Abstract
Communication of antibiotic resistance among bacteria via small molecules is implicated in transient reduction of bacterial susceptibility to antibiotics, which could lead to therapeutic failures aggravating the problem of antibiotic resistance. Released putrescine from the extremely antibiotic-resistant bacterium Burkholderia cenocepacia protects less-resistant cells from different species against the antimicrobial peptide polymyxin B (PmB). Exposure of B. cenocepacia to sublethal concentrations of PmB and other bactericidal antibiotics induces reactive oxygen species (ROS) production and expression of the oxidative stress response regulator OxyR. We evaluated whether putrescine alleviates antibiotic-induced oxidative stress. The accumulation of intracellular ROS, such as superoxide ion and hydrogen peroxide, was assessed fluorometrically with dichlorofluorescein diacetate, while the expression of OxyR and putrescine synthesis enzymes was determined in luciferase assays using chromosomal promoter-lux reporter system fusions. We evaluated wild-type and isogenic deletion mutant strains with defects in putrescine biosynthesis after exposure to sublethal concentrations of PmB and other bactericidal antibiotics. Exogenous putrescine protected against oxidative stress induced by PmB and other antibiotics, whereas reduced putrescine synthesis resulted in increased ROS generation and a parallel increased sensitivity to PmB. Of the 3 B. cenocepacia putrescine-synthesizing enzymes, PmB induced only BCAL2641, an ornithine decarboxylase. This study reveals BCAL2641 as a critical component of the putrescine-mediated communication of antibiotic resistance and as a plausible target for designing inhibitors that would block the communication of such resistance among different bacteria, ultimately reducing the window of therapeutic failure in treating bacterial infections.
INTRODUCTION
The relentless increase in multidrug resistance, particularly intrinsic high-level resistance, undermines new treatments improving health and extending the life of patients, especially those with chronic conditions (1). For example, respiratory failure secondary to chronic pulmonary bacterial infection in patients with cystic fibrosis (CF) hinders the dramatic improvements in survival achieved over the last several decades and remains the primary cause of death (2). The emergence of growing numbers of cystic fibrosis pathogens with intrinsic multidrug resistance, such as Burkholderia cepacia complex, Stenotrophomonas maltophilia, Achromobacter xylosoxidans, and nontuberculous mycobacteria creates a further need for novel therapies (2). We investigate the mechanisms of high-level intrinsic multidrug resistance using Burkholderia cenocepacia as a model bacterium. B. cenocepacia is an environmental, opportunistic pathogen that belongs to the B. cepacia complex and causes serious respiratory infections in CF patients (3). These infections are associated with faster decline in lung function, debilitating exacerbations, and ultimately death (4–6), and they also reduce the survival of CF patients after lung transplant (7).
While genetic mechanisms are considered the quintessential means of transfer of antibiotic resistance traits among bacteria, small molecules are also capable of modulating the antibiotic response of bacteria (8). The clinical outcome of antibiotic treatment does not always correlate with the expectations based on in vitro susceptibility testing performed on individual clinical isolates (9). Due to the polymicrobial nature of many infections (10), cross talk between the different bacterial species is likely to occur during infection. Such chemical communication of antibiotic resistance among bacteria may aggravate the problem of antibiotic resistance by potentially causing transient reduction in the susceptibility to antibiotics, leading to therapeutic failures. For example, a transient increase in resistance to antimicrobial peptides by exposure to host polyamines was shown for the urogenital pathogen Neisseria gonorrhoeae (11). Identification of chemical communicators of antibiotic resistance and their mechanism of protection would provide another avenue for intervention to combat the increase and spread of antimicrobial resistance. Recently, we demonstrated that B. cenocepacia exhibits a nongenetic mechanism to reduce antibiotic susceptibility that is chemically mediated by putrescine and YceI, a small secreted protein of unknown function that is highly conserved in bacteria (12). Putrescine is a polyamine produced by almost all living organisms (13). When released from B. cenocepacia, putrescine protects less-resistant cells from the same and different species from the antimicrobial peptide polymyxin B (PmB) (12).
The mechanism of protection is partly due to the ability of putrescine to compete with PmB for binding to the surface of B. cenocepacia (12). However, polyamines can also quench oxidative species (14) and protect membranes from lipid peroxidation (15). Various classes of antibiotics induce oxidative stress and increased production of reactive oxygen species (ROS) (16–19). Although the specific lethal role of ROS generated in response to antibiotics remains under discussion (16, 20, 21), oxidative stress constitutes a burden on the bacterial cells (22). Conceivably, protection from oxidative stress accompanying antibiotic exposure would improve the bacterial response to antibiotics, thus increasing resistance.
