Abstract
The emergence of antibiotic resistance in recent years has radically reduced the clinical efficacy of many antibacterial treatments and now poses a significant threat to public health. One of the earliest studied well-validated targets for antimicrobial discovery is the bacterial cell wall. The essential nature of this pathway, its conservation among bacterial pathogens, and its absence in human biology have made cell wall synthesis an attractive pathway for new antibiotic drug discovery. Herein, we describe a highly sensitive screening methodology for identifying chemical agents that perturb cell wall synthesis, using the model of the Gram-positive bacterium Bacillus subtilis. We report on a cell-based pilot screen of 26,000 small molecules to look for cell wall-active chemicals in real time using an autonomous luminescence gene cluster driven by the promoter of ywaC, which encodes a guanosine tetra(penta)phosphate synthetase that is expressed under cell wall stress. The promoter-reporter system was generally much more sensitive than growth inhibition testing and responded almost exclusively to cell wall-active antibiotics. Follow-up testing of the compounds from the pilot screen with secondary assays to verify the mechanism of action led to the discovery of 9 novel cell wall-active compounds.
INTRODUCTION
The emergence and spread of antibiotic resistance in recent years have reduced the clinical efficacy of many antibacterial treatments, posing a significant threat to public health. With the 1929 discovery of penicillin, the first antibacterial agent to be widely used, the cell wall was one of the earliest targets exploited in antibacterial drug discovery (1). Indeed, cell wall-active agents have been a rich source of efficacious antibiotics. These drugs are typically bactericidal in nature and target extracytoplasmic functions in bacterial cells, eliminating the requirement for cell penetration. Although intensively explored, the cell wall still has many untapped targets and provides new opportunities for the discovery of novel antibacterial chemical agents (2–4). Indeed, nearly all cell wall-active agents have been discovered using screens for growth inhibition, followed by cumbersome secondary screens for activity directed at the cell wall. What is lacking are selective primary screening assays for the sensitive detection of cell wall-active compounds, avoiding off-target and nuisance compounds.
The bacterial cell wall is an essential macromolecular structure with key roles in numerous functions, including maintenance of cell shape, growth, and division (5). The Gram-positive cell wall is composed of two main components, i.e., peptidoglycan (PG) and wall teichoic acid (WTA), in roughly equal quantities. Both are synthesized on the lipid carrier undecaprenol phosphate (6). Existing antibacterials targeting the cell wall are largely inhibitors of PG synthesis; however, WTA and undecaprenol synthesis are emerging as promising new targets (7–10).
Previous work from our laboratory identified upregulation of the ywaC gene upon inhibition of WTA synthesis (11, 12). After that identification, we constructed a promoter-reporter system by fusing the promoter of ywaC to the lux genes, providing a real-time luminescence signal. The system was tested against an extensive panel of antibiotics and showed exquisite selectivity for antibiotics targeting cell wall biosynthesis (11). Thus, the ywaC promoter-reporter system was able to sense cell wall stress in Bacillus subtilis resulting from the perturbation of PG, WTA, and undecaprenol synthesis. The ywaC gene encodes a largely uncharacterized protein that, in recent years, has been shown to have GTP pyrophosphokinase activity (13). This class of enzyme synthesizes guanosine tetra(penta)phosphate [(p)ppGpp], a well-known bacterial “alarmone” that influences gene expression during cell stress. In addition, studies using promoter census searches have revealed that ywaC is a member of the σW regulon, which includes genes that are upregulated in response to cell wall-active antibiotics (14). Indeed, other promoter-reporter-based systems have been used, in several bacterial hosts, to detect the presence of cell wall inhibitors (15–22). For the most part, these systems respond only to compounds that block PG synthesis and thus are insensitive to, for example, WTA and undecaprenol synthesis.
Herein we describe the development and implementation of a kinetic whole-cell screening method using the PywaC-lux promoter-reporter system. Using a library of ∼26,000 diverse synthetic compounds, we show that the real-time assay is robust, highly sensitive, and amenable to high-throughput screening (HTS). Furthermore, the cell-based screen was extremely effective in enriching for cell wall-active chemicals. Secondary assays of the active compounds derived from this screen identified 9 novel compounds that target cell wall biogenesis in the Gram-positive B. subtilis model (Fig. 1).
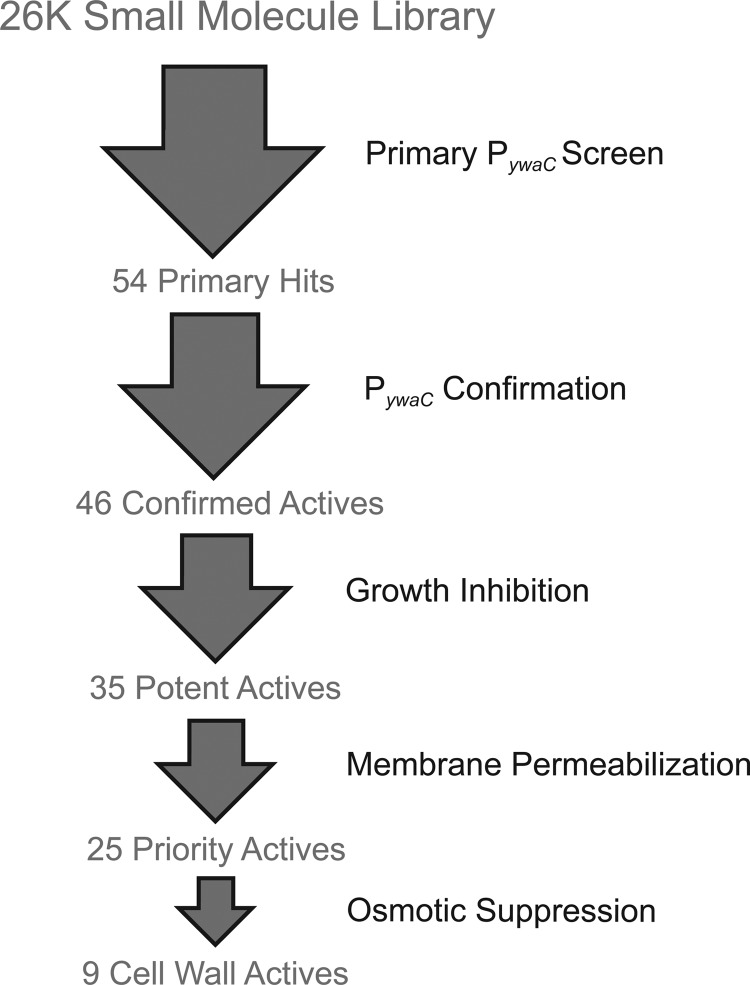
FIG 1.
