Abstract
The respiratory syncytial virus (RSV) L protein is a viral RNA-dependent RNA polymerase that contains multiple enzyme activities required for RSV replication. The RSV L inhibitors described in literature are limited by their cytotoxicity or the lack of RSV B subtype coverage. Here, we characterize a new RSV L inhibitor with strong antiviral activity against both RSV A and B subtypes and no detectable cytotoxicity. This compound, AZ-27, was equally active against RSV live viruses and subgenomic replicons and demonstrated advantages over other classes of RSV inhibitors in time-of-addition and cell line dependency studies. Resistance studies identified a dominant mutation in the putative capping enzyme domain of L protein, which conferred strong resistance to the AZ-27 series but not other classes of RSV inhibitors, supporting RSV L protein as the direct target for AZ-27. This novel and broad-spectrum RSV L polymerase inhibitor may pave the way toward an efficacious RSV therapeutic and provide a new tool for interrogation of the L protein function.
INTRODUCTION
Respiratory syncytial virus (RSV) is an enveloped, nonsegmented negative-sense RNA virus in the Paramyxoviridae family. RSV infection is ubiquitous in that virtually everyone is infected by the age of 2 years and reinfection occurs throughout all ages. It is the leading cause of acute lower respiratory tract infections in young children, the elderly, and immunosuppressed patients (1). Progress has been made toward vaccine development, but many challenges remain, as highlighted by the short-lived natural immune response against RSV with high reinfection rate, the difficulty in eliciting a protective immune response in neonates, and the unexpected enhancement of disease by RSV vaccination observed in the formalin-inactivated RSV vaccine trial (2). Immunoprophylaxis with RSV-neutralizing antibodies has been successful in protecting high-risk infants and children. However, there is no RSV-specific therapy available for postinfection treatment, and RSV continues to be the number one reason for infant hospitalization (3). The only approved treatment for RSV is ribavirin, which has limited clinical utility due to its high toxicity and controversial efficacy (4). Therefore, finding an effective treatment for RSV infection remains an important public health priority.
The limited understanding of the molecular mechanisms of RSV replication and pathogenesis has hampered the development of RSV therapeutics (5). RSV replication requires the viral RNA genome, mRNAs, 11 viral proteins, and many host factors, all of which are potential targets for therapeutic intervention. Targeting host factors holds the promise of broader-spectrum coverage and a potentially higher barrier to resistance. However, there may also be on-target toxicity, the adverse pharmacologic effect of interfering with a cellular target important for host function, which would be of particular concern in treating young infants, the main population affected by severe RSV diseases. Antivirals directly targeting viral proteins with no close human homolog may prove advantageous in mitigating this safety risk. Most previous development of RSV drugs has been focused on RSV fusion inhibitors and failed to progress beyond phase I-II clinical trials (4). A small interfering RNA (siRNA) agent targeting RSV nucleoprotein (N) mRNA was recently advanced to phase II trials; however, it did not meet the primary clinical endpoint of reduced bronchiolitis obliterans (4). Development of the RSV inhibitor RSV604, targeting N protein at postentry steps, was halted after a phase II clinical trial for undisclosed reasons. In vitro data demonstrated that RSV604 maintained potency across a wider range of times of addition relative to infection than did known fusion inhibitors (6), raising the question of whether targeting viral replication may be more advantageous for the short treatment window associated with acute viral respiratory diseases.
A functional RSV RNA replication complex requires four viral proteins: the large protein (L), phosphoprotein (P), matrix 2-1 (M2-1), and N. The required enzymatic activities are primarily associated with L, making it an attractive drug target (7). L functions as the RNA-dependent RNA polymerase to replicate the viral RNA genome and transcribe mRNAs, the capping enzyme to cap the mRNA 5′ end, and the methylase to methylate the cap. Six conserved regions in L have been identified across the nonsegmented negative-sense RNA virus family and were implicated in the individual enzymatic activities (7). The predicted structural and functional domains of RSV L have not been directly demonstrated due to the challenges of recombinant protein production and biochemical assay development for this very large protein (250 kDa) (8). Recent progress has been made with developing an in vitro RSV L polymerase assay but not yet with the assay for the L capping enzyme (9), which is particularly complex and understudied. It is likely to be a polyribonucleotidyltransferase, similar to that found in vesicular stomatitis virus, which mediates unusual capping. Whether it also possesses RNA triphosphatase and guanylyltransferase activities to form a eukaryote-like cap, as reported for the rinderpest virus, remains to be determined (10). Novel L inhibitors could serve as useful chemical biology tools to dissect the domains and functions of L protein.
Viral RNA and DNA polymerases are among the most common and successful targets for many antiviral therapies. Three classes of RSV L inhibitors have been reported to date. Ribavirin is a nucleoside analog that has been suggested to inhibit the polymerases of many DNA/RNA viruses, including RSV (4). The Boehringer Ingelheim benzimidazole series of L inhibitors, exemplified by compound D (BI cpd D), was identified through a novel RSV poly(A) capture screen and demonstrated potent inhibition of RSV in vitro and in the RSV mouse model (11). In vitro characterization suggested that it functioned through interfering with the viral mRNA capping (12). However, the observed cytotoxicity of this compound may limit its therapeutic index (11). Benzazepine YM-53403 is another RSV L inhibitor that was identified through a compound library screen (13). Serial passaging of virus in the presence of BI cpd D and YM-53403 generated resistance mutations in the RSV L coding region, providing the main evidence linking these two classes of inhibitors to the L protein target.
