Abstract
This study critically evaluated the new European Committee for Antimicrobial Susceptibility Testing (EUCAST) antibiotic susceptibility testing guidelines on the basis of a large set of disk diffusion diameters determined for clinical isolates. We report several paradigmatic problems that illustrate key issues in the selection of clinical susceptibility breakpoints, which are of general importance not only for EUCAST but for all guidelines systems, i.e., (i) the need for species-specific determinations of clinical breakpoints/epidemiological cutoffs (ECOFFs), (ii) problems arising from pooling data from various sources, and (iii) the importance of the antibiotic disk content for separating non-wild-type and wild-type populations.
INTRODUCTION
Since 2010, the European Committee for Antimicrobial Susceptibility Testing (EUCAST) has issued regularly updated antibiotic susceptibility testing (AST) guidelines, including clinical susceptibility breakpoints (CBPs), which are determined mainly on the basis of epidemiological cutoffs (ECOFFs), pharmacokinetic/pharmacodynamic (PK/PD) parameters and, in part, clinical outcome data (1–3). EUCAST aims at full transparency in the still-complex, mostly consensus-driven process of CBP determinations by open source publication of documents for diameter/MIC distributions and ECOFF data (4). Diameter and MIC distributions may readily be retrieved from the EUCAST webpage, http://www.eucast.org/zone_diameter_distributions. The Clinical and Laboratory Standards Institute (CLSI) uses a similar process for establishing clinical breakpoints (5).
In 2010, the Institute of Medical Microbiology (IMM), University of Zürich, changed its guidelines from those of CLSI to the EUCAST guidelines. During this shift, we established a database containing nonduplicate disk diffusion diameters of >30,000 clinical isolates with the view to validate EUCAST AST guidelines as part of the quality control management in our clinical microbiology laboratory (6, 7). Despite our data being largely consistent with those of EUCAST, we encountered some problems and inconsistencies in EUCAST AST guidelines that exemplified several important key issues in setting CBPs that, incidentally, also apply to other systems, such as that of CLSI (8): (i) the need for species-specific determinations of CBPs/ECOFFs, (ii) problems arising from pooling data from various sources, and (iii) the importance of the antibiotic disk content for the separation of resistant populations from the wild-type population.
MATERIALS AND METHODS
Clinical isolates.
All nonduplicate clinical isolates included in this study were isolated over a 3-year period from 2010 until 2012 in the clinical microbiology laboratory of the Institute for Medical Microbiology, University of Zürich, which mainly serves a 750-bed tertiary-care hospital (University Hospital of Zürich). Isolates of the same species were considered duplicates if they (i) originated from the same patient, and (ii) showed one major and two minor differences in AST interpretation at the most. All duplicates were excluded from the analysis, resulting in a reduction of included isolates by 33% on average (species-specific reductions: Staphylococcus aureus, 39%; coagulase-negative staphylococci [CoNS], 32%; Enterococcus faecalis, 23%; Enterobacter cloacae, 31%; Klebsiella pneumoniae, 30%). Only isolates considered clinically relevant were included. The absolute numbers of species/drug combinations for Staphylococcus aureus, CoNS, Enterococcus faecalis, Enterobacter cloacae, and Klebsiella pneumoniae are shown below in Fig. 1 and 2.
FIG 1.
