Abstract
The type VI secretion system (T6SS) is the most prevalent bacterial secretion system and an important virulence mechanism utilized by Gram-negative bacteria, either to target eukaryotic cells or to combat other microbes. The components show much variability, but some appear essential for the function, and two homologues, denoted VipA and VipB in Vibrio cholerae, have been identified in all T6SSs described so far. Secretion is dependent on binding of an α-helical region of VipA to VipB, and in the absence of this binding, both components are degraded within minutes and secretion is ceased. The aim of the study was to investigate if this interaction could be blocked, and we hypothesized that such inhibition would lead to abrogation of T6S. A library of 9,600 small-molecule compounds was screened for their ability to block the binding of VipA-VipB in a bacterial two-hybrid system (B2H). After excluding compounds that showed cytotoxicity toward eukaryotic cells, that inhibited growth of Vibrio, or that inhibited an unrelated B2H interaction, 34 compounds were further investigated for effects on the T6SS-dependent secretion of hemolysin-coregulated protein (Hcp) or of phospholipase A1 activity. Two compounds, KS100 and KS200, showed intermediate or strong effects in both assays. Analogues were obtained, and compounds with potent inhibitory effects in the assays and desirable physicochemical properties as predicted by in silico analysis were identified. Since the compounds specifically target a virulence mechanism without affecting bacterial replication, they have the potential to mitigate the virulence with minimal risk for development of resistance.
INTRODUCTION
Vibrio cholerae causes the devastating disease cholera, which is endemic in several Southeast Asian and African countries and in Haiti. It also causes extensive epidemics. The disease is a major public health concern in countries where it is endemic and causes more than 500,000 cases and many deaths annually (1). The principal means of treatment is hydration, but antibiotic treatment is recommended for severely ill patients; however, a caveat is that drug-resistant strains are becoming increasingly common. Several cholera vaccines exist, but their protective efficacy is limited and of rather short duration (1). Thus, to successfully control cholera, there is need for additional therapies. The severe form of cholera is caused by serogroups O1 and O139, and their expression of the cholera toxin is essential for the devastating diarrhea. There are also toxin-negative variants of V. cholerae, and although they do not cause cholera, they are emerging extraintestinal pathogens. Common to all V. cholerae strains is the possession of a type VI secretion system (T6SS). Although it is a recently described secretion system, T6SSs are the most common form of bacterial secretion systems and present in many important Gram-negative pathogens (2–5). T6SSs constitute four major phylogenetic groups, and, intriguingly, representatives of several of these groups may be present in a given genome, e.g., six T6SS clusters are present in Burkholderia pseudomallei (6). This emphasizes the heterogeneity of T6SSs and indicates that the functions are diverse and likely specialized. As other secretion systems, T6SSs perform key roles to target eukaryotic cells and facilitate virulence in eukaryotes (6–12); however, unlike other secretion systems, the T6SS also plays a central role in interbacterial competition, a potentially very important virulence mechanism in bacterial communities such as the gastrointestinal tract (13–22).
T6SSs are encoded by 15 to 20 genes, but the genetic contents and organization vary considerably (2). Approximately a dozen T6SS proteins appear to constitute the core subunits, and two of these, VipA and VipB, are highly conserved and homologues are present in all T6SS clusters identified so far (2, 23–25). The two proteins interact, and this binding is essential for type VI secretion (T6S) (4, 26). This interaction has been shown to occur in all tested, clinically important pathogens, such as V. cholerae, Francisella tularensis, Escherichia coli, Pseudomonas aeruginosa, and Yersinia pseudotuberculosis (4). Several of the Vip homologues also bind to the heterologous partners from other bacteria, and, thus, the mechanism behind the complex formation appears to be highly conserved (4). The interaction is highly specific, since even single amino acid substitutions in the α-helical region of VipA abrogate binding to VipB (26).
VipA-VipB complexes form filaments that structurally resemble the bacteriophage T4 contractile tail sheath, and they show a regular movement and cycle between assembly, quick contraction, disassembly, and reassembly (27). The filaments are quickly disassembled by ClpV, an ATPase family protein (21, 27–29). Thus, the coordinated functions of VipA, VipB, and ClpV are needed for active secretion. In most T6SSs, hemolysin-coregulated protein (Hcp) and valine-glycine repeat protein G (VgrG) are exported by the secretion machinery, but most likely numerous other proteins are secreted as well (30–36). The well-characterized Hcp protein is not only secreted but also suggested to be a chaperone that binds to cognate T6SS effector molecules, and it forms a hexameric pore that is required for secretion of diverse effectors encompassing several enzymatic classes (37–39). It has been recently demonstrated that a family of phospholipases are common T6SS-dependent effectors, and they degrade phosphatidylethanolamine, leading to bacterial lysis (13, 14, 40). Also, it has been found that the T6SS is triggered in response to bacterium-bacterium contact and suggested that this is not only an “offensive” weapon but also a preemptive mechanism which supports the cooperation of entire bacterial populations (41). In addition, T6SS has been found to be activated by means of membrane-disrupting products, such as polymyxin B and the mating pair formation system of T4SS (42), indicating that it may be a means of protecting the integrity of the cell wall.
In view of all of the available evidence that demonstrates strict molecular constraints and high interspecies conservation of the VipA-VipB interaction, we hypothesized that it would be possible to inhibit the interaction by use of small-molecule compounds and that such inhibitors would block T6S and thereby impede virulence of Vibrio and other important Gram-negative pathogens.
MATERIALS AND METHODS
Plasmids, strains, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. All bacterial strains were grown in Luria-Bertani (LB) broth or agar (BD, Sparks, MD, USA) at 37°C. When needed, antibiotics were used at the following concentrations: carbenicillin, 50 μg/ml; kanamycin, 50 μg/ml; rifampin, 100 μg/ml; and tetracycline, 10 μg/ml.
Compounds.
The small-molecular-weight compound collection was purchased from ChemBridge (San Diego, CA) and consisted of 9,600 compounds. The compounds were dissolved in dimethyl sulfoxide (DMSO) at 5 mM and stored in 96-well plates at room temperature (RT) in a desiccator at Laboratories of Chemical Biology Umeå. Analogues of KS100 and KS200 were purchased separately from ChemBridge, Sigma-Aldrich (St. Louis, MO), Specs (Delft, the Netherlands), and Chemdiv (San Diego, CA).
DNA manipulations.