Here we show that when present at sublethal concentrations, PmB and other bactericidal antibiotics induce oxidative stress in B. cenocepacia. Our findings revealed that exogenous and endogenous putrescine protects against antibiotic-mediated oxidative stress. This work exposes another mechanism of putrescine-mediated protection from antibiotics alongside protection of the cell surface from binding of PmB as previously described (12). By examining the expression patterns of the different putrescine-synthesizing enzymes in response to antibiotics, we discovered that the ornithine decarboxylase BCAL2641 is a plausible target for designing inhibitors that would block putrescine-mediated communication of antibiotic resistance among different bacteria, ultimately reducing the window of therapeutic failure in treating bacterial infections.
MATERIALS AND METHODS
Strains and reagents.
Table 1 lists the bacteria and plasmids used in this study. Bacteria grew in LB at 37°C. Antibiotics (Sigma, St. Louis, MO) were diluted in water, except for PmB, which was diluted in 0.2% bovine serum albumin–0.01% glacial acetic acid buffer. Rifampin was dissolved in dimethyl sulfoxide (DMSO).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference(s) |
|---|---|---|
| Strains | ||
| Burkholderia cenocepacia | ||
| K56-2 | ET12 clone related to J2315 (CF clinical isolate, BCRRC) | 43 |
| OME11 | K56-2 ΔBCAL2641 | 12 |
| OME12 | K56-2 ΔBCAM1111-1112 | 12 |
| OME49 | OME12 Prha::BCAL2641 | This study |
| OME50 | K56-2 PBCAL2641::pGSVTp-luxCDABE; Tpr | This study |
| OME51 | OME12 PBCAL2641::pGSVTp-luxCDABE; Tpr | This study |
| OME52 | K56-2 PBCAM1111::pGSVTp-luxCDABE; Tpr | This study |
| OME53 | OME11 PBCAM1111::pGSVTp-luxCDABE; Tpr | This study |
| OME54 | K56-2 PBCAM1112::pGSVTp-luxCDABE; Tpr | This study |
| OME55 | OME11 PBCAM1112::pGSVTp-luxCDABE; Tpr | This study |
| OME56 | K56-2 PoxyR::pGSVTp-luxCDABE; Tpr | This study |
| OME57 | OME11 PoxyR::pGSVTp-luxCDABE; Tpr | This study |
| OME58 | OME12 PoxyR::pGSVTp-luxCDABE; Tpr | This study |
| Escherichia coli | ||
| DH5α | F− ϕ80lacZ M15 endA1 recA1 supE44 hsdR17(rK− mK+) deoR thi-1 nupG supE44 gyrA96 relA1 Δ(lacZYA-argF)U169 λ− | Laboratory stock |
| GT115 | F− mcrAΔ(mrr-hsdRMS-mcrBC) ϕ80ΔlacZΔM15 ΔlacX74 recA1 rpsL(StrA) endA1 Δdcm uidA(ΔMluI)::pir-116 ΔsbcC-sbcD | InvivoGen |
| Plasmids | ||
| pRK2013 | oricolE1, RK2 derivative; Kanr mob+ tra+ | 26 |
| pGSVTp-lux | Mobilizable suicide vector containing the lux operon, derivative from pGSV3-lux; OriT Tpr | 44, 45 |
| pSC200 | oriR6K, PRhaB rhamnose-inducible promoter, Tpr mob+ | 28 |
| pOE14 | PBCAL2641::luxCDABE transcriptional fusion in pGSVTp-lux | This study |
| pOE17 | Prha::BCAL2641 in pSC200 | This study |
| pOE18 | PBCAM1111::luxCDABE transcriptional fusion in pGSVTp-lux | This study |
| pOE19 | PBCAM1112::luxCDABE transcriptional fusion in pGSVTp-lux | This study |
| pOE20 | PoxyR::luxCDABE transcriptional fusion in pGSVTp-lux | This study |
Tpr, trimethoprim resistance; Kanr, kanamycin resistance; Tetr, tetracycline resistance; BCRRC, B. cepacia Research and Referral Repository for Canadian CF Clinics.
General molecular techniques.
DNA manipulations were performed as previously described (23). T4 DNA ligase (Roche Diagnostics, Laval, Quebec, Canada), Antarctic phosphatase (New England BioLabs, Pickering, Ontario, Canada) and restriction endonucleases (Roche or New England BioLabs) were used as recommended by the manufacturers. Transformation of Escherichia coli GT115 was performed using the calcium chloride method (24). Mobilization of plasmids into B. cenocepacia was conducted by triparental mating (25), using E. coli DH5α carrying the helper plasmid pRK2013 (26). DNA amplification by PCR was performed using a C1000 thermal cycler (Bio-Rad Laboratories, Ltd., Mississauga, Ontario, Canada) with Taq or HotStar HiFidelity DNA polymerases (Qiagen, Mississauga, Ontario, Canada) and optimized for each primer pair. DNA sequencing was carried out at the DNA sequencing Facility of York University, Toronto, Canada, or at Eurofins MWG Operon, Huntsville, AL. The DNA sequences were analyzed with the BLAST computer program and compared to the sequenced genome of B. cenocepacia strain J2315.