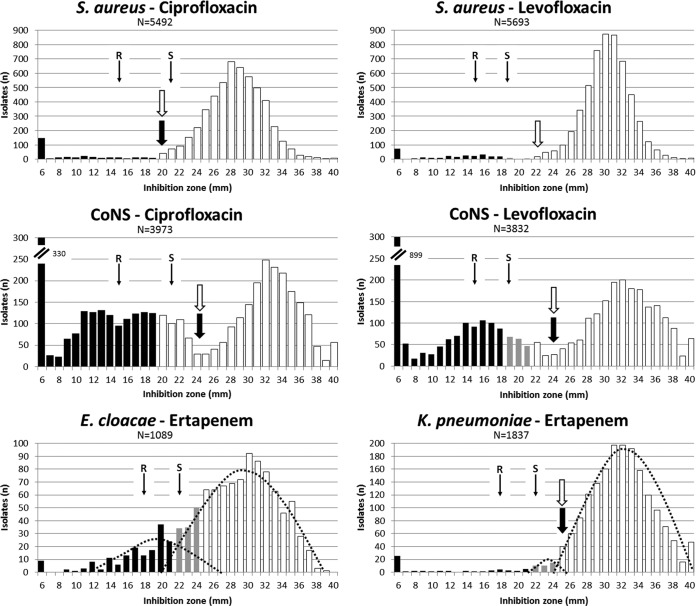
Species-specific breakpoints and overlapping populations. The graphs show inhibition zone diameter distributions of isolates tested in the clinical laboratory of the Institute of Medical Microbiology, University of Zürich. Values were determined by the EUCAST-recommended disk diffusion procedure. Categorization according to EUCAST clinical breakpoints is illustrated with the following symbols: white bars, susceptible; gray bars, intermediate; black bars, resistant; black arrow, EUCAST ECOFF, as listed in EUCAST diameter distributions (13); white arrow, IMM ECOFF (present study); thin arrows, CLSI clinical breakpoints; dotted lines, predicted population distributions; CoNS, coagulase-negative staphylococci. N, number of isolates; R, resistant; S, susceptible.
FIG 2.
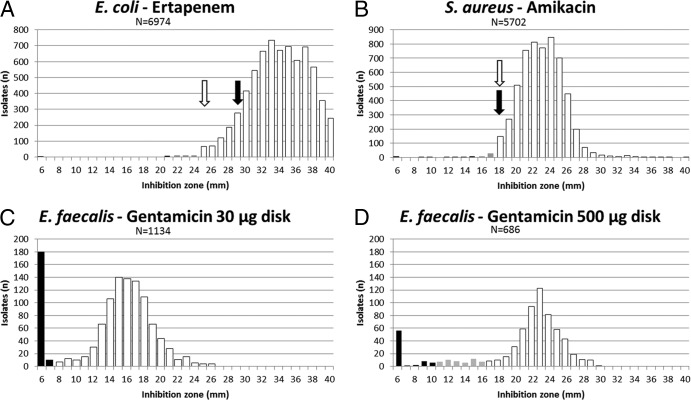
Diameter distributions, results of data pooling from various sources, and importance of antibiotic disk contents. Diameter distributions originated from isolates tested in the clinical laboratory of the Institute of Medical Microbiology, University of Zürich. Values were determined by the EUCAST-recommended disk diffusion procedure. White bars, EUCAST susceptible category; gray bars, EUCAST intermediate category; black bars, EUCAST resistant category; black arrows, EUCAST ECOFFs as listed in EUCAST diameter distributions (13); white arrows, IMM ECOFFs (present study). The 500-μg gentamicin disk content was recommended by CA-SFM and in accordance with CBPs as shown. N, number of isolates.
Susceptibility testing.
For susceptibility testing, the disk diffusion method (with disks from i2a, Montpellier, France) was carried out using Mueller-Hinton agar (Becton, Dickinson, Franklin Lakes, NJ) and a McFarland 0.5 dilution from overnight cultures as an inoculum, followed by incubation at 35°C for 16 to 18 h according to EUCAST recommendations (9). Inhibition zone diameters were determined and recorded using the semiautomated Sirweb/Sirscan system (i2a).
ECOFF determinations.
ECOFFs were determined by visual inspection of diameter distributions (the consensus of five experienced persons, i.e., the “eyeball method”) (8). EUCAST does not recommend a method for ECOFF determinations, nor does it indicate the method used for determination of reported EUCAST ECOFFs.
Software.
Calculations were performed using Microsoft Excel 2010 software (Microsoft Corporation, Redmond, WA).
RESULTS AND DISCUSSION
Reliable CBPs/ECOFFs require species-specific determinations.
Inhibition zone diameters and MIC distributions are species specific in terms of ranges, means, and standard deviations (8). Thus, ECOFFs and CBPs, by definition, need to be determined on a species-specific basis, a principle acknowledged by EUCAST and CLSI (10, 11). To avoid increased guideline complexity, group-specific CBPs have been suggested, e.g., the majority of staphylococcal CBPs are defined at the genus level, and Enterobacteriaceae CBPs are applicable at the group level (3, 5). However, there is agreement that CBPs must never split a wild-type population due to biological variation within wild-type isolates (11). A practical solution to reduce the highly complex process of setting CBPs for a large number of drugs is the use of drug class representatives with use of AST extrapolations for other class members (3, 5).