Plasmids used in this study are listed in Table S1 in the supplemental material. Primers and restriction enzymes used to generate the plasmids are listed in Table S2 in the supplemental material. All the DNA fragments were amplified with the Expand Long-Range dNTPack (Roche, Mannheim, Germany) and were initially cloned into the pCR4-TOPO TA cloning vector (Invitrogen, Carlsbad, CA, USA) in recipient strain E. coli Top10. Chromosomal DNA of E. coli 536 and P. aeruginosa PAK was used as the templates for bacterial two-hybrid constructs of AaiA-AaiB (ECP_0238-ECP_0237) and HsiB2-HsiC2 (PA1657-PA1658). The hsic2 allele was generated by overlap PCR to mutate the intrinsic NotI site. All the DNA fragments were confirmed by sequencing (Eurofins, Ebersberg, Germany) and were first introduced into NdeI/NotI-digested pACTR-AP-Zif or pBRGPω and then into E. coli DH5αF′IQ. For use in the bacterial two-hybrid assay, the plasmids were cotransformed into E. coli KDZif1ΔZ by electroporation.
Bacterial two-hybrid screening assay.
E. coli KDZif1ΔZ expressing the previously described VCA0107-VCA0108 (4), the AaiA-AaiB, or the HsiB2-HsiC2 construct was grown at 37°C in LB with the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) at 0.4 mM. Bacterial cells were permeabilized with 13% of SDS-CHCl3 and assayed for β-galactosidase activity after 5 min or longer as described previously (30, 43, 44). Screening assays were performed in duplicate and repeated once, and values were expressed as averages based on one experiment; duplicate measurements differed by less than 10%. Bacteria treated with 1% DMSO were used as the control, and their β-galactosidase activity was defined as 100%. The activity of samples was presented as the percentage of the control value.
In silico calculations of compound properties.
Properties were calculated using QikProp (45, 46) on low-energy three-dimensional (3D) conformations (i.e., the global minimum) of the molecules generated by the Macromodel (47) using the OPLS2005 force field (48) and a water model (49). The conformational search was a Monte Carlo-based torsional sampling of the molecules with the Polak-Ribière conjugate gradient energy minimization method (50). Calculated properties included the total solvent accessible surface area in Å2 (SASA), the logarithmic octanol/water partition coefficient (logP), the logarithmic aqueous solubility in mol/dm3 (logS), apparent (i.e., the calculated value of the composite effects of traversing all permeation pathways in the cell system) Caco-2 cell permeability in nm/s (Caco-2), and apparent Madin-Darby canine kidney (MDCK) cell permeability in nm/s, where the assessment uses a knowledge-based set of rules, including checking for suitable number of rotatable bonds, logP, solubility, and cell permeability.
Cytotoxicity test.
Cytotoxicity of the compounds was essentially tested as described previously (51). The compounds were added at 50 μM to 1 × 105 cells/well of J774A.1 macrophages (ATCC TIB-67) cultured in advanced Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) (GIBCO, New York, USA) at 37°C in 5% CO2. After 24 h of incubation, lactate dehydrogenase (LDH) released in supernatant was measured by using a CytoTox 96 nonradioactive cytotoxicity assay kit (Promega, Madison, WI, USA), and cells were monitored under a microscope. The value of cells treated with DMSO only and lysed by the addition of phosphate-buffered saline (PBS) with 0.1% deoxycholate was set as 100% (30, 52). Values of samples were shown as the percentage of the control value.
Growth inhibition test.
The compounds were added at 50 μM to bacterial strains cultured in LB broth. The A600 at the 4-, 8-, 12-, and 18-h time points was measured. The bacteria were in stationary phase beyond the 8-h time point. Bacteria incubated with 2% DMSO were used as a control.
Hcp secretion assay.
V. cholerae, 2 × 108 cells/ml, was incubated with compounds at an indicated concentration for 4 h in LB broth at 37°C in 5% of CO2. Bacterial cells were pelleted at 16,100 × g for 5 min, and proteins in supernatants were precipitated with 12% trichloroacetic acid. Proteins were subjected to Western blot analysis and probed with rabbit antiserum specific to Hcp, a kind gift from Sun Nyunt Wai, Department of Molecular Biology, Umeå University (53). Bacteria incubated with 2% DMSO only were used as a control.
Interbacterial competition assay.
The assay was performed according to a protocol previously described (16). V. cholerae V52 and E. coli 363 (a kind gift from Anders Johansson, Department of Clinical Microbiology, Umeå University) were mixed at a ratio of 10:1 in PBS. Compounds dissolved in DMSO were added to the mixture at a concentration of 200 μM, and DMSO only was used as a control. The mixture of bacteria (109 bacterial cells/ml) and compounds or DMSO only was spotted on LB agar plates and incubated at 37°C for 4 h. The bacterial spots were harvested, diluted in PBS, and spotted on LB agar plates containing antibiotics for the selection and quantification of V. cholerae or E. coli. After 14 to 18 h of incubation at 37°C, the density of the spots was determined visually.
Enzymatic assay of phospholipase activity.
V. cholerae, 2 × 108 cells/ml, was incubated with compounds at a concentration of 50 μM for 4 h in LB at 37°C in 5% of CO2. Proteins in supernatants were examined by the phospholipase A1 assay as previously described (40). Briefly, the assay was performed by incubating supernatants with phospholipase A1 substrate PED-A1 (Invitrogen, Eugene, OR, USA) for 30 min at RT and measuring the fluorescence intensity with a microplate reader equipped for excitation in the range of 450 to 490 nm and fluorescence emission around 515 nm. Samples with bacteria treated with DMSO were used as controls, and their phospholipase activity was arbitrarily set as 100%. The activity of samples was shown as the percentage inhibition versus the control sample.
Statistical evaluation.
Student's two-tailed t test was used to compare means between groups. All statistical analysis was conducted using SPSS, version 20.
Nucleotide sequence accession no.
Sequences have been deposited in GenBank under accession numbers KJ735588 (PA1657) and KJ735589 (PA1658).
RESULTS
Identification of candidate compounds.
