Abstract
CBR703 was reported to inhibit bacterial RNA polymerase (RNAP) and biofilm formation, considering it to be a good candidate for further optimization. While synthesized derivatives of CBR703 did not result in more-active RNAP inhibitors, we observed promising antibacterial activities. These again correlated with a significant cytotoxicity toward mammalian cells. Furthermore, we suspect the promising effects on biofilm formation to be artifacts. Consequently, this class of compounds can be considered unattractive as antibacterial agents.
TEXT
Bacterial RNA polymerase (RNAP) is essential for bacterial growth and survival and is thus an attractive target for drug development (1, 2). Along with the recently FDA-approved fidaxomycin (3), the rifamycins, applied as first-line antituberculosis drugs, are the only RNAP inhibitors that are in clinical use (2). However, similarly to other anti-infectives, the use of rifamycins resulted in the occurrence of resistant bacterial strains (1, 4–7), which represents a remarkable threat to public health (8, 9). Consequently, there is a need to focus on novel promising inhibitors. Recently, interesting peptidic and peptidomimetic (10–12) as well as nonpeptidic (13–18) small-molecule RNAP inhibitors have been described. Another example is CBR703 (Fig. 1), whose mechanism of action is reported to be different from that of the rifamycins (19, 20). This compound has been identified in a high-throughput screening searching for small-molecule inhibitors of RNAP (19). Two more-potent analogs in that report reveal the potential of optimizing CBR703 by structural enlargement. Furthermore, pursuing the hypothesis that RNAP is of particular importance for bacterial survival in biofilms, Villain-Guillot et al. showed CBR703 to significantly reduce Staphylococcus epidermidis biofilm mass (21). We therefore considered CBR703 to be a promising starting point for drug development. Consequently, we focused on CBR703 to perform systematic modifications on its core structure, aiming to obtain a more appropriate starting point for further structural optimization.
FIG 1.
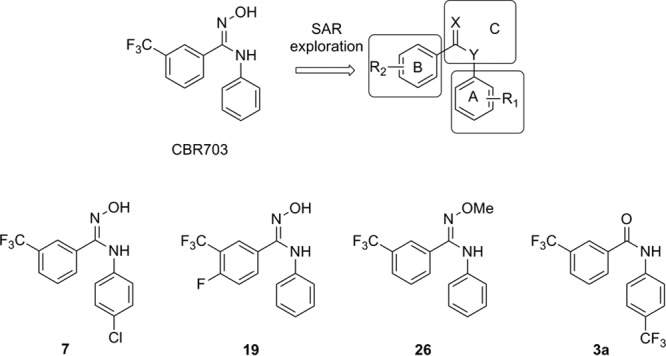
CBR703 and the most potent compounds in different classes: compound 7, best compound against E. coli TolC bearing an amidoxime group; compound 19, most RNAP-inhibitory derivative; compound 26, the only RNAP inhibitor after replacement of the amidoxime linker; compound 3a, most active against E. coli TolC. SAR, structure activity relationship.
Detailed information concerning the materials and methods used in synthesis and biology can be found in the supplemental material.
