Abstract
Natural resistance to lincosamides and streptogramins A (LSA), which is a species characteristic of Bacillus subtilis and Enterococcus faecalis, has never been documented in the Staphylococcus genus. We investigate here the molecular basis of the LSA phenotype exhibited by seven reference strains of Staphylococcus sciuri, including the type strains of the three described subspecies. By whole-genome sequencing of strain ATCC 29059, we identified a candidate gene that encodes an ATP-binding cassette protein similar to the Lsa and VmlR resistance determinants. Isolation and reverse transcription-quantitative PCR (qRT-PCR) expression studies confirmed that Sal(A) can confer a moderate resistance to lincosamides (8 times the MIC of lincomycin) and a high-level resistance to streptogramins A (64 times the MIC of pristinamycin II). The chromosomal location of sal(A) between two housekeeping genes of the staphylococcal core genome supports the gene's ancient origins and thus innate resistance to these antimicrobials within S. sciuri subspecies.
INTRODUCTION
Macrolides, lincosamides, and streptogramins (MLS) constitute the group of antibiotics employed to treat staphylococcal and streptococcal infections in human and veterinary medicine. Resistance to these antibiotics is increasing among clinical isolates, mainly as a result of acquisition of exogenous genes carried by mobile elements, plasmids, and/or transposons. The validated list of MLS resistance (MLS-R) determinants includes rRNA methylases, efflux pumps, and inactivating enzymes (1).
Efflux-mediated resistance to MLS antibiotics in staphylococci relies on the ATPase activity of a very special kind of ATP-binding cassette (ABC) protein (2–4). These MLS-R ABC proteins form a robust cluster that corresponds to the antibiotic resistance (ARE) protein subfamily, closely related to, but distinct from, the regulatory (REG) subfamily in the phylogenetic and functional classification of ABC systems (5). Their protein designations depend on both the resistance phenotype and the bacterial origin of the genes. Two resistance spectra have been reported in the literature for ABC-based MLS efflux among Gram-positive cocci; the first one is encoded by msr genes and leads to macrolide and sometimes streptogramin B (M/MSB) resistance, while the other is encoded by lsa and vga genes and leads to resistance to lincosamides and streptogramin A (LSA). Thus far, three classes of msr, five classes of lsa, and five classes of vga genes have been shown to be clinically relevant because of their detection in pathogens such as Staphylococcus, Streptococcus, and Enterococcus strains (1, 6, 7). One of these genes is intrinsic to Enterococcus faecalis; this gene is lsa(A), whose integrity and constitutive expression have been linked to the LSA phenotype, useful as a species identification marker in enterococci (8).
Staphylococci can acquire the LSA phenotype via mobile genetic elements. Though lsa(B)- and lsa(E)-carrying plasmids have been described, vga genes, especially vga(A), carried by plasmids and/or transposons, have been found predominantly among clinical strains isolated in human medicine (9). Our most recent study failed to identify the resistance gene responsible for the LSA phenotype within a Staphylococcus sciuri isolate (10). We report here the nucleotide sequence, genome localization, and antibiotic-inducible expression of sal(A), a new category of ARE-encoding genes that we found in all our reference strains belonging to the three subspecies described for the S. sciuri species.
(Part of this work was presented at the 52nd Interscience Conference on Antimicrobial Agents and Chemotherapy [ICAAC], San Francisco, CA, 2012 [11].)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Seven reference strains of S. sciuri were included in this study. They were previously assigned by ribotype delineation to the three validly described subspecies: S. sciuri subsp. sciuri, S. sciuri subsp. carnaticus, and S. sciuri subsp. rodentium (12, 13). Four strains were listed by the World Federation of Culture Collections: ATCC 29059 (CIP 105824) and type strain ATCC 29062 (CIP 81.62T), which belong to S. sciuri subsp. sciuri, type strain ATCC 700058 (CIP 105826T) of S. sciuri subsp. carnaticus, and type strain ATCC 700061 (CIP 105829T) of S. sciuri subsp. rodentium. The three other strains, SS226, BL2, and SVv1, originally described by Wesley Kloos and which belong to S. sciuri subsp. sciuri, S. sciuri subsp. carnaticus, and S. sciuri subsp. rodentium, respectively, have since obtained the designations BM12487, BM12489, and BM12494, respectively. Escherichia coli strain Top10 was used as a cloning host for all plasmid constructs, and Staphylococcus aureus strain RN4220 was used as a recipient for plasmid electrotransformation. Staphylococcus saprophyticus strain ATCC 15305 (CIP 76.125T) was used as a control. When required, ampicillin at 100 μg/ml or chloramphenicol at 10 μg/ml was added to the culture media (Becton, Dickinson, Le Pont de Claix, France) for plasmid maintenance in E. coli or S. aureus, respectively. These antibiotic compounds, as well as lincomycin (Lc), were purchased from Sigma (Saint-Quentin Fallavier, France), while the two others, rifampin and pristinamycin IIA (PIIA), were kindly provided by Sanofi-Aventis (Croix-de-Berny, Antony, France). Bacterial strains were grown at 37°C in Luria-Bertani medium for E. coli or in brain heart infusion for staphylococci. Broth cultures were vigorously shaken, and cell density was monitored by measuring the absorbance at 600 nm.