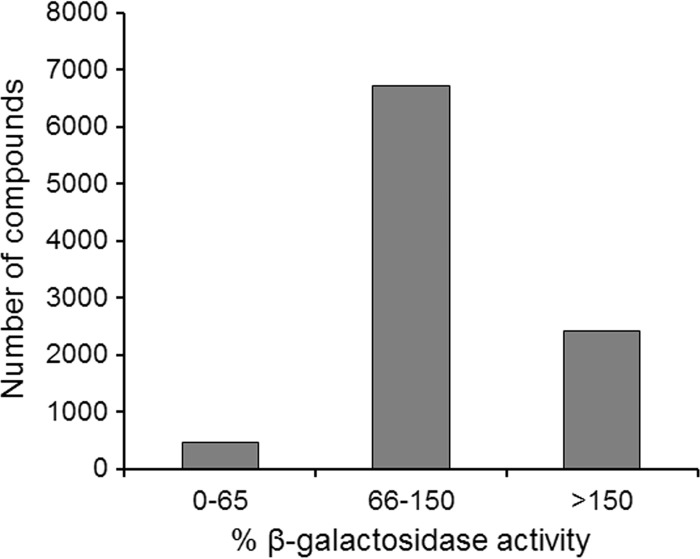
The screening strategy was based on determination of β-galactosidase activity in a bacterial 2-hybrid interaction assay, where the VipA-VipB binding was analyzed (44, 54). A 9,600-compound library was screened at the concentration of 50 μM for the ability to inhibit β-galactosidase activity (54). The primary hits of the screening were 470 compounds that showed more than a 35% inhibition of the β-galactosidase activity (Fig. 1), representing a hit rate of approximately 4.9%.
FIG 1.
Relative effects on the VipA-VipB interaction of the 9,600 compounds analyzed. The activity of the samples was assayed using the B2H assay and expressed as percentages relative to the samples with DMSO only (100%).
Down-selection of candidate compounds.
The 470 compounds were screened for inhibitory activity on the growth of V. cholerae. Moreover, the effect of the compounds on the unrelated B2H interaction of the Francisella tularensis proteins SspA/MglA (44) was investigated to remove compounds with nonspecific inhibition of the B2H assay. This down-selection led to the identification of 34 hits that inhibited the VipA-VipB B2H interaction but showed no effects in the other assays.
Cytotoxic effects exerted by down-selected compounds.
The down-selected compounds were further analyzed for cytotoxicity when added to the macrophage-like cell line J774, since it was hypothesized that marked cell cytotoxicity would be an undesirable feature. Addition of the selected compounds to cultures of J774 macrophages was performed, and LDH release was measured to evaluate compound cytotoxicity. No marked differences were observed after 24 h of incubation between J774 macrophages treated with KS100 and the effect of DMSO only, whereas KS200 resulted in somewhat elevated LDH levels (see Fig. S1 in the supplemental material). Microscopic examination of the morphology of the treated cells was also performed, and no or only slightly deviating morphology was observed in relation to the cells with the addition of DMSO only (see Table S3 in the supplemental material).
Growth inhibition effects on Gram-negative bacteria.
To investigate whether KS100 or KS200 (50 μM) showed any effects on bacterial growth, they were added to 11 Gram-negative bacterial strains, each representing different species, and were examined in broth. No significant growth inhibitory effects were observed at the 18-h time point (P > 0.05) compared to the control with DMSO only (see Fig. S2 in the supplemental material).
Based on the aforementioned criteria, the KS100 and KS200 compounds were identified as the most useful in view of their potent VipA-VipB binding inhibition, no nonspecific B2H inhibitory activity, lack of bactericidal activity and growth inhibition, as well as low cytotoxic properties. It should be noted that KS200 induced intermediate levels of cytotoxicity; however, since it showed very potent inhibition of T6SS effector functions, it was still selected, as we hypothesized that analogues could be identified that still showed potent T6S inhibition but were less cytotoxic.
Selection of analogues to KS100 and KS200.
Based on the structures of the down-selected compounds KS100 and KS200, commercially available structural analogues were obtained and evaluated with regard to physicochemical properties and behavior in several biological assays. Analogues were selected based on their structural similarity to KS100 with regard to the heterocyclic dibenzothiophene sulfone parental scaffold. Analogue structural variations included hetero atom variation in the scaffold, different oxidation states of the scaffold sulfur, and substituents on the dibenzothiophene of various size and electronic properties. Analogues selected for KS200 all had the central benzoxazole scaffold with various phenyl-substituents in the 2 position and mono- and biphenylic substituents on the imine in position 5. The original imine was exchanged for an amine in some of the KS200 analogues. In total, 17 analogues of the KS100 series and 12 of the KS200 series were obtained. All compounds were characterized by the calculation of physicochemical properties such as lipophilicity (hydrophobicity), solubility, molecular weight, and permeability, and they basically followed the rules of Lipinski et al. and Veber et al. (55, 56).
According to in silico calculations, the octanol/water partition coefficients (logP) of a subset of the compounds KS100, KS103, KS105, KS107, KS108, KS112, and KS116 were in the ideal range with a logP of <3, i.e., low lipophilicity (see Table S3 in the supplemental material). The most water-soluble compounds, i.e., logS of >−4, were KS103, KS105, KS108, KS110, KS111, KS112, KS115, and KS116, most of which also showed low logP values (see Table S3). The values for membrane permeability in Caco-2 and MDCK cells showed high correlation, and all of the analogues in the KS200 series showed very high permeability values, >2,400, whereas values for the KS100 series analogues were much lower (see Table S3). Still, nine of them showed high membrane permeability values, >500, and KS107, KS111, K115, and KS116 all had values of >1,500 for both Caco-2 and MDCK permeability. All of the analogues had substantial solvent accessible surface areas (SASA), between 430 and 930 (see Table S3).
In summary, the analogues showed markedly distinct predicted physicochemical properties since several of the KS100 analogues showed low logP and high logS values, whereas many of them had rather low membrane permeability values. There were some exceptions, however, and, e.g., KS116 was among the compounds with low logP and high logS values but still very high permeability values. In contrast, none of the compounds of the KS200 series showed low logP and high logS values, but all had very high permeability values.
Cytotoxic properties and growth inhibition effects exerted by the KS100 and KS200 analogues.
The effects on eukaryotic cells of the compounds were tested by adding them to J774 cells and following the release of LDH and cell morphology as a measure of cell cytotoxicity. Among the analogues, addition of KS102, KS110, KS203, KS205, KS207, and KS208 resulted in high values, >29.5%, of released LDH compared to that for DMSO only, 12.6% (see Fig. S1 and Table S3 in the supplemental material; P < 0.05), whereas the addition of KS101, KS103, KS104, KS105, KS106, KS107, KS109, and KS114 did not result in any significant differences compared to cultures with DMSO only, and the LDH levels were <15% but in abnormal morphologies (i.e., rounding up) of the cells as judged by microscopy (see Table S3). In contrast, the addition of KS108, KS111, KS112, KS113, KS115, KS116, KS117, KS209, or KS210 resulted in low LDH release, <15% (see Fig. S1 and Table S3) and no obvious microscopic effect on J774 cells (see Table S3). The effects on growth of 12 Gram-negative strains, each representing a unique species of the analogues, were also examined. None of the compounds induced any significant growth inhibition at the 18-h time point (see Fig. S2) or at any earlier time point.