Screening workflow. A diverse collection of 26,000 synthetic compounds was screened using the PywaC reporter system, which generated 54 active compounds. Activity was confirmed for 46 compounds upon retesting. Confirmed active compounds were subjected to growth inhibition studies using B. subtilis, which revealed 35 potent active compounds. Subsequently, a membrane-permeabilization counterscreen was used to exclude membrane-active compounds, which yielded 25 priority active compounds. For 9 of these compounds, the activity phenocopied known cell wall-active antibiotics, as PywaC activity could be suppressed with the inclusion of osmoprotectants (MSM) in growth medium, and produced morphological defects in B. subtilis; these constituted novel cell wall-active compounds.
MATERIALS AND METHODS
Solid agar-based PywaC screening methodology.
Screens of PywaC transcriptional activity were performed in 96-well microtiter plates (catalog no. 6005688; PerkinElmer), in duplicate, using a stand-alone Biomek FX workstation (Beckman Coulter Inc.). The night before screening, a single colony of strain EB1385 (11) was grown in 5 ml of Luria-Bertani (LB) broth supplemented with erythromycin (0.5 μg/ml). On the day of screening, cells were harvested at an optical density at 600 nm (OD600) of 3.0. The cells were then diluted into fresh LB broth supplemented with antibiotic, to a final OD600 of 0.02. To each well of a 96-well microtiter plate, 1 μl of compound (1 mM in 100% dimethyl sulfoxide [DMSO]) was added; 99 μl of liquid (warmed) 0.5% LB agar supplemented with antibiotic was subsequently transferred to the 96-well microtiter plates containing compound. The liquid LB agar was allowed to solidify at room temperature for 2 h. After 2 h at room temperature, 20 μl of cells at a final OD600 of 0.02 was spotted on the surface of the solid LB agar. Plates were allowed to dry for approximately 1 h before the start of the assay. Assay luminescence was read using an EnVision Multilabel plate reader (PerkinElmer), with an emission wavelength of 492 nm, over the course of 19 h. Fosmidomycin (10 μM) was used as the high control, and neat DMSO was used as the low control. In instances in which osmotic suppression assays were performed, LB medium was supplemented with 20 mM MgCl2-0.5 M sucrose-20 mM maleic acid (MSM). Calculation of the fold increase in luminescence was determined for each sample by dividing the luminescence values of the sample by the average of the values for the low controls on the same assay plate at the same time point.
MIC determinations.
The MIC defines the lowest concentration of a compound required to inhibit the growth of a particular strain. A single colony of wild-type (WT) B. subtilis strain 168 (EB6) was grown in 5 ml of LB broth. Following 16 h of growth, the overnight culture was diluted 100-fold into fresh LB broth. Once the cells reached an OD600 of 0.3 to 0.5, the cells were diluted 10,000-fold into fresh LB broth. The MICs of various antibacterial agents were determined for each of the strains by adding 198 μl of the diluted cells to wells of a clear 96-well microtiter plate (catalog no. 3370; Corning Costar) containing the compound at 2-fold dilutions. The plates were incubated at 30°C for 16 h, with shaking at 250 rpm. Optical density was measured at 600 nm with a Spectra-max Plus instrument (Molecular Devices).
EC50 determinations.
A dose-response assessment (50% effective concentration [EC50]) was determined for each priority active compound obtained from the primary screen by using a cell-based assay for growth inhibition. The assay was conducted as described for the MIC determinations in liquid LB medium. Inhibitors were added at half-logarithmic dilutions from 100 μM to 0.001 μM. EC50s were determined by the use of nonlinear least-squares regression using GraFit Workspace (Erithacus Software Ltd.).
Cytoplasmic membrane permeability assay.
Cytoplasmic membrane permeabilization was determined by using the membrane-potential-sensitive cyanine dye DiSC3(5) (23). Strain EB6 was grown at 30°C, with shaking, to mid-logarithmic phase (OD600, 0.5 to 0.6). Cells were harvested by centrifugation at 16,000 × g, washed once with buffer (150 mM Tris-HCl, pH 7.2), and resuspended in assay buffer (10 mM Tris-HCl, pH 7.2) to an OD600 of 0.04. A 100-μl cell suspension was placed in each well of a clear 96-well microtiter plate (catalog no. 3370; Corning Costar). The cell suspension was incubated with 0.4 μM DiSC3(5) until DiSC3(5) uptake was maximal (as indicated by a stable reduction in fluorescence due to fluorescence quenching as the dye became concentrated in the cell by the membrane potential). The desired concentration of test compound was subsequently added, and the fluorescence reading was monitored using a Synergy HT multimode microplate reader (BioTek) at an excitation wavelength of 622 nm and an emission wavelength of 670 nm. The maximal fluorescence increase due to disruption of the cytoplasmic membrane was recorded. A blank with only cells and the dye was used for background subtraction.
Morphological studies of B. subtilis.
Cultures were prepared as described previously for MIC determination and then chemically fixed in a 1:10 (vol/vol) culture/20 mM HEPES (pH 6.8)-1.5% glutaraldehyde solution. Samples were fixed at 4°C overnight, to limit de novo cell wall biosynthesis during fixation. Samples were then negatively stained with 1.5% nigrosin, flushed with N2 gas to remove bubbles, and gently heated at 55°C to bring cells to a common focal plane. Samples were visualized using bright-field microscopy with an inverted Nikon Eclipse TE200 microscope at ×1,000 magnification, with a Plan Fluor Apo 100× objective. Micrographs were acquired using the open-source microscopy package Micro Manager (24).
RESULTS
Amenability of the PywaC reporter system to high-throughput screening.