Here, we describe the characterization of a novel RSV L inhibitor designed based on the YM-53403 series (14). This inhibitor achieved significantly improved potency and RSV subtype coverage and demonstrated a desirable antiviral profile and a resistance pattern linking its mechanism of action (MoA) to the L protein.
MATERIALS AND METHODS
Cells and viruses.
HEp-2, BHK-21, A549, and HEK293 cells (ATCC) were cultured according to ATCC instructions. The RSV replicon cell lines (Apath) were cultured as previously described (15, 16). Differentiated human bronchial epithelial cell (HBEC) air-liquid interface cultures (HBEC-ALI) (MatTek) were cultured and maintained according to the manufacturer's instructions.
RSV A2 (ATCC), B-Washington B-9320 (B-WST) (ATCC), and clinical isolates (kindly provided by MedImmune) (17) were propagated, and their titers were determined by a 50% tissue culture infective dose (TCID50) assay in HEp-2 cells as previously reported (17).
Compounds.
RSV inhibitors AZ-27, AZD4316 (patent WO 2010/103306 A1 [see Fig. S1 in the supplemental material]), YM-53403, and BI cpd D were synthesized in-house (11, 13, 14). Ribavirin was purchased from EMD Chemicals. Compounds were solubilized in dimethyl sulfoxide (DMSO) to a 10 mM concentration and serially diluted to the desired compound concentrations with a final DMSO concentration of 0.1% (vol/vol) in assay medium for testing.
RSV ELISA.
An RSV enzyme-linked immunosorbent assay (ELISA) was adapted from the RSV microneutralization assay (18). HEp-2 cells were used in all RSV ELISAs except for the cell type dependency analysis, in which A549, HEK293, and BHK-21 cells were also tested in parallel. Cells were seeded in 96-well plates (5 × 103 to 10 × 103 cells/well) and incubated overnight. Cultures were preincubated with compounds for 1 h at 37°C, followed by RSV infection at a multiplicity of infection (MOI) of 0.1 (or otherwise specified) and cultured for 3 days (A2) or 4 days (B-WST and clinical isolates). RSV replication level was determined through quantitation of expressed RSV F protein by ELISA. Briefly, cells were fixed with 80% acetone for 20 min at 4°C and blocked with casein block–phosphate-buffered saline (PBS) (Thermo) for 1 h, followed by incubation with mouse anti-RSV F monoclonal antibody (1:4,000) (Millipore) in washing buffer (PBS with 0.1% Tween 20) at 37°C for 1 h. The samples were then washed 3 times and incubated at 37°C for 1 h with peroxidase-conjugated goat anti-mouse IgG (1:8,000) (Millipore), followed by an additional 3 washes before the addition of SureBlue tetramethylbenzidine (TMB) peroxidase substrate. The reaction was stopped with 0.2 M sulfuric acid, and absorbance was read at 450 nm.
The compound 50% effective concentration (EC50) for all assays was calculated as previously described using the percent activity of the maximal signal (virus-only infection or replicon only) at multiple compound concentrations by the XLFit model (16).
Time-of-addition studies were performed using the RSV ELISA described above, with the exception that two different MOIs (0.02 and 1) of RSV A2 virus were used and compounds were added to cells at either 2 h preinfection or 6, 12, and 24 h postinfection (hpi).
RSV replicon assay.
The anti-RSV activity of compounds was assessed in the BHK-21-based RSV replicon cell line as previously described (16). Briefly, APC126-E cells were plated 4 h prior to compound addition; EnduRen substrate was added after 48 h of culture to measure replicon luciferase reporter signal through luminescence detection.
The effect of compound on the RSV replicon-driven green fluorescent protein (GFP) expression was also monitored by addition of compounds to HeLa-based RSV replicon cells seeded overnight, followed by an additional 48-h culture and examination of GFP expression using a FLoid Cell Imaging Station (Invitrogen) (15).
RSV reverse transcription-quantitative PCR (qRT-PCR) assay in HBEC-ALI.
Compounds were added to the basal medium of HBEC-ALI transwell cultures 1 h prior to RSV A2 infection (MOI, 0.1) through the apical side of the culture. Virus inoculum was removed after 2 h of incubation at 37°C, followed by apical wash with medium twice and 72 h of culture. The basal medium was replaced every 24 h with fresh medium containing compound. HBEC-ALI samples were homogenized in lysis buffer for RNA extraction using a Maxwell 16 total RNA purification kit (Promega). RSV RNA was quantified using the TaqMan RNA-to-CT 1-Step kit (Invitrogen) with the following primers and probe recognizing RSV N RNA sequence: RSVNA2_Forward, 5′-AGATCAACTTCTGTCATCCAGCAA-3′; RSVNA2_Reverse, 5′-TTCTGCACATCATAATTAGGAGTATCAAT-3′; RSVNA2_Probe, 5′–6-carboxyfluorescein (FAM)–CACCATCCAACGGAGCACAGGAGAT-6-carboxytetramethylrhodamine (TAMRA)-3′. PCR cycles consisted of 48°C for 15 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. In vitro-transcribed RSV N RNA was used to generate a standard curve for quantitation.
Cytotoxicity assay.