Effects of compounds on the VipA-VipB interaction of V. cholerae, the AaiA-AaiB interaction of E. coli, and the HsiB2-HsiC2 interaction of P. aeruginosa as assessed by the B2H assay.
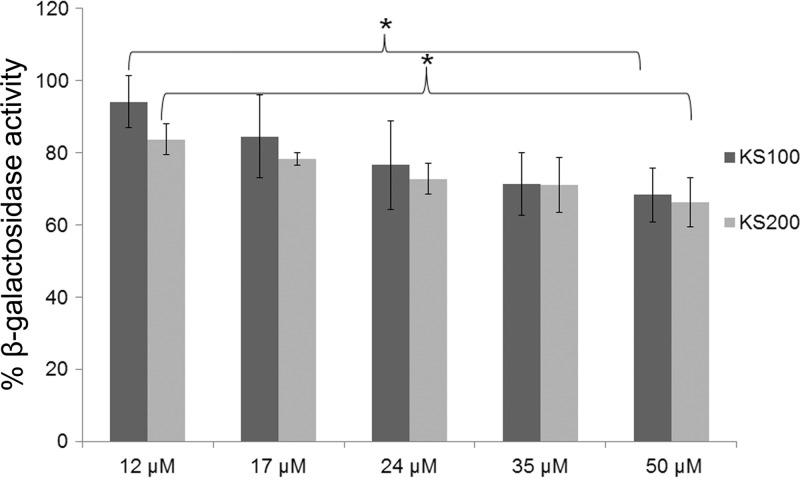
To investigate whether the VipA-VipB interaction was affected in a dose-dependent fashion by the down-selected compounds KS100 and KS200, they were serially diluted and the β-galactosidase activity of the B2H was measured (Fig. 2). A significant difference in β-galactosidase activity was observed between the samples with the lowest concentration, 12 μM, versus the highest, 50 μM (P < 0.05). The values recorded were somewhat higher than those observed in the original screening procedure (Fig. 1). The analogues were also included in the B2H analysis and tested at a concentration of 50 μM. Most of the compounds showed values in the range of 60 to 90%, although K206 and KS208 showed values of approximately 50%, similar to those of KS100 and KS200 (see Fig. S3). However, some analogues showed distinctly higher values of >90%, i.e., little or no inhibition, in particular KS108 but also KS110, KS111, KS112, KS209, KS210, KS211, and KS212.
FIG 2.
Dose-dependent inhibition of the VipA-VipB interaction. Serial dilutions of KS100 and KS200 were assayed in the B2H system, and values of β-galactosidase activity are shown as percentages relative to samples with DMSO only. Data are presented as means ± standard deviation (SD) from three wells from one representative experiment of three. *, P < 0.05.
To analyze if the compounds affected also homologous B2H interactions, the effects on the interaction of the VipA-VipB analogues, AaiA-AaiB of E. coli and HsiB2-HsiC2 of P. aeruginosa, were investigated. The compounds showed variable effects on the B2H interactions, and only KS104 showed a significant inhibition of all three B2H assays at a concentration of 50 μM, P < 0.05 (see Fig. S3).
Effects on T6SS-dependent Hcp secretion, phospholipase activity, and the interbacterial competition assay exerted by the KS100 and KS200 compounds and analogues.
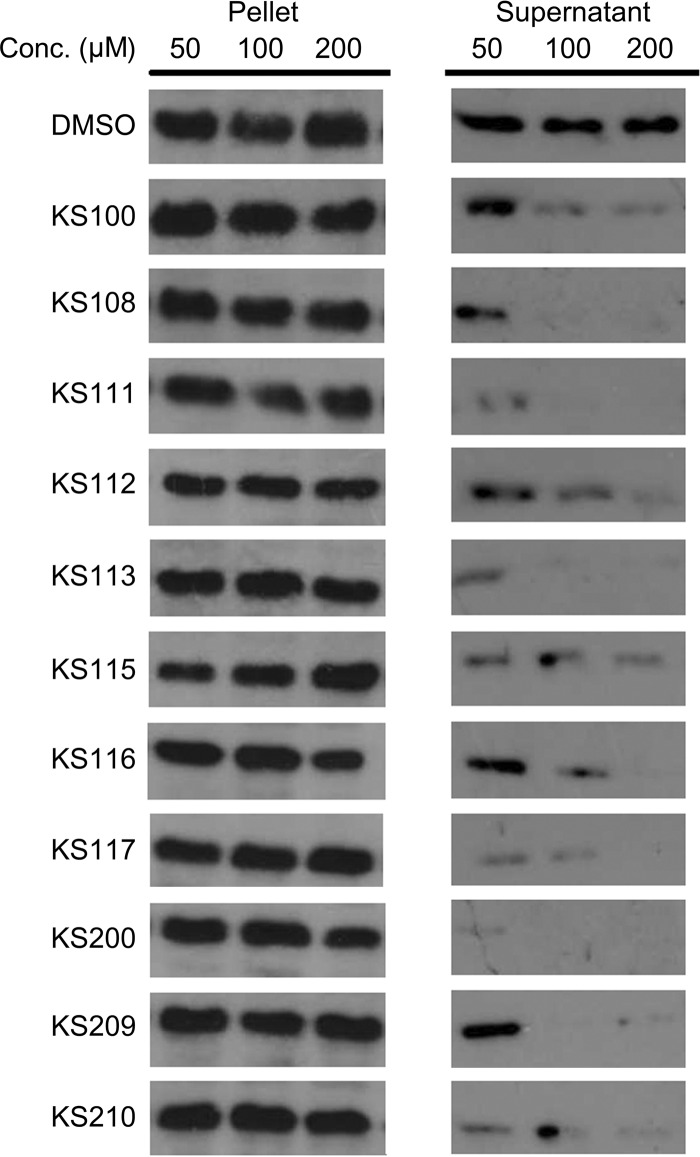
The selected compounds were next tested to determine how they affected T6SS functions. To this end, the secretion of the very-well-characterized T6SS effector, Hcp, was analyzed. A normal VipA-VipB interaction is an absolute requirement for Hcp secretion since VipA variants that are unable to bind to VipB efficiently do not support secretion of Hcp (26, 34). KS100, KS200, and the analogues were serially diluted to final concentrations of 50 μM, 100 μM, or 200 μM and incubated with V. cholerae V52 for 4 h. Extracts from the cells and culture supernatants were examined by Western blotting, and a dose-dependent inhibition of Hcp secretion was observed (Fig. 3). At 200 μM, the Hcp secretion was essentially eliminated by many compounds (see Table S3). KS100, KS200, and the low cytotoxic analogues all showed complete inhibition at 200 μM and marked inhibition at 100 μM (Fig. 3).
