Abstract
The molecular epidemiology and mechanisms of resistance of carbapenem-resistant Enterobacteriaceae (CRE) were determined in hospitals in the countries of the Gulf Cooperation Council (GCC), namely, Saudi Arabia, United Arab Emirates, Oman, Qatar, Bahrain, and Kuwait. Isolates were subjected to PCR-based detection of antibiotic-resistant genes and repetitive sequence-based PCR (rep-PCR) assessments of clonality. Sixty-two isolates which screened positive for potential carbapenemase production were assessed, and 45 were found to produce carbapenemase. The most common carbapenemases were of the OXA-48 (35 isolates) and NDM (16 isolates) types; 6 isolates were found to coproduce the OXA-48 and NDM types. No KPC-type, VIM-type, or IMP-type producers were detected. Multiple clones were detected with seven clusters of clonally related Klebsiella pneumoniae. Awareness of CRE in GCC countries has important implications for controlling the spread of CRE in the Middle East and in hospitals accommodating patients transferred from the region.
INTRODUCTION
International travel is a major mode of the spread of multiresistant Gram-negative bacilli, including carbapenem-resistant Enterobacteriaceae (CRE) (1). The countries of the Gulf Cooperation Council (GCC) (Saudi Arabia, United Arab Emirates [UAE], Oman, Kuwait, Qatar, and Bahrain) exemplify the potential for international travel as a significant issue: large numbers of citizens seek medical care in specialized centers in the United States and Europe, substantial proportions of the population are migrant workers from the Indian subcontinent, and millions visit the region annually for the Hajj and other religious events (2). As one of many desperately needed first steps to control the spread of CRE, we aimed to determine in this collaborative work the molecular genetics of CRE in the countries of the GCC. To our knowledge there has been no surveillance on the molecular genetics of CRE in this region in the past. For this reason, we have performed a “snapshot” assessment of the molecular epidemiology of CRE in the countries of the Gulf Cooperation Council.
MATERIALS AND METHODS
Bacterial isolates.
Between July 2011 and January 2013, 413 clinical Escherichia coli and Klebsiella pneumoniae isolates were collected from six participating institutes across the GCC states (one hospital each from Saudi Arabia, United Arab Emirates [UAE], Kuwait, Qatar, Oman, and Bahrain) (Table 1), as part of a region-wide collaborative study on multidrug-resistant Gram-negative bacilli. E. coli and Klebsiella spp. were identified and tested for their susceptibility to a panel of antimicrobials using semiautomated systems in each clinical microbiology laboratory (Table 1). Isolates were included on the basis of showing decreased susceptibility to cefotaxime (MIC, ≥2 μg/ml), ceftriaxone (MIC, ≥2 μg/ml), ceftazidime (MIC, ≥8 μg/ml), cefepime (MIC, ≥16 μg/ml), imipenem (MIC, ≥2 μg/ml), or meropenem (MIC, ≥2 μg/ml). Only one isolate per patient was included.
TABLE 1.