Our laboratory originally reported on studies of the PywaC reporter to identify interactions among WTA, PG, and undecaprenol synthesis in B. subtilis 168 while helping to explain the complexity of teichoic acid gene dispensability (11). Here, we have optimized the PywaC reporter system for high-throughput screening (HTS), harnessing its capacity to enrich for perturbants of cell wall synthesis. In modifying the previously reported method, a kinetic assay was developed in which luminescence was measured continuously for 19 h on solid LB agar containing compounds at a desired screening concentration of 10 μM. As an obligate aerobe, our engineered B. subtilis strain posed an interesting screening challenge. Typically, HTS assays of bacteria are performed in liquid medium; however, shaking is required for reproducible growth of our reporter strain under such conditions. Furthermore, the kinetic nature of our assay meant that multiple readings were required. Because multimode readers are commonly integrated with nonshaking incubation chambers, we needed an alternative solution. We found that solid agar provided ideal growth conditions and generated a robust reporter signal that was highly amenable to kinetic luminescence detection. In further optimizing the assay, three key parameters were evaluated, i.e., the cell culture inoculum, the temporal window in which luminescence was measured, and the screening concentration of the compounds. Different inocula were tested, to identify an optimal starting density at which most cell wall-active compounds upregulated the transcriptional response of PywaC. At 6 × 106 CFU ml−1, all cell wall inhibitors tested yielded at least a 2.1-fold increase.
To probe the sensitivity of our assay system, we turned to conventional cell wall-active compounds of diverse mechanisms, for example, targeting both the membrane and extracellular steps in PG synthesis, as well as the 1-deoxy-d-xylulose-5-diphosphate (DOXP) isoprenoid biosynthesis pathway (Table 1). In addition to cell wall-active compounds, we found that the reporter system was sensitive to membrane-active chemicals. The sensitivity of the PywaC promoter-reporter was defined by the minimum activating concentration (MAC), which was compared with the MIC for known antibiotics. MAC values were evaluated based on a statistical increase of 4 standard deviations (2.1-fold increase) over the value for the low control (1% DMSO). Nearly all molecules had MIC/MAC ratios of >1, demonstrating that the reporter system was capable of detecting activity at concentrations below the MICs for most compounds. Particularly noteworthy was the high sensitivity for vancomycin, fosmidomycin, and various cephalosporins, for which ratios of >10 were seen. Nearly all of the aforementioned compounds showed meaningful activation of the reporter at ≤10 μM. Thus, we chose this concentration for our high-throughput screen.
TABLE 1.
Evaluation of the sensitivity of the HTS-optimized PywaC reporter system with select cell wall-associated and membrane-perturbing compoundsa
| Compound | Mode of action | MAC (μM) | MIC (μM) | MIC/MAC ratio |
|---|---|---|---|---|
| Carbenicillin | Transpeptidase inhibition | 0.21 | 1.65 | 7.86 |
| Nafcillin | Transpeptidase inhibition | 0.75 | 1.51 | 2.01 |
| Cephalothin | Transpeptidase inhibition | 1.58 | 1.58 | 1.00 |
| Cefoxitin | Transpeptidase inhibition | 1.46 | 5.85 | 4.01 |
| Cefuroxime | Transpeptidase inhibition | 0.74 | 23.60 | 31.90 |
| Ceftazidime | Transpeptidase inhibition | 9.15 | 29.3 | 3.20 |
| Cefotaxime | Transpeptidase inhibition | 1.37 | 10.9 | 7.96 |
| Ceftriaxone | Transpeptidase inhibition | 0.56 | 18.0 | 32.14 |
| Vancomycin | Transpeptidase inhibition | 0.01 | 0.42 | 42.00 |
| Fosfomycin | MurA inhibition | 145.00 | 145.00 | 1.00 |
| d-Cycloserine | Alanine racemase inhibition | 7.19 | 3.59 | 0.50 |
| Tunicamycin | MraY and TagO inhibition | 0.12 | 0.24 | 2.00 |
| Bacitracin | Undecaprenol recycling | 3.56 | 112.4 | 31.57 |
| Fosmidomycin | DOXP reductoisomerase | 6.81 | 109.00 | 16.01 |
| Daptomycin | Membrane potential, lipoteichoic acid | 1.24 | 0.39 | 0.31 |
| Polymyxin B | Membrane permeability | 3.84 | 7.68 | 2.00 |
| EDTA | Membrane permeability | 548.00 | 2190.00 | 4.00 |
The sensitivities of the PywaC reporter system were determined with select compounds by evaluating the MIC and minimum activating concentration (MAC) values and generating MIC/MAC ratios. A high MIC/MAC ratio represents sensitivity of the reporter below the MIC for the compound.
Kinetic screening of a 26,000-member small-molecule library for PywaC activation.
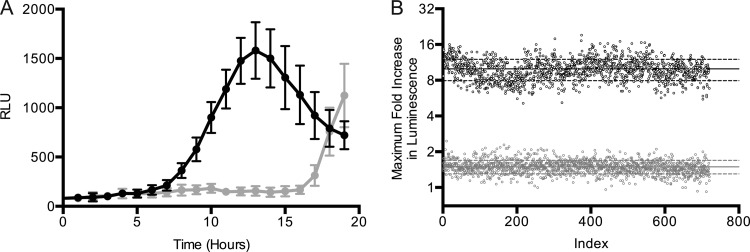
A diverse collection of ∼26,000 synthetic compounds, commercially obtained from Chembridge and Maybridge Chemicals, was screened against the HTS-optimized PywaC reporter system at concentrations of 10 μM, in duplicate, for 19 h, with luminescence readings being obtained every 60 min (see the supplemental material for the full data set). There was no theoretical ceiling for the luminescence resulting from reporter gene activation; however, fosmidomycin (10 μM) provided practical control for our screen. Fosmidomycin targets the DOXP pathway, specifically the reductoisomerase IspC. The latter is essential for the production of undecaprenol phosphate, the lipid species that supports both PG and WTA biosynthesis. DMSO at 1% was chosen as a low control, as this was the final concentration of DMSO present in each well containing a compound for analysis. Figure 2A shows an example of the time course for reporter luminescence in response to fosmidomycin and illustrates a strong broad peak of luminescence that is characteristic of the reporter system. The increase in luminescence signal in response to fosmidomycin correlates with growth. A maximum is reached as cells begin entering the stationary phase. Interestingly, at this point the negative control (1% DMSO) shows an increase in luminescence. This tailing effect can be attributed to nutrient starvation, as YwaC is part of a class of enzymes that synthesize (p)ppGpp, a well-known bacterial alarmone that is produced during cell stress, including amino acid starvation. In our analysis of compound data, the maximum relative light unit (RLU) values were recorded and normalized to the values for the DMSO controls on the same screening plate. This generated values for fold increases, corresponding to the activation of the compound relative to that recorded for the 1% DMSO control. The inclusion of fosmidomycin as a control allowed us to monitor the robustness of the reporter throughout the screening procedure (Fig. 2B).