FIG 3.
Dose-dependent inhibition of the Hcp secretion. The V. cholerae strain V52 was incubated with either DMSO as the control or selected compounds at 50, 100, or 200 μM. Extracts from the cells and culture supernatants were separated by SDS-PAGE and examined by Western blotting using rabbit antiserum against Hcp.
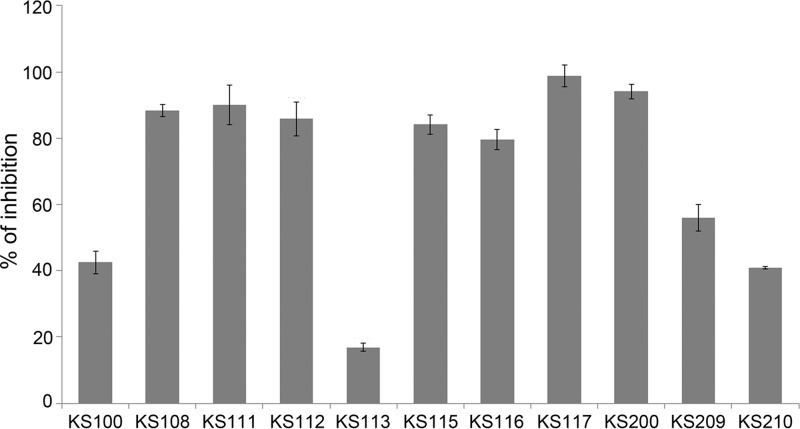
Phospholipase A1 function, another T6SS-dependent effector mechanism, was also investigated as a parameter of the activity of the compounds by measuring its activity in the supernatant (40). KS100 showed significant inhibition, 42.5%, and KS200 very potent inhibition, 94.1%, compared with that of the DMSO control (Fig. 4; see also Table S3 in the supplemental material). Among the analogues, a majority showed >50% inhibition, and they were particularly common among the KS100 analogues, and, notably, KS101, KS102, KS104, KS108, KS110, KS111, KS112, KS115, KS116, and KS117 all showed values of >80%. The effects of the low cytotoxic compounds are presented in Fig. 4. Among these, KS108, KS111, KS112, KS115, KS116, and KS117 showed marked inhibition of both phospholipase A1 activity and Hcp secretion.
FIG 4.
Inhibition of phospholipase A1 secretion. Selected compounds at a concentration of 50 μM were tested for inhibition of phospholipase A1 activity and expressed as percentage inhibition relative to samples with DMSO only. Experiments were carried out in triplicates, and the means ± SD are shown from one representative experiment of three. The P values were all <0.001.
The compounds were also investigated for their ability to interfere in an interbacterial competition assay, killing of E. coli effectuated by V. cholerae. The killing was assessed by estimating the density of E. coli bacteria growing on the antibiotic-selective agar plates. None of the compounds significantly affected the killing of E. coli (data not shown).
SAR of KS100 and KS200 analogues.
To evaluate the effects of the down-selected compounds and the analogues on T6SS, the structure-activity relationship (SAR) was focused on the most widely used assay with regard to T6S function, Hcp secretion. In the KS100 set of analogues, smaller and less lipophilic compounds generally showed prominent inhibition of Hcp secretion, and three compounds, i.e., KS103, KS105, and KS108, displayed very low logP and a stronger inhibition than the parental compound KS100, and two, KS112 and KS116, showed a logP of <3.0 and an inhibition as strong as that of the parental compound (see Table S3). Although no consistent conclusions could be drawn regarding the optimal substitution pattern on the parental scaffold from this limited set of analogues, some trends could be identified. An oxidization of the sulfur appeared to be slightly preferable over nonoxidization in dibenzothiophenes with phenylic substituents (cf. KS105 and KS109). There were no dramatic differences in activity between the 3,7- and 2,8-substituted dibenzothiophene scaffolds, and various substitution patterns on the parental heterocyclic dibenzothiophene sulfone framework (KS105, KS109, KS113, KS115, and KS117), as well as on dibenzofuran scaffolds (KS112 and KS116) and carbazole scaffolds (KS110 and KS111), resulted in active compounds. This feature gives flexibility to tune properties like solubility, toxicity, and membrane permeability in a future potential lead optimization effort. It was also of interest that the symmetry of the KS100 class was not essential for greater activity, as evidenced by the potent inhibition displayed by the asymmetrical compounds KS111, KS115, KS116, and KS117. Notably, asymmetrical compounds displayed lower cell toxicity than most symmetrical compounds.
The KS200 analogues generally displayed a lower inhibitory capacity of the Hcp secretion than the KS100 analogues. Exceptions were compounds KS210 and KS212, although the former compound showed substantial cytotoxicity. The SAR analysis of the KS200 compounds indicated that there is an intricate relation between activity and cytotoxicity among these compounds. Most of the least toxic compounds, i.e., KS209, KS210, and KS211, have a common chemical structure with variation in the position of the substituent on the phenyl in the 2 position on the benzoxazole scaffold. Here, a meta-substituent, i.e., m-methyl in KS210, seemed to exert more potent inhibition. The other meta-substituent compound, KS204 (m-iodo), also showed some inhibitory effect but increased cytotoxicity compared to that of KS210, which we hypothesize is due to the missing o-chloro on the benzoate moiety. KS210 showed good cell permeability capacity but poor water solubility according to the in silico calculations.
DISCUSSION
The role of T6SS for cholera pathogenesis is intriguing and by far not fully explored. Although the cholera toxin is the principal pathogenetic mechanism and essential for the clinical manifestations of classical cholera (57, 58), there is accumulating evidence that T6SS is an important adjunct virulence mechanism. For example, secretion of Hcp and VgrG contributes to the diarrhea and facilitates bacterial replication in a mouse model of cholera (39). The contribution of T6SS in this regard may be dependent on its central role in interbacterial competition, since this should be a very important virulence mechanism in complex bacterial communities, such as the gastrointestinal tract (41). In addition, there is evidence that T6SS targets the intestinal epithelium and thereby modulates the inflammatory response such that bacterial survival will be favored (33). Thus, inhibition of T6SS may adversely affect the virulence of gastrointestinal pathogens by impairing their interbacterial competitiveness as well as their ability to modulate the local immune response.