Summary of CRE clinical isolates in the GCC states
| Location | Hospital name | Hospital type | Hospital capacity (no. of beds) | Semiautomated system used for species identification and antibiotic sensitivity | Study isolates received (no.) | No. (%) of isolates with reduced susceptibility to ertapenem | No. (%) of carbapenemase and CTX-M-15-type genes |
No. (%) of plasmid replicon typinga |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NDM type | OXA-48 type | KPC type | IMP type | VIM type | CTX-M-15 type | IncL/M | IncA/C | |||||||
| Riyadh, Saudi Arabia | King Abdulaziz Medical City | Tertiary and academic | 1,000 | Vitek II, bioMérieux | E. coli (151) | 2 (1.3) | 1 (50) | 0 | 0 | 0 | 0 | 2 (100) | NTb | NT |
| K. pneumoniae (77) | 40 (52) | 10 (25) | 31 (77.5)c | 0 | 0 | 0 | 28 (70) | 23 (74) | 1 (3) | |||||
| Abu Dhabi, United Arab Emirates | Sheikh Zayed Military Hospital | Tertiary | 365 | Vitek II, bioMérieux | E. coli (29) | 1 (3) | 0 | 0 | 0 | 0 | 0 | 1 (100) | NT | NT |
| K. pneumoniae (16) | 4 (25) | 3 (75) | 0 | 0 | 0 | 0 | 4 (100) | NT | NT | |||||
| Kuwait, Kuwait | Al-Ameri Hospital | Tertiary | 398 | Vitek II, bioMérieux | E. coli (18) | 0 | NAd | NA | NA | NA | NA | NA | NT | NT |
| K. pneumoniae (13) | 0 | NA | NA | NA | NA | NA | NA | NT | NT | |||||
| Muscat, Oman | The Royal Hospital | Teaching tertiary | 750 | Phoenix, Becton, Dickinson | E. coli (23) | 2 (9) | 0 | 0 | 0 | 0 | 0 | 1 (50) | NT | NT |
| K. pneumoniae (14) | 3 (21) | 1 (33) | 1 (33) | 0 | 0 | 0 | 3 (100) | 0 | 0 | |||||
| Doha, Qatar | Hamad Medical Cooperation | Tertiary | >1,300 | Phoenix, Becton, Dickinson | E. coli (23) | 4 (17) | 0 | 1 (25) | 0 | 0 | 0 | 3 (75) | 0 | 0 |
| K. pneumoniae (16) | 5 (31) | 1 (20) | 2 (40) | 0 | 0 | 0 | 5 (100) | 0 | 1 (8) | |||||
| Manama, Bahrain | Samlaniya Medical Complex | Tertiary and teaching | 1,000 | Phoenix, Becton, Dickinson | E. coli (22) | 0 | NA | NA | NA | NA | NA | NA | NT | NT |
| K. pneumoniae (11) | 1 (0.9) | 0 | 0 | 0 | 0 | 0 | 1 (100) | NT | NT | |||||
| Total | ||||||||||||||
| E. coli | 266 (64) | 9 (3.4) | 1 (6) | 1 (6.4) | 0 | 0 | 0 | 7 (78) | 0 | 0 | ||||
| K. pneumoniae | 147 (36) | 53 (36) | 15 (28) | 34 (63)c | 0 | 0 | 0 | 41 (77) | 23 (68) | 2 (5.9) | ||||
| 413 | 62 (15) | 16 (22.5) | 35 (49) | 0 | 0 | 0 | 48 (77) | 23 (66) | 2 (5.7) | |||||
Only on OXA-48-type-positive isolates.
NT, not tested because they were OXA-48-type negative.
Six isolates coharbored genes of the blaOXA-48 and blaNDM types.
NA, not applicable for further testing because isolates were susceptible to ertapenem.
Isolates were sent to the research laboratory at the University of Queensland Centre for Clinical Research (UQCCR). Bacterial species were confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) on a Microflex platform (Bruker Daltonics, Inc.). All 413 isolates underwent initial screening for carbapenem resistance as determined by reduced susceptibility to ertapenem by disk diffusion; ertapenem was chosen as the screening carbapenem based on its ability to detect NDM-1, KPC, and low-level carbapenemase producers (3).
PCR for carbapenemase genes and CTX-M-15 ESBL.