FIG 2.
Assessment of the robustness of the PywaC reporter HTS assay. (A) A representative time course of the luminescence response (in relative light units [RLU]) for the positive control compound fosmidomycin at a concentration of 10 μM is shown (black circles). Also shown is the luminescence with the addition of 1% DMSO (gray circles). (B) Fosmidomycin was used a control compound on each screening plate. Shown is the maximal PywaC response recorded in response to 10 μM fosmidomycin for each of the control wells (black circles) relative to that of 1% DMSO treatment (gray circles). Solid black and gray horizontal lines correspond to the averages for fosmidomycin and DMSO activation, respectively. Dashed black and gray horizontal lines correspond to ±1 standard deviation from the mean of activation due to fosmidomycin and DMSO, respectively.
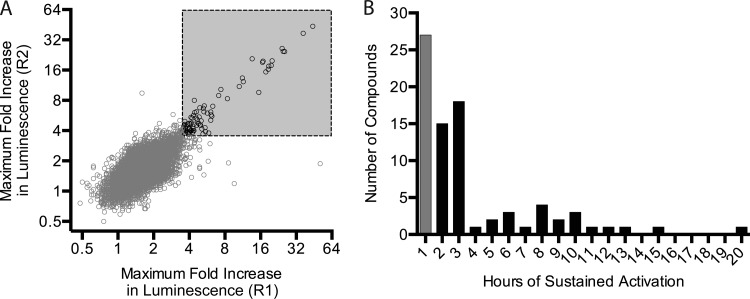
Figure 3A shows a plot of the data for the 26,000 compounds in which the fold increases in luminescence of replicates are plotted against one another, revealing strong reproducibility of the assay duplicates. A statistical analysis of the data revealed a standard deviation of 0.57 about the mean for the 1% DMSO control; instead of a statistical cutoff value, we elected to choose a threshold (3.5-fold increase) that made for a manageable number of active compounds. The threshold of a 3.5-fold increase led to 81 active compounds in the primary screen. Figure 3B shows an analysis of the duration of luminescence increases of ≥3.5-fold and reveals that several of our active compounds resulted from a single spike in luminescence. Where this behavior was quite different from, for example, that of our control compound fosmidomycin, we suspected that these spikes might be fleeting luminescence artifacts. To guard against possible artifacts, the 81 primary active compounds were further evaluated to ensure that the increases were sustained for at least 2 consecutive time points (2 h); that analysis led to 54 active compounds. These compounds were subjected to retesting under primary screening conditions, which yielded 46 confirmed active compounds.
FIG 3.
Replica plot and hit selection for the primary PywaC kinetic screen of 26,000 small molecules. (A) Duplicate samples from the primary screen describe the maximum fold increases in luminescence achieved during the 19-hour kinetic reading. With an arbitrary cutoff value of ≥3.5-fold increases for both replicates, the data shown as black circles correspond to 81 compounds designated active compounds, while compounds shown as gray circles are inactive. The gray box is the zone of the replica plot in which the primary hits are found, i.e., >3.5-fold in both replicate 1 and replicate 2. (B) The plot shows the duration of activation for the 81 molecules (PywaC activity above a 3.5-fold increase). Fifty-four compounds (black bars) showed sustained activity for at least two sequential kinetic readings. The remaining 27 compounds (gray bar) showed activity for only one time point and were deprioritized for further testing.
EC50 and membrane permeabilization studies.
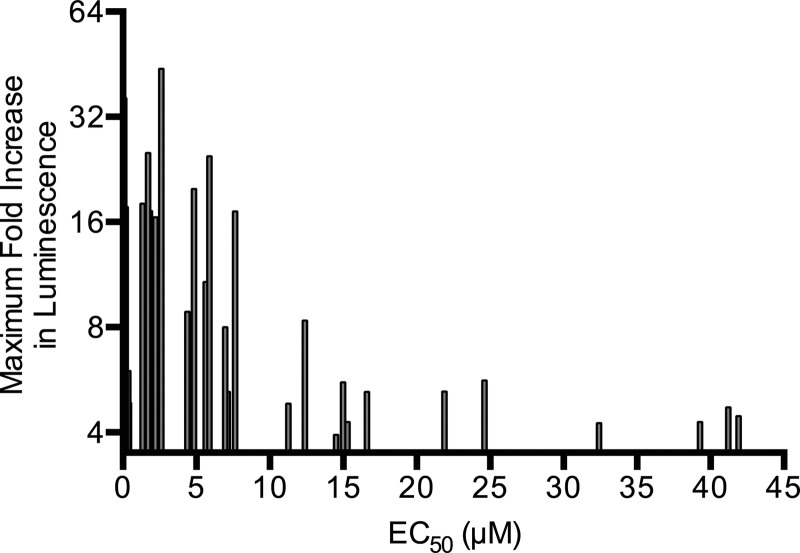
Studies of dose-dependent growth inhibition were conducted with the 46 confirmed active compounds using wild-type B. subtilis 168. Approximately 35 compounds showed clear growth inhibition, with EC50s between 0.05 and 80 μM (see Table S1 in the supplemental material). Potency showed modest correlation with PywaC responses seen in the primary screen, i.e., molecules with potent EC50s (0 to 10 μM) showed the strongest PywaC transcriptional activity (Fig. 4).
FIG 4.
Correlation of potency and PywaC activity. EC50 data on the inhibition of B. subtilis growth for 35 potent active compounds (x axis) are plotted against maximal PywaC activities (y axis).
As membrane-active compounds were shown previously to increase PywaC transcriptional activity, a membrane permeabilization counterscreen was performed with the 35 potent active compounds using the membrane potential-sensitive cyanine dye DiSC3(5). In this assay, membrane-active compounds lead to an increase or decrease in steady-state fluorescence through perturbation of the proton motive force (25). Of the 35 potent active compounds tested, 8 molecules were membrane active (see Table S1 in the supplemental material). With two compounds unavailable for reorder, this left us with 25 priority active compounds for follow-up testing to confirm activity on the cell wall.
Suppression of PywaC transcriptional activity of cell wall-active antibiotics by osmoprotectants.