For sufficient statistical power, an analysis should include a minimum of 30 isolates to form a Gaussian distribution suitable for the ECOFF determination (central limit theorem) (12). The 21,434 Enterobacteriaceae isolates tested for antibiotic susceptibility to 27 individual drugs in the IMM clinical laboratory from 2010 to 2012 comprised 11,597 (54%) E. coli, 3,016 (14%) K. pneumoniae, 1,685 (8%) Enterobacter cloacae, 1,089 (5%) Proteus mirabilis, 905 (4.2%) Klebsiella oxytoca, 735 (3.4%) Serratia marcescens, 535 (2.5%) Citrobacter koseri, 481 (2.2%) Enterobacter aerogenes, 454 (2.1%) Citrobacter freundii, 404 (1.9%) Morganella morganii, 276 (1.3%) Proteus vulgaris, 121 (0.6%) Hafnia alvei, 50 (0.23%) Providencia rettgeri, 37 (0.17%) Serratia liquefaciens, and 33 (0.15%) Pantoea agglomerans isolates. For all other Enterobacteriaceae species, the IMM database contained less than 30 isolates. The species listed above accounted for >99.5% of all Enterobacteriaceae isolated in our clinical laboratory. Given these numbers, which can serve as an approximation for other laboratories, sufficient AST data would be available for >99.5% of clinical Enterobacteriaceae isolates. E. coli, K. pneumoniae, and E. cloacae alone, as the three most frequently isolated species, accounted for 76.1% of all isolates. Automated diameter reading and documentation systems can facilitate the collection of comparably high numbers of diameter values.
When evaluating our clinical isolate database, we observed that genus- and group-specific CBPs resulted in several problems, including potential interpretation errors. This situation is best exemplified by fluoroquinolones for staphylococci and by ertapenem for E. cloacae and K. pneumoniae.
The ciprofloxacin EUCAST ECOFFs for S. aureus (20 mm) and for CoNS (24 mm) are consistent with both the EUCAST and IMM diameter distributions (Fig. 1A and B) (13). Likewise, the EUCAST levofloxacin ECOFF of 24 mm for CoNS is consistent with the IMM diameter distribution (Fig. 1D). EUCAST does not define an S. aureus levofloxacin ECOFF, most likely due to the low number of available data points (n = 107). Despite different S. aureus and CoNS ciprofloxacin ECOFFs, and a missing levofloxacin ECOFF for S. aureus, both EUCAST and CLSI CBP tables define uniform staphylococcal CBPs for fluoroquinolones, which do not differentiate S. aureus from non-S. aureus staphylococci, e.g., there are uniform staphylococcal ciprofloxacin CBPs for susceptibility at ≥20 mm and resistance at <20 mm (EUCAST) or ≥21 mm for susceptibility and resistance at ≤15 mm (CLSI), and there are uniform staphylococcal levofloxacin CBPs for susceptibility at ≥22 mm and resistance at <19 mm (EUCAST) or at ≥19 mm for susceptibility and resistance at ≤15 mm (CLSI) (3, 5). The IMM S. aureus ciprofloxacin ECOFF (20 mm; n = 5,492 isolates) (Fig. 1A) correlates with EUCAST and CLSI susceptible breakpoints. However, both EUCAST and CLSI susceptible ciprofloxacin CBPs applied to CoNS split the “out-of-ECOFF” population (usually considered “non-wild type”) to different interpretative categories (Fig. 1B). Similarly, the uniform staphylococcal EUCAST and CLSI levofloxacin CBPs split a homogeneously distributed “out-of-ECOFF” (non-wild-type) CoNS population (see IMM distribution; n = 3,832 isolates tested with levofloxacin) (Fig. 1D) between all three interpretative categories (susceptible, intermediate, and resistant). In contrast, the susceptible EUCAST levofloxacin CBP (≥22 mm) and the susceptible CLSI CBP (≥19 mm) were largely adequate for S. aureus, based on assigning only the wild-type population to the susceptible category (Fig. 1C). We assume that the uniform EUCAST staphylococcal ciprofloxacin and levofloxacin CBPs were extrapolated from S. aureus data, a procedure apparently inappropriate for CoNS. Consequently, fluoroquinolone CBPs for staphylococci must be defined either at the group level (coagulase-negative staphylococci) or in a species-specific fashion, as done by EUCAST and CLSI for cefoxitin for S. aureus and Staphylococcus lugdunensis or, most recently, by EUCAST for Staphylococcus saprophyticus and Staphylococcus pseudointermedius (3, 5).