Fluorometric determination of ROS.
Overnight cultures of the parental B. cenocepacia strain K56-2 and the appropriate mutants in LB medium were diluted to an optical density at 600 nm (OD600) of 0.1 in fresh medium. Five-milliliter aliquots were incubated at 37°C for 3 h at 200 rpm. Antibiotics and/or putrescine was added at the specified concentrations, and the cultures were further incubated at 37°C for 2 h at 200 rpm. After incubation, the OD600 was measured, and aliquots containing cells equivalent to an OD600 of 0.4 were pelleted, washed with phosphate-buffered saline (PBS), and resuspended in 1 ml of PBS. Superoxide radicals and other ROS were determined by diluting the obtained suspension 100-fold in 1 ml PBS and adding 2′,7′-dichlorofluorescein diacetate (DCF) to a final concentration of 2 μM. The reaction mixture was then incubated at 37°C for 30 min with rotation. After incubation, the fluorescence was measured in 200-μl aliquots placed into 96-well white plates (Microfluor-2 White; Thermo Scientific) at λ excitation (λex) of 480 nm and λ emission (λem) of 521 nm, using a Cary Eclipse fluorescence spectrophotometer (Varian, Inc., Mississauga, Ontario, Canada). In addition, the OD600s of the same suspensions were measured and used to normalize the fluorescence values. Hydroxyl radical production was determined in 600-μl bacterial suspensions without dilution using 3′-(p-hydroxyphenyl) fluorescein (HPF) at a final concentration of 5 μM. Fluorescence was measured at λex of 495 nm and λem of 530 nm in 200-μl aliquots placed into 96-well white plates. Background fluorescence of each probe in buffer control was subtracted. Autofluorescence of the bacterial suspensions, without adding the probes, was measured and corrected for by subtraction from the fluorescence signals. Data were normalized to the OD600 of the bacterial suspensions. The suspensions were protected from light throughout the assays to avoid photo-oxidation.
Antibiotic susceptibility testing.
Overnight cultures of the parental B. cenocepacia strain K56-2 and the appropriate mutants in LB medium were diluted to an optical density at 600 nm (OD600) of 0.0008 (low inoculum) or 0.04 (high inoculum) in fresh LB medium and 0.04 in fresh M9 minimal medium with or without the antibiotic and incubated at 37°C with medium continuous shaking in a Bioscreen C automated growth curve analyzer (MTX Lab Systems, Vienna, VA). Bacterial growth was assessed turbidimetrically at 600 nm.
In vitro antioxidant activity assay.
The ability of putrescine to scavenge free radicals was determined using a system of in vitro generation of superoxide radicals containing phenazine methosulfate (PMS)-NADH as previously described (27). Briefly, the reaction mixture consisted of 21 mM phosphate buffer (pH 8.3), 0.7 mM NADH, 17 μM Nitro Blue Tetrazolium, and the corresponding quantity of putrescine. The reaction was initiated by adding 4 μM PMS. The reaction mixtures were mixed, and the amount of formazan formation was measured immediately using the spectrophotometer at 560 nm. The percentage of inhibition of formazan formation by putrescine was calculated relative to the control lacking putrescine.
Transcriptional fusions to luxCDABE.
The promoter regions from BCAL2641, BCAM1111, BCAM1112, and OxyR were PCR amplified. The PCR products were digested with EcoRI and cloned into the EcoRI-digested and dephosphorylated pGSVTp-lux plasmid. The orientation of the promoter region was checked by PCR and luminescence of E. coli GT115 colonies carrying the plasmids. The resulting plasmids contained the promoter region of the genes of interest fused to the luxCDABE reporter system. The plasmids were mobilized into K56-2 and the appropriate mutants by triparental mating. Transconjugants (carrying the chromosomal promoter-reporter fusions) were selected on LB agar plates containing 100 μg/ml trimethoprim (Tp), 200 μg/ml ampicillin, and 10 μg/ml gentamicin.
Luminescence expression assays.