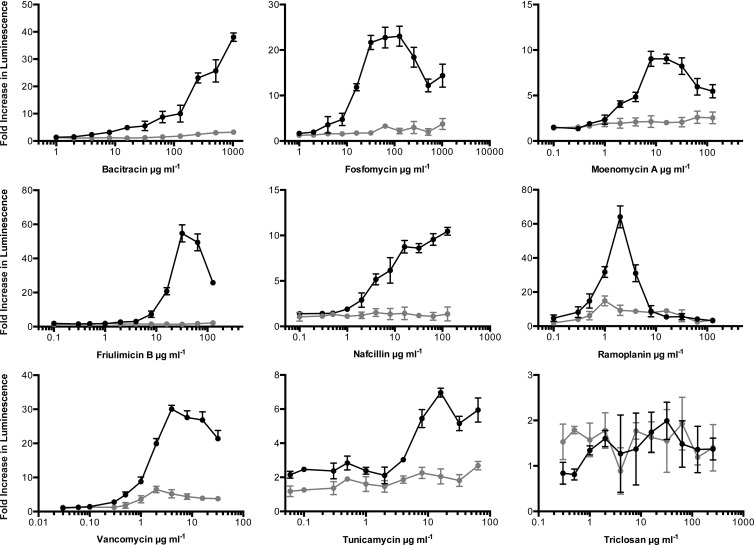
To determine whether the transcriptional responses of PywaC could be suppressed by osmoprotectants when exposed to cell wall-active agents, a panel of known cell wall-active antibiotics was tested in rich medium in a dose-dependent manner, in the presence and absence of 20 mM MgCl2, 0.5 M sucrose, and 20 mM maleic acid (26). Responses were monitored for compounds with diverse targets, including transpeptidation (vancomycin and nafcillin), cytoplasmic synthesis of precursors (fosfomycin), and membrane-associated steps (bacitracin, friulimicin B, ramoplanin, moenomycin A, and tunicamycin). In the presence of osmoprotectants, the reporter was suppressed for all cell wall-active compounds tested (Fig. 5).
FIG 5.
PywaC promoter-reporter responses to a panel of antibiotics in the presence and absence of osmoprotectants. Peak responses of the PywaC reporter were measured in the presence (black circles) and absence (gray circles) of osmoprotectants (MSM) for a panel of largely cell wall-active antibiotics at a range of concentrations. Fold increases in luminescence were calculated as the normalized luminescence (RLU/OD600) value of the reporter strain divided by the normalized luminescence value without antibiotic at the same time point.
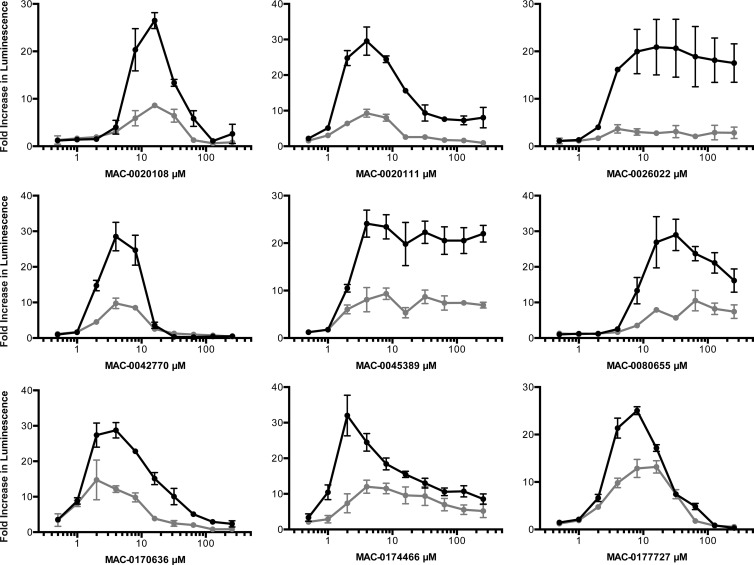
The 25 priority active compounds were reordered, and liquid chromatography-mass spectrometry confirmed the purity (>80%) and identity of each. Having demonstrated the suppression of reporter luminescence for known cell wall-active compounds, we endeavored to explore this phenotype for the priority active compounds. PywaC reporter activity was monitored in a dose-dependent manner at 0.5 to 256 μM, for 19 h, on solid LB agar in the presence and absence of osmoprotectants (MSM). Of the 25 compounds, 9 showed signature luminescence responses that were concentration dependent and could be at least partially suppressed by osmoprotectants (Fig. 6), suggesting that these compounds are active against the bacterial cell wall biosynthetic machinery (Table 2).
FIG 6.
Dose-response curves for the PywaC promoter-reporter in the absence and presence of osmoprotectants, leading to 9 novel cell wall-active inhibitors. Peak responses of the PywaC reporter were measured in the presence (black circles) and absence (gray circles) of osmoprotectants (MSM) for 25 priority active compounds, of which 9 phenocopied known cell wall-active antibiotics, identifying them as cell wall-active compounds. Fold increases in luminescence were calculated by dividing the RLU value in the presence of inhibitor by the value for the 1% DMSO control.
TABLE 2.