The effect of compounds on cell viability was measured by the standard format previously described (16). Briefly, cells were seeded and incubated with compound under the assay conditions described above, followed by CellTiter-Glo substrate (Promega) addition and luminescence detection. The 50% cytotoxic concentration (CC50) of the compound was calculated as previously described using the percent activity of the maximal signal (cells without compound) at multiple compound concentrations by the XLFit model (16).
RSV resistance selection. (i) Resistant replicon.
RSV replicon cells (APC126) were continuously cultured for 4 weeks in the presence of 0.2% DMSO (control) or 0.06 μM or 0.3 μM AZ-27 and passaged once when 90% confluence was reached at the end of week 2. Cells with diminished replicon expression resulting from AZ-27 inhibition were eliminated by the presence of blasticidin in the medium. The surviving AZ-27-resistant cells were pooled at the end of week 4 and analyzed for a resistance phenotype.
(ii) Resistant viruses.
RSV A2 virus was cultured for 7 passages in the presence of 0.2% DMSO (control) or AZ-27 (0.06 μM or 0.3 μM) to select for resistant viruses. Briefly, HEp-2 cells were pretreated with AZ-27 for 1 h before RSV infection (MOI of 0.2). After 3 to 4 days of culture, both intracellular virus and virus released to the culture medium were collected (by freeze-thawing the cells twice) and passaged to fresh HEp-2 cells in the presence of AZ-27. The resistance phenotype of these viruses was analyzed by RSV ELISA.
Sequence analysis.
RNA was isolated from RSV replicon and virus-infected cells using the RNeasy RNA extraction kit (Qiagen) and then in vitro transcribed to cDNA using the SuperScript III First Strand Synthesis system (Invitrogen) with primer 5′-GTGTCAAAAACTAATATCTCGT-3′. RSV cDNA was amplified by PCR with primers F1 (5′-GATAAGTACCACTTAAATTTAACTCCCTTGG-3′) and R1 (5′-GCATTACACTCTGAGAAAGAGATAACACC-3′) for the NS1 to M2-2 region and F2 (5′-GTGTCATAACACTCAATTCTAACACTCACC-3′) and R2 (5′-CCTCCAAGATTAAAATGATAACTTTAGGATTAG-3′) for the M2-2 to trailer region.
Nextera XT libraries were generated from 1.5 ng of PCR product (Illumina) and sequenced on a MiSeq V2 sequencer (Illumina) according to the manufacturer's instructions. The resulting reads were assembled and analyzed using CLCBio Genomics Workbench v6.5 (Qiagen). Quality-based single-nucleotide polymorphisms (SNPs)/indels were identified by mapping to a control reference assembly (DMSO treatment samples) at a minimum frequency of 20% with coverage of >100-fold.
Representative L protein sequences for the paramyxovirus family members measles virus (MeV), Nipah virus (NiV), human parainfluenza viruses 2 and 4b (hPiV2 and hPiV4b, respectively), mumps virus (MuV), and Newcastle disease virus (NDV) and consensus sequences of RSV A and B subtypes were obtained from GenBank sequence databases for standard alignment analysis.
RESULTS
Anti-RSV activity of AZ-27.
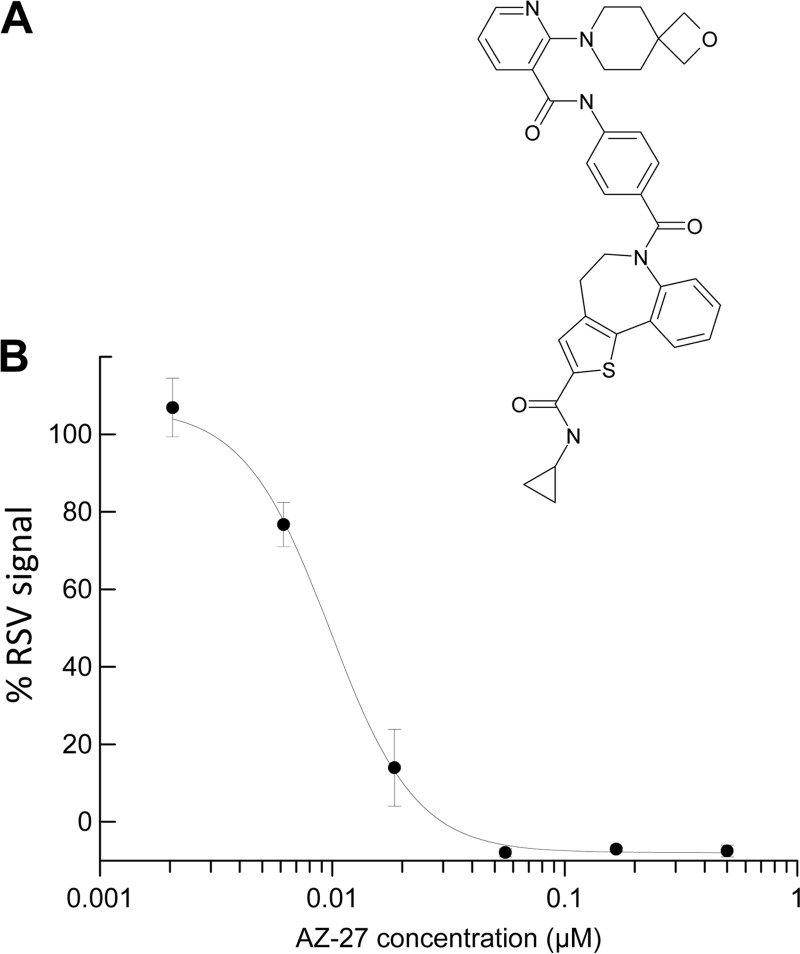
AZ-27 was discovered through scaffold modification of the previously reported RSV L inhibitor YM-53403 (Fig. 1A) (13, 14). This novel molecule demonstrated nanomolar-concentration activity against RSV A2 in the RSV ELISA following a 3-day infection (Fig. 1B), with an ∼75-fold improvement over the parent compound as measured by EC50 (Table 1). AZ-27 showed no cytotoxicity when we examined compound interference of cell proliferation at the highest concentration tested (100 μM), similar to YM-53403 and superior to the RSV L inhibitor BI cpd D (Fig. 1B and Table 1) (11, 13).