Antimicrobial susceptibility assays.
MLS resistance patterns of bacteria were determined on Mueller-Hinton agar using antibiotic disks purchased from Bio-Rad (Marnes-la-Coquette, France) or loaded manually with 20 μg PIIA. MICs of Lc and PIIA were obtained with serial 1:2 dilutions of the antibiotics in Mueller-Hinton agar using 104 CFU per spot (2). Each MIC determination was carried out in triplicate from two independent series of experiments, with plates incubated for 18 h at 37°C. Reference breakpoints were those published in the Communiqués CASFM, which are annually revised and updated (see http://www.sfm-microbiologie.org for details). The control strain was S. aureus RN4220.
DNA manipulations.
Genomic DNA of S. sciuri ATCC 29059 was extracted and purified by using the QIAamp DNA minikit (Qiagen GmbH, Hilden, Germany) with lysostaphin (Sigma) added to the lysis buffer. Plasmid DNA was isolated from E. coli using the QIAprep spin system (Qiagen). Gene cloning in pRB474 (14) was achieved in successive steps, starting from a blunt-ended DNA fragment obtained by PCR and then purified by the QIAquick PCR purification kit (Qiagen) before being inserted into the pCR4-Blunt Topo vector (Invitrogen, Carlsbad, CA). Bacterial DNA was amplified using PfuUltra II fusion high-fidelity DNA polymerase (Agilent Technologies, Santa Clara, CA) from boiled colonies resuspended in sterile water, of which 5 μl was incorporated into 50-μl reaction mixtures. PCR was performed in an Eppendorf vapo.protect Mastercycler pro system using primer pairs described in Table 1 and classic thermal cycling parameters, such as 1 min at 52°C for the annealing segment and 2 min at 72°C for the elongation segment. The sizes of the PCR products were estimated by ethidium bromide staining after 1%, wt/vol, agarose gel electrophoresis using a 1-kb DNA ladder (Fermentas, Vilnius, Lithuania) as a double-stranded DNA standard. The clean PCR products were either directly sequenced or cloned into pCR4 for sequencing, and the PstI-EcoRI DNA fragment was later subcloned into a pRB474 shuttle vector (14) using the instructions, reagents, and enzymes contained in a rapid DNA dephosphorylation and ligation kit (Roche Applied Science, Indianapolis, IN). The resulting recombinant plasmid and the empty shuttle vector were introduced into S. aureus by electroporation as previously described (2). Each of the two categories of transformants was analyzed by antimicrobial susceptibility assays, as described above.
TABLE 1.
List of primers used for this study
| Expt usage | Target | Primer pair | Primer sequence from 5′ to 3′ |
|---|---|---|---|
| Gene cloning | sal(A) | SalA-P7 | GCGTCTGCAGGTGTAATACATGGGTAGa |
| SalA-P8 | AAAGACTAGAACTCTGTTTCACG | ||
| Gene detection | sal(A) | SalA-P1 | CTATTAATCGATGAACCAACAAACC |
| SalA-P2 | TTGATTTACCTGTACCATTTCTGC | ||
| Control gene primers for qRT-PCR | rrs | 16SF-qRTPCR | ACGTGGGTAACCTACCTATAAGACTGGAT |
| 16SR-qRTPCR | TACCTTACCAACTAGCTAATACG | ||
| Gene-specific primers for qRT-PCR | sal(A) | SalAF-qRTPCR | CGATGAACCAACAAACCACA |
| SalAR-qRTPCR | GCCCCATCAAGACTCAATTC |
Nucleotides in boldface correspond to the PstI site used for sal(A) gene cloning into pRB474.
RMAs.
The purified genomic DNA of S. sciuri ATCC 29059 was amplified, fragmented, and labeled using the GeneChip resequencing assay kit (Affymetrix Inc., Santa Clara, CA). The labeled sample was loaded onto a preheated PathogenID v.1 microarray cartridge (Affymetrix) containing 250 antibiotic resistance genes (15). Table S1 in the supplemental material lists the nature of the genes tiled on the microarray. MLS-R genes represent 26% of the total gene number (66 of the 250 nonredundant targeted sequences). After an overnight hybridization at 45°C, the GeneChip was washed, stained, and scanned according to Affymetrix's instructions. The raw image file (.DAT) obtained after the scan was transformed into a .CEL file. Nucleotide sequences were delivered in FASTA format by GeneChip sequence analysis software (GSEQ v4.1), which uses a derivative of the ABACUS base-calling algorithm (16). This algorithm consists of an automated statistical method that analyzes raw microarray assay (RMA) hybridization data and optimizes the base-calling process. For each base call, a quality score is defined. If this score is below a chosen threshold, an undetermined base, N, rather than A, T, C, or G is displayed. Sequence information is then analyzed manually by using the continuous or discontinuous stretch of resolved bases as a query for database similarity searches.
WGS.