Nine cell wall-active lead compoundsa
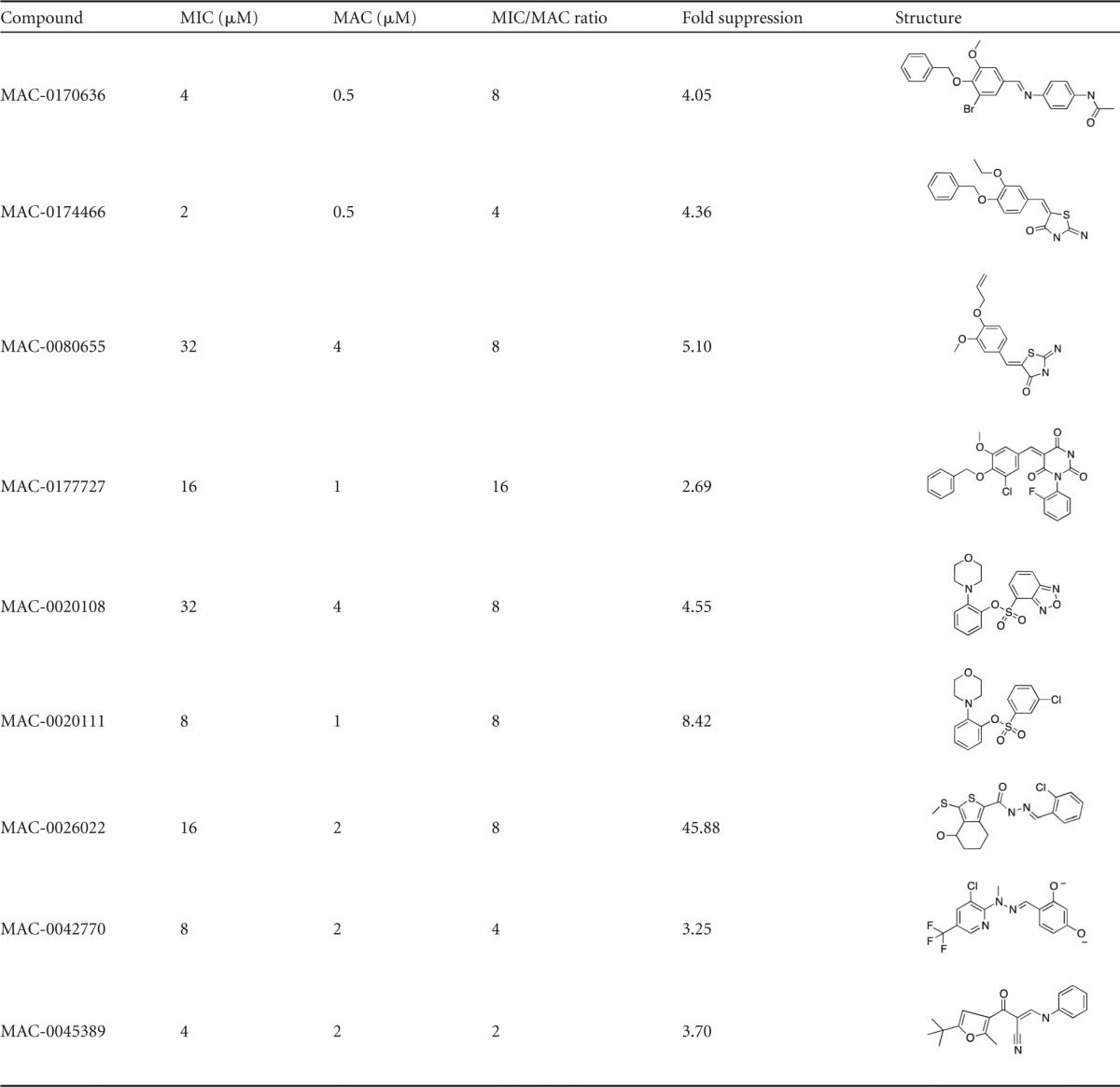
For each compound, the MIC against B. subtilis 168 and the minimum activating concentration (MAC) for the PywaC reporter were recorded. MIC/MAC ratios of >1 demonstrate sensitivity of the reporter below the MIC. Fold suppression of PywaC activity in response to the compound with osmoprotectants was calculated by dividing the increase in luminescence in the absence of osmoprotectants by the increase in luminescence in the presence of osmoprotectants. High fold suppression values indicate suppression of the activity of the molecule against cell wall biosynthesis in the presence of osmoprotectants.
Morphological effects of PywaC-active compounds against B. subtilis 168 at sub-MIC levels.
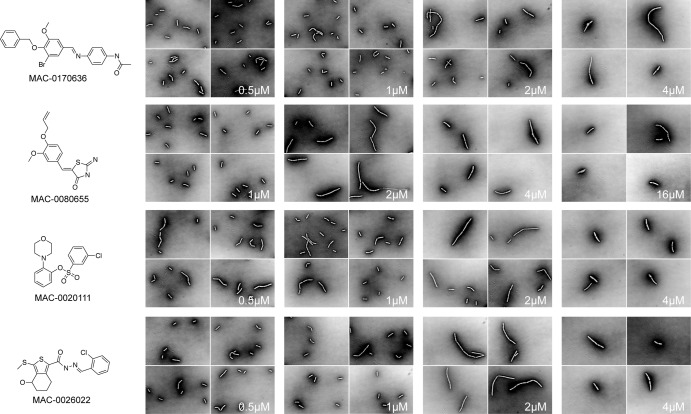
With the 9 compounds phenocopying known cell wall-active antibiotics, we sought further confirmation through microscopic examination. Dose-dependent studies were performed with each of the 9 compounds at sub-MIC through MIC levels. The fractions of abnormal cells in treated populations were observed to be inversely proportional to the cell culture density, demonstrating dose-dependent filamentous morphology characteristic of cell wall-active molecules (see Fig. S2 in the supplemental material). All 9 of the compounds showed induction of morphological changes at sub-MIC levels, in a dose-dependent manner (see Fig. S1 in the supplemental material), and four are highlighted in Fig. 7. We conclude that these 9 compounds are likely cell wall active.
FIG 7.
Morphological studies of B. subtilis 168 in the presence of 9 novel cell wall-active compounds. B. subtilis 168 was grown to midexponential phase and visualized in the presence of 9 novel cell wall-active compounds, 4 of which are shown here (for a full set of micrographs, see Fig. S1 in the supplemental material), at sub-MIC levels in a dose-dependent manner.
DISCUSSION
In the work presented here, we have made use of the specific responses of the B. subtilis ywaC promoter to cell wall-active antibiotics to develop a real-time high-throughput assay for novel chemical agents that affect cell wall biosynthesis. Optimized for solid growth medium, the PywaC-lux promoter-reporter system was a robust sensitive reporter of the action of known cell wall-active antibiotics with various mechanisms of action, including both intracellular and extracellular steps in PG synthesis and the DOXP pathway for isoprenoid biosynthesis. The sensitivity of the reporter was highlighted by comparing MIC and MAC values. Exquisite sensitivity for some cell wall-active antibiotics was seen, in some cases at concentrations 30-fold lower than the MIC.
A pilot-scale screen of 26,000 diverse synthetic compounds using the PywaC-lux promoter-reporter system revealed 54 active compounds, 46 of which were confirmed on retesting. Approximately 35 of these had potent whole-cell activity against B. subtilis and were further evaluated with a counterscreen for off-target membrane-active compounds using DiSC3(5), a fluorescence reporter of membrane polarization in bacteria (25). This left 25 priority active compounds, which were reordered, checked for quality and identity, and tested for dose-dependent responses of the PywaC-lux promoter-reporter system in the presence and absence of osmoprotectants. Nine of the 25 compounds showed signature luminescence responses that were concentration dependent and suppressed by osmoprotectants. All 9 of these chemicals are most likely novel cell wall-active compounds.
