Abstract
New Delhi metallo-β-lactamase-3 (NDM-3) was identified in a multidrug-resistant Escherichia coli isolate, NCGM77, obtained from the feces of a patient in Japan. The enzymatic activities of NDM-3 against β-lactams were similar to those of NDM-1, although NDM-3 showed slightly lower kcat/Km ratios for all the β-lactams tested except for doripenem. The genetic context for blaNDM-3 was tnpA-blaNDM-3-bleMBL-trpF-dsbC-tnpA-sulI-qacEdeltaI-aadA2-dfrA1, which was present on an approximately 250-kb plasmid.
TEXT
Metallo-β-lactamases (MBLs) are produced by many species of Gram-negative bacteria and some species of Gram-positive bacteria, including Bacillus spp. (1, 2). MBLs can confer resistance or reduced susceptibility to carbapenems and, usually, to cephalosporins and to penicillins except for monobactams (3). New Delhi metallo-β-lactamase-1 (NDM-1), a recently discovered MBL, was initially found in Sweden from Klebsiella pneumoniae and Escherichia coli isolates that originated from India (4). Subsequently, at least 10 NDM variants (see www.lahey.org/studies) have been reported in several different countries (5–17).
Escherichia coli NCGM77 was isolated from the feces of a patient in a medical setting in Japan in 2013. The isolate was phenotypically identified, and species identification was confirmed by 16S rRNA sequencing (18). MICs were determined using the broth microdilution method as recommended by the Clinical and Laboratory Standards Institute (19). E. coli NCGM77 was resistant to various tested β-lactams (Table 1); the MICs of the other antibiotics were 32 μg/ml (amikacin), 8 μg/ml (arbekacin), 256 μg/ml (ciprofloxacin), <0.25 μg/ml (colistin), >1,024 μg/ml (fosfomycin), 64 μg/ml (gentamicin), 512 μg/ml (kanamycin), 32 μg/ml (levofloxacin), 1 μg/ml (minocycline), <0.25 μg/ml (tigecycline), and 64 μg/ml (tobramycin). PCR analysis for MBL genes of blaDIM, blaGIM, blaIMP, blaNDM, blaSIM, blaSPM, and blaVIM was performed (20, 21). On the basis of the PCR results, the isolate was positive for blaNDM. The DNA sequence of the PCR product revealed that the isolate had the blaNDM-3 gene (9). Multilocus sequence typing (MLST) of NCGM77 found it to be sequence type 88 (ST88) (E. coli MLST database [see http://www.pasteur.fr/recherche/genopole/PF8/mlst/EColi.html]). Pseudomonas aeruginosa IOMTU9 (17) was used as a source of the blaNDM-1 gene.
TABLE 1.
MICs of various β-lactams for E. coli strains NCGM77 and DH5α transformed with plasmids carrying NDM-1 or NDM-3
| Antibiotic | MIC (μg/ml) for: |
|||
|---|---|---|---|---|
| NCGM77 | pHSG398/NDM-3 | pHSG398/NDM-1 | pHSG398 | |
| Ampicillin | >1,024 | 256 | 256 | 4 |
| Ampicillin-sulbactam | >1,024 | 128 | 128 | 2 |
| Aztreonam | >1,024 | 0.063 | 0.063 | 0.063 |
| Cefepime | >1,024 | 0.25 | 0.5 | 0.063 |
| Cefoselis | —a | 2 | 4 | 0.031 |
| Cefotaxime | >1,024 | 4 | 8 | 0.031 |
| Cefoxitin | >1,024 | 32 | 64 | 4 |
| Cefpirome | —a | 1 | 0.5 | 0.015 |
| Ceftazidime | >1,024 | 256 | 256 | 0.25 |
| Ceftriaxone | —a | 8 | 8 | 0.031 |
| Cephradine | >1,024 | 256 | 256 | 16 |
| Doripenem | 32 | 0.125 | 0.125 | 0.031 |
| Imipenem | 16 | 0.25 | 0.5 | 0.063 |
| Meropenem | 32 | 0.25 | 0.5 | <0.015 |
| Moxalactam | —a | 8 | 8 | 0.125 |
| Penicillin G | >1,024 | 128 | 256 | 32 |
—, MICs of cefoselis, cefpirome, ceftriaxone, and moxalactam for the NCGM77 strain were not determined.