FIG 1.
Discovery of a novel RSV inhibitor. (A) AZ-27 chemical structure. (B) Inhibition of RSV A2 replication by AZ-27. Data shown are percentage of RSV signal (mean ± standard deviation of triplicates) following 3-day infection in HEp-2 cells in the presence of AZ-27 from a representative RSV ELISA.
TABLE 1.
Anti-RSV activity of RSV L inhibitors
| Compound | Mean ± SD (n = 3 to 16) |
Selective index, CC50/EC50d | |||
|---|---|---|---|---|---|
| RSV EC50 (μM)a |
Replicon assay, EC50 (μM)b | Cytotoxicity assay, CC50 (μM)c | |||
| A2 | B-WST | ||||
| YM-53403 | 0.75 ± 0.31 | >20 | 1.2 ± 0.93 | >50 | NA |
| BI cpd D | 0.32 ± 0.04 | 0.25 ± 0.004 | 0.77 ± 0.21 | 9.3 ± 2.2 | 29 |
| AZ-27 | 0.01 ± 0.004 | 1.3 ± 0.89 | 0.05 ± 0.02 | >100 | >79 |
EC50 measured by RSV ELISA in HEp-2 cells following 3-day (A2) or 4-day (B-WST) RSV infection in the presence of compound.
EC50 measured by RSV replicon luciferase assay following 2-day compound treatment of BHK-based RSV A2 replicon cells.
CC50 measured by cytotoxicity assay in parallel with the EC50 assays.
Selective index calculated as the ratio between CC50 and EC50 of the A2 or B-WST ELISA; NA, not applicable.
Mechanism of inhibition.
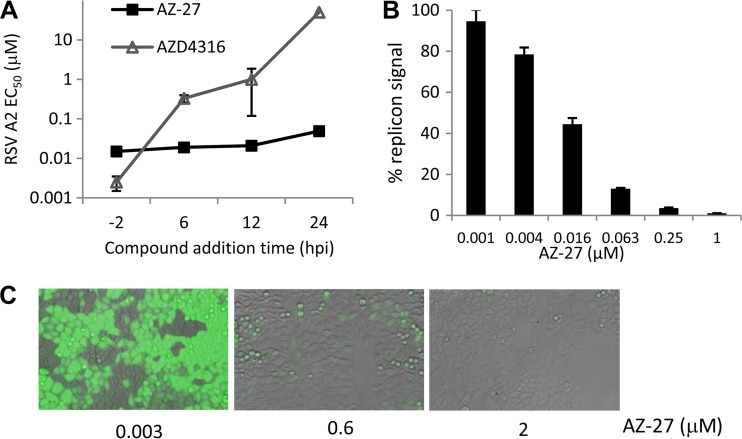
To evaluate the mechanism of inhibition for AZ-27, a time-of-addition study was carried out to compare the activities of inhibitors when added before and after RSV infection. AZ-27 remained potent when added up to 24 h postinfection (hpi), in contrast to the fusion inhibitor AZD4316, which showed a loss in activity when added after 6 hpi (Fig. 2A). This result indicated that AZ-27 is targeting a postentry step(s) in the viral life cycle. The activity of AZ-27 was also more consistent than that of the fusion inhibitor when the effects of viral titer in the assay were compared. This was most evident with compound addition at 6 and 12 hpi, where the MOI increase from 0.02 to 1 resulted in only a severalfold EC50 shift for AZ-27, in contrast to the over-100-fold shift observed for the fusion inhibitor (Table 2).
FIG 2.
AZ-27 targeting RSV replication. (A) Comparison of the AZ-27 and fusion inhibitor AZD4316 activities in a time-of-addition study. EC50s were measured by RSV ELISA in HEp-2 cells following a 3-day infection by RSV A2 at an MOI of 0.02 with compounds added before (−2 h) or after (+6 to 24 h) the infection. Data are means ± standard deviations from triplicates of a representative experiment with EC50s of >50 μM depicted as 50 μM. (B) Inhibition of BHK-21 cell-based RSV replicon following 2-day AZ-27 treatment in a representative replicon luciferase assay. Mean replicon luciferase signals ± standard deviations from triplicates are depicted. (C) Inhibition of HeLa cell-based RSV replicon following 2-day AZ-27 treatment as visualized by microscopy of the replicon GFP reporter expression.
TABLE 2.