Crude genomic DNA for PCR was extracted from the isolates using the heat lysis method. The presence of genes of the blaNDM and blaOXA-48 types (4, 5) (Table 2) was sought on all isolates with reduced susceptibility to ertapenem, using a multiplex PCR with GoTaq green master mix. PCR was performed with 0.4 μM each primer and 0.75 μl of DNA template. The PCR conditions were as follows: initial denaturation at 95°C for 2 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 45 s, and extension at 70°C for 60 s, and a final extension at 70°C for 5 min. PCR products of the blaOXA-48-type gene found in 10 different K. pneumoniae isolates representing different clones were sequenced to identify the blaOXA-48-type variants. All isolates with a reduced susceptibility to ertapenem were also tested in a singleplex reaction for the other major carbapenemase groups that confer clinically relevant resistance to carbapenems, the blaKPC, blaVIM, and blaIMP types (4–8) (Table 2). Isolates with negative PCR results for the tested carbapenemase genes were subjected to the Carba NP test, as described previously (9). PCR for blaCTX-M-15-type genes was also performed on the isolates to check if they were coharboring this pandemic extended-spectrum beta-lactamase (ESBL) type (10) (Table 2).
TABLE 2.
Oligonucleotides used to amplify selected beta-lactamase genes
| Primer name | Target | Sequence (5′ → 3′) | Size (bp) | Annealing temp (°C) | Reference |
|---|---|---|---|---|---|
| CTX-M-15-F | blaCTX-M-15 | CACACGTGGAATTTAGGGACT | 996 | 55 | 10 |
| CTX-M-15-R | GCCGTCTAAGGCGATAAACA | ||||
| IMP-F | blaIMP | CTACCGCAGCAGAGTCTTTGC | 591 | 58 | 7 |
| IMP-R | GAACAACCAGTTTTGCCTTACC | ||||
| KPC-F | blaKPC | ATCTGACAACAGGCATGACG | 452 | 55 | 4 |
| KPC-R | GACGGCCAACACAATAGGTG | ||||
| NDM-F | blaNDM | GCAGGTTGATCTCCTGCTTG | 203 | 55 | 4 |
| NDM-R | ACGGTTTGGCGATCTGGT | ||||
| OXA-48-F | blaOXA-48 | GCGTGGTTAAGGATGAACAC | 438 | 55 | 5 |
| OXA-48-R | CATCAAGTTCAACCCAACCG | ||||
| VIM-F | blaVIM | GATGGTGTTTGGTCGCATA | 390 | 55 | 8 |
| VIM-R | CGAATGCGCAGCACCAG |
Plasmid typing.
Isolates found carrying genes of the blaOXA-48 type were subjected to PCR-based replicon typing analysis (PBRT), as described by Carattoli et al. (11), to determine plasmid incompatibility types. The primer pairs targeting IncL/M were used, and isolates negative for IncL/M were subsequently screened for IncA/C (Table 1). These two plasmid replicon types were selected based on reports suggesting dissemination of blaOXA-48 in IncL/M- and IncA/C-type plasmids (12, 13).
Clonal analysis of NDM- or/and OXA-48-producing Klebsiella pneumoniae by rep-PCR.
The genetic relatedness among K. pneumoniae isolates from the GCC was determined by repetitive sequence-based PCR (rep-PCR) typing using the DiversiLab system (bioMérieux, Oakleigh, Australia). The DNA fragment patterns were analyzed by the appropriate software using Pearson correlation coefficient pairwise pattern matching to determine the clonal relationships and to create dendrograms. A cluster of closely related isolates was defined as isolates sharing ≥95% similarity and indistinguishable isolates of ≥97% (14).
Human ethics.
The University of Queensland granted human ethics clearance to conduct this project (no. 2011000474). Permission from King Abdulaziz Medical City, Saudi Arabia, was granted to conduct the region-wide collaborative study on multidrug resistant Gram-negative bacilli (reference no. IRBC/193/12).
RESULTS
Bacterial isolates and ertapenem susceptibility.
Of the 413 isolates assessed, a total of 62 nonrepetitive isolates that were not susceptible to ertapenem were subjected to further analysis; 53 were K. pneumoniae, and 9 were E. coli (Table 1).
Antibiotic resistance genes.