Overnight cultures in LB containing 100 μg/ml Tp were diluted into fresh LB medium to an OD600 of 0.04. After addition of the antibiotics and/or putrescine, 300 μl of sample was loaded in triplicate, for each time point, in a 100-well honeycomb microtiter plate. The plates were incubated at 37°C with continuous shaking in a Bioscreen C automated growth curve analyzer (MTX Lab Systems, Vienna, VA). Growth was monitored by measuring the OD600 at 37°C every 30 min. At predetermined time points postinoculation, the Bioscreen was paused, and three 200-μl aliquots for each condition tested were transferred into a flat-bottom 96-well microtiter plate (Microfluor-2 White; Thermo Scientific), and luminescence (in relative light units [RLU]) was measured using a Fluoroskan Ascent FL microplate fluorometer and luminometer (Thermo Scientific, Ottawa, Ontario, Canada). Expression levels of each gene of interest in the different strain backgrounds were calculated as RLU/OD600 for each time point.
Construction of a conditional mutant.
A fragment (∼300 bp) spanning the 5′ region of BCAL2641 was PCR amplified, digested by NdeI and XbaI, and cloned into the NdeI- and XbaI-digested and dephosphorylated pSC200 plasmid. The plasmids were mobilized into OME12 (ΔBCAM1111-BCAM1112 [hereafter, ΔBCAM1111-1112]) by triparental mating. Transconjugants were selected on LB agar plates containing 100 μg/ml of trimethoprim (Tp), 200 μg/ml ampicillin, 10 μg/ml gentamicin, and 0.5% (wt/vol) rhamnose. This strategy creates conditional mutants in which the expression of the targeted gene depended on the rhamnose concentration in the medium (28).
TLC analyses of polyamines.
The conditional mutant and the wild type were grown at 37°C in M9 minimal medium supplemented with final concentrations of 100 μg/ml Tp and 0.4% (wt/vol) rhamnose (the permissive condition of expression). An aliquot of an overnight culture in M9 medium with rhamnose was spun down and washed three times with sterile phosphate-buffered saline (PBS), resuspended in PBS, and adjusted to an OD600 of 1. Drops (10 μl) of undiluted suspension and 10-fold serial dilutions were plated onto M9 agar plates supplemented with 0.4% (wt/vol) glucose and incubated at 37°C (the nonpermissive condition of expression). Bacteria growing on the plates were collected and suspended in sterile PBS, and the OD600 was adjusted to 0.1. Polyamines were extracted, derivatized to their dansyl derivatives, sequentially separated on thin-layer chromatography (TLC) silica gel plates (20 by 20 cm) (Merck, Darmstadt, Germany) in two solvent systems—(i) benzene-triethylamine (20:2 [vol/vol]) and (ii) benzene-methanol (10:0.45 [vol/vol])—and visualized under UV light as previously described (12). Standard solutions of putrescine, cadaverine, spermidine, and spermine (0.2 mM each) were treated similarly and included as controls.
Catalase enzyme activity assay.
Overnight cultures of the wild-type B. cenocepacia strain K56-2 in LB were diluted to an OD600 of 0.04 in 30 ml fresh LB medium, with or without antibiotics, and incubated at 37°C at 200 rpm for 16 h. Bacterial cells were pelleted, washed with sterile PBS, and resuspended in 300 μl PBS (or less if necessary, depending on bacterial inhibition of growth by antibiotics). The OD600 of the bacterial suspensions was measured. The catalase enzyme activity was evaluated by the method described by Iwase et al. (29). Briefly, 100 μl of bacterial suspension or bovine liver catalase solution at different concentrations was added in a glass tube followed by the addition of 100 μl of 1% Triton X-100. Finally, 100 μl of undiluted hydrogen peroxide (30%) was added to the solutions, mixed thoroughly, and incubated at room temperature. The height of O2-forming foam that remained constant for 15 min in the test tube was finally measured using a ruler. The catalase activity of bacterial suspensions was determined using calibration curves constructed using the standard catalase solutions with different concentrations and normalized to the OD600 of the tested suspensions.
Statistical analyses.
Unpaired Student's t tests were conducted with GraphPad Prism 5.0.
RESULTS AND DISCUSSION
Putrescine reduces ROS production induced by PmB.