Impact of viral titers and time of compound addition on compound EC50
| Compound | RSV A2 MOI | EC50 (μM)a |
EC50 fold shift |
||||||
|---|---|---|---|---|---|---|---|---|---|
| −2 h | +6 h | +12 h | +24 h | −2 h | +6 h | +12 h | +24 h | ||
| AZ-27 | 0.02 | 0.015 ± 0.0004b | 0.019 ± 0.002 | 0.021 ± 0.005 | 0.049 ± 0.002 | 1 | 1 | 1 | 3 |
| 1 | 0.052 ± 0.005 | 0.062 ± 0.004 | 0.07 ± 0.002 | 28 ± 19 | 3 | 4 | 5 | 1,867 | |
| AZD4316 | 0.02 | 0.0025 ± 0.001b | 0.33 ± 0.07 | 1.0 ± 0.88 | >50 | 1 | 132 | 412 | >20,000 |
| 1 | 0.095 ± 0.02 | >50 | >50 | >50 | 38 | >20,000 | >20,000 | >20,000 | |
Compound EC50s were measured by RSV ELISA following 3-day RSV A2 infection in HEp-2 cells, with compounds added before (−2 h) or after (+6, +12, or +24 h) RSV infection at 0 h. EC50 data are means ± standard deviations from triplicate results.
Baseline for EC50 fold shift calculation.
The subgenomic RSV replicon system was used to determine whether AZ-27 targets RSV RNA replication (15, 16). The expression of luciferase and GFP reporters in this system is driven by the viral RNA replication process; the viral entry, assembly, and release steps of the viral life cycle were omitted as a result of the F, G, and SH gene deletions from the RSV replicon. AZ-27 demonstrated similar potencies in both the replicon and live virus assays, indicating that viral replication is the target of this compound (Table 1). Studies using BHK or HeLa cell-based RSV replicons and luciferase or GFP reporter readout gave comparable results (Fig. 2B and C).
Spectrum and selectivity.
AZ-27 was tested against a panel of nine RSV A and four RSV B laboratory and clinical strains to assess the spectrum of its antiviral activity. AZ-27 inhibited all the strains tested, with higher potency observed against the RSV A subtype (average EC50 = 24 ± 9 nM) than against the B subtype (average EC50 = 1.0 ± 0.28 μM). This is a significant improvement compared to YM-53403, which was not active against the B strains tested (Table 1). AZ-27 was RSV specific, with no activity (EC50, >100 μM) against other RNA and DNA viruses tested, including human metapneumovirus, influenza virus A, human rhinovirus, and cytomegalovirus (data not shown).
To evaluate possible cell type dependency for AZ-27 activity, several immortalized cell lines and differentiated HBEC cultures (HBEC-ALI) were infected with RSV A2 in the presence of compounds; viral inhibition was measured by RSV ELISA or qRT-PCR assay. AZ-27 was equally potent in all host cell types tested (Table 3). In contrast, the RSV N inhibitor RSV604 showed a cell-type-dependent activity and was inactive against RSV infection in BHK-21 cells.
TABLE 3.
AZ-27 activity against RSV infection in different cell types
| Compound | RSV A2 EC50 (μM)c |
||||
|---|---|---|---|---|---|
| HEp-2a | A549a | HEK293a | BHK-21a | HBEC-ALIb | |
| RSV604 | 1.6 ± 0.26 | 1.2 ± 0.14 | 0.77 ± 0.17 | >50 | 0.44 ± 0.30 |
| AZ-27 | 0.01 ± 0.004 | 0.03 ± 0.006 | 0.02 ± 0.003 | 0.05 ± 0.03 | 0.07 ± 0.03 |
EC50 measured by RSV ELISA in the indicated cell types following 3-day infection and compound treatment.
EC50 measured by qRT-PCR quantitation of viral RNAs in HBEC-ALI following 3-day infection and compound treatment.
Data are means ± standard deviations (n = 2 to 5).
Resistance.
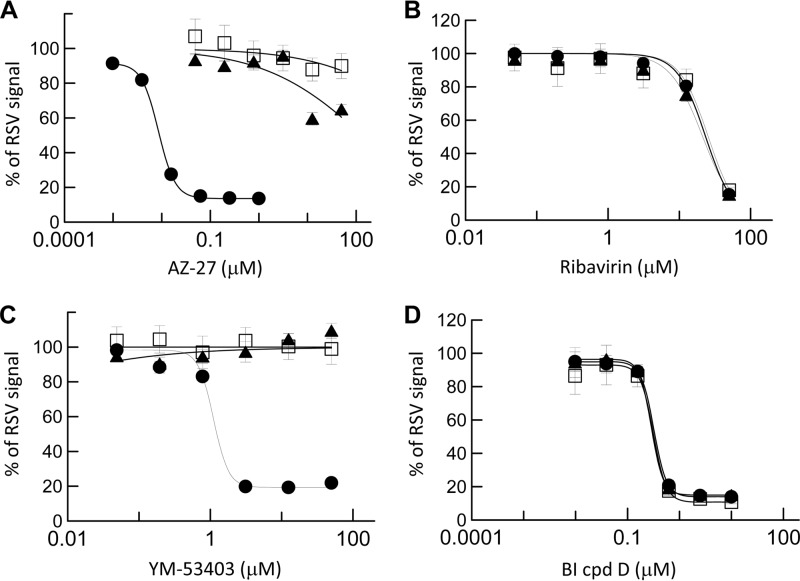
To profile viral resistance to AZ-27, resistant RSV A2 viruses were selected by continuous passage of the virus in HEp-2 cells in the presence of AZ-27. After 7 passages at 3- to 4-day intervals over 4 weeks, viruses cultured under fixed concentrations of 0.06 or 0.3 μM AZ-27 reached titers similar to those of the control wild-type virus (2 × 107 to 9 × 107 TCID50/ml) upon propagation. These viruses were tested for susceptibility to AZ-27 and other RSV inhibitors in the RSV ELISA. As shown in the viral inhibition dose-response curves, the AZ-27-selected viruses exhibited strong resistance toward AZ-27 and YM-53403 while maintaining sensitivity to other classes of inhibitors tested, including ribavirin and BI cpd D (Fig. 3). A greater-than-5,000-fold increase in EC50 against AZ-27 was observed in the resistant virus (Table 4).