A total of 35 (49%) isolates (34 K. pneumoniae and 1 E. coli) were OXA-48-type producers and a total of 16 (23%) (15 K. pneumoniae and 1 E. coli) were NDM-type producers. Six of these isolates coproduced the NDM type with the OXA-48 type, and all were K. pneumoniae isolates from Saudi Arabia (Table 1). Sequencing results of the blaOXA-48-type gene carried by representative K. pneumoniae isolates from each clone showed that all were carrying blaOXA-48 except one isolate from Qatar that produced OXA-181 (Fig. 1). All 62 isolates were negative for genes of the blaKPC, blaVIM, and blaIMP types. A total of 17 (27%) isolates (7 E. coli and 10 K. pneumoniae) were negative for all the tested carbapenemase genes. These isolates were also negative for carbapenemase production, as shown by the Carba NP test.
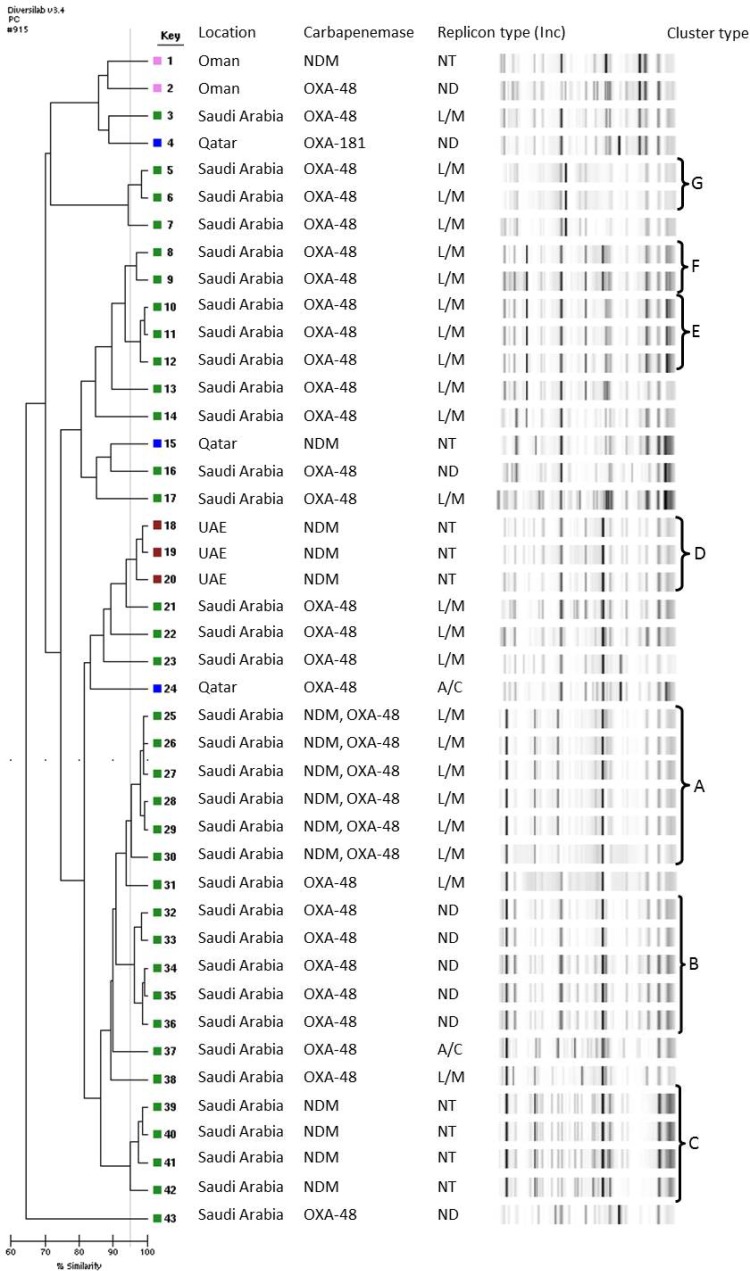
FIG 1.