EUCAST does not define an ertapenem ECOFF for E. cloacae. Most probably, this is due to a shoulder in the population caused by a resistant subpopulation that overlaps with the wild type, which becomes clearly visible in the E. cloacae/ertapenem IMM diameter distribution, compared to available EUCAST data (1,089 isolates with IMM [Fig. 1E] versus 109 isolates in the EUCAST distribution [13]). Despite the lack of an ECOFF for ertapenem and E. cloacae, EUCAST recommends uniform susceptible (≥25 mm)/resistant (<22 mm) ertapenem CBPs for all Enterobacteriaceae species, as does CLSI (susceptible and resistant CBPs of ≥22 mm, and ≤18 mm, respectively) (3, 5). However, testing ertapenem susceptibility in E. cloacae appears to be unreliable in terms of clinical categorization: wild-type and resistant populations cannot safely be distinguished by inhibition zone diameter determinations alone (Fig. 1E). Ertapenem AST for E. cloacae may, consequently, be discouraged, or isolates should be tested for the presence of resistance mechanisms.
For K. pneumoniae, the EUCAST and IMM ertapenem ECOFFs are identical (25 mm) (Fig. 1F). The ECOFF matches the EUCAST susceptible Enterobacteriaceae CBP of ≥25 mm, appropriately assigning wild-type isolates to the susceptible category (13). IMM distributions show a minor non-wild-type population, which overlaps the wild type, although the situation is less pronounced than with E. cloacae (Fig. 1F, compare with E). The minor K. pneumoniae non-wild-type population is categorized intermediate by EUCAST Enterobacteriaceae CBPs, which seems reasonable. According to the susceptible ertapenem CLSI CBP (≥22 mm), this minor non-wild-type population is categorized as susceptible. Clinical and PK/PD data, and also molecular characterization for resistance mechanisms, are mandatory to decide if such isolates constitute a distinct population and can be reliably treated.
For both E. cloacae and K. pneumoniae, the overlapping non-wild-type populations are, most likely, caused by increased beta-lactamase production combined with an outer membrane porin deficiency causing reduced permeability. This combination of resistance mechanisms is commonly found in E. cloacae and K. pneumoniae but rarely in E. coli (14–16).
The above examples indicate that CBPs are preferably set on a species-specific basis, to prevent erroneous treatment recommendations. Combinations of resistance mechanisms can result in complex population diameter distributions that lack a clear discrimination between wild-type and non-wild-type populations. This phenotypic indifference may warrant determination of resistance mechanisms (biochemically or genotypically) rather than mere assessment of MIC/diameter CBPs. The reduced discriminative power of CBPs becomes even more limiting when one considers measurement inaccuracies, which further hamper correct assignment of isolates to interpretative clinical categories (17). The unavoidable increase in complexity of species-specific CBPs may prompt the use of software systems for AST categorization, e.g., zone diameter classifications (18).
Pooling data from various sources may lower population discrimination.
The EUCAST E. coli ertapenem ECOFF was set at 29 mm with a minor population between 27 and 29 mm, which was obviously considered non-wild type (13). This decision was made on the basis of 579 data points from 2 sources. The IMM distribution comprised 6,974 ertapenem diameter values of clinically relevant, nonduplicate E. coli isolates. Our data suggest an ECOFF of 25 mm to appropriately assign wild-type and non-wild-type isolates (Fig. 2A), as an ECOFF of 29 mm would split the Gaussian distribution of wild-type isolates.
The S. aureus amikacin EUCAST ECOFF is 18 mm (13). However, the EUCAST distribution (841 data points from 4 sources) seems to contain a resistant S. aureus subpopulation that overlaps with the wild type, which is split by the amikacin CBPs (susceptible and resistant CBPs of ≥18 mm and <16 mm, respectively) (13). This overlapping resistant population is missing in the IMM distribution (5,702 data points) (Fig. 2B).