The blaNDM-3 and blaNDM-1 genes were cloned into the corresponding sites of pHSG398 (TaKaRa, Shiga, Japan) using the primer set EcoRI-NDM-F (5′-GGGAATTCATGGAATTGCCCAATATTATG-3′) and PstI-NDM-R (5′-AACTGCAGTCAGCGCAGCTTGTCGGCCAT-3′). The Escherichia coli DH5α strain was transformed with pHSG398-NDM-3 or pHSG398-NDM-1 to determine the MICs of β-lactams.
The open reading frames of NDM-1 and NDM-3 without signal peptide regions were cloned into the pET28a expression vector (Novagen, Inc., Madison, WI) using the primer set BamHI-TEV-NDM-F (5′-ATGGATCCGAAAACCTGTATTTCCAAGGCCAGCAAATGGAAACTGGCGAC-3′) and XhoI-NDM-R (5′-ATCTCGAGTCAGCGCAGCTTGTCGGCCATG-3′). Plasmids were transformed into E. coli BL21-CodonPlus (DE3)-RIP (Agilent Technologies, Santa Clara, CA). Recombinant NDM proteins were purified using nickel-nitrilotriacetic acid (Ni-NTA) agarose according to the manufacturer's instructions (Qiagen, Hilden, Germany). His tags were removed by digestion with TurboTEV protease (Accelagen, San Diego, CA), and untagged proteins were purified by an additional passage over the Ni-NTA agarose. The purities of NDM-1 and NDM-3 were >90%, as estimated by SDS-PAGE. During the purification procedure, the presence of β-lactamase activity was monitored using nitrocefin (Oxoid Ltd., Basingstoke, United Kingdom). Initial hydrolysis rates were determined in 50 mM phosphate buffer (pH 7.0) containing 5 μM Zn(NO3)2 at 37°C, using a UV-visible spectrophotometer (V-530; Jasco, Tokyo, Japan). The Km and kcat values and the kcat/Km ratios were determined by analyzing β-lactam hydrolysis using a Lineweaver-Burk plot. Wavelengths and extinction coefficients for β-lactam substrates have been reported previously (22–24). Three individual experiments were performed to determine the Km and kcat values.
All cloned genes in the pHSG398 and pQE2 vectors were sequenced using an ABI PRISM 3130 sequencer (Applied Biosystems, Foster City, CA).
The plasmid harboring blaNDM-3 was extracted (25) and sequenced by MiSeq (Illumina, San Diego, CA). The size of the plasmid harboring blaNDM-3 was determined using pulsed-field gel electrophoresis (PFGE) and Southern hybridization (12). A probe for blaNDM-3 was amplified by PCR by using the EcoRI-NDM-F and the PstI-NDM-R primers. Signal detection was carried out using the DIG High Prime DNA labeling and detection starter kit II (Roche Applied Science, Indianapolis, IN).
The blaNDM-3 probe hybridized to a 250-kb plasmid (Fig. 1). The sequence surrounding blaNDM-3 was tnpA-blaNDM-3-bleMBL-trpF-dsbC-tnpA-sulI-qacEdeltaI-aadA2-dfrA1. This plasmid showed more than 99.9% identity at the nucleotide sequence level from 69,229 to 78,275 bp of the pGUE plasmid (GenBank accession no. JQ364967) from the E. coli strain GUE, which was isolated in India (26). The entire sequence of the plasmid was not determined with the sequence data generated by MiSeq (Illumina).
FIG 1.
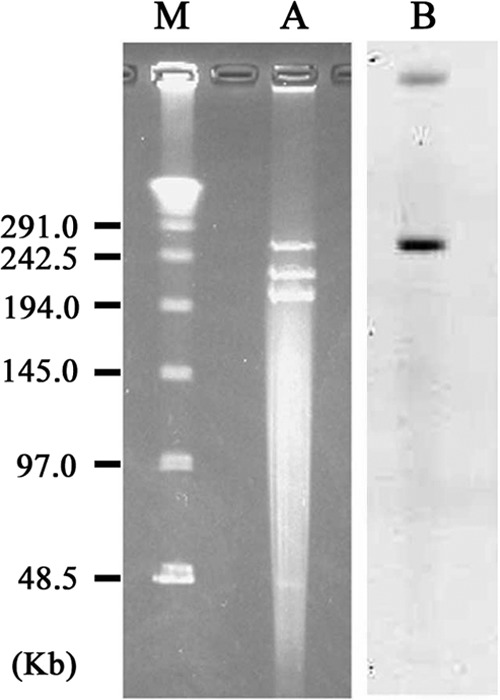
Localization of the blaNDM-3 gene on the plasmid of E. coli strain NCGM77 separated by PFGE. Lane M, midrange PFG marker (New England BioLabs, Tokyo, Japan); lane A, plasmids of E. coli strain NCGM77; lane B, hybridization of the plasmid with a probe specific for the blaNDM-3 gene.