Dendrogram illustrating the genetic relationship and clusters of 43 NDM- or OXA-48-producing K. pneumoniae isolates from the GCC states. ND, not determined; NT, not tested.
Genes of the blaCTX-M-15 type were detected in 7 (78%) E. coli isolates and in 41 (77%) K. pneumoniae isolates. Among the E. coli isolates, the blaCTX-M-15-type ESBL gene was coharbored with the blaNDM-type gene in a single isolate. Coproduction of the CTX-M-15 type in K. pneumoniae with NDM occurred in 13 isolates and coproduction with the OXA-48 type in 23 isolates. Five K. pneumoniae isolates carried all 3 (blaCTX-M-15-type, blaNDM-type, and blaOXA-48) genes. Of all isolates that tested negative for carbapenemases (n = 17), 94% produced CTX-M-15-type ESBL.
Plasmid replicon typing.
Among the 35 OXA-48-type-producing isolates, 23 (62%) were plasmid replicon type IncL/M, and all were from Saudi Arabia. Of the IncL/M-negative isolates (n = 14), 2 were positive for IncA/C.
Clonal analysis of carbapenem-resistant Klebsiella pneumoniae by rep-PCR.
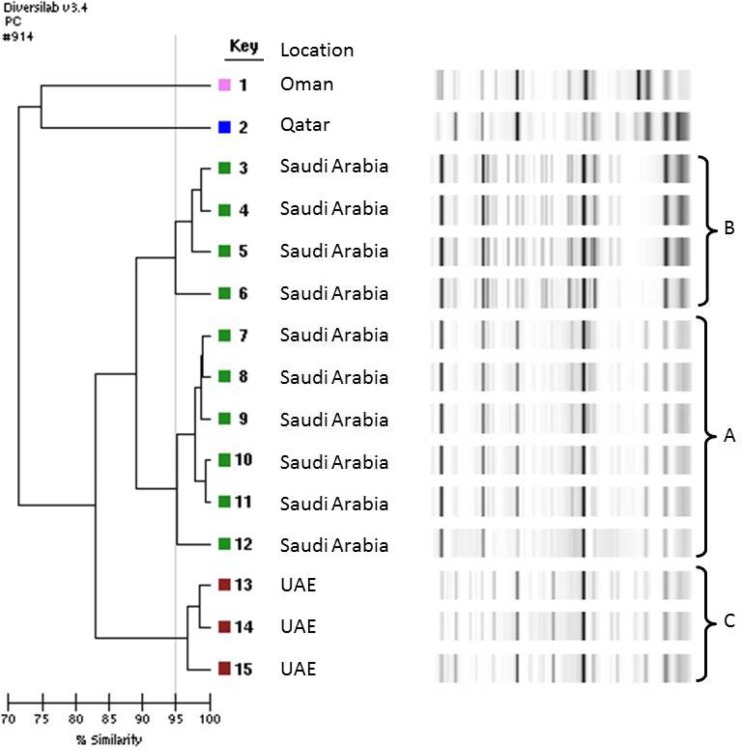
Clonal analysis was performed on all carbapenemase-producing K. pneumoniae isolates from different countries within the GCC. The rep-PCR results reveal seven well-defined clusters (Fig. 1). The main cluster (cluster A) represented six blaNDM-type- and blaOXA-48-carrying K. pneumoniae isolates from Saudi Arabia. Two smaller clusters (cluster B and C), representing four NDM-type-positive isolates and five OXA-48-positive isolates, respectively, were also observed from Saudi Arabia. Additional clusters of NDM-type-positive isolates from the UAE (cluster D), and OXA-48-positive isolates from Saudi Arabia (clusters E, F, and G), demonstrated high genetic relatedness (≥97% similarity). NDM-type-producing isolates from Qatar and Oman were genetically unrelated to all other NDM-type-positive strains isolated from the region (Fig. 2). Analysis by the DiversiLab system demonstrated good correlation between the isolates and their country of origin. Figure 2 highlights the three well-defined clusters (A and B from Saudi Arabia and C from UAE) of NDM-type-producing K. pneumoniae and demonstrates the high genetic relatedness of isolates from individual hospitals.