Diameter and MIC values are influenced by the method used for their determinations (19). EUCAST distribution data represent a compilation from numerous sources (13). This raises some concerns about data consistency and bias due to the various methods used and the different measurement precisions in individual laboratories. Technical variations between individual laboratories may decrease the discriminative power of diameter distributions due to a higher spread of distribution curves and, therefore, could hamper clear differentiation of wild-type and non-wild-type populations. On the other hand, analyzing data from multiple sources is required in order to limit bias due to local epidemiology and to make AST data more robust. The zone diameters presented in this study were determined by using an automated reading system (Sirscan). Reading zone diameters produced by using automats is officially approved by EUCAST and CLSI (5, 9). A weekly quality control is performed in our laboratory. Using EUCAST-recommended quality control isolates, we found that median diameter values, standard deviations, and diameter ranges closely matched EUCAST criteria (see Fig. S1 in the supplemental material), indicating that a comparison of IMM and EUCAST data is appropriate. In addition, we recently showed that diameter readings by the Sirscan zone reader could be reliably compared to the EUCAST-recommended manual method (20).
Possible reasons for the discrepancies observed include different epidemiologies, a lower number of data points in the EUCAST distribution, pooling results from various sources, bias due to a low number of laboratories contributing data (a single source for our data and 2 and 4 sources for the EUCAST data), and differences in ECOFF determination methods. In our study, we used the eyeball method for ECOFF determinations, i.e., a visual consensus value (8). EUCAST does not officially recommend an ECOFF method, nor does it provide information on how reported EUCAST ECOFFs were determined. The eyeball method is, however, frequently used and can at least provide a reasonable ECOFF estimation. Next to the visual inspection of data distributions, several statistical methods have been described to determine ECOFFs in order to deal with statistical problems such as skewed and/or outlier distributions, but no single method has been commonly accepted as the gold standard to date (21–25).
The data used in this study were limited, as they originated from a single source (the clinical laboratory of the IMM Zürich) and, thus, represent the local epidemiology for the organisms. While the problems described for our setting may not be encountered by all clinical laboratories, the CBP inconsistencies we observed were of a more general character.
Antibiotic disk content is critical for delineating distinct non-wild-type and wild-type populations.
For determination of high-level aminoglycoside resistance in enterococci, antibiotic disks with high drug loads have long been recommended by CLSI and the Comité de l'Antibiogramme de la Societe Francaise de Microbiologie (CA-SFM), i.e., a load of gentamicin of 100 μg/disk is recommended by CLSI and 500 μg/disk is recommended by the CA-SFM (5, 26). EUCAST recommends a lower disk content of 30 μg/disk in order to enhance the sensitivity of high-level resistance detection (3). This approach can cause practical problems in separating resistant and wild-type populations, as illustrated here with E. faecalis and gentamicin: the IMM diameter distribution for the 30-μg disk (Fig. 2C) showed a wild-type population that was left-shifted, close to the resistant population. Reliable assignment of isolates to the resistant or wild-type population is hampered due to measurement inaccuracies, as seen with the small separation between the wild-type and non-wild-type populations. As a consequence, interpretation errors will inevitably occur due to technical measurement inaccuracies (17). It may be advisable to either increase the disk content, thereby increasing the distance/separation between wild-type and high-level-resistant populations, or to introduce a gray, or “buffer” zone surrounding the ECOFF. Figure 2D shows the distribution data for E. faecalis and the 500-μg gentamicin disk according to the 2013 CA-SFM CBPs. Clearly, the wild-type population (deemed susceptible) can be separated from high-level-resistant isolates. Wild-type and high-level-resistant populations are separated by a transitional population of uncertain assignment (deemed intermediate). In addition, to assign such transitional populations to the susceptible or resistant category, guideline-issuing organizations, such as CLSI and EUCAST, are increasingly advocating use of determination of resistance mechanisms to validate the phenotypic interpretive criteria (27).