There are several features that make the VipA-VipB interaction of T6SS an attractive target for antimicrobial therapy. One is the presence of the two genes in all T6SS clusters discovered so far, and another one is the high degree of conservation in their interaction, as evidenced by the effective binding of several of the homologous proteins with cognate partners from other bacterial species (2, 4, 26). Thus, there is reason to believe that compounds that inhibit the interaction in one species may also confer effects on the interaction in other species. Moreover, the available evidence indicates that the interaction is an essential prerequisite for the T6SS effector mechanisms. Thus, effective inhibition of the interaction of VipA-VipB and their homologues has the potential to abrogate T6SS functions in a wide variety of pathogenic bacteria.
In the present study, we developed a high-throughput screening assay for small-molecule inhibitors of VipA-VipB binding in V. cholerae based on a B2H protein-protein interaction assay. The strategy to use B2H as a screening method in this context has only rarely been implemented (59), but, if successful, it should result in the identification of highly specific compounds in view of the strict prerequisites of the assay. It was anticipated that the strategy would be affected by several caveats; protein-protein interactions are generally highly specific, and, therefore, there will be very strict requirements for small-molecule inhibitors. Further, since it is assumed that there is an intracellular localization of the proteins, the inhibitors need to be highly membrane diffusible. As a reflection of these caveats, even though our original screen identified some 5% of the compounds as inhibitory, the subsequent analysis excluded a high proportion of these, and only a few compounds were finally down-selected. In conjunction to the screening procedure, we also tested if the identified compounds prevented bacterial replication of V. cholerae and other clinically important Gram-negative bacteria, and those that showed inhibition were excluded from further characterization. Thereby, at least from a theoretical standpoint, this will minimize the risk of development of antibiotic resistance against such compounds.
The VipA-VipB interaction is an absolute prerequisite for the function of the T6S, and, thus, it would be postulated that the inhibition of the interaction will lead to the abrogation of Hcp secretion or phospholipase activity, both shown to be important T6SS effector mechanisms (28, 34). In fact, numerous studies have demonstrated not only a close link between T6SS and bacterial secretion, necessary for virulence, but also the structural roles of VipA, VipB, and Hcp in the needle-like structure required for secretion (2, 4, 13, 14, 27–29, 34, 39, 53, 60). Our work based on the analogues of the two parental compounds demonstrated, however, that some preferentially inhibited one or the other of the assays. Thus, even if the VipA-VipB interaction without doubt is required for T6S, it is possible that less potent inhibitors of the interaction may still allow sufficient function for the subsequent activation of the machinery and, hence, Hcp secretion or phospholipase activity. Moreover, we also observed that the compounds showed variable effects on the VipA-VipB homologous interactions of AaiA-AaiB and HsiB2-HsiC2, indicating that the interactions are to some degree distinct. The two parental compounds KS100 and KS200 were selected since they showed potent inhibition of both VipA-VipB interaction as well as Hcp secretion. The B2H assay was performed using an E. coli strain, thereby minimizing the possibility that the compounds identified would exert activities that target other essential functions in V. cholerae.
The physical properties of the compounds resulted in some constraints when performing the assays; in particular, the limited solubility prevented the possibility to perform testing of the potency of inhibition in the B2H assay over a wide range of concentrations and, therefore, the half maximal inhibitory concentration could not be calculated. Our data showed, however, that the inhibitory effects of these compounds on the VipA-VipB B2H assay were dose dependent below 100 μM and on Hcp secretion below 500 μM.
Our analysis of the structure and activity relationship of the parental compounds, KS100 and KS200, and their analogues allowed us to make certain inferences regarding the structural constraints of the biological activities. Of the KS100 analogues, smaller and less lipophilic compounds showed the most prominent Hcp secretion inhibition. Notably, the dibenzothiophene sulfone structure of KS100 was possible to modify in multiple ways, including the addition of heteroaromatic moieties like dibenzothiophenes, dibenzofurans, and carbazoles, with preserved activity, and it was observed that asymmetrical compounds displayed lower cell toxicity than symmetrical ones. Fewer inferences were possible to make regarding the KS200 analogues, but the important, albeit unwanted, characteristic of cell cytotoxicity was minimized when compounds had a common 2-methoxy-4-[(1-oxa-5-indenylimino)methyl]phenyl 2,4-dichlorobenzoate structure, despite that their substituents varied.
The KS200 analogues, besides that a majority showed marked cytotoxicity, displayed high membrane permeability and were markedly hydrophobic and showed intermediate inhibition of the Hcp secretion and phospholipase activity. The KS100 analogues, in contrast, showed much more diverse activity spectra. A majority showed minimal cytotoxicity, and this was correlated to intermediate or low hydrophobic properties. Many of these displayed prominent inhibition of Hcp secretion and phospholipase activity, and some, e.g., KS112 and KS116, also showed high membrane permeability.
In summary, we established a high-throughput screening assay for small-molecule inhibitors targeting the interaction of VipA and VipB, which is essential for the T6SS of V. cholerae, and we demonstrated that KS100, KS200, and some of their analogues are effective to inhibit V. cholerae T6SS effector mechanisms. We were able to make certain inferences regarding the structure-activity relationship of the compounds; however, the exact molecular prerequisites of these inhibitors and how the effector functions are inhibited will require additional structural analyses.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants 2006-3426, 2006-2877, 2009-5026, and 2013-4581 from the Swedish Research Council and a stipend from the Knut and Alice Wallenberg foundation to K.S. and a grant from the Medical Faculty, Umeå University, Umeå, Sweden. Support was also obtained from the Chemical Biology Consortium of Sweden. The work was performed in part at the Umeå Centre for Microbial Research (UCMR).
We thank Sun Nyunt Wai, Department of Molecular Biology, Umeå University, for advice regarding the phospholipase A1 assay.
Footnotes
Published ahead of print 5 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02819-13.