FIG 2.
Dendrogram of 15 NDM-producing K. pneumoniae isolates from the GCC states.
DISCUSSION
We have described the molecular genetics of recent isolates of CRE from patients in selected GCC hospitals. We had several major findings. First, we have found that the OXA-48-type carbapenemase was the dominant mechanism responsible for CRE in this study, and it has been identified for the first time in K. pneumoniae and E. coli isolates from Qatar. Genes of the OXA-48 type and related OXA enzymes have been found to be widely prevalent in North Africa, the Middle East, and the Indian subcontinent (15), and, more importantly, large numbers of outbreaks have occurred in regions such as Europe and Australia, where CRE is not endemic, as a result of international transfer of patients (12, 16). In turn, this has resulted in endemicity in hospitals in countries such as France (12). OXA-48 producers have recently been reported for the first time in the United States (17). Importantly, the first reported case in the United States was a patient who had been recently hospitalized in Saudi Arabia (17). There is clearly a need to consider colonization of OXA-48 producers in patients transferred from the Middle East to the United States or Europe for medical care.
Second, we have found that multiple clones of OXA-48-type-producing K. pneumoniae are circulating within hospitals in the Gulf Cooperation Council (Fig. 1). This finding suggests that OXA-48-type producers have been prevalent in hospitals in the region for a prolonged period of time. The finding of isolates sharing common rep-PCR profiles reemphasizes the need for optimized infection control in hospitals in the region, especially given the previous findings of clusters of multiresistant Acinetobacter baumannii in hospitals in the region (18).
Third, we have detected NDM-type producers in several countries across the GCC. Although there was a high diversity of rep-PCR profiles in different isolates from different countries, common profiles were found within individual hospitals (Fig. 2). NDM producers have been previously identified in countries of the Gulf Cooperation Council (2, 19), and international transfer of NDM-1 producers from patients previously hospitalized in the Middle East has been reported (20). However, this is the first report describing the identification of NDM-type-producing K. pneumoniae in Qatar and isolates that are coharboring NDM-type producers with OXA-48-type producers in Saudi Arabia. Index patients in outbreaks of NDM-1 producers occurring in North America and Europe have typically had previous hospitalizations in overseas countries (21), emphasizing the need for preemptive contact isolation precautions in patients with previous overseas health care contacts (1).
No isolate was found to produce the KPC, VIM, or IMP beta-lactamase, although these carbapenemases have been previously detected in Saudi Arabia and Kuwait (2, 22–24). For 17 isolates, no carbapenemase activity or carbapenemase genes were identified, suggesting a noncarbapenemase-related resistance mechanism. These isolates produced genes of the CTX-M-15 type and most likely had extended-spectrum beta-lactamase production associated with decreased permeability of the outer membrane (25).
In summary, we have evaluated CRE in hospitals from across the Gulf Cooperation Council. Although this is not a formal surveillance study, it is the first “snapshot” study to determine the molecular epidemiology of CRE in the region. Our findings of multiple clusters of OXA-48-type- and NDM-type-producing K. pneumoniae have important implications for control of spread of CRE both in the Middle East and in hospitals accommodating patients transferred from the region. Additionally, attention to hospital antibiotic stewardship, the availability of over-the-counter antibiotics, and agricultural use of antibiotics all have relevance to control of CRE in the region.
ACKNOWLEDGMENTS
The “Surveillance of Antibiotic Resistant Gram Negative Bacilli in Saudi Arabia and the Gulf States” (project no. IRBC/193/12) is supported by The Ministry of National Guard, Health Affairs, King Abdullah International Medical Research Centre, Saudi Arabia. H.M.Z. is academically sponsored by the government of Saudi Arabia to pursue postgraduate studies in the field of clinical microbiology and infectious diseases. T.R.W. is funded by the HEFC, British government.