FIG 3.
Susceptibility of AZ-27-resistant virus to RSV inhibitors. RSV A2 virus was cultured in HEp-2 cells in the presence of DMSO (●; control) or AZ-27 (▲, 0.06 μM; □, 0.3 μM) for 4 weeks. The titers of the surviving viruses were determined, and the viruses were tested in a 3-day RSV ELISA in HEp-2 cells to determine their susceptibility to AZ-27 (A), ribavirin (B), YM-53403 (C), and BI cpd D (D). The data shown are means ± standard deviations of triplicates from a representative experiment.
TABLE 4.
Compound susceptibility of AZ-27-resistant RSV and replicon
| Compound | EC50 (μM), mean ± SDd |
|||||
|---|---|---|---|---|---|---|
| RSV A2a |
RSV repliconb |
|||||
| Control virus | AZ-27-resistant virus | EC50 fold shiftc | Control replicon | AZ-27-resistant replicon | EC50 fold shiftc | |
| YM-53403 | 1.7 ± 0.03 | >50 | >29 | 3.8 ± 0.8 | >50 | >13 |
| AZ-27 | 0.01 ± 0.0005 | >50 | >5,000 | 0.01 ± 0.004 | 4.7 ± 0.74 | 470 |
| BI cpd D | 0.37 ± 0.01 | 0.33 ± 0.02 | 1 | 0.77 ± 0.27 | 0.65 ± 0.1 | 1 |
| Ribavirin | 23 ± 1.2 | 25 ± 0.48 | 1 | 5.6 ± 2.5 | 6.0 ± 0.68 | 1 |
EC50 measured by RSV ELISA in HEp-2 cells following 3-day infection of control or resistant virus in the presence of compounds.
EC50 measured by RSV replicon luciferase assay following 2-day compound treatment of control or resistant replicon cells.
Ratio of EC50 between resistant and control virus or replicon.
Data from triplicate results.
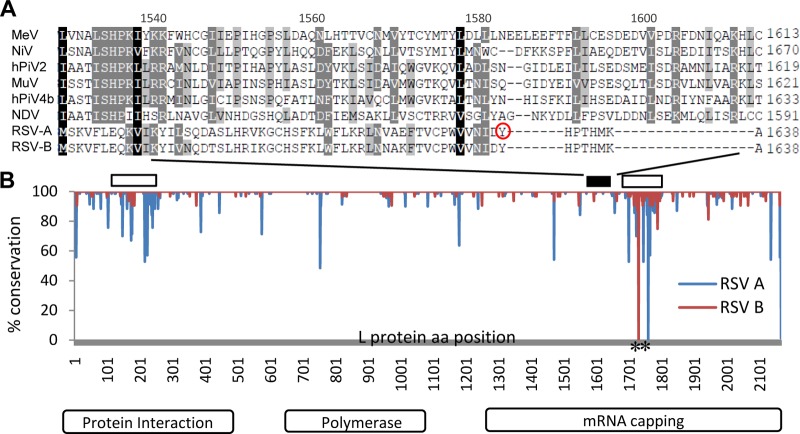
Full-genome sequence analysis of the resistant viruses was carried out, and a single dominant mutation site was identified. One hundred percent of the viral stocks from independent resistance selections all contained a Y1631H variant in the L protein, which was also identified as the substitution responsible for YM-53403 resistance (13). This result supports L protein as the target of AZ-27, and a single mutation is sufficient to confer strong resistance toward AZ-27. L domain analysis for paramyxoviruses suggested that Y1631 is located around the capping enzyme domain of this multifunctional protein (Fig. 4) (8). Y1631 and the nearby amino acid sequence are 100% conserved in the L protein across RSV A and B isolates, while differing from those observed in other paramyxoviruses analyzed.
FIG 4.
Conservation of RSV L domain conferring resistance to AZ-27. (A) Sequence alignment of the L protein fragments from RSV and other paramyxoviruses overlapping the AZ-27 resistance site Y1631 (circle). Amino acid position numbers of MeV L protein are shown on the top. (B) Conservation of the RSV L protein sequences. One hundred two full-length RSV L protein sequences available were compared to the RSV L consensus sequence. The level of conservation at each amino acid (aa) position for RSV A (blue line) and B (red line) isolates is depicted as the percentage of isolates sharing the consensus sequence. The open boxes on top mark the two variable regions, and the black box highlights the location of the fragment depicted in panel A. The two alignment gaps are labeled with asterisks.