In total, 30 final compounds and 24 intermediates were obtained and tested for Escherichia coli RNAP inhibition and their ability to inhibit the growth of E. coli TolC (see Table S1 to S3 in the supplemental material). According to their structures, the synthesized derivatives can be divided into three groups with modifications in part A, B, or C (Fig. 1). Compounds 1 to 25 (see Scheme S1 in the supplemental material) with introduction of substituents into the aromatic moieties (part A or B) were prepared by condensation of an intermediate amide with hydroxylamine (22, 23). In order to ensure an appropriate coverage of lipophilic and electronic properties, the substituents were chosen rationally from all quadrants of a Craig plot (e.g., Hansch-Fujita π versus σ constant) (24). The results (see Table S1) showed that compounds 1 to 25 display decreased RNAP inhibition compared to CBR703, with the exception of two compounds (18 and 19) with similar activities (50% inhibitory concentrations [IC50s] in the range of 20 μM). As previously reported (19), there were two more-potent CBR703 analogs with a larger size, one of which was optimized by replacing the linker amidoxime with a pyrazole system. To investigate this structural modification, compounds 26 to 30 with a different linking part (part C) have been synthesized (see Table S2). Remarkably, in our case, replacement of the amidoxime moiety by other functional groups, including N-heterocycles, led to a decrease in or complete loss of activity. Additionally, all amide intermediates turned out to be inactive against RNAP (see Table S3). Surprisingly, 11 compounds, including intermediates with little or even no RNAP inhibition, showed stronger antibacterial potency in E. coli TolC than CBR703. Compound 3a with a MIC of 2 μg/ml was even more potent than rifampin. The fact that no correlation between RNAP inhibition and antibacterial activity (see Table S1 to S3) could be observed led us to the conclusion that additional mechanisms besides RNAP inhibition must be responsible for the antibacterial activity.
To obtain further information about the antibacterial profiles, four compounds (Fig. 1) were selected based on the results of the previous experiments (see Table S1 to S3 in the supplemental material) and compared with reference compounds. In a first step, the effects of these compounds on the growth of E. coli K-12, Pseudomonas aeruginosa PAO1, Bacillus subtilis, and Staphylococcus aureus were investigated (Table 1). Notably, compounds 7 (the best compound against E. coli TolC bearing an amidoxime group) and 19 (the most active RNAP inhibitor) showed only moderate activity against B. subtilis. Compound 3a (the most active compound against E. coli TolC) exhibited rather potent activities against B. subtilis and S. aureus. For compound 26 (the only compound with RNAP inhibition after replacement of the amidoxime linker), we observed no detectable activities against Gram-positive species. None of the compounds inhibited the growth of Gram-negative strains K-12 and PAO1. In addition, the toxicity of the inhibitors toward mammalian cells was tested using the human embryonic kidney 293 (HEK293) cell line. Interestingly, the most active compound in the MIC experiment, compound 3a, showed significant cytotoxicity and the other tested compounds were also at least moderately toxic (Table 2). As it is known that lipophilic compounds bind to serum proteins, which were also present in our MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay as a component of fetal calf serum (FCS), we added the same amount of FCS (10%) to the bacterial growth medium and performed the MIC determinations in E. coli TolC. Surprisingly, the antibacterial activity of the tested compounds was abolished or drastically reduced (see Table S4). This finding led to the assumption that the cytotoxicities of our compounds are even more pronounced in the absence of serum.
TABLE 1.
RNAP inhibition and antibacterial profile of selected compoundsa
| Compound | % inhibition of E. coli RNAP (at 50 μM) or IC50 value | MIC (μg/ml)b |
||||
|---|---|---|---|---|---|---|
| Gram-negative bacteria |
Gram-positive bacteria |
|||||
| E. coli TolC | E. coli K-12 | P. aeruginosa PAO1 | B. subtilis | S. aureus | ||
| CBR703 | 18 μMc | 14 | >25 | >25 | >25 | >25 |
| 7 | 35 | 9 | >25 | >25 | 23 | >25 |
| 19 | 19 μMc | 21 | >50 | >50 | 43 | >50 |
| 26 | 29 | 24 | >25 | >25 | >25 | >25 |
| 3a | n.i | 2 | >25 | >25 | 5 | 11 |
| Rifampin | 24 nMc | 6 | 7 | 13 | 5 | 0.02 |
No correlation between RNAP inhibition and antibacterial activities was observed, suggesting that the antibacterial activity was due to a mechanism other than RNAP inhibition. The standard deviations (SD) in these experiments were <25% (in most cases, <15%). n.i, no inhibition (<10% inhibition).
>25 and >50, MIC determinations were limited due to insufficient solubility of the compound.