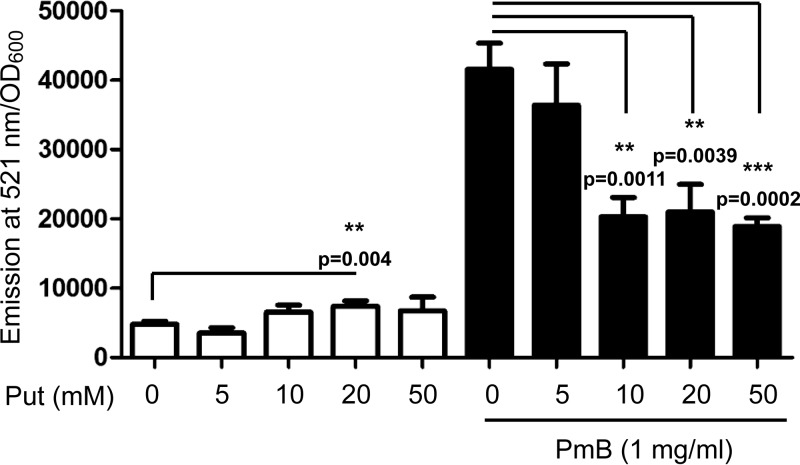
Treatment of B. cenocepacia K56-2 with 1 mg/ml PmB led to significantly increased production of intracellular ROS, as detected by 2′,7′-dichlorofluorescein diacetate (DCF) (Fig. 1). DCF is a colorless, nonfluorescent fluorescein derivative that passively diffuses into cells, where the two acetate groups are cleaved by intracellular esterases to yield the non-cell-permeable 2′,7′-dichlorofluorescein (30). This cleaved product becomes trapped within the cells and becomes oxidized by intracellular ROS, resulting in the formation of a highly fluorescent product; hence, it is a measure of generalized oxidant production rather than that of any particular reactive species (30). Lower concentrations of PmB (0.5 mg/ml or less) did not alter the intracellular DCF-detectable ROS pool (data not shown), whereas due to its reduced solubility in the culture medium, higher concentrations of PmB could not be reliably tested. Since putrescine protects B. cenocepacia from PmB (12), we assessed whether it also alleviates PmB-induced ROS production. Compared to control cells, exogenous putrescine reduced DCF-detectable ROS generation in PmB-treated bacteria (Fig. 1). This effect was assessed at 2 h of incubation with PmB and/or putrescine to avoid potential interference from putrescine degradation or metabolic by-products at prolonged incubation times. It should be noted that putrescine did not decrease the background ROS levels produced by bacterial cells not exposed to PmB but rather caused a slight but significant increase in DCF-detected ROS levels compared to the level in control cells at 20 mM (Fig. 1, white bars). We attributed these results to polyamine catabolism, which also generates ROS (31).
FIG 1.
Putrescine reduces ROS production induced by PmB in B. cenocepacia K56-2. ROS were detected by DCF. n = 6 from 2 independent experiments. **, P < 0.01; ***, P < 0.001.
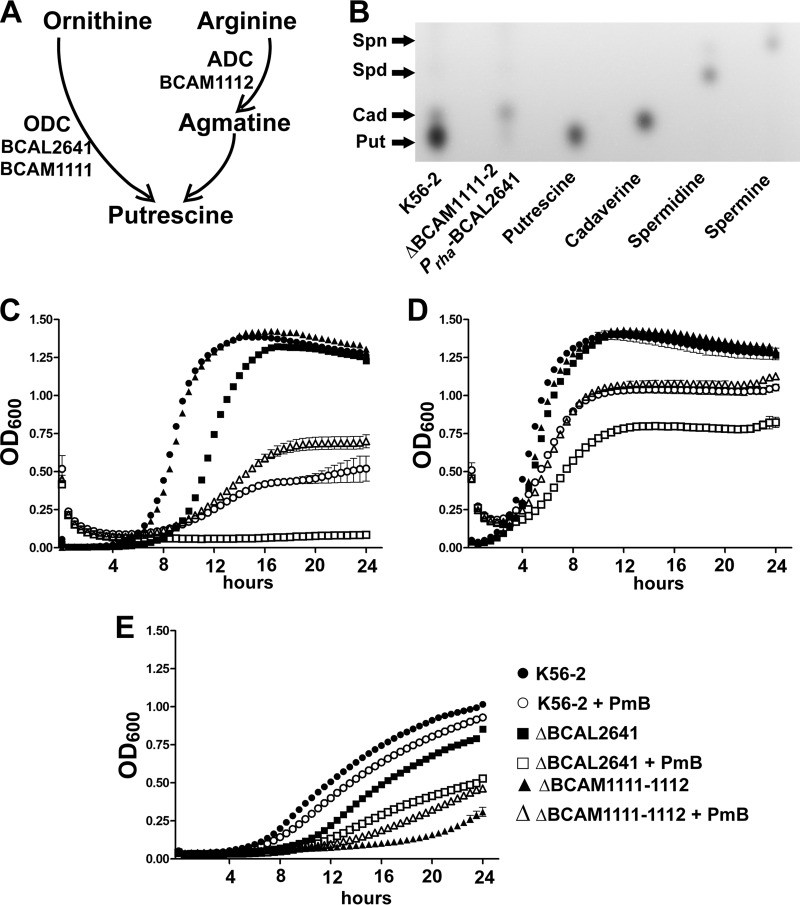
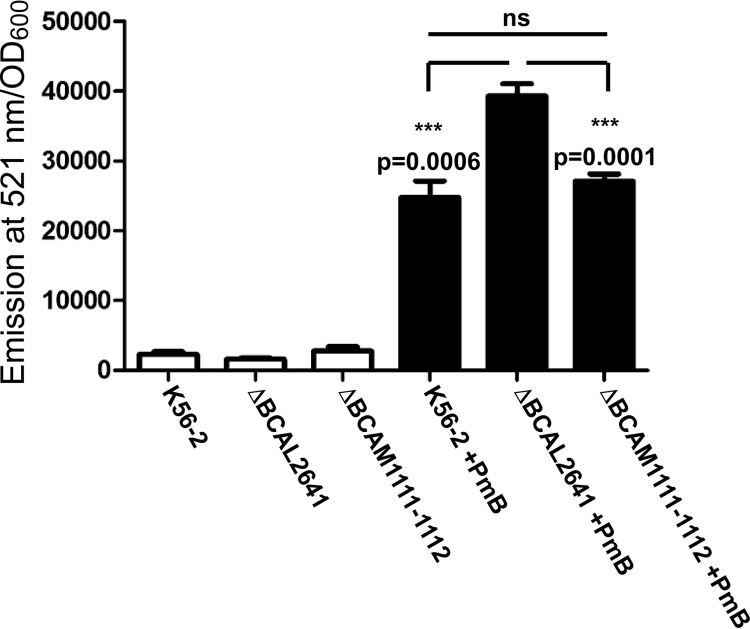
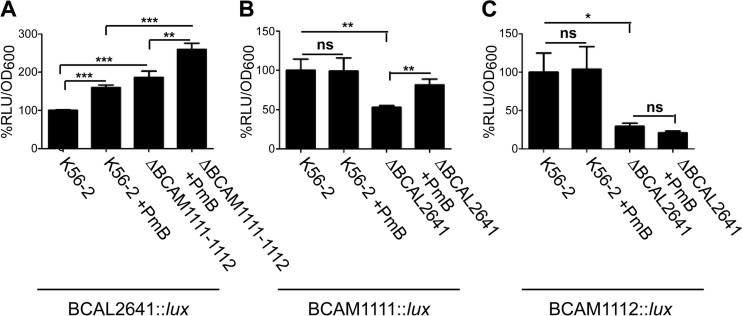
To assess whether endogenous putrescine also has the ability to reduce ROS levels in PmB-treated B. cenocepacia, we employed deletion mutants in the putrescine biosynthesis pathway. Putrescine can arise through the action of either ornithine decarboxylase or arginine decarboxylase (12). B. cenocepacia has two ornithine decarboxylase homologues, BCAL2641 and BCAM1111, and one arginine decarboxylase protein, BCAM1112 (Fig. 2A). The ornithine decarboxylase BCAL2641 is encoded by a gene located on chromosome 1 of B. cenocepacia; whereas both the ornithine decarboxylase BCAM1111 and the