We thank all the staff from the collaborating clinical microbiology laboratories across the GCC states and the active role of the GCC Center for Infection Control under the umbrella of the National Guard Health Affairs. We also thank Wan Keat Yam and Moongaambikai Thangaveloo for helping to prepare the transport media.
D.L.P. has received honoraria for advisory board participation from AstraZeneca, Pfizer, and Merck, not relating to this work.
Footnotes
Published ahead of print 17 March 2014
REFERENCES
- 1.Rogers BA, Aminzadeh Z, Hayashi Y, Paterson DL. 2011. Country-to-country transfer of patients and the risk of multi-resistant bacterial infection. Clin. Infect. Dis. 53:49–56. 10.1093/cid/cir273 [DOI] [PubMed] [Google Scholar]
- 2.Zowawi HM, Balkhy HH, Walsh TR, Paterson DL. 2013. β-Lactamase production in key Gram-negative pathogen isolates from the Arabian Peninsula. Clin. Microbiol. Rev. 26:361–380. 10.1128/CMR.00096-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nordmann P, Poirel L, Carrer A, Toleman MA, Walsh TR. 2011. How to detect NDM-1 producers. J. Clin. Microbiol. 49:718–721. 10.1128/JCM.01773-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim S. 2011. Development of multiplex PCR: β-lactamase genes and virulence determinants in E. coli. University of Queensland Centre for Clinical Research, Brisbane St Lucia, Australia [Google Scholar]
- 5.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70:119–123. 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 6.Sidjabat H, Nimmo GR, Walsh TR, Binotto E, Htin A, Hayashi Y, Li J, Nation RL, George N, Paterson DL. 2011. Carbapenem resistance in Klebsiella pneumoniae due to the New Delhi metallo-β-lactamase. Clin. Infect. Dis. 52:481–484. 10.1093/cid/ciq178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirel L, Naas T, Nicolas D, Collet L, Bellais S, Cavallo JD, Nordmann P. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891–897. 10.1128/AAC.44.4.891-897.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellington MJ, Kistler J, Livermore DM, Woodford N. 2007. Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J. Antimicrob. Chemother. 59:321–322. 10.1093/jac/dkl481 [DOI] [PubMed] [Google Scholar]
- 9.Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 18:1503–1507. 10.3201/eid1809.120355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muzaheed Doi Y, Adams-Haduch JM, Endimiani A, Sidjabat HE, Gaddad SM, Paterson DL. 2008. High prevalence of CTX-M-15-producing Klebsiella pneumoniae among inpatients and outpatients with urinary tract infection in Southern India. J. Antimicrob. Chemother. 61:1393-1394. 10.1093/jac/dkn109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- 12.Potron A, Poirel L, Rondinaud E, Nordmann P. 2013. Intercontinental spread of OXA-48 β-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro Surveill. 18(31). pii: 20549 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20549 [DOI] [PubMed] [Google Scholar]
- 13.Potron A, Poirel L, Nordmann P. 2014. Derepressed transfer properties leading to the efficient spread of the plasmid encoding carbapenemase OXA-48. Antimicrob. Agents Chemother. 58:467–471. 10.1128/AAC.01344-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brolund A, Haeggman S, Edquist PJ, Gezelius L, Olsson-Liljequist B, Wisell KT, Giske CG. 2010. The DiversiLab system versus pulsed-field gel electrophoresis: characterisation of extended spectrum beta-lactamase producing Escherichia coli and Klebsiella pneumoniae. J. Microbiol. Methods 83:224–230. 10.1016/j.mimet.2010.09.004 [DOI] [PubMed] [Google Scholar]