High-throughput whole-genome sequencing (WGS) of the genomic DNA of S. sciuri ATCC 29059 was performed using Roche 454 GS Titanium chemistry (GATC Biotech, Constance, Germany). The 83,000 reads were assembled by CLC Workbench 5.2 software, which generated 371 contigs. The nucleotide and deduced protein sequences were analyzed with the tBLASTn and BLASTp programs available at the Institut Pasteur Biology IT Center through the Mobyle platform (17). Search for a candidate S. sciuri ARE protein(s) was conducted using Vga(A) and VmlR as queries (GenBank accession numbers AF117259 and BSU05610, respectively). The start codon of the presumptive gene and its putative Shine-Dalgarno sequence were determined by visual inspection.
Multiple-sequence alignments.
Nucleotide and amino acid polymorphisms were analyzed using the Clustal X v2.1 package (18). Comparison of a set of 25 representative ARE proteins with a confirmed or likely role in MLS resistance was performed using the Muscle algorithm (19). Phylogenetic analysis was done using PhyML 3.0 software, and the resulting tree was displayed with FigTree.
Induction tests and qRT-PCR.
The impact of 3-h sub-MIC treatments on resistance properties of S. sciuri ATCC 29059 was tested twice in broth before plating. Antibiotic was added at an optical density at 600 nm (OD600) of 0.1. Total RNA of bacterial cells was extracted at an OD600 of 0.6 with TRIzol reagent (Invitrogen) as previously described (20) and then treated with the Turbo DNA-free kit (Invitrogen) to remove any trace of DNA contamination. Quantitation of target gene expression was performed in relation to 16S rRNA expression using reagents of the LightCycler RNA amplification kit SYBR green I (Roche) according to the manufacturer's instructions. The amount of nucleic acid material was calculated by measuring the absorbance at 260 nm of the RNA samples. Each of the two independent series of experiments was analyzed in quadruplicate using 50 ng or 1 ng of input RNA and a 0.5 μM concentration of gene-specific primers (Table 1). Reverse transcription was carried out at 55°C for 10 min. PCR mixtures were subjected to 1 cycle at 95°C for 30 s and 45 cycles at 95°C for 5 s, 54°C for 10 s, and 72°C for 20 s. Identification of the PCR products was carried out by DNA melting profiling; profiles were automatically converted into melting peaks by Light Cycler instrument software (Roche).
Nucleotide sequence accession number.
The nucleotide sequence of sal(A) found within contig 252 of the CLC workbench assembly of the WGS of S. sciuri strain ATCC 29059 has been deposited into the GenBank database under accession no. KC693025.
RESULTS
LSA resistance is intrinsic to S. sciuri species.
Antimicrobial susceptibility testing was performed on the seven S. sciuri strains using the disk diffusion technique. S. saprophyticus strain ATCC 15305 was MLS susceptible, like the control strain RN4220. All S. sciuri strains exhibited an unambiguous LSA phenotype, with growth up to the PIIA-containing disk and a zone inhibition diameter around the Lc disk ranging from 12 to 16 mm. Thus, they were all categorized as resistant to both antibiotics. They all showed intermediate susceptibility to clindamycin, with diameters of the inhibition zones ranging from 19 and 21 mm, and full susceptibility to pristinamycin, which is comprised of a mixture of 70% SA and 30% SB compounds. The MICs of Lc and PIIA for strain ATCC 29059 were 16 μg/ml and 32 μg/ml, respectively. Precultivation of the strain for 3 h at one-eighth of the MIC of Lc led to a 2-fold increase in both values. This strain was chosen for an in-depth genetic study in order to characterize the molecular basis of LSA resistance in S. sciuri.
No LSA resistance gene was identified by RMA.
A total of 250 resistance genes were selected from the GenBank database to be represented in the PathogenID v.1 microarray (see Table S1 in the supplemental material). A set of 28 sequences coding for ARE proteins which can be either intrinsic or acquired among Gram-positive bacteria were tiled on the array. Only the sequence that codes for VmlR yielded a short fingerprint of 30 nucleotides with the labeled and fragmented DNA of strain ATCC 29059. When translated, this region of homology provided the peptide signature of the conserved H-loop motif shared by some categories of ABC proteins, including those of the ARE subfamily. Thus, neither the length of the determined nucleotide sequence (Fig. S1) nor the discriminative power of the deduced peptide stretch (IVVSHDR) was strongly indicative of the presence of an ARE-encoding gene in the genome of S. sciuri ATCC 29059. Indeed, there are at least three ABC proteins of the REG subfamily coded by the staphylococcal core genome: SACOL2036 (homologous to the YdiF protein of Bacillus subtilis), SACOL1427 (homologous to YkpA of B. subtilis), and SACOL0779 (homologous to YfmR of B. subtilis). None of these three ubiquitous proteins have a demonstrated role in MLS resistance. Rather, due to their similarity to the Saccharomyces cerevisiae regulatory protein GCN20, they might be involved in global gene regulation networks (21). Only fine-tuned analysis of full-length polypeptides would distinguish ARE from REG members. The data sequences recovered here by RMA for ARE scanning were not large enough for such a precise characterization. This was in sharp contrast to those obtained for other gene targets, such as mecA, rpoB, gyrA, or rrs, with which the accuracy of the identification process was optimal (data not shown). It was therefore anticipated that either a shotgun cloning or a complete genome sequencing might decipher the nature of the LSA resistance mechanism in strain ATCC 29059 and, more broadly, in S. sciuri species.