arginine decarboxylase BCAM1112 are encoded by genes located adjacent to each other, but in the opposite orientation, on chromosome 2. In a previous study, we showed that the ΔBCAL2641 mutant had a greater reduction in the amount of secreted putrescine compared with the wild type than the ΔBCAM1111-1112 mutant (12). Here, we confirmed that these three enzymes are the only contributors to putrescine production in B. cenocepacia. A conditional mutant of BCAL2641 in the ΔBCAM1111-1112 background did not produce detectable levels of putrescine under the nonpermissive conditions of expression compared to the wild-type strain (Fig. 2B). With respect to the response to PmB, the ornithine decarboxylase BCAL2641 was the only enzyme, among the 3 putrescine synthesis enzymes, involved in resistance against PmB. The ΔBCAL2641 mutant, but not the ΔBCAM1111-1112 mutant, had increased susceptibility to PmB compared to the wild type when tested in LB medium (Fig. 2C and D) or M9 medium (Fig. 2E). Although the growth of the ΔBCAM1111-1112 mutant was not impaired in LB medium regardless of the initial inoculum size (Fig. 2C and D), it exhibited significant reduction in growth compared to the wild-type cells in M9 medium (Fig. 2E). Nevertheless, this mutant did not show increased susceptibility to PmB in M9 medium, in which its growth was retarded (Fig. 2E). On the contrary, the ΔBCAL2641 mutant showed slight reduction in growth in LB medium only at a low inoculum size (Fig. 2C) but not at a high inoculum size (Fig. 2D) or in M9 medium (Fig. 2E). This suggests that these genes involved in putrescine synthesis are not functionally redundant; they seem to be stimulated under different conditions and regulated differently, with BCAL2641 only involved in resistance to antibiotics. Next, detection of ROS by DCF was assessed after incubation of the ΔBCAL2641 and ΔBCAM1111-1112 mutants with PmB for 16 h to allow the different enzymes to reach their maximum expression levels, which occurred at about 12 h in the luminescence expression assays (not shown). No differences were observed in PmB-untreated cells between the wild type and the deletion mutants (Fig. 3, white bars). In contrast, the ΔBCAL2641 mutant exhibited a significant increase in levels of superoxide and other ROS detected by DCF in response to PmB compared to the wild type, whereas the ΔBCAM1111-1112 mutant produced the same level as the parental strain (Fig. 3). Together, these results support the notion that putrescine reduces the level of PmB-induced ROS production, and this reduction contributes to protection of bacteria from the bactericidal effects of PmB.