- 15.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J. Antimicrob. Chemother. 67:1597–1606. 10.1093/jac/dks121 [DOI] [PubMed] [Google Scholar]
- 16.Espedido BA, Steen JA, Ziochos H, Grimmond SM, Cooper MA, Gosbell IB, van Hal SJ, Jensen SO. 2013. Whole genome sequence analysis of the first Australian OXA-48-producing outbreak-associated Klebsiella pneumoniae isolates: the resistome and in vivo evolution. PLoS One 8:e59920. 10.1371/journal.pone.0059920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathers AJ, Hazen KC, Carroll J, Yeh AJ, Cox HL, Bonomo RA, Sifri CD. 2013. First clinical cases of OXA-48-producing carbapenem-resistant Klebsiella pneumoniae in the United States: the “menace” arrives in the new world. J. Clin. Microbiol. 51:680–683. 10.1128/JCM.02580-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balkhy HH, Bawazeer MS, Kattan RF, Tamim HM, Al Johani SM, Aldughashem FA, Al Alem HA, Adlan A, Herwaldt LA. 2012. Epidemiology of Acinetobacter spp.-associated healthcare infections and colonization among children at a tertiary-care hospital in Saudi Arabia: a 6-year retrospective cohort study. Eur. J. Clin. Microbiol. Infect. Dis. 31:2645–2651. 10.1007/s10096-012-1608-8 [DOI] [PubMed] [Google Scholar]
- 19.Ghazawi A, Sonnevend A, Bonnin RA, Poirel L, Nordmann P, Hashmey R, Rizvi TA, Hamadeh MB, Pál M. 2012. NDM-2 carbapenemase-producing Acinetobacter baumannii in the United Arab Emirates. Clin. Microbiol. Infect. 18:E34–E36. 10.1111/j.1469-0691.2011.03726.x [DOI] [PubMed] [Google Scholar]
- 20.Birgy A, Doit C, Mariani-Kurkdjian P, Genel N, Faye A, Arlet G, Bingen E. 2011. Early detection of colonization by VIM-1-producing Klebsiella pneumoniae and NDM-1-producing Escherichia coli in two children returning to France. J. Clin. Microbiol. 49:3085-3087. 10.1128/JCM.00540-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowe CF, Kus JV, Salt N, Callery S, Louie L, Khan MA, Vearncombe M, Simor AE. 2013. Nosocomial transmission of New Delhi metallo-β-lactamase-1-producing Klebsiella pneumoniae in Toronto, Canada. Infect. Control Hosp. Epidemiol. 34:49–55. 10.1086/668778 [DOI] [PubMed] [Google Scholar]
- 22.Al-Qadheeb NS, Althawadi S, Alkhalaf A, Hosaini S, Alrajhi AA. 2010. Evolution of tigecycline resistance in Klebsiella pneumoniae in a single patient. Ann. Saudi Med. 30:404–407. 10.4103/0256-4947.67087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dashti AA, Vali L, Jadaon MM, El-Shazly S, Amyes SG. 2011. The emergence of carbapenem resistance in ESBL-producing Escherichia coli O25B-ST131 strain from community acquired infection in Kuwait. In 1st International Conference on Prevention and Infection Control. International Conference on Prevention and Infection Control, Geneva, Switzerland [Google Scholar]
- 24.Jamal W, Rotimi VO, Albert MJ, Khodakhast F, Nordmann P, Poirel L. 2013. High prevalence of VIM-4 and NDM-1 metallo-β-lactamase among carbapenem-resistant Enterobacteriaceae. J. Med. Microbiol. 62:1239–1244. 10.1099/jmm.0.059915-0 [DOI] [PubMed] [Google Scholar]
- 25.Dortet L, Poirel L, Al Yaqoubi F, Nordmann P. 2012. NDM-1, OXA-48 and OXA-181 carbapenemase-producing Enterobacteriaceae in Sultanate of Oman. Clin. Microbiol. Infect. 18:E144–E148. 10.1111/j.1469-0691.2012.03796.x [DOI] [PubMed] [Google Scholar]