One candidate ARE gene was identified by WGS.
From de novo assembly of the entire genome of strain ATCC 29059, a tBLASTn search identified a putative ARE protein encoded by contig 252. The encoding gene had 1,626 bp and appeared to be inserted between two housekeeping genes, iscS and mnmA, of the staphylococcal core genome (Fig. 1). All features of a class 2 ABC protein were found in the deduced protein sequence (Fig. 2): two Walker A motifs (positions 35 to 43 and 347 to 355), two Walker B motifs (positions 151 to 155 and 451 to 455), two ABC signatures (positions 131 to 136 and 431 to 436), and two H-loop switches (positions 185 and 485). The closest NCBI matches were ≤70% amino acid identity with two other hypothetical proteins of unassembled staphylococcal genomes recently released in databases: WP_016910816 of Staphylococcus vitulinus and WP_016998509 of Staphylococcus lentus. The highest scores of amino acid identity with referenced members of the ABCISSE database (http://www.pasteur.fr/abcisse) were 28% with VmlR, formerly ExpZ, 27% with Vga(A), and 24% with Lsa(A). Such high-scoring results with ARE proteins conferring an LSA phenotype strongly suggested that the candidate gene found within contig 252 was responsible for the LSA resistance properties observed in strain ATCC 29059 and possibly in the other tested S. sciuri strains. In contrast, none of the other assembled staphylococcal genomes at the same chromosomal location contained an ARE-encoding gene. Only the genome of S. saprophyticus, strain ATCC 15305, harbors a similar genetic structure at this locus (Fig. 1). However, the presence of a stop codon at the end of the first quarter of the presumptive resistance gene would disrupt the full-length reading frame. Without such a mutation, the deduced polypeptide from this pseudogene would have 27% and 25% amino acid identity with Vga(A) and VmlR, respectively (Fig. 2). As mentioned above, strain ATCC 15305 is unambiguously susceptible to all MLS antibiotics.
FIG 1.
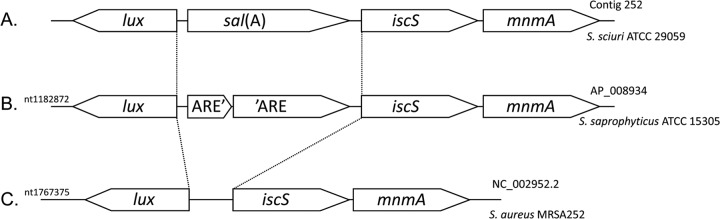
Genomic organization of the sal(A) gene locus in Staphylococceae. Only three organizations were found by reciprocal BLAST analysis with the available full-length genomes of the Staphylococceae family. Investigated species were Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus saprophyticus, Staphylococcus carnosus, Staphylococcus pseudintermedius, Staphylococcus lugdunensis, and Macrococcus caseolyticus. (A) Gene order of S. sciuri where sal(A) is present and expressed; (B) gene order of S. saprophyticus where there is a disrupted ARE gene orthologous to sal(A); (C) gene order of all other genomes where sal(A) is absent. No residual traces of sal(A) gene sequences were detected in these genomes. nt, nucleotide.
FIG 2.
Multiple alignment of Sal(A)-related proteins. Analysis was carried out with Sal(A) from S. sciuri (KC693025) (lane 1), VmlR from B. subtilis (BSU05610) (lane 3), Vga(A) from S. aureus (M90056) (lane 4), and the reconstructed ARE protein from S. saprophyticus ATCC 15305 that may arise from the fusion of two contiguous coding sequences (SSP1137 plus SSP1138) (lane 2). The two copies of the Walker A and Walker B motifs, as well as ABC signatures and H-loops, are indicated in boldface and underlined. Identical and similar amino acids are marked by asterisks (identity), colons (strong similarity), and dots (family similarity). The boldface letter X denotes the stop codon of SSP1137 of S. saprophyticus.
Characterization of the novel sal(A) gene.