FIG 2.
BCAL2641 is the only putrescine synthesis enzyme in B. cenocepacia involved in reduced susceptibility to PmB. (A) Putrescine synthesis pathway in B. cenocepacia K56-2 together with the enzymes involved. ADC, arginine decarboxylase; ODC, ornithine decarboxylase. (B) TLC plate showing the lack of production of putrescine in the ΔBCAM1111-1112 Prha-BCAL2641 conditional mutant under nonpermissive conditions. Put, putrescine; Cad, cadaverine; Spd, spermidine; Spn, spermine. (C to E) Sensitivity of the wild-type and ΔBCAL2641 (OME11) and ΔBCAM1111-1112 (OME12) putrescine synthesis mutants to 2,048 μg/ml PmB as determined turbidimetrically. n = 3 from a representative experiment. (C) Low initial inoculum in LB medium; (D) high initial inoculum in LB medium; (E) M9 minimal medium.
FIG 3.
BCAL2641 is the main ornithine decarboxylase responsible for reduction of ROS accumulation. ROS production in response to 1 mg/ml PmB in wild-type strain K56-2 compared to the ΔBCAL2641 (OME11) and ΔBCAM1111-1112 (OME12) putrescine synthesis mutants detected by DCF. n = 6 from 2 independent experiments. ns, not significant. ***, P < 0.001.
Hydroxyl radical is another ROS that may be produced upon oxidative stress. Others have used hydroxyphenyl fluorescein (HPF) to fluorometrically detect hydroxyl radicals upon antibiotic stress (16). Using HPF in experiments similar to those described above, we found a comparable pattern of reduction of PmB-induced ROS by putrescine (data not shown). However, the fluorescence signal detected by HPF was too low compared to that detected by DCF and required a 100-fold-higher inoculum than that for the DCF experiments to detect signal above the background noise of fluorescence. Such a high inoculum of cells led to high autofluorescence compared to the actual fluorescence signal detected upon addition of HPF, which was not the case with the DCF assays (see Fig. S1 in the supplemental material). Thus, we disregarded the results of HPF assays. Similar criticism about the use of HPF was raised recently concerning the interference between the autofluorescence of cells with the actual fluorescence in the presence of the probe, especially upon antibiotic treatment (32).
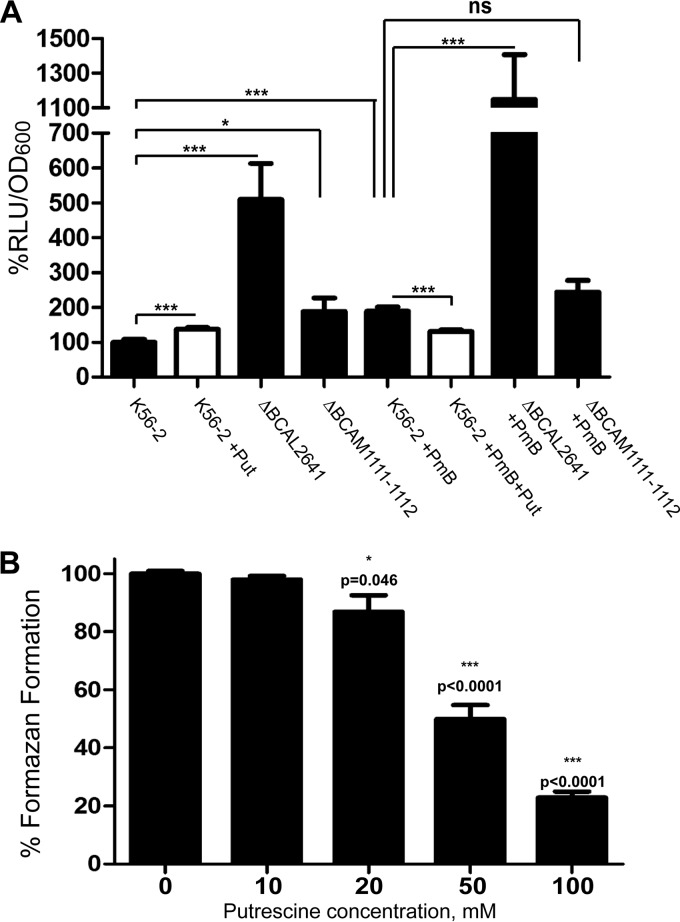
Although the DCF fluorometric assay is a well-established method and has many advantages over other techniques developed for measurement of intracellular ROS (30), the probe may be nonselective in reacting with other oxidants, such as hydroxyl radicals and lipid peroxides (33). Hence, to provide additional evidence supporting the DCF fluorometric assays results, we measured the expression of OxyR as an independent indicator of oxidative stress. OxyR belongs to the LysR family of transcription factors whose regulon is involved in the cellular response to oxidative stress (22). OxyR is very sensitive to ROS concentrations; it is activated at very low hydrogen peroxide concentrations, leading to upregulation to its regulon (34). Moreover, expression of oxyR is upregulated in response to hydrogen peroxide using LacZ promoter fusions (35); similarly, another LysR-type transcription regulator involved in the response to oxidative stress is overexpressed in response to ROS (36). Therefore, we constructed derivatives of wild-type and mutant strains carrying an oxyR::lux promoter fusion to measure oxyR gene expression at chromosomal levels. PmB stimulated the oxyR expression (Fig. 4A), which was consistent with the induction