In contrast to the ECOFF definition, the setting of CBPs cannot be based on microbiological data alone but needs to consider clinical and PK/PD data as mandatory sources of information. However, microbiological data should not be assigned less weight than PK/PD data in making such determinations. Preliminary CBPs should preferably be consistent with both systems and should ultimately be validated by clinical outcome studies. In conclusion, our data illustrate some issues of general importance in determining CBPs: (i) reliable CBPs/ECOFFs require species-specific criteria, (ii) pooling data from different sources may influence diameter distributions, and (iii) the antibiotic disk content is critical for separating resistant and wild-type populations.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the laboratory technicians of the Institute of Medical Microbiology, University of Zürich, for their dedicated help, and to Reinhard Zbinden, Guido Bloemberg, Vanessa Deggim, Florian Maurer, Vera Rüegger, and Andrea Zbinden for valuable discussions.
There are no conflicts of interest to declare.
This work was supported by the University of Zürich.
Footnotes
Published ahead of print 28 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02489-13.
REFERENCES
- 1.Courvalin P. 2008. Can pharmacokinetic-pharmacodynamic parameters provide dosing regimens that are less vulnerable to resistance? Clin. Microbiol. Infect. 14:989–994. 10.1111/j.1469-0691.2008.02081.x [DOI] [PubMed] [Google Scholar]
- 2.European Committee on Antimicrobial Susceptibility Testing. 2009. Breakpoint tables for interpretation of MICs and zone diameters, version 1.0. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/EUCAST_breakpoints_v1.0_20091221.pdf Accessed 18 January 2014 [Google Scholar]
- 3.European Committee on Antimicrobial Susceptibility Testing. 2014. Breakpoint tables for interpretation of MICs and zone diameters, version 4.0. http://www.eucast.org/clinical_breakpoints/ Accessed 18 January 2014 [Google Scholar]
- 4.European Committee on Antimicrobial Susceptibility Testing. 2014. Rationale documents. http://www.eucast.org/documents/rd/ Accessed 18 January 2014 [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. CLSI document M 100-S 24. CLSI, Wayne, PA [Google Scholar]
- 6.Hombach M, Bloemberg GV, Bottger EC. 2012. Effects of clinical breakpoint changes in CLSI guidelines 2010/2011 and EUCAST guidelines 2011 on antibiotic susceptibility test reporting of Gram-negative bacilli. J. Antimicrob. Chemother. 67:622–632. 10.1093/jac/dkr524 [DOI] [PubMed] [Google Scholar]
- 7.Hombach M, Wolfensberger A, Kuster SP, Bottger EC. 2013. Influence of clinical breakpoint changes from CLSI 2009 to EUCAST 2011 AST guidelines on multidrug resistance rates of Gram-negative rods. J. Clin. Microbiol. 51:2385–2387. 10.1128/JCM.00921-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnidge J, Paterson DL. 2007. Setting and revising antibacterial susceptibility breakpoints. Clin. Microbiol. Rev. 20:391–408. 10.1128/CMR.00047-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.European Committee on Antimicrobial Susceptibility Testing. 2013. Disk diffusion manual, v. 3.0. http://www.eucast.org/antimicrobial_susceptibility_testing/disk_diffusion_methodology/ Accessed 18 January 2014 [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2008. Development of in vitro susceptibility testing criteria and quality control parameters; approved guideline, 3rd ed. CLSI document M23–A3. CLSI, Wayne, PA [Google Scholar]
- 11.European Committee on Antimicrobial Susceptibility Testing. 2010. Setting breakpoints for new antimicrobial agents, EUCAST SOP 1.0. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/EUCAST_SOPs/EUCAST_SOP_1.0_Setting_breakpoints_new_agents_20100926.pdf Accessed 18 January 2014 [Google Scholar]
- 12.Horng-Jinh C, Kuo-Chang H, Chao-Hsien W. 2006. Determination of sample size in using central limit theorem for Weibull distribution. Inf. Manage. Sci. 17:31–46 [Google Scholar]
- 13.European Committee on Antimicrobial Susceptibility Testing. 2014. Zone diameter distributions. http://www.eucast.org/zone_diameter_distributions/ Accessed 18 January 2014 [Google Scholar]