The latter assay was inspired by evidence dating back more than 40 years that established that cell wall-deficient cells (induced by genetic mutation or chemical perturbation) could be rescued with osmoprotectants and divalent cations (27–32). In recent years, it was shown that a stable L-form of B. subtilis lacking a cell wall could be generated by depletion of the murE operon and a single mutation in ispA when cultured in rich medium supplemented with high concentrations of sucrose and Mg2+ (26). We hypothesized that the activity of PywaC induced by cell wall-active compounds could be suppressed in the presence of osmoprotectants, similar to findings for chemical or genetic blocks in cell wall synthesis. Such suppression phenotypes were used to prioritize novel compounds for studies of the cell wall mode of action. In examining known cell wall-directed antibiotics in dose and temporal studies, we found that the transcriptional activity of the PywaC reporter system could indeed be suppressed in the presence of osmoprotectants.
Morphological changes of various bacterial species due to exposure to cell wall-active agents have been well documented over the years and show varied phenotypes, including blebbing, rounding, and long filamentation, as well as increases in division septa (33–37). To determine whether the 9 compounds thought to be selectively targeting cell wall biosynthesis showed similar phenotypes, imaging studies for these compounds were performed at sub-MIC and MIC levels. All 9 of the compounds showed induction of morphological changes at sub-MIC levels, in a dose-dependent manner. Interestingly, MAC-0080655 gives rise to long filamentous cells at low concentrations; when the MIC is approached, however, cells become shorter and spheroplasts begin to appear. Such morphological dependence on concentration was described previously for β-lactams in Escherichia coli (37).
Other cell wall reporter-based strains that respond to inhibition of essential steps in PG synthesis, including a vanH promoter-reporter fused to lacZ, have previously been developed. However, such systems report on only a subset of cell wall targets, such as inhibition of transglycosylation (17). In contrast, the PywaC reporter described herein has a nearly comprehensive response to cell wall synthesis inhibition, with demonstrated sensitivity for multiple pathways that converge on cell wall assembly, including PG, isoprenoid, and WTA biosynthesis.
The PywaC reporter screen takes a whole-cell approach, with a unique ability to report on biological activity at sub-MIC levels. Thus, the assay has the potential to discover cell wall-active compounds that would otherwise be discarded in conventional screens for viability. While the PywaC promoter-reporter could be a valuable follow-up tool to enrich for cell wall-targeted chemicals among whole-cell-active collections, it is arguably best used as a primary screening tool, with the rare capacity to report on previously untapped chemical areas. Such sensitivity could be particularly useful in, for example, the search for novel natural-product inhibitors, where high-throughput efforts typically use crude or semipure extracts that may contain molecules of interest at subinhibitory concentrations. In addition, unlike typical whole-cell screening approaches, the PywaC promoter-reporter system eliminates the need for follow-up testing of uninteresting and generally reactive nuisance compounds while reporting specifically on inhibitors that perturb cell wall biosynthesis. Our efforts have generated 9 novel cell wall-active compounds for further mode-of-action studies to elucidate their specific targets in the context of cell wall assembly.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Canada Research Chair salary award to E.D.B. and by an operating grant from the Canadian Institutes of Health Research (grant MOP-15496).
Footnotes
Published ahead of print 31 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02352-14.
REFERENCES
- 1.Anonymous. 1943. Penicillin. Science 98:192. 10.1126/science.98.2539.192-b [DOI] [PubMed] [Google Scholar]
- 2.Brown ED, Wright GD. 2005. New targets and screening approaches in antimicrobial drug discovery. Chem. Rev. 105:759–774. 10.1021/cr030116o [DOI] [PubMed] [Google Scholar]
- 3.Falconer SB, Brown ED. 2009. New screens and targets in antibacterial drug discovery. Curr. Opin. Microbiol. 12:497–504. 10.1016/j.mib.2009.07.001 [DOI] [PubMed] [Google Scholar]
- 4.Sewell EW, Brown ED. 2014. Taking aim at wall teichoic acid synthesis: new biology and new leads for antibiotics. J. Antibiot. (Tokyo) 67:43–51. 10.1038/ja.2013.100 [DOI] [PubMed] [Google Scholar]
- 5.Hancock IC. 1997. Bacterial cell surface carbohydrates: structure and assembly. Biochem. Soc. Trans. 25:183–187 [DOI] [PubMed] [Google Scholar]
- 6.Bhavsar AP, Brown ED. 2006. Cell wall assembly in Bacillus subtilis: how spirals and spaces challenge paradigms. Mol. Microbiol. 60:1077–1090. 10.1111/j.1365-2958.2006.05169.x [DOI] [PubMed] [Google Scholar]
- 7.Farha MA, Leung A, Sewell EW, D'Elia MA, Allison SE, Ejim L, Pereira PM, Pinho MG, Wright GD, Brown ED. 2013. Inhibition of WTA synthesis blocks the cooperative action of PBPs and sensitizes MRSA to β-lactams. ACS Chem. Biol. 8:226–233. 10.1021/cb300413m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Gill CJ, Lee SH, Mann P, Zuck P, Meredith TC, Murgolo N, She X, Kales S, Liang L, Liu J, Wu J, Santa Maria J, Su J, Pan J, Hailey J, Mcguinness D, Tan CM, Flattery A, Walker S, Black T, Roemer T. 2013. Discovery of wall teichoic acid inhibitors as potential anti-MRSA β-lactam combination agents. Chem. Biol. 20:272–284. 10.1016/j.chembiol.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swoboda JG, Meredith TC, Campbell J, Brown S, Suzuki T, Bollenbach T, Malhowski AJ, Kishony R, Gilmore MS, Walker S. 2009. Discovery of a small molecule that blocks wall teichoic acid biosynthesis in Staphylococcus aureus. ACS Chem. Biol. 4:875–883. 10.1021/cb900151k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tidten-Luksch N, Grimaldi R, Torrie LS, Frearson JA, Hunter WN, Brenk R. 2012. IspE inhibitors identified by a combination of in silico and in vitro high-throughput screening. PLoS One 7:e35792. 10.1371/journal.pone.0035792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Elia MA, Millar KE, Bhavsar AP, Tomljenovic AM, Hutter B, Schaab C, Moreno-Hagelsieb G, Brown ED. 2009. Probing teichoic acid genetics with bioactive molecules reveals new interactions among diverse processes in bacterial cell wall biogenesis. Chem. Biol. 16:548–556. 10.1016/j.chembiol.2009.04.009 [DOI] [PubMed] [Google Scholar]