To confirm the AZ-27 resistance profile and evaluate the RSV replicon as a tool for resistance studies, the blasticidin-resistant RSV A2 replicon cells were cultured in the presence of AZ-27 and blasticidin for 4 weeks. The resultant replicon demonstrated strong resistance toward AZ-27 and YM-53403 while remaining sensitive to ribavirin and BI cpd D, similar to the results from the resistant virus (4). The EC50 shift against AZ-27 in the RSV replicon luciferase assay was over 400-fold. Full replicon sequence analysis revealed that L protein Y1631 was also the major resistance site, with 58% of sequence reads from the pooled replicon population harboring the mutation. Interestingly, Y1631C was the amino acid substitution observed at this site, as opposed to the Y1631H variant observed in the resistant virus. These data suggested that the RSV replicon can be used for compound resistance studies and further implicated Y1631 as the key mutation site for AZ-27 resistance.
DISCUSSION
We demonstrated AZ-27 as the most potent RSV A replication inhibitor reported to date. Time-of-addition, MOI, and cell type dependency analysis revealed the strength of this novel inhibitor in maintaining potency compared to several other classes of RSV inhibitors tested. AZ-27 showed significant improvement in RSV spectrum coverage compared to similar compounds in this class. A single mutation in the L protein capping enzyme domain conferred strong resistance toward AZ-27, supporting L as the direct target of this compound. This report also demonstrated for the first time that the RSV replicon system can be a useful tool for compound mechanism-of-action and resistance studies.
Even with the improved RSV A and B subtype coverage, the activity of AZ-27 against the RSV B subtype (EC50, ∼1 μM) remained weaker than that against the A subtype strains (EC50, 10 to 40 nM). The mechanism behind this difference is yet to be understood. The lack of biochemical assays and structure information for the RSV L capping enzyme has limited the means to address this question. Interestingly, the site of the L mutation leading to AZ-27 resistance, as well as the nearby amino acid residues, is identical among RSV A and B strains, suggesting that the feature responsible for generating AZ-27 resistance may be separated from what drives the subtype-dependent susceptibility toward this compound (Fig. 4). A similar phenomenon was also observed with other unrelated chemical series sharing the same MoA for RSV inhibition (unpublished data). Mapping out the L sequence/region that determines this RSV subtype susceptibility profile may help in better understanding and overcoming the compound potency limitations. Since the clinical prevalences and disease severities of RSV A and B are similar, it is important to develop efficacious therapeutics that cover both subtypes (19). The physical properties of this series also require improvement to become a more drug-like molecule (14). AZ-27 represents significant progress in this inhibitor class and provides a foundation for further optimization. AZ-27 may also serve as a useful tool compound to investigate the RSV subtype differences and functions of the L domain.
Acute viral respiratory diseases, including RSV, are typically associated with a short treatment window and high viral titers in patients. The past development of RSV treatments has been focused primarily on entry inhibitors, with most programs failing to go beyond phase II clinical trials. Compared to compounds targeting RSV entry, RSV L inhibitors of different scaffolds all demonstrated a longer window of potency in time-of-addition and MOI studies (data not shown). This in vitro observation merits further in vivo and clinical investigations to determine whether viral replication is a more attractive target for delivering efficacious therapies for respiratory virus infection.
The L protein Y1631 variants were not found in any of the currently deposited RSV sequences in GenBank, suggesting that preexisting resistance toward AZ-27 may not be a major concern. However, as a single mutation is sufficient to confer strong resistance to AZ-27, we cannot rule out the possibility of a rapid emergence of resistant viruses upon use of this inhibitor class as a monotherapy. As demonstrated by antivirals against other viruses, combination therapy with compounds of different mechanisms of inhibition is a desirable long-term approach to increase the resistance barrier and address this risk.
Supplementary Material
ACKNOWLEDGMENTS
We thank Melinda Foulk for compound synthesis; James Whiteaker and Kathy MacCormack for sequencing sample preparation; Nancy Ulbrandt, Qing Zhu, Hong Ji, Josie McAuliffe, Nicole Kallewaard-LeLay, Kelly Huang, Leslie Wachter, Subramaniam Krishnan, and Catherine Svabek for RSV, human rhinovirus, and human metapneumovirus isolates, assay protocols, HBEC-ALI, and other technical support; WuXi AppTec for virology assay support; and Giuseppe Ciaramella, Scott Butler, and Paul Miller for scientific discussions.
Footnotes
Published ahead of print 28 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02540-14.