Expression of the blaNDM-3 and blaNDM-1 genes in E. coli DH5α conferred resistance or reduced susceptibility to all cephalosporins, moxalactam, and carbapenems (Table 1). The MIC of cefpirome was 2-fold higher for E. coli expressing NDM-3 than for E. coli expressing NDM-1. In contrast, those of cefepime, cefoselis, cefotaxime, cefoxitin, imipenem, meropenem, and penicillin G were 2-fold lower for NDM-3 than for NDM-1.
As shown in Table 2, recombinant NDM-3 and NDM-1 hydrolyzed all β-lactams tested except for aztreonam. The profiles of the enzymatic activities of NDM-3 against β-lactams tested were similar to those of NDM-1, although NDM-3 had slightly but significantly lower kcat/Km ratios for all β-lactams tested except for doripenem. The lower kcat/Km ratios were mostly caused by the lower kcat values of NDM-3 compared with those of NDM-1, i.e., the values of NDM-3 were 19.0 to 47.5% of those of NDM-1 (Table 2). In fact, the substitution from Asp to Asn at position 95 of NDM appeared to decrease the hydrolysis rate of all β-lactams tested except for doripenem (Table 2). An amino acid substitution at position 95 from Asp to Asn decreased the kcat values of NDM-3 compared to those of NDM-1 (Table 2). Residue 95 is in α1, which is located on the protein surface. The crystal structure of NDM-1 revealed that the active site of NDM-1 is located at the bottom of a shallow groove enclosed by 2 important loops, L3 and L10 (27–30). Residue 95 in α1, however, was not located in these loops. This residue may indirectly affect the interaction of the substrate with the active site. Among all 9 NDM variants, amino acid substitutions were identified at 7 positions (28, 88, 95, 130, 152, 154, and 233). It remains unclear which position(s) plays a critical role in the enzymatic activities. Relative to IMP-1 and VIM-2, NDM-1 does not bind to carbapenems as tightly, but it turns over carbapenems at a rate similar to that of VIM-2 (4). NDM-4 with an amino acid substitution at position 154 (Met to Leu) showed greater hydrolytic activity toward carbapenems, cephalotin, cefotaxime, and ceftazidime than that shown by NDM-1 (12). NDM-5 with substitutions at positions 88 (Val to Leu) and 154 (Met to Leu) resulted in reduced susceptibilities of E. coli transformants to cephalosporins and carbapenems (12). NDM-8 with substitutions at positions 130 (Asp to Gly) and 154 (Met to Leu) resulted in enzymatic activities against β-lactams that were similar to those of NDM-1 (17). The drug susceptibilities of E. coli transformants with blaNDM-2, blaNDM-3, blaNDM-6, blaNDM-7, and blaNDM-9 have not been reported.
TABLE 2.
Kinetic parameters of NDM-3 and NDM-1 enzymesa
| β-Lactam | NDM-3 |
NDM-1 |
||||
|---|---|---|---|---|---|---|
| Kmb (μM) | kcatb (s−1) | kcat/Km (μM−1 s−1) | Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) | |
| Ampicillin | 228 ± 35 | 73 ± 7 | 0.32 | 500 ± 14 | 255 ± 8 | 0.51 |
| Aztreonam | NHc | NHc | NHc | NHc | NHc | NHc |
| Cefepime | 103 ± 10 | 17 ± 1 | 0.16 | 147 ± 18 | 41 ± 3 | 0.28 |
| Cefotaxime | 28 ± 3 | 24 ± 1 | 0.9 | 36 ± 4 | 63 ± 2 | 1.7 |
| Cefoxitin | 17 ± 1 | 1.6 ± 0.2 | 0.10 | 20 ± 3 | 4.4 ± 0.2 | 0.22 |
| Ceftazidime | 64 ± 9 | 11 ± 1 | 0.17 | 233 ± 35 | 58 ± 5 | 0.25 |
| Cephradine | 12 ± 3 | 26 ± 1 | 2.2 | 13 ± 2 | 66 ± 1 | 5.0 |
| Doripenem | 92 ± 2 | 34 ± 1 | 0.37 | 116 ± 18 | 41 ± 4 | 0.35 |
| Imipenem | 148 ± 13 | 25 ± 1 | 0.17 | 123 ± 21 | 59 ± 3 | 0.48 |
| Meropenem | 81 ± 4 | 32 ± 1 | 0.40 | 78 ± 6 | 74 ± 2 | 0.95 |
| Penicillin G | 42 ± 3 | 47 ± 1 | 1.1 | 24 ± 4 | 99 ± 4 | 4.3 |
The proteins were initially modified by a His tag, which was removed after purification.