IC50 value.
TABLE 2.
Investigation of cytotoxicity in HEK293 cellsa
| Compound | LD50 at 24 h (μM) | LD50 at 72 h (μM) |
|---|---|---|
| CBR703 | 58 | 52 |
| 7 | 43 | 40 |
| 19 | 34 | 33 |
| 26 | 25 | 38 |
| 3a | 15 | 13 |
| Rifampin | >100 | 81 |
| Doxorubicin | 5 | 0.3 |
The most potent antibacterial compound, compound 3a, was also the most toxic one. Rifampin, negative control; doxorubicin, positive control; LD50, 50% lethal dose.
As it had been shown that CBR703 efficiently eradicated biofilm-embedded bacteria (21), we considered that this effect could be due to Fe(III) chelation (25, 26). The fact that the amidoxime moiety plays a prominent role in the activity in our compounds and the well-known property of amidoxime functional groups to form Fe(III) complexes gave rise to the presumption that the amidoximes display their antibacterial effect due to such a complexation (27, 28). Consequently, we examined this hypothesis. First, the ability of CBR703 to form Fe(III) complexes was confirmed by a color change reaction (29). After addition of a CBR703 solution, the brown red FeCl3 solution turned to blue whereas this change was not observed after addition of compound 26 (see Table S5 in the supplemental material). In a following step, the complex stability constants were determined by potentiometric titration. Thereby, it was uncovered that formation of Fe(OH)3 was observed even under acidic conditions (pH = 4). This means that, under physiological conditions, CBR703 cannot form stable Fe(III) complexes. These results were supported by biological tests which were performed in parallel. Indeed, addition of Fe(III) had an effect on the anti-TolC activity of the positive control deferoxamine mesylate (DFO)—a known iron chelator with antibacterial activity (30)—but not on that of CBR703, leading to our conclusion that the antibacterial effects of CBR703 are not attributable to iron complexation (see Fig. S1). Interestingly, each of the three most antibacterial compounds (compounds 3a, 10a, and 21a) possesses two strong electron-withdrawing (leading to a polarity decrease) and highly lipophilic CF3 groups which might be the reason for their antibacterial potency. Such properties could facilitate cell penetration and could furthermore result in nonspecific inhibition of a variety of other enzymes.
During the determination of MIC values, we found that CBR703 showed slight precipitation at 100 μg/ml whereas its MIC was determined to be 100 μg/ml in the literature (21). Beyond that, a significant effect on Staphylococcus epidermidis biofilm was reported at concentrations between 100 and 400 μg/ml. At these concentrations, we observed strong and concentration-dependent precipitation of CBR703 and selected derivatives in Mueller-Hinton broth (MHB) (Fig. 2A; see also Fig. S2 in the supplemental material), the medium used in the literature (21). Nevertheless, we evaluated all compounds on S. aureus biofilms with concentrations in a soluble range but without observing an effect. At higher concentrations (100 to 400 μg/ml), CBR703 and its derivatives (e.g., compounds 7 and 19) showed a clear reduction in biofilm formation (Fig. 2B), indicating a correlation between antibiofilm activity and precipitation.
FIG 2.