- 14.Garcia-Sureda L, Domenech-Sanchez A, Barbier M, Juan C, Gasco J, Alberti S. 2011. OmpK26, a novel porin associated with carbapenem resistance in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 55:4742–4747. 10.1128/AAC.00309-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang XD, Cai JC, Zhou HW, Zhang R, Chen GX. 2009. Reduced susceptibility to carbapenems in Klebsiella pneumoniae clinical isolates associated with plasmid-mediated beta-lactamase production and OmpK36 porin deficiency. J. Med. Microbiol. 58:1196–1202. 10.1099/jmm.0.008094-0 [DOI] [PubMed] [Google Scholar]
- 16.Woodford N, Dallow JW, Hill RL, Palepou MF, Pike R, Ward ME, Warner M, Livermore DM. 2007. Ertapenem resistance among Klebsiella and Enterobacter submitted in the UK to a reference laboratory. Int. J. Antimicrob. Agents 29:456–459. 10.1016/j.ijantimicag.2006.11.020 [DOI] [PubMed] [Google Scholar]
- 17.Hombach M, Bottger EC, Roos M. 2013. The critical influence of the intermediate category on interpretation errors in revised EUCAST and CLSI antimicrobial susceptibility testing guidelines. Clin. Microbiol. Infect. 19:E59–E71. 10.1111/1469-0691.12090 [DOI] [PubMed] [Google Scholar]
- 18.Winstanley T, Courvalin P. 2011. Expert systems in clinical microbiology. Clin. Microbiol. Rev. 24:515–556. 10.1128/CMR.00061-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koeth LM, King A, Knight H, May J, Miller LA, Phillips I, Poupard JA. 2000. Comparison of cation-adjusted Mueller-Hinton broth with Iso-Sensitest broth for the NCCLS broth microdilution method. J. Antimicrob. Chemother. 46:369–376. 10.1093/jac/46.3.369 [DOI] [PubMed] [Google Scholar]
- 20.Hombach M, Zbinden R, Bottger EC. 2013. Standardisation of disk diffusion results for antibiotic susceptibility testing using the sirscan automated zone reader. BMC Microbiol. 13:225. 10.1186/1471-2180-13-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canton E, Peman J, Hervas D, Iniguez C, Navarro D, Echeverria J, Martinez-Alarcon J, Fontanals D, Gomila-Sard B, Buendia B, Torroba L, Ayats J, Bratos A, Sanchez-Reus F, Fernandez-Natal I, FUNGEMYCA Study Group 2012. Comparison of three statistical methods for establishing tentative wild-type population and epidemiological cutoff values for echinocandins, amphotericin B, flucytosine, and six Candida species as determined by the colorimetric Sensititre YeastOne method. J. Clin. Microbiol. 50:3921–3926. 10.1128/JCM.01730-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kronvall G, Kahlmeter G, Myhre E, Galas MF. 2003. A new method for normalized interpretation of antimicrobial resistance from disk test results for comparative purposes. Clin. Microbiol. Infect. 9:120–132. 10.1046/j.1469-0691.2003.00546.x [DOI] [PubMed] [Google Scholar]
- 23.Meletiadis J, Mavridou E, Melchers WJ, Mouton JW, Verweij PE. 2012. Epidemiological cutoff values for azoles and Aspergillus fumigatus based on a novel mathematical approach incorporating cyp51A sequence analysis. Antimicrob. Agents Chemother. 56:2524–2529. 10.1128/AAC.05959-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaller MA, Boyken L, Hollis RJ, Kroeger J, Messer SA, Tendolkar S, Diekema DJ. 2011. Wild-type MIC distributions and epidemiological cutoff values for posaconazole and voriconazole and Candida spp. as determined by 24-hour CLSI broth microdilution. J. Clin. Microbiol. 49:630–637. 10.1128/JCM.02161-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turnidge J, Kahlmeter G, Kronvall G. 2006. Statistical characterisation of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin. Microbiol. Infect. 12:418–425. 10.1111/j.1469-0691.2006.01377.x [DOI] [PubMed] [Google Scholar]
- 26.Comité de l'Antibiogramme de la Societe Francaise de Microbiologie. 2013. Recommandations. http://www.sfm-microbiologie.org/ Accessed 18 January 2014 [Google Scholar]
- 27.European Committee on Antimicrobial Susceptibility Testing. 2014. EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. http://www.eucast.org/resistance_mechanisms/ Accessed 18 January 2014 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.