Km and kcat values represent the means of 3 independent experiments ± the standard deviations.
NH, no hydrolysis was detected under conditions with substrate concentrations up to 1 mM and enzyme concentrations up to 700 nM.
This is the first report describing NDM-3-producing Gram-negative pathogens in Japan. It appears that NDMs have recently begun evolving; therefore, careful monitoring of NDM-producing pathogens is required.
Plasmid sequence accession number.
The plasmid sequence including the blaNDM-3 gene has been deposited in GenBank under the accession number AB898038.
ACKNOWLEDGMENTS
This study was supported by International Health Cooperation Research (grant 23-A-301), the Ministry of Health, Labor and Welfare of Japan (grant H24-Shinko-Ippan-010), and the JSPS KAKENHI (grant 24790432).
Footnotes
Published ahead of print 31 March 2014
REFERENCES
- 1.Queenan AM, Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20:440–458. 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlesinger SR, Kim SG, Lee JS, Kim SK. 2011. Purification development and characterization of the zinc-dependent metallo-beta-lactamase from Bacillus anthracis. Biotechnol. Lett. 33:1417–1422. 10.1007/s10529-011-0569-9 [DOI] [PubMed] [Google Scholar]
- 3.Bush K. 2001. New beta-lactamases in Gram-negative bacteria: diversity and impact on the selection of antimicrobial therapy. Clin. Infect. Dis. 32:1085–1089. 10.1086/319610 [DOI] [PubMed] [Google Scholar]
- 4.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-beta-lactamases: a last frontier for beta-lactams? Lancet Infect. Dis. 11:381–393. 10.1016/S1473-3099(11)70056-1 [DOI] [PubMed] [Google Scholar]
- 6.Pillai DR, McGeer A, Low DE. 2011. New Delhi metallo-beta-lactamase-1 in Enterobacteriaceae: emerging resistance. CMAJ 183:59–64. 10.1503/cmaj.101487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaase M, Nordmann P, Wichelhaus TA, Gatermann SG, Bonnin RA, Poirel L. 2011. NDM-2 carbapenemase in Acinetobacter baumannii from Egypt. J. Antimicrob. Chemother. 66:1260–1262. 10.1093/jac/dkr135 [DOI] [PubMed] [Google Scholar]
- 8.Espinal P, Fugazza G, Lopez Y, Kasma M, Lerman Y, Malhotra-Kumar S, Goossens H, Carmeli Y, Vila J. 2011. Dissemination of an NDM-2-producing Acinetobacter baumannii clone in an Israeli rehabilitation center. Antimicrob. Agents Chemother. 55:5396–5398. 10.1128/AAC.00679-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poirel L, Bonnin RA, Boulanger A, Schrenzel J, Kaase M, Nordmann P. 2012. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56:1087–1089. 10.1128/AAC.05620-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghazawi A, Sonnevend A, Bonnin RA, Poirel L, Nordmann P, Hashmey R, Rizvi TA, Hamadeh MB, Pal T. 2012. NDM-2 carbapenemase-producing Acinetobacter baumannii in the United Arab Emirates. Clin. Microbiol. Infect. 18:E34–E36. 10.1111/j.1469-0691.2011.03726.x [DOI] [PubMed] [Google Scholar]
- 11.Rogers BA, Sidjabat HE, Silvey A, Anderson TL, Perera S, Li J, Paterson DL. 2013. Treatment options for New Delhi metallo-beta-lactamase-harboring Enterobacteriaceae. Microb. Drug Resist. 19:100–103. 10.1089/mdr.2012.0063 [DOI] [PubMed] [Google Scholar]
- 12.Nordmann P, Boulanger AE, Poirel L. 2012. NDM-4 metallo-beta-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob. Agents Chemother. 56:2184–2186. 10.1128/AAC.05961-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hornsey M, Phee L, Wareham DW. 2011. A novel variant, NDM-5, of the New Delhi metallo-beta-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 55:5952–5954. 10.1128/AAC.05108-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williamson DA, Sidjabat HE, Freeman JT, Roberts SA, Silvey A, Woodhouse R, Mowat E, Dyet K, Paterson DL, Blackmore T, Burns A, Heffernan H. 2012. Identification and molecular characterisation of New Delhi metallo-beta-lactamase-1 (NDM-1)- and NDM-6-producing Enterobacteriaceae from New Zealand hospitals. Int. J. Antimicrob. Agents 39:529–533. 10.1016/j.ijantimicag.2012.02.017 [DOI] [PubMed] [Google Scholar]