REFERENCES
- 1.WHO. 2010. Cholera fact sheet. WHO, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs107/en/index.htm [Google Scholar]
- 2.Bingle LE, Bailey CM, Pallen MJ. 2008. Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 11:3–8. 10.1016/j.mib.2008.01.006 [DOI] [PubMed] [Google Scholar]
- 3.Wang M, Luo Z, Du H, Xu S, Ni B, Zhang H, Sheng X, Xu H, Huang X. 2011. Molecular characterization of a functional type VI secretion system in Salmonella enterica serovar Typhi. Curr. Microbiol. 63:22–31. 10.1007/s00284-011-9935-z [DOI] [PubMed] [Google Scholar]
- 4.Bröms JE, Lavander M, Sjöstedt A. 2009. A conserved alpha-helix essential for a type VI secretion-like system of Francisella tularensis. J. Bacteriol. 191:2431–2446. 10.1128/JB.01759-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blondel CJ, Jimenez JC, Contreras I, Santiviago CA. 2009. Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics 10:354. 10.1186/1471-2164-10-354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burtnick MN, Brett PJ, Harding SV, Ngugi SA, Ribot WJ, Chantratita N, Scorpio A, Milne TS, Dean RE, Fritz DL, Peacock SJ, Prior JL, Atkins TP, Deshazer D. 2011. The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect. Immun. 79:1512–1525. 10.1128/IAI.01218-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blondel CJ, Jimenez JC, Leiva LE, Alvarez SA, Pinto BI, Contreras F, Pezoa D, Santiviago CA, Contreras I. 2013. The type VI secretion system encoded in Salmonella pathogenicity island 19 is required for Salmonella enterica serotype Gallinarum survival within infected macrophages. Infect. Immun. 81:1207–1220. 10.1128/IAI.01165-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cascales E. 2008. The type VI secretion toolkit. EMBO Rep. 9:735–741. 10.1038/embor.2008.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blondel CJ, Yang HJ, Castro B, Chiang S, Toro CS, Zaldivar M, Contreras I, Andrews-Polymenis HL, Santiviago CA. 2010. Contribution of the type VI secretion system encoded in SPI-19 to chicken colonization by Salmonella enterica serotypes Gallinarum and Enteritidis. PLoS One 5:e11724. 10.1371/journal.pone.0011724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Pace F, Nakazato G, Pacheco A, de Paiva JB, Sperandio V, da Silveira WD. 2010. The type VI secretion system plays a role in type 1 fimbria expression and pathogenesis of an avian pathogenic Escherichia coli strain. Infect. Immun. 78:4990–4998. 10.1128/IAI.00531-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H, Coulthurst SJ, Pritchard L, Hedley PE, Ravensdale M, Humphris S, Burr T, Takle G, Brurberg MB, Birch PR, Salmond GP, Toth IK. 2008. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog. 4:e1000093. 10.1371/journal.ppat.1000093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng J, Leung KY. 2007. Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 66:1192–1206. 10.1111/j.1365-2958.2007.05993.x [DOI] [PubMed] [Google Scholar]
- 13.Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37. 10.1016/j.chom.2009.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell AB, Singh P, Brittnacher M, Bui NK, Hood RD, Carl MA, Agnello DM, Schwarz S, Goodlett DR, Vollmer W, Mougous JD. 2012. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 11:538–549. 10.1016/j.chom.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng J, Ho B, Mekalanos JJ. 2011. Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One 6:e23876. 10.1371/journal.pone.0023876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. 2010. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. U. S. A. 107:19520–19524. 10.1073/pnas.1012931107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. 2010. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 6:e1001068. 10.1371/journal.ppat.1001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coulthurst SJ. 2013. The Type VI secretion system—a widespread and versatile cell targeting system. Res. Microbiol. 6:640–654. 10.1016/j.resmic.2013.03.017 [DOI] [PubMed] [Google Scholar]
- 19.Schwarz S, Hood RD, Mougous JD. 2010. What is type VI secretion doing in all those bugs? Trends Microbiol. 18:531-537. 10.1016/j.tim.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, Coulthurst SJ. 2011. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J. Bacteriol. 193:6057–6069. 10.1128/JB.05671-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapitein N, Bönemann G, Pietrosiuk A, Seyffer F, Hausser I, Locker JK, Mogk A. 2013. ClpV recycles VipA/VipB tubules and prevents nonproductive tubule formation to ensure efficient type VI protein secretion. Mol. Microbiol. 87:1013–1028. 10.1111/mmi.12147 [DOI] [PubMed] [Google Scholar]
- 22.Jani AJ, Cotter PA. 2010. Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe 8:2–6. 10.1016/j.chom.2010.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lossi NS, Manoli E, Forster A, Dajani R, Pape T, Freemont P, Filloux A. 2013. The HsiB1C1 (TssB-TssC) complex of the Pseudomonas aeruginosa type VI secretion system forms a bacteriophage tail sheathlike structure. J. Biol. Chem. 288:7536–7548. 10.1074/jbc.M112.439273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Bruin OM, Duplantis BN, Ludu JS, Hare RF, Nix EB, Schmerk CL, Robb CS, Boraston AB, Hueffer K, Nano FE. 2011. The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins IglA, IglB, IglC, PdpB and DotU suggest roles in type VI secretion. Microbiology 157:3483–3491. 10.1099/mic.0.052308-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bröms JE, Sjöstedt A, Lavander M. 2010. The role of the Francisella tularensis pathogenicity island in type VI secretion, intracellular survival, and modulation of host cell signaling. Front. Microbiol. 1:136. 10.3389/fmicb.2010.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bröms JE, Ishikawa T, Wai SN, Sjöstedt A. 2013. A functional VipA-VipB interaction is required for the type VI secretion system activity of Vibrio cholerae O1 strain A1552. BMC Microbiol. 13:96. 10.1186/1471-2180-13-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186. 10.1038/nature10846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bönemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. 2009. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 28:315–325. 10.1038/emboj.2008.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pietrosiuk A, Lenherr ED, Falk S, Bönemann G, Kopp J, Zentgraf H, Sinning I, Mogk A. 2011. Molecular basis for the unique role of the AAA+ chaperone ClpV in type VI protein secretion. J. Biol. Chem. 286:30010–30021. 10.1074/jbc.M111.253377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bröms JE, Meyer L, Lavander M, Larsson P, Sjöstedt A. 2012. DotU and VgrG, core components of type VI secretion systems, are essential for Francisella tularensis LVS pathogenicity. PLoS One 7:e34639. 10.1371/journal.pone.0034639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Maayer P, Venter SN, Kamber T, Duffy B, Coutinho TA, Smits TH. 2011. Comparative genomics of the type VI secretion systems of Pantoea and Erwinia species reveals the presence of putative effector islands that may be translocated by the VgrG and Hcp proteins. BMC Genomics 12:576. 10.1186/1471-2164-12-576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hachani A, Lossi NS, Hamilton A, Jones C, Bleves S, Albesa-Jove D, Filloux A. 2011. Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. J. Biol. Chem. 286:12317–12327. 10.1074/jbc.M110.193045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartonickova L, Sterzenbach T, Nell S, Kops F, Schulze J, Venzke A, Brenneke B, Bader S, Gruber AD, Suerbaum S, Josenhans C. 2013. Hcp and VgrG1 are secreted components of the Helicobacter hepaticus type VI secretion system and VgrG1 increases the bacterial colitogenic potential. Cell Microbiol. 15:992–1011. 10.1111/cmi.12094 [DOI] [PubMed] [Google Scholar]