REFERENCES
- 1.Collins PL, Fearns R, Graham BS. 2013. Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease. Curr. Top. Microbiol. Immunol. 372:3–38. 10.1007/978-3-642-38919-1_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins PL, Melero JA. 2011. Progress in understanding and controlling respiratory syncytial virus: still crazy after all these years. Virus Res. 162:80–99. 10.1016/j.virusres.2011.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borchers AT, Chang C, Gershwin ME, Gershwin LJ. 2013. Respiratory syncytial virus—a comprehensive review. Clin. Rev. Allergy Immunol. 45:331–379. 10.1007/s12016-013-8368-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roymans D, Koul A. 2011. Treatment of respiratory syncytial virus infection: past, present and future. In Resch B. (ed), Human respiratory syncytial virus infection. InTech, Rijeka, Croatia. 10.5772/26528 [DOI] [Google Scholar]
- 5.Lay MK, Gonzalez PA, Leon MA, Cespedes PF, Bueno SM, Riedel CA, Kalergis AM. 2013. Advances in understanding respiratory syncytial virus infection in airway epithelial cells and consequential effects on the immune response. Microbes Infect. 15:230–242. 10.1016/j.micinf.2012.11.012 [DOI] [PubMed] [Google Scholar]
- 6.Chapman J, Abbott E, Alber DG, Baxter RC, Bithell SK, Henderson EA, Carter MC, Chambers P, Chubb A, Cockerill GS, Collins PL, Dowdell VC, Keegan SJ, Kelsey RD, Lockyer MJ, Luongo C, Najarro P, Pickles RJ, Simmonds M, Taylor D, Tyms S, Wilson LJ, Powell KL. 2007. RSV604, a novel inhibitor of respiratory syncytial virus replication. Antimicrob. Agents Chemother. 51:3346–3353. 10.1128/AAC.00211-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morin B, Kranzusch PJ, Rahmeh AA, Whelan SP. 2013. The polymerase of negative-stranded RNA viruses. Curr. Opin. Virol. 3:103–110. 10.1016/j.coviro.2013.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dochow M, Krumm SA, Crowe JE, Jr, Moore ML, Plemper RK. 2012. Independent structural domains in paramyxovirus polymerase protein. J. Biol. Chem. 287:6878–6891. 10.1074/jbc.M111.325258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tremaglio CZ, Noton SL, Deflube LR, Fearns R. 2013. Respiratory syncytial virus polymerase can initiate transcription from position 3 of the leader promoter. J. Virol. 87:3196–3207. 10.1128/JVI.02862-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gopinath M, Shaila MS. 2009. RNA triphosphatase and guanylyl transferase activities are associated with the RNA polymerase protein L of rinderpest virus. J. Gen. Virol. 90:1748–1756. 10.1099/vir.0.010975-0 [DOI] [PubMed] [Google Scholar]
- 11.Mason SW, Lawetz C, Gaudette Y, Do F, Scouten E, Lagace L, Simoneau B, Liuzzi M. 2004. Polyadenylation-dependent screening assay for respiratory syncytial virus RNA transcriptase activity and identification of an inhibitor. Nucleic Acids Res. 32:4758–4767. 10.1093/nar/gkh809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liuzzi M, Mason SW, Cartier M, Lawetz C, McCollum RS, Dansereau N, Bolger G, Lapeyre N, Gaudette Y, Lagacé L, Massariol MJ, Dô F, Whitehead P, Lamarre L, Scouten E, Bordeleau J, Landry S, Rancourt J, Fazal G, Simoneau B. 2005. Inhibitors of respiratory syncytial virus replication target cotranscriptional mRNA guanylylation by viral RNA-dependent RNA polymerase. J. Virol. 79:13105–13115. 10.1128/JVI.79.20.13105-13115.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sudo K, Miyazaki Y, Kojima N, Kobayashi M, Suzuki H, Shintani M, Shimizu Y. 2005. YM-53403, a unique anti-respiratory syncytial virus agent with a novel mechanism of action. Antiviral Res. 65:125–131. 10.1016/j.antiviral.2004.12.002 [DOI] [PubMed] [Google Scholar]
- 14.Xiong H, Foulk M, Aschenbrenner L, Fan J, Tiong-Yip CL, Johnson KD, Moustakas D, Fleming PR, Brown DG, Zhang M, Ferguson D, Wu D, Yu Q. 2013. Discovery of a potent respiratory syncytial virus RNA polymerase inhibitor. Bioorg. Med. Chem. Lett. 23:6789–6793. 10.1016/j.bmcl.2013.10.018 [DOI] [PubMed] [Google Scholar]
- 15.Malykhina O, Yednak MA, Collins PL, Olivo PD, Peeples ME. 2011. A respiratory syncytial virus replicon that is noncytotoxic and capable of long-term foreign gene expression. J. Virol. 85:4792–4801. 10.1128/JVI.02399-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tiong-Yip CL, Plant H, Sharpe P, Fan J, Rich K, Gorseth E, Yu Q. 2014. Development of a high-throughput replicon assay for the identification of respiratory syncytial virus inhibitors. Antiviral Res. 101:75–81. 10.1016/j.antiviral.2013.11.003 [DOI] [PubMed] [Google Scholar]
- 17.Zhu Q, Patel NK, McAuliffe JM, Zhu W, Wachter L, McCarthy MP, Suzich JA. 2012. Natural polymorphisms and resistance-associated mutations in the fusion protein of respiratory syncytial virus (RSV): effects on RSV susceptibility to palivizumab. J. Infect. Dis. 205:635–638. 10.1093/infdis/jir790 [DOI] [PubMed] [Google Scholar]
- 18.Wu H, Pfarr DS, Tang Y, An LL, Patel NK, Watkins JD, Huse WD, Kiener PA, Young JF. 2005. Ultra-potent antibodies against respiratory syncytial virus: effects of binding kinetics and binding valence on viral neutralization. J. Mol. Biol. 350:126–144. 10.1016/j.jmb.2005.04.049 [DOI] [PubMed] [Google Scholar]
- 19.Jafri HS, Wu X, Makari D, Henrickson KJ. 2013. Distribution of respiratory syncytial virus subtypes A and B among infants presenting to the emergency department with lower respiratory tract infection or apnea. Pediatr. Infect. Dis. J. 32:335–340. 10.1097/INF.0b013e318282603a [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.