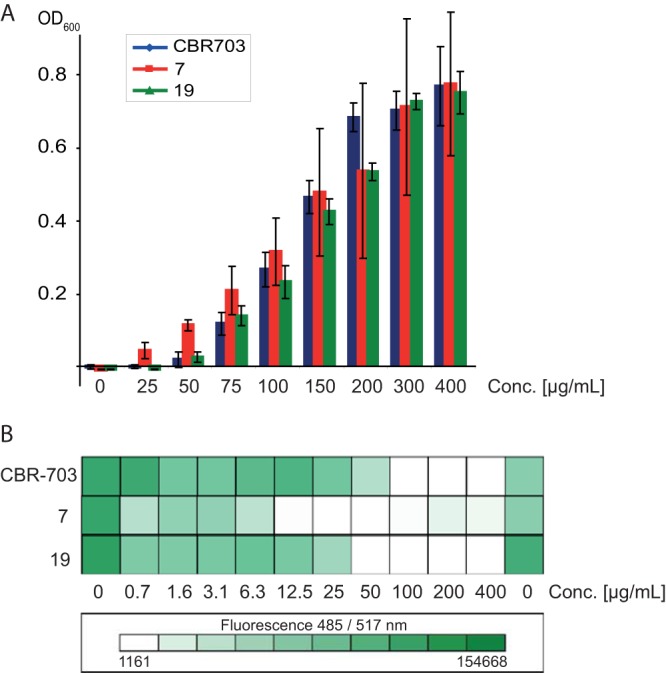
Correlation between precipitation and biofilm mass. (A) Concentration-dependent precipitation of CBR703 and compounds 7 and 19 in MHB. (B) Quantification of the washed biofilm mass. A complete biofilm reduction can be observed only at concentrations at which precipitates have formed. The most prominent effect can be observed for compound 7.
In this work, we designed and synthesized derivatives of CBR703 as a follow-up to a published paper (19), aiming to optimize their promising biological effects by modifying the core structure. However, no compound showed enhanced RNAP inhibition. Nevertheless, in some cases we observed promising antibacterial activities. These again turned out to correlate with a significant cytotoxicity toward HEK293 cells. Furthermore, the reported effects on biofilm formation, which were among the main reasons for choosing CBR703 as a starting point, were suspected to be artifacts due to compound precipitation. This finding should be a reminder to the scientific community to be cautious with published data, as they could include such artifacts (31). Consequently, we rank this class of compounds as unattractive for development as antibacterial agents.
Supplementary Material
ACKNOWLEDGMENTS
W. Zhu thanks China Scholarship Council for her Ph.D. fellowship.
We thank Jeannine Jung and Jannine Ludwig for technical assistance, Kaspar Hegetschweiler and Bernd Morgenstern of the Department of Chemistry, Saarland University, Germany, for the determination of complex stability constants, Werner Tegge of the Helmholtz Centre for Infection Research, Braunschweig, Germany, for supporting M. Fountain's biofilm tests, and Wolfgang Witte of the Robert Koch Institute, Wernigerode, Germany, for kindly providing the MSSA ST30 strain.
Footnotes
Published ahead of print 12 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02600-14.
REFERENCES
- 1.Chopra I. 2007. Bacterial RNA polymerase: a promising target for the discovery of new antimicrobial agents. Curr. Opin. Investig. Drugs 8:600–607 [PubMed] [Google Scholar]
- 2.Mariani R, Maffioli SI. 2009. Bacterial RNA polymerase inhibitors: an organized overview of their structure, derivatives, biological activity and current clinical development status. Curr. Med. Chem. 16:430–454. 10.2174/092986709787315559 [DOI] [PubMed] [Google Scholar]
- 3.Talpaert M, Campagnari F, Clerici L. 1975. Lipiarmycin: an antibiotic inhibiting nucleic acid polymerases. Biochem. Biophys. Res. Commun. 63:328–334. 10.1016/S0006-291X(75)80047-7 [DOI] [PubMed] [Google Scholar]
- 4.Darst SA. 2004. New inhibitors targeting bacterial RNA polymerase. Trends Biochem. Sci. 29:159–162. 10.1016/j.tibs.2004.02.005 [DOI] [PubMed] [Google Scholar]
- 5.Villain-Guillot P, Bastide L, Gualtieri M, Leonetti JP. 2007. Progress in targeting bacterial transcription. Drug Discov. Today 12:200–208. 10.1016/j.drudis.2007.01.005 [DOI] [PubMed] [Google Scholar]
- 6.Bryskier A. 2005. Anti-MRSA agents: under investigation, in the exploratory phase and clinically available. Expert Rev. Anti Infect. Ther. 3:505–553. 10.1586/14787210.3.4.505 [DOI] [PubMed] [Google Scholar]