- 34.Ishikawa T, Sabharwal D, Bröms J, Milton DL, Sjöstedt A, Uhlin BE, Wai SN. 2012. Pathoadaptive conditional regulation of the type VI secretion system in Vibrio cholerae O1 strains. Infect. Immun. 80:575–584. 10.1128/IAI.05510-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U. S. A. 104:15508–15513. 10.1073/pnas.0706532104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. 2009. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U. S. A. 106:4154–4159. 10.1073/pnas.0813360106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530. 10.1126/science.1128393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silverman JM, Agnello DM, Zheng H, Andrews BT, Li M, Catalano CE, Gonen T, Mougous JD. 2013. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol. Cell 51:584–593. 10.1016/j.molcel.2013.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pukatzki S, McAuley SB, Miyata ST. 2009. The type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12:11–17. 10.1016/j.mib.2008.11.010 [DOI] [PubMed] [Google Scholar]
- 40.Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, Mougous JD. 2013. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496:508–512. 10.1038/nature12074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alteri CJ, Himpsl SD, Pickens SR, Lindner JR, Zora JS, Miller JE, Arno PD, Straight SW, Mobley HL. 2013. Multicellular bacteria deploy the type VI secretion system to preemptively strike neighboring cells. PLoS Pathog. 9:e1003608. 10.1371/journal.ppat.1003608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho BT, Basler M, Mekalanos JJ. 2013. Type 6 secretion system-mediated immunity to type 4 secretion system-mediated gene transfer. Science 342:250–253. 10.1126/science.1243745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vallet-Gely I, Donovan KE, Fang R, Joung JK, Dove SL. 2005. Repression of phase-variable cup gene expression by H-NS-like proteins in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 102:11082–11087. 10.1073/pnas.0502663102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Charity JC, Costante-Hamm MM, Balon EL, Boyd DH, Rubin EJ, Dove SL. 2007. Twin RNA polymerase-associated proteins control virulence gene expression in Francisella tularensis. PLoS Pathog. 3:e84. 10.1371/journal.ppat.0030084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jorgensen WL, Duffy EM. 2000. Prediction of drug solubility from Monte Carlo simulations. Bioorg. Med. Chem. Lett. 10:1155–1158. 10.1016/S0960-894X(00)00172-4 [DOI] [PubMed] [Google Scholar]
- 46.Duffy EM, Jorgensen WL. 2000. Prediction of properties from simulations: free energies of solvation in hexadecane, octanol, and water. J. Am. Chem. Soc. 122:2878–2888. 10.1021/ja993663t [DOI] [Google Scholar]
- 47.Mohamadi F, Richards NGJ, Guida WC, Liskamp R, Lipton M, Caufield C, Chang G, Hendrickson T, Still WC. 1990. Macromodel—an integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J. Comp. Chem. 11:440–467. 10.1002/jcc.540110405 [DOI] [Google Scholar]
- 48.Banks JL, Beard HS, Cao Y, Cho AE, Damm W, Farid R, Felts AK, Halgren TA, Mainz DT, Maple JR, Murphy R, Philipp DM, Repasky MP, Zhang LY, Berne BJ, Friesner RA, Gallicchio E, Levy RM. 2005. Integrated modeling program, applied chemical theory (IMPACT). J. Comput. Chem. 26:1752–1780. 10.1002/jcc.20292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ooi T, Oobatake M, Nemethy G, Scheraga HA. 1987. Accessible surface areas as a measure of the thermodynamic parameters of hydration of peptides. Proc. Natl. Acad. Sci. U. S. A. 84:3086–3090. 10.1073/pnas.84.10.3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polak E, Ribiere G. 1969. Note sur la convergence de méthodes de directions conjuguées. ESAIM 3:35–43 [Google Scholar]
- 51.Andersson EK, Strand M, Edlund K, Lindman K, Enquist PA, Spjut S, Allard A, Elofsson M, Mei YF, Wadell G. 2010. Small-molecule screening using a whole-cell viral replication reporter gene assay identifies 2-{[2-(benzoylamino)benzoyl]amino}-benzoic acid as a novel antiadenoviral compound. Antimicrob. Agents Chemother. 54:3871–3877. 10.1128/AAC.00203-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bröms JE, Meyer L, Sun K, Lavander M, Sjöstedt A. 2012. Unique substrates secreted by the type VI secretion system of Francisella tularensis during intramacrophage infection. PLoS One 7:e50473. 10.1371/journal.pone.0050473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ishikawa T, Rompikuntal PK, Lindmark B, Milton DL, Wai SN. 2009. Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS One 4:e6734. 10.1371/journal.pone.0006734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dove SL, Hochschild A. 2004. A bacterial two-hybrid system based on transcription activation. Methods Mol. Biol. 261:231–246. 10.1385/1-59259-762-9:231 [DOI] [PubMed] [Google Scholar]
- 55.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. 2001. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46:3–26. 10.1016/S0169-409X(00)00129-0 [DOI] [PubMed] [Google Scholar]
- 56.Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. 2002. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 45:2615–2623. 10.1021/jm020017n [DOI] [PubMed] [Google Scholar]
- 57.Kaper JB, Morris JG, Jr, Levine MM. 1995. Cholera. Clin. Microbiol. Rev. 8:48–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanchez J, Holmgren J. 2011. Cholera toxin—a foe & a friend. Indian J. Med. Res. 133:153–163 [PMC free article] [PubMed] [Google Scholar]
- 59.Wrench AP, Gardner CL, Siegel SD, Pagliai FA, Malekiha M, Gonzalez CF, Lorca GL. 2013. MglA/SspA complex interactions are modulated by inorganic polyphosphate. PLoS One 8:e76428. 10.1371/journal.pone.0076428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U. S. A. 103:1528–1533. 10.1073/pnas.0510322103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.