- 15.Cuzon G, Bonnin RA, Nordmann P. 2013. First identification of novel NDM carbapenemase, NDM-7, in Escherichia coli in France. PLoS One. 8:e61322. 10.1371/journal.pone.0061322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottig S, Hamprecht AG, Christ S, Kempf VA, Wichelhaus TA. 2013. Detection of NDM-7 in Germany, a new variant of the New Delhi metallo-beta-lactamase with increased carbapenemase activity. J. Antimicrob. Chemother. 68:1737–1740. 10.1093/jac/dkt088 [DOI] [PubMed] [Google Scholar]
- 17.Tada T, Miyoshi-Akiyama T, Dahal RK, Sah MK, Ohara H, Kirikae T, Pokhrel BM. 2013. NDM-8 metallo-beta-lactamase in a multidrug-resistant Escherichia coli strain isolated in Nepal. Antimicrob. Agents Chemother. 57:2394–2396. 10.1128/AAC.02553-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki MT, Taylor LT, DeLong EF. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605–4614. 10.1128/AEM.66.11.4605-4614.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 9th ed. Approved standard M07-A9 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 20.Ellington MJ, Kistler J, Livermore DM, Woodford N. 2007. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J. Antimicrob. Chemother. 59:321–322. 10.1093/jac/dkl481 [DOI] [PubMed] [Google Scholar]
- 21.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70:119–123. 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 22.Boschi L, Mercuri PS, Riccio ML, Amicosante G, Galleni M, Frere JM, Rossolini GM. 2000. The Legionella (Fluoribacter) gormanii metallo-beta-lactamase: a new member of the highly divergent lineage of molecular-subclass B3 beta-lactamases. Antimicrob. Agents Chemother. 44:1538–1543. 10.1128/AAC.44.6.1538-1543.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crowder MW, Walsh TR, Banovic L, Pettit M, Spencer J. 1998. Overexpression, purification, and characterization of the cloned metallo-beta-lactamase L1 from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 42:921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Queenan AM, Shang W, Flamm R, Bush K. 2010. Hydrolysis and inhibition profiles of beta-lactamases from molecular classes A to D with doripenem, imipenem, and meropenem. Antimicrob. Agents Chemother. 54:565–569. 10.1128/AAC.01004-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarczynski MC, Meyer WJ, Min JJ, Wood KA, Hellwig RJ. 1994. Two-minute miniprep method for plasmid DNA isolation. Biotechniques 16:514–519 [PubMed] [Google Scholar]
- 26.Bonnin RA, Poirel L, Carattoli A, Nordmann P. 2012. Characterization of an IncFII plasmid encoding NDM-1 from Escherichia coli ST131. PLoS One 7:e34752. 10.1371/journal.pone.0034752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green VL, Verma A, Owens RJ, Phillips SE, Carr SB. 2011. Structure of New Delhi metallo-beta-lactamase 1 (NDM-1). Acta Crystallogr. F Struct. Biol. Cryst. Commun. 67:1160–1164. 10.1107/S1744309111029654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y, Tesar C, Mire J, Jedrzejczak R, Binkowski A, Babnigg G, Sacchettini J, Joachimiak A. 2011. Structure of apo- and monometalated forms of NDM-1—a highly potent carbapenem-hydrolyzing metallo-beta-lactamase. PLoS One 6:e24621. 10.1371/journal.pone.0024621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King D, Strynadka N. 2011. Crystal structure of New Delhi metallo-beta-lactamase reveals molecular basis for antibiotic resistance. Protein Sci. 20:1484–1491. 10.1002/pro.697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Hao Q. 2011. Crystal structure of NDM-1 reveals a common beta-lactam hydrolysis mechanism. FASEB J. 25:2574–2582. 10.1096/fj.11-184036 [DOI] [PubMed] [Google Scholar]