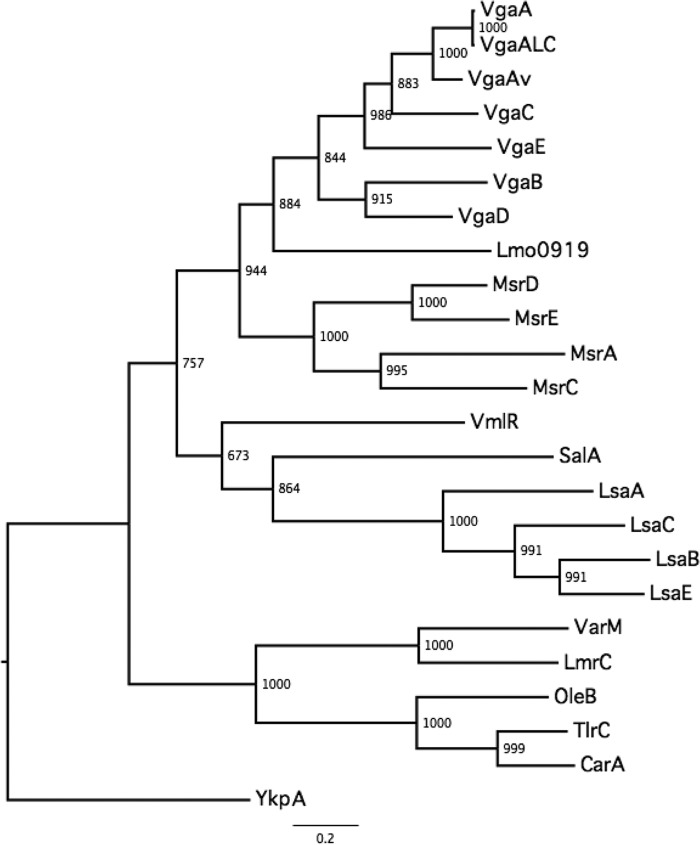
The putative ARE-encoding gene of strain ATCC 29059 was amplified using primers P7 and P8 (Table 1) and cloned into shuttle vector pRB474. The recombinant plasmid was electroporated into S. aureus RN4220. In comparison with the recipient carrying the empty plasmid, S. aureus RN4220 transformants carrying the S. sciuri gene exhibited 8-fold- and 64-fold-increased MIC values of Lc (8 μg/ml) and PIIA (64 μg/ml), respectively. Such a result showed that the cloned gene was capable of conferring an LSA phenotype. In multiple-sequence alignment, the novel characterized ARE protein of S. sciuri was confirmed to be similar to the VmlR determinant of B. subtilis, and it was found to be much more closely related to the Lsa group than to the Vga group of proteins on the phylogenetic tree (Fig. 3). On the basis of these functional data and this clustering analysis, the novel ARE member received the designation Sal(A) from the MLS resistance gene nomenclature center (http://faculty.washington.edu/marilynr/).
FIG 3.
Phylogenetic relatedness among the experimentally tested ARE proteins. The maximum likelihood tree, using YkpA as an outgroup REG protein, was built by PhyML using the best-fitting model, which consists of WAG plus I (0.05) plus G (2.54), and 1,000 bootstraps. The basal group corresponds to ARE proteins from the MLS producers. Three main ARE groups were recovered; they correspond to Lsa proteins, Msr proteins, and Vga proteins. Sal(A) is closer to Lsa than to Vga.
Expression and distribution of sal(A).
To study sal(A) gene expression in strain ATCC 29059, a series of qRT-PCR experiments were conducted with and without antibiotic exposure at one-eighth the MICs of Lc and PIIA. Both antibiotics were potent inducers of the targeted RNA transcript(s), since Lc and PIIA led to an 8-fold and a 6-fold increase in sal(A) gene expression, respectively. Specificity of induction was tested with chloramphenicol and rifampin under the same conditions, and no change in sal(A) expression was observed. Standard PCR experiments checked that sal(A) could be amplified from all other studied S. sciuri strains (see Fig. S2 in the supplemental material).
DISCUSSION
The sal(A) gene characterized in this study was found to be highly divergent from all other thus-far-described ARE-encoding genes. It had not been detected either previously by PCR (10) nor here by RMA. Only WGS and BLAST analyses against sequences in ABCISSE, a relational database displaying the properties and total subunit compositions of experimentally investigated ABC systems (21), pointed out its likely involvement in the LSA resistance phenotype of S. sciuri strain ATCC 29059. This assumption was confirmed by its subsequent cloning into S. aureus strain RN4220. Based on the gain in MIC values of Lc and PIIA obtained by use of this reference strain, the novel gene has been definitively included in the long list of MLS resistance determinants (22).
Despite its demonstrated inducibility in strain ATCC 29059, the sal(A) gene appears to be loosely regulated by the presence of lincosamide or SA antibiotics in culture medium. Neither the mechanism of gene induction nor the determination of the transcription start site(s) was investigated here. Indeed, that type of analysis was not considered of high clinical relevance, mainly because the LSA resistance trait could be easily evidenced in classical antibiograms without preincubation treatment for all tested S. sciuri strains. Since PIIA-loaded disks are not commercially available, the use of Lc disks would be the best way to detect the LSA phenotype. While lnu (formerly lin) resistance genes confer high-level resistance to lincomycin in staphylococci (up to a MIC of 128 μg/ml) and no detectable resistance to clindamycin, the sal(A), vga, and lsa genes confer low-level resistance to both antibiotics (23, 24). We speculate that pleuromutilin antibiotics, such as tiamulin, preferentially used in veterinary medicine but not tested here because of their unavailability, would be good alternatives to screen for the LSA and pleuromutilin resistance phenotype. In light of the present study, it can be anticipated that the high-level resistance to tiamulin exhibited by the recently collected lnu(A)-carrying S. sciuri clinical strain (23) resulted from the indigenous sal(A) gene. This assumption needs to be further investigated.