- 12.Bhavsar AP, Beveridge TJ, Brown ED. 2001. Precise deletion of tagD and controlled depletion of its product, glycerol 3-phosphate cytidylyltransferase, leads to irregular morphology and lysis of Bacillus subtilis grown at physiological temperature. J. Bacteriol. 183:6688–6693. 10.1128/JB.183.22.6688-6693.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nanamiya H, Kasai K, Nozawa A, Yun C-S, Narisawa T, Murakami K, Natori Y, Kawamura F, Tozawa Y. 2008. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol. Microbiol. 67:291–304. 10.1111/j.1365-2958.2007.06018.x [DOI] [PubMed] [Google Scholar]
- 14.Cao M, Kobel PA, Morshedi MM, Wu MFW, Paddon C, Helmann JD. 2002. Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316:443–457. 10.1006/jmbi.2001.5372 [DOI] [PubMed] [Google Scholar]
- 15.Lai MH, Kirsch DR. 1996. Induction signals for vancomycin resistance encoded by the vanA gene cluster in Enterococcus faecium. Antimicrob. Agents Chemother. 40:1645–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ulijasz AT, Grenader A, Weisblum B. 1996. A vancomycin-inducible lacZ reporter system in Bacillus subtilis: induction by antibiotics that inhibit cell wall synthesis and by lysozyme. J. Bacteriol. 178:6305–6309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mani N, Sancheti P, Jiang ZD, McNaney C, DeCenzo M, Knight B, Stankis M, Kuranda M, Rothstein DM, Sanchet P, Knighti B. 1998. Screening systems for detecting inhibitors of cell wall transglycosylation in Enterococcus: cell wall transglycosylation inhibitors in Enterococcus. J. Antibiot. (Tokyo) 51:471–479 [DOI] [PubMed] [Google Scholar]
- 18.Cao M, Wang T, Ye R, Helmann JD. 2002. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis σW and σM regulons. Mol. Microbiol. 45:1267–1276. 10.1046/j.1365-2958.2002.03050.x [DOI] [PubMed] [Google Scholar]
- 19.Hong H-J, Paget MSB, Buttner MJ. 2002. A signal transduction system in Streptomyces coelicolor that activates the expression of a putative cell wall glycan operon in response to vancomycin and other cell wall-specific antibiotics. Mol. Microbiol. 44:1199–1211. 10.1046/j.1365-2958.2002.02960.x [DOI] [PubMed] [Google Scholar]
- 20.Mascher T, Zimmer SL, Smith T-A, Helmann JD. 2004. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48:2888–2896. 10.1128/AAC.48.8.2888-2896.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lacriola CJ, Falk SP, Weisblum B. 2013. Screen for agents that induce autolysis in Bacillus subtilis. Antimicrob. Agents Chemother. 57:229–234. 10.1128/AAC.00741-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urban A, Eckermann S, Fast B, Metzger S. 2007. Novel whole-cell antibiotic biosensors for compound discovery. Appl. Environ. Microbiol. 73:6436–6443. 10.1128/AEM.00586-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sims PJ, Waggoner AS, Wang CH, Hoffman JF. 1974. Studies on the mechanism by which cyanine dyes measure membrane potential in red blood cells and phosphatidylcholine vesicles. Biochemistry 13:3315–3330. 10.1021/bi00713a022 [DOI] [PubMed] [Google Scholar]
- 24.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. 2010. Computer control of microscopes using μManager. Curr. Protoc. Mol. Biol. Unit 14.20. 10.1002/0471142727.mb1420s92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farha MA, Verschoor CP, Bowdish D, Brown ED. 2013. Collapsing the proton motive force to identify synergistic combinations against Staphylococcus aureus. Chem. Biol. 20:1168–1178. 10.1016/j.chembiol.2013.07.006 [DOI] [PubMed] [Google Scholar]
- 26.Leaver M, Domínguez-Cuevas P, Coxhead JM, Daniel RA, Errington J. 2009. Life without a wall or division machine in Bacillus subtilis. Nature 457:849–853. 10.1038/nature07742 [DOI] [PubMed] [Google Scholar]
- 27.Burmeister HR, Hesseltine CW. 1968. Induction and propagation of a Bacillus subtilis L form in natural and synthetic media. J. Bacteriol. 95:1857–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chatterjee AN, Young FE. 1972. Regulation of the bacterial cell wall: isolation and characterization of peptidoglycan mutants of Staphylococcus aureus. J. Bacteriol. 111:220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilpin RW. 1976. Time-lapse photography of Bacillus subtilis L-forms replicating in liquid medium. J. Bacteriol. 127:1018–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilpin RW, Young FE, Chatterjee AN. 1973. Characterization of a stable L-form of Bacillus subtilis 168. J. Bacteriol. 113:486–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray T, Popham DL, Setlow P. 1998. Bacillus subtilis cells lacking penicillin-binding protein 1 require increased levels of divalent cations for growth. J. Bacteriol. 180:4555–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young FE, Haywood P, Pollock M. 1970. Isolation of L-forms of Bacillus subtilis which grow in liquid medium. J. Bacteriol. 102:867–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhanel GG, Hoban DJ, Harding GK. 1992. Subinhibitory antimicrobial concentrations: a review of in vitro and in vivo data. Can. J. Infect. Dis. 3:193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorian V. 1975. Some effects of subinhibitory concentrations of antibiotics on bacteria. Bull. N. Y. Acad. Med. 51:1046–1055 [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmerman SB, Stapley EO. 1976. Relative morphological effects induced by cefoxitin and other β-lactam antibiotics in vitro. Antimicrob. Agents Chemother. 9:318–326. 10.1128/AAC.9.2.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amini B. 2009. Effect of different sub MIC concentrations of penicillin, vancomycin and ceftazidime on morphology and some biochemical properties of Staphylococcus aureus and Pseudomonas aeruginosa isolates. Iranian J. Microbiol. 1:43–47 [Google Scholar]
- 37.Greenwood D, O'Grady F. 1973. Comparison of the responses of Escherichia coli and Proteus mirabilis to seven β-lactam antibodies. J. Infect. Dis. 128:211–222. 10.1093/infdis/128.2.211 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.