of intracellular ROS detected by DCF (Fig. 1 and 3). Likewise, catalase activity, regulated by OxyR (22), increased in response to PmB (Table 2). This further confirms the induction of intracellular ROS in response to PmB and validates the findings of DCF fluorometric and oxyR expression assays as measures of intracellular ROS. oxyR expression was significantly higher in the ΔBCAL2641 mutant than in the parental strain in the presence or absence of PmB. In contrast, no difference in oxyR expression between the wild type and the ΔBCAM1111-1112 mutant was detected in response to PmB (Fig. 4A). No differences in the growth rates of the different strains were noted in the absence of PmB, whereas the ΔBCAL2641 mutant was more susceptible to PmB than the wild type and the ΔBCAM1111-1112 mutant (see Fig. S2 in the supplemental material). These results follow the same pattern of ROS generated in response to PmB in the mutants compared to the wild-type strain (Fig. 3).
FIG 4.
(A) Induction of OxyR expression as an indicator of ROS accumulation in the wild type (OME56) compared to the ΔBCAL2641 (OME57) and ΔBCAM1111-1112 (OME58) putrescine synthesis mutants in response to 500 μg/ml PmB with or without 10 mM putrescine determined by the luciferase expression assay at 3 h. Results are shown as percentage of relative light units (RLU)/OD600 relative to the OME56 control (K56-2 background). The mean RLU/OD600 of the control is 0.09567. The percentages of OD600 are shown in Fig. S2 in the supplemental material. n = 9 from 3 different clones. ns, not significant. *, P < 0.05; and ***, P < 0.001. (B) In vitro antioxidant activity of putrescine. n = 6 from 2 independent experiments.
TABLE 2.
Catalase enzyme activities in this study
| Antibiotic (concn, μg/ml) | Catalase activity, % U/OD600 (SEM)a | P value for difference from controlb |
|---|---|---|
| None | 100 (1.3) | NA |
| Polymyxin B (500) | 120.1 (4.2) | 0.0002 |
| Norfloxacin (8) | 110.6 (1.2) | 0.0012 |
| Rifampin (16) | 134.5 (5.2) | <0.0001 |
| Ceftazidime (32) | 99.8 (12.4) | 0.978 (NS) |
| Gentamicin (1,000) | 48.0 (7.1) | <0.0001 |
, standard error of the mean. Results from 2 independent experiments are shown (n = 6). The r2 values of the calibration curves are 0.9644 and 0.9544.
NA, not applicable; NS, not significant.
Next, we investigated the mechanism by which putrescine protects from oxidative stress. Putrescine stimulated the expression of oxyR (Fig. 4A), probably as a result of a slight induction of ROS accumulation as detected by DCF (Fig. 1). However, putrescine alleviated the increase in oxyR expression in response to PmB (Fig. 4A), suggesting a protective effect against ROS. Nevertheless, putrescine did not induce a statistically significant difference in growth of the wild type in the presence or absence of PmB at this early time point of incubation (3 h) under the conditions of this test (see Fig. S2 in the supplemental material). Supporting the protective role of putrescine from oxidative stress, we confirmed the antioxidant properties of putrescine by demonstrating that it could scavenge superoxide radicals generated in vitro from a phenazine methosulfate-NADH system in a concentration-dependent manner (Fig. 4B). Together, the results of this section reveal a link between reduced susceptibility to PmB, induction of ROS production, and expression of OxyR with the intracellular level of putrescine, which can be attributed to the antioxidant properties of this polyamine.