As judged by its genetic environment in strain ATCC 29059, sal(A) cannot be easily disseminated by horizontal gene transfer, and its localization between two housekeeping genes of the staphylococcal core genome is a rather limited way of spreading, especially within the genus Staphylococcus, where generalized transduction and transformation events are scarce for most of the studied chromosomal markers (25, 26). Moreover, the presence of sal(A) in the chromosome of strain ATCC 29059, a strain of S. sciuri isolated from the healthy skin of a Virginia opossum in 1972 (27), long before the intensive use of antimicrobials in medicine and agriculture practices, lends support to the hypothesis of a natural resistance trait. Since sal(A) was successfully amplified by the use of external gene primers for the three S. sciuri subspecies and since this detection is correlated with both the LSA phenotype of the strains and the evolution time of the species, stable inheritance of the resistance gene from a common ancestor is likely. Such a progenitor might have belonged to the Gram-positive bacterial lineage, prior to the separation of the Firmicutes and Actinobacteria phyla (28). The latter phylum is recognized as the source of more than two-thirds of clinically useful antibiotics, and each MLS producer strain was found to have at least one ARE protein encoded by its genome in the vicinity of antibiotic biosynthesis genes (29, 30, 38). In these MLS-producing strains, ARE proteins confer self-resistance or natural immunity against the antibiotic that they synthesize. It might be speculated, in light of recently reported findings, that a ubiquitous microorganism like S. sciuri, which has been isolated in soil, sand, and marsh grass (31), has maintained an indigenous Sal(A) determinant to survive within lincosamide-containing environments. This is probably what also happens with many other species of the Bacilli class, as recently exemplified by molecular characterization of VmlR in B. subtilis (32) and Lsa(A) in E. faecalis (8), which confer the same intrinsic resistance properties, the LSA resistance phenotype, upon the bacteria in which they reside.
In contrast to lsa(A), from which were derived the lsa(B) (33), lsa(C) (34), and lsa(E) (7) genes, neither sal(A)- nor vmlR-related genes have been detected to date outside their linked species, and the present study did not find a reservoir of LSA resistance genes in staphylococci. The origin of vga variant genes remains obscure at this time. According to us, it is now clear that most staphylococcal species lost their ancestral ARE-encoding gene, represented by sal(A) in S. sciuri, during their species evolution; bit by bit, they became highly restricted to the skin and mucous membranes of their mammal, fish, reptile, and bird hosts. Staphylococcal strains, currently confronted with much MLS use in a wide variety of human processes, have developed various successful strategies for acquiring a whole range of MLS resistance genes from the environment (35). While S. sciuri and sal(A) cannot be seen as immediate threats to public health, both should be included in the monitoring memos for MLS antibiotic resistance surveys. In addition, the report of the presence of a sal(A)-related pseudogene in S. saprophyticus might be of epidemiological concern, especially since none of the thus-far-described resistance genes can be found in an ML-resistant strain (36). Indeed, it was recently reported that only one nucleotide mutation within a resident cryptic ARE-encoding gene of the Enterococcus faecium genome would be enough to generate an LSA-resistant strain from a susceptible one (37). With respect to these documented examples, ABC-mediated MLS-R clinical cases may have been underestimated in the literature.
Supplementary Material
ACKNOWLEDGMENTS
N.B. was supported by the Académie Nationale de Medecine under an Elisabeth Taub award.
C.H. and O.C. are indebted to Cécile Wandersman for her constant support and interest during this work. We are grateful to the Antibacterial Agents Unit and the Genotyping Platform, both located at the Institut Pasteur, for great flexibility in allowing us to use the Roche LightCycler sequence detection instrument and the Affymetrix station, respectively. We thank Olivier Poupel and Nicolas Rosenfeld for help in qRT-PCR experiments and Elie Dassa for critical reading of the final draft document.
Footnotes
Published ahead of print 31 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02797-13.
