and its application for protein sequencing by electrospray and matrix-assisted laser desorption/ionization mass spectrometry. Electrophoresis 17:945–953. 10.1002/elps.1150170517 [DOI] [PubMed] [Google Scholar]
- 33.Sulakvelidze A, Alavidze Z, Morris JG., Jr 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649–659. 10.1128/AAC.45.3.649-659.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischetti VA, Nelson D, Schuch R. 2006. Reinventing phage therapy: are the parts greater than the sum? Nat. Biotechnol. 24:1508–1511. 10.1038/nbt1206-1508 [DOI] [PubMed] [Google Scholar]
- 35.Low LY, Yang C, Perego M, Osterman A, Liddington R. 2011. Role of net charge on catalytic domain and influence of cell wall binding domain on bactericidal activity, specificity, and host range of phage lysins. J. Biol. Chem. 286:34391–34403. 10.1074/jbc.M111.244160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simmonds RS, Pearson L, Kennedy RC, Tagg JR. 1996. Mode of action of a lysostaphin-like bacteriolytic agent produced by Streptococcus zooepidemicus 4881. Appl. Environ. Microbiol. 62:4536–4541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia E, Garcia JL, Garcia P, Arraras A, Sanchez-Puelles JM, Lopez R. 1988. Molecular evolution of lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Proc. Natl. Acad. Sci. U. S. A. 85:914–918. 10.1073/pnas.85.3.914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoopes JT, Stark CJ, Kim HA, Sussman DJ, Donovan DM, Nelson DC. 2009. Use of a bacteriophage lysin, PlyC, as an enzyme disinfectant against Streptococcus equi. Appl. Environ. Microbiol. 75:1388–1394. 10.1128/AEM.02195-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ribelles P, Rodriguez I, Suarez JE. 2012. LysA2, the Lactobacillus casei bacteriophage A2 lysin is an endopeptidase active on a wide spectrum of lactic acid bacteria. Appl. Microbiol. Biotechnol. 94:101–110. 10.1007/s00253-011-3588-5 [DOI] [PubMed] [Google Scholar]
- 40.Schleifer KH, Kandler O. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, Ricklefs SM, Sturdevant DE, Graham MR, Vuopio-Varkila J, Hoe NP, Musser JM. 2005. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J. Infect. Dis. 192:771–782. 10.1086/432514 [DOI] [PubMed] [Google Scholar]
- 42.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. 2006. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2:e5. 10.1371/journal.ppat.0020005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bustamante N, Campillo NE, Garcia E, Gallego C, Pera B, Diakun GP, Saiz JL, Garcia P, Diaz JF, Menendez M. 2010. Cpl-7, a lysozyme encoded by a pneumococcal bacteriophage with a novel cell wall-binding motif. J. Biol. Chem. 285:33184–33196. 10.1074/jbc.M110.154559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loeffler JM, Djurkovic S, Fischetti VA. 2003. Phage lytic enzyme Cpl-1 as a novel antimicrobial for pneumococcal bacteremia. Infect. Immun. 71:6199–6204. 10.1128/IAI.71.11.6199-6204.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng Q, Nelson D, Zhu S, Fischetti VA. 2005. Removal of group B streptococci colonizing the vagina and oropharynx of mice with a bacteriophage lytic enzyme. Antimicrob. Agents Chemother. 49:111–117. 10.1128/AAC.49.1.111-117.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson D, Loomis L, Fischetti VA. 2001. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. U. S. A. 98:4107–4112. 10.1073/pnas.061038398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Sun JH, Lu CP. 2009. Purified recombinant phage lysin LySMP: an extensive spectrum of lytic activity for swine streptococci. Curr. Microbiol. 58:609–615. 10.1007/s00284-009-9379-x [DOI] [PubMed] [Google Scholar]
- 48.Yoong P, Schuch R, Nelson D, Fischetti VA. 2006. PlyPH, a bacteriolytic enzyme with a broad pH range of activity and lytic action against Bacillus anthracis. J. Bacteriol. 188:2711–2714. 10.1128/JB.188.7.2711-2714.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zimmer M, Vukov N, Scherer S, Loessner MJ. 2002. The murein hydrolase of the bacteriophage phi3626 dual lysis system is active against all tested Clostridium perfringens strains. Appl. Environ. Microbiol. 68:5311–5317. 10.1128/AEM.68.11.5311-5317.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Donovan DM, Lardeo M, Foster-Frey J. 2006. Lysis of staphylococcal mastitis pathogens by bacteriophage phi11 endolysin. FEMS Microbiol. Lett. 265:133–139. 10.1111/j.1574-6968.2006.00483.x [DOI] [PubMed] [Google Scholar]