- 7.Floss HG, Yu TW. 2005. Rifamycin—mode of action, resistance, and biosynthesis. Chem. Rev. 105:621–632. 10.1021/cr030112j [DOI] [PubMed] [Google Scholar]
- 8.Coates ARM, Halls G, Hu Y. 2011. Novel classes of antibiotics or more of the same? Br. J. Pharmacol. 163:184–194. 10.1111/j.1476-5381.2011.01250.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walsh C. 2003. Where will new antibiotics come from? Nat. Rev. Microbiol. 1:65–70. 10.1038/nrmicro727 [DOI] [PubMed] [Google Scholar]
- 10.Hüsecken K, Negri M, Fruth M, Boettcher S, Hartmann RW, Haupenthal J. 2013. Peptide-based investigation of the Escherichia coli RNA polymerase σ(70):core interface as target site. ACS Chem. Biol. 8:758–766. 10.1021/cb3005758 [DOI] [PubMed] [Google Scholar]
- 11.Kuznedelov K, Semenova E, Knappe TA, Mukhamedyarov D, Srivastava A, Chatterjee S, Ebright RH, Marahiel MA, Severinov K. 2011. The antibacterial threaded-lasso peptide capistruin inhibits bacterial RNA polymerase. J. Mol. Biol. 412:842–848. 10.1016/j.jmb.2011.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma C, Yang X, Kandemir H, Mielczarek M, Johnston EB, Griffith R, Kumar N, Lewis PJ. 2013. Inhibitors of bacterial transcription initiation complex formation. ACS Chem. Biol. 8:1972–1980. 10.1021/cb400231p [DOI] [PubMed] [Google Scholar]
- 13.Sahner JH, Groh M, Negri M, Haupenthal J, Hartmann RW. 2013. Novel small molecule inhibitors targeting the “switch region” of bacterial RNAP: structure-based optimization of a virtual screening hit. Eur. J. Med. Chem. 65:223–231. 10.1016/j.ejmech.2013.04.060 [DOI] [PubMed] [Google Scholar]
- 14.Hinsberger S, Hüsecken K, Groh M, Negri M, Haupenthal J, Hartmann RW. 2013. Discovery of novel bacterial RNA polymerase inhibitors: pharmacophore-based virtual screening and hit optimization. J. Med. Chem. 56:8332–8338. 10.1021/jm400485e [DOI] [PubMed] [Google Scholar]
- 15.André E, Bastide L, Michaux-Charachon S, Gouby A, Villain-Guillot P, Latouche J, Bouchet A, Gualtieri M, Leonetti JP. 2006. Novel synthetic molecules targeting the bacterial RNA polymerase assembly. J. Antimicrob. Chemother. 57:245–251. 10.1093/jac/dki426 [DOI] [PubMed] [Google Scholar]
- 16.Arhin F, Belanger O, Ciblat S, Dehbi M, Delorme D, Dietrich E, Dixit D, Lafontaine Y, Lehoux D, Liu J, McKay GA, Moeck G, Reddy R, Rose Y, Srikumar R, Tanaka KS, Williams DM, Gros P, Pelletier J, Parr TRJ, Far AR. 2006. A new class of small molecule RNA polymerase inhibitors with activity against rifampicin-resistant Staphylococcus aureus. Bioorg. Med. Chem. 14:5812–5832. 10.1016/j.bmc.2006.05.035 [DOI] [PubMed] [Google Scholar]
- 17.Buurman EdT, Foulk MA, Gao N, Laganas VA, McKinney DC, Moustakas DT, Rose JA, Shapiro AB, Fleming PR. 2012. Novel rapidly diversifiable antimicrobial RNA polymerase switch region inhibitors with confirmed mode of action in Haemophilus influenzae. J. Bacteriol. 194:5504–5512. 10.1128/JB.01103-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elgaher WAM, Fruth M, Groh M, Haupenthal J, Hartmann RW. 2014. Expanding the scaffold for bacterial RNA polymerase inhibitors: design, synthesis and structure activity relationships of ureido-heterocyclic-carboxylic acids. RSC Adv. 4:2177–2194. 10.1039/c3ra45820b [DOI] [Google Scholar]