REFERENCES
- 1.Roberts MC. 2008. Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol. Lett. 282:147–159. 10.1111/j.1574-6968.2008.01145.x [DOI] [PubMed] [Google Scholar]
- 2.Chesneau O, Ligeret H, Hosan-Aghaie N, Morvan A, Dassa E. 2005. Molecular analysis of resistance to streptogramin A compounds conferred by the Vga proteins of staphylococci. Antimicrob. Agents Chemother. 49:973–980. 10.1128/AAC.49.3.973-980.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacquet E, Girard JM, Ramaen O, Pamlard O, Lévaique H, Betton JM, Dassa E, Chesneau O. 2008. ATP hydrolysis and pristinamycin IIA inhibition of the Staphylococcus aureus Vga(A), a dual ABC protein involved in streptogramin A resistance. J. Biol. Chem. 283:25332–25339. 10.1074/jbc.M800418200 [DOI] [PubMed] [Google Scholar]
- 4.Novotna G, Janata J. 2006. A new evolutionary variant of the streptogramin A resistance protein, Vga(A)LC, from Staphylococcus haemolyticus with shifted substrate specificity towards lincosamides. Antimicrob. Agents Chemother. 50:4070–4076. 10.1128/AAC.00799-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson AL, Dassa E, Orelle C, Chen J. 2008. Structure, function, and evolution of bacterial ATP-binding cassette systems. Mol. Biol. Rev. 72:317–364. 10.1128/MMBR.00031-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwendener S, Perreten V. 2011. New transposon Tn6133 in methicillin-resistant Staphylococcus aureus ST398 contains vga(E), a novel streptogramin A, pleuromutilin, and lincosamide resistance gene. Antimicrob. Agents Chemother. 55:4900–4904. 10.1128/AAC.00528-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wendlandt S, Lozano C, Kadlec K, Gomez-Sanz E, Zarazaga M, Torres M, Schwarz S. 2013. The enterococcal ABC transporter gene lsa(E) confers combined resistance to lincosamides, pleuromutilins and streptogramin A antibiotics in methicillin-susceptible and methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 68:473–475. 10.1093/jac/dks398 [DOI] [PubMed] [Google Scholar]
- 8.Chowdhury SA, Arias CA, Nallapareddy SR, Reyes J, Willems RJL, Murray BE. 2009. A trilocus sequence typing scheme for hospital epidemiology and subspecies differentiation of an important nosocomial pathogen, Enterococcus faecalis. J. Clin. Microbiol. 47:2713–2719. 10.1128/JCM.00667-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lozano C, Aspiroz C, Rezusta A, Gomez-Sanz Simon C, Gomez P, Ortega C, Revillo MJ, Zarazaga M, Torres C. 2012. Identification of novel vga(A)-carrying plasmids and a Tn5406-like transposon in methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis of human and animal origin. Int. J. Antimicrob. Agents 40:306–312. 10.1016/j.ijantimicag.2012.06.009 [DOI] [PubMed] [Google Scholar]
- 10.Tessé S, Trueba F, Berthet N, Hot C, Chesneau O. 2013. Resistance genes underlying the LSA phenotype of French staphylococcal isolates. Antimicrob. Agents Chemother. 57:4543–4546. 10.1128/AAC.00259-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hot C, Berthet N, Chesneau O.Abstr. 52nd Intersci. Conf. Antimicrob. Agents Chemother, abstr. C1–1349.2012. [Google Scholar]
- 12.Chesneau O, Morvan A, Aubert S, El Solh N. 2000. The value of rRNA gene restriction site polymorphism analysis for delineating taxa in the genus Staphylococcus. Int. J. Syst. Evol. Microbiol. 50:689–697. 10.1099/00207713-50-2-689 [DOI] [PubMed] [Google Scholar]
- 13.Kloos WE, Ballard DN, Webster JA, Hubner RJ, Tomasz A, Couto I, Sloan GL, Dehart HP, Fiedler F, Schubert K, de Lancastre H, Santos Sanches I, Heath HE, Leblanc PA, Ljungh A. 1997. Ribotype delineation and description of Staphylococcus sciuri subspecies and their potential as reservoirs of methicillin resistance and staphylolytic enzyme genes. Int. J. Syst. Bacteriol. 47:313–323. 10.1099/00207713-47-2-313 [DOI] [PubMed] [Google Scholar]
- 14.Bruckner R. 1992. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene 122:187–192. 10.1016/0378-1119(92)90048-T [DOI] [PubMed] [Google Scholar]
- 15.Berthet N, Dickinson P, Filliol I, Reinhardt AK, Batejat C, Vallaeys T, Kong KA, Davies C, Lee W, Zhang S, Turpaz Y, Heym B, Coralie G, Dacheux L, Burguière AM, Bourhy H, Old IG, Manuguerra JC, Cole ST, Kennedy GC. 2008. Massively parallel pathogen identification using high-density microarrays. Microb. Biotechnol. 1:79–86. 10.1111/j.1751-7915.2007.00012.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cutler DJ, Zwick ME, Carrasquillo MM, Yohn CT, Tobin KP, Kashuk C, Mathews DJ, Shah NA, Eichler EE, Warrington JA, Chakravarti A. 2001. High-throughput variation detection and genotyping using microarrays. Genome Res. 11:1913–1925. 10.1101/gr.197201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Néron B, Ménager H, Maufrais C, Joly N, Maupetit J, Letort S, Carrere Tuffery SP, Letondal C. 2009. Mobyle: a new full web bioinformatics framework. Bioinformatics 25:3005–3011. 10.1093/bioinformatics/btp493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 19.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Even S, Burguière P, Auger S, Soutourina O, Danchin A, Martin-Verstraete I. 2006. Global control of cysteine metabolism by CymR in Bacillus subtilis. J. Bacteriol. 188:2184–2197. 10.1128/JB.188.6.2184-2197.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dassa E. 2011. Natural history of ABC systems: not only transporters. Essays Biochem. 50:19–42. 10.1042/bse0500019 [DOI] [PubMed] [Google Scholar]