- 51.Rashel M, Uchiyama J, Ujihara T, Uehara Y, Kuramoto S, Sugihara S, Yagyu K, Muraoka A, Sugai M, Hiramatsu K, Honke K, Matsuzaki S. 2007. Efficient elimination of multidrug-resistant Staphylococcus aureus by cloned lysin derived from bacteriophage phi MR11. J. Infect. Dis. 196:1237–1247. 10.1086/521305 [DOI] [PubMed] [Google Scholar]
- 52.Obeso JM, Martinez B, Rodriguez A, Garcia P. 2008. Lytic activity of the recombinant staphylococcal bacteriophage phiH5 endolysin active against Staphylococcus aureus in milk. Int. J. Food Microbiol. 128:212–218. 10.1016/j.ijfoodmicro.2008.08.010 [DOI] [PubMed] [Google Scholar]
- 53.Yokoi KJ, Kawahigashi N, Uchida M, Sugahara K, Shinohara M, Kawasaki K, Nakamura S, Taketo A, Kodaira K. 2005. The two-component cell lysis genes holWMY and lysWMY of the Staphylococcus warneri M phage varphiWMY: cloning, sequencing, expression, and mutational analysis in Escherichia coli. Gene 351:97–108. 10.1016/j.gene.2005.03.006 [DOI] [PubMed] [Google Scholar]
- 54.Takac M, Witte A, Blasi U. 2005. Functional analysis of the lysis genes of Staphylococcus aureus phage P68 in Escherichia coli. Microbiology 151:2331–2342. 10.1099/mic.0.27937-0 [DOI] [PubMed] [Google Scholar]
- 55.Mayer MJ, Narbad A, Gasson MJ. 2008. Molecular characterization of a Clostridium difficile bacteriophage and its cloned biologically active endolysin. J. Bacteriol. 190:6734–6740. 10.1128/JB.00686-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gu J, Xu W, Lei L, Huang J, Feng X, Sun C, Du C, Zuo J, Li Y, Du T, Li L, Han W. 2011. LysGH15, a novel bacteriophage lysin, protects a murine bacteremia model efficiently against lethal methicillin-resistant Staphylococcus aureus infection. J. Clin. Microbiol. 49:111–117. 10.1128/JCM.01144-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loessner MJ, Kramer K, Ebel F, Scherer S. 2002. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44:335–349. 10.1046/j.1365-2958.2002.02889.x [DOI] [PubMed] [Google Scholar]
- 58.O'Flaherty S, Coffey A, Meaney W, Fitzgerald GF, Ross RP. 2005. The recombinant phage lysin LysK has a broad spectrum of lytic activity against clinically relevant staphylococci, including methicillin-resistant Staphylococcus aureus. J. Bacteriol. 187:7161–7164. 10.1128/JB.187.20.7161-7164.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaeng S, Scherer S, Neve H, Loessner MJ. 2000. Gene cloning and expression and secretion of Listeria monocytogenes bacteriophage-lytic enzymes in Lactococcus lactis. Appl. Environ. Microbiol. 66:2951–2958. 10.1128/AEM.66.7.2951-2958.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tipper DJ, Berman MF. 1969. Structures of the cell wall peptidoglycans of Staphylococcus epidermidis Texas 26 and Staphylococcus aureus Copenhagen. I. Chain length and average sequence of cross-bridge peptides. Biochemistry 8:2183–2192 [DOI] [PubMed] [Google Scholar]
- 61.Severin A, Tomasz A. 1996. Naturally occurring peptidoglycan variants of Streptococcus pneumoniae. J. Bacteriol. 178:168–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schleifer KH, Kilpper-Balz R, Kraus J, Gehring F. 1984. Relatedness and classification of Streptococcus mutans and “mutans-like” streptococci. J. Dent. Res. 63:1047–1050. 10.1177/00220345840630080701 [DOI] [PubMed] [Google Scholar]
- 63.Fittipaldi N, Sekizaki T, Takamatsu D, de la Cruz Dominguez-Punaro M, Harel J, Bui NK, Vollmer W, Gottschalk M. 2008. Significant contribution of the pgdA gene to the virulence of Streptococcus suis. Mol. Microbiol. 70:1120–1135. 10.1111/j.1365-2958.2008.06463.x [DOI] [PubMed] [Google Scholar]
- 64.Wood BJB, Holzapfel WH. 1992. The lactic acid bacteria, 1st ed. Elsevier Applied Science, London, United Kingdom [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.