- 19.Artsimovitch I, Chu C, Lynch AS, Landick R. 2003. A new class of bacterial RNA polymerase inhibitor affects nucleotide addition. Science 302:650–654. 10.1126/science.1087526 [DOI] [PubMed] [Google Scholar]
- 20.Malinen AM, Nandymazumdar M, Turtola M, Malmi H, Grocholski T, Artsimovitch I, Belogurov GA. 2014. CBR antimicrobials alter coupling between the bridge helix and the β subunit in RNA polymerase. Nat. Commun. 5:3408. 10.1038/ncomms4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villain-Guillot P, Gualtieri M, Bastide L, Leonetti JP. 2007. In vitro activities of different inhibitors of bacterial transcription against Staphylococcus epidermidis biofilm. Antimicrob. Agents Chemother. 51:3117–3121. 10.1128/AAC.00343-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Chen X, Fan P, Mihalic JT, Cutler ST. 2001. Synthesis, antibacterial activity and RNA polymerase inhibition of phenylamidine derivatives. PCT Int. Appl. WO 2001051456 A2 20010719 [Google Scholar]
- 23.Krajete A, Steiner G, Kopacka H, Ongania KH, Wurst K, Kristen MO, Preishuber-Pflugl P, Bildstein B. 2004. Iminohydroxamato early and late transition metal halide complexes—new precatalysts for aluminoxane-cocatalyzed olefin insertion polymerization. Eur. J. Inorg. Chem. 8:1740–1752. 10.1002/ejic.200300405 [DOI] [Google Scholar]
- 24.Patrick GL. 1995. Chapter 9.5, the Craig plot, p 143 In An introduction to medicinal chemistry, 1st ed. Oxford University Press, New York, NY [Google Scholar]
- 25.Singh PK, Parsek MR, Greenberg EP, Welsh MJ. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417:552–555. 10.1038/417552a [DOI] [PubMed] [Google Scholar]
- 26.Ardehali R, Shi L, Janatova J, Mohammad SF, Burns GL. 2002. The effect of apo-transferrin on bacterial adhesion to biomaterials. Artif. Organs 26:512–520. 10.1046/j.1525-1594.2002.06923.x [DOI] [PubMed] [Google Scholar]
- 27.Thompson MG, Corey BW, Si Y, Craft DW, Zurawski DV. 2012. Antibacterial activities of iron chelators against common nosocomial pathogens. Antimicrob. Agents Chemother. 56:5419–5421. 10.1128/AAC.01197-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Léséleuc L, Harris G, Kuo LR, Chen W. 2012. In vitro and in vivo biological activities of iron chelators and gallium nitrate against Acinetobacter baumannii. Antimicrob. Agents Chemother. 56:5397–5400. 10.1128/AAC.00778-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith PAS. 1966. The chemistry of open-chain organic nitrogen compounds, vol 2, p 73–76 Benjamin Publishers, New York, NY [Google Scholar]
- 30.Flournoy DJ. 1991. In vitro antimicrobial properties of deferoxamine mesylate. Eur. J. Clin. Microbiol. Infect. Dis. 10:597–598. 10.1007/BF01967285 [DOI] [PubMed] [Google Scholar]
- 31.Zhu W, Groh M, Haupenthal J, Hartmann RW. 2013. A detective story in drug discovery: elucidation of a screening artifact reveals polymeric carboxylic acids as potent inhibitors of RNA polymerase. Chem. Eur. J. 19:8397–8400. 10.1002/chem.201301289 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