- 22.Roberts MC. 2011. Environmental macrolide-lincosamide-streptogramin, and tetracycline resistance genes. Front. Microbiol. 2:40. 10.3389/fmicb.2011.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lozano C, Aspiroz C, Saenz Y, Ruiz-Garcia M, Royo-Garcia G, Gomez-Sanz E, Ruiz-Larrea F, Zarazaga M, Torres C. 2012. Genetic environment and location of the lnu(A) and lnu(B) genes in methicillin-resistant Staphylococcus aureus and other staphylococci of animal and human origin. J. Antimicrob. Chemother. 67:2804–2808. 10.1093/jac/dks320 [DOI] [PubMed] [Google Scholar]
- 24.Lüthje P, von Köckritz-Blickwede M, Schwarz S. 2007. Identification and characterization of nine novel types of small staphylococcal plasmids carrying the lincosamide nucleotidyltransferase gene lnu(A). J. Antimicrob. Chemother. 59:600–606. 10.1093/jac/dkm008 [DOI] [PubMed] [Google Scholar]
- 25.Maslanova I, Doskar J, Varga M, Kuntova L, Muzik J, Maluskova D, Ruzickova V, Pantucek R. 2013. Bacteriophages of Staphylococcus aureus efficiently package various bacterial genes and mobile genetic elements including SCCmec with different frequencies. Environ. Microbiol. Rep. 5:66–73. 10.1111/j.1758-2229.2012.00378.x [DOI] [PubMed] [Google Scholar]
- 26.Morikawa K, Takemura AJ, Inose Y, Tsai M, Nguyen Thi LT, Ohta T, Msadek T. 2012. Expression of a cryptic secondary sigma factor gene unveils natural competence for DNA transformation in Staphylococcus aureus. PLoS Pathog. 8:e1003003. 10.1371/journal.ppat.1003003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kloos WE, Schleifer KH, Smith RF. 1976. Characterization of Staphylococcus sciuri sp. nov. and its subspecies. Int. J. Syst. Bacteriol. 26:22–37. 10.1099/00207713-26-1-22 [DOI] [Google Scholar]
- 28.Cavalier-Smith T. 2010. Deep phylogeny, ancestral groups and the four ages of life. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365:111–132. 10.1098/rstb.2009.0161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peschke U, Schmidt H, Zhang HZ, Piepersberg W. 1995. Molecular characterization of the lincomycin-production gene cluster of Streptomyces lincolnensis 78-11. Mol. Microbiol. 16:1137–1156. 10.1111/j.1365-2958.1995.tb02338.x [DOI] [PubMed] [Google Scholar]
- 30.Pulsawat N, Kitani S, Nihira T. 2007. Characterization of biosynthetic gene cluster for the production of virginiamycin M, a streptogramin type A antibiotic, in Streptomyces virginiae. Gene 393:31–42. 10.1016/j.gene.2006.12.035 [DOI] [PubMed] [Google Scholar]
- 31.Dakic I, Morrison D, Vukovic D, Savic B, Shittu A, Jezek P, Hauschild T, Stepanovic S. 2005. Isolation and molecular characterization of Staphylococcus sciuri in the hospital environment. J. Clin. Microbiol. 43:2782–2785. 10.1128/JCM.43.6.2782-2785.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohki R, Tateno K, Takizawa T, Aiso T, Murata M. 2005. Transcriptional termination control of a novel ABC transporter gene involved in antibiotic resistance in Bacillus subtilis. J. Bacteriol. 187:5946–5954. 10.1128/JB.187.17.5946-5954.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kehrenberg C, Ojo KK, Schwarz S. 2004. Nucleotide sequence and organization of the multiresistance plasmid pSCFS1 from Staphylococcus sciuri. J. Antimicrob. Chemother. 54:936–939. 10.1093/jac/dkh457 [DOI] [PubMed] [Google Scholar]
- 34.Malbruny B, Werno AM, Murdoch DR, Leclercq R, Cattoir V. 2011. Cross-resistance to lincosamides, streptogramins A, and pleuromutilins due to the lsa(C) gene in Streptococcus agalactiae UCN70. Antimicrob. Agents Chemother. 55:1470–1474. 10.1128/AAC.01068-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wendlandt S, Fessler AT, Monecke S, Ehricht R, Schwarz S, Kadlec K. 2013. The diversity of antimicrobial resistance genes among staphylococci of animal origin. Int. J. Med. Microbiol. 303:338–349. 10.1016/j.ijmm.2013.02.006 [DOI] [PubMed] [Google Scholar]
- 36.Le Bouter A, Leclerq R, Cattoir V. 2011. Molecular basis of resistance to macrolides, lincosamides and streptogramins in Staphylococcus saprophyticus clinical isolates. Int. J. Antimicrob. Agents 37:118–123. 10.1016/j.ijantimicag.2010.10.008 [DOI] [PubMed] [Google Scholar]
- 37.Isnard C, Malbruny B, Leclercq R, Cattoir V. 2013. Genetic basis for in vitro and in vivo resistance to lincosamides, streptogramins A, and pleuromutilins (LSAP phenotype) in Enterococcus faecium. Antimicrob. Agents Chemother. 57:4463–4469. 10.1128/AAC.01030-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karray F, Darbon E, Oestreicher N, Dominguez H, Tuphile K, Gagnat J, Blondelet-Rouault MH, Gerbaud C, Pernodet JL. 2007. Organization of the biosynthetic gene cluster of the macrolide antibiotic spiramycin in Streptomyces ambofaciens. Microbiology 153:4111–4122. 10.1099/mic.0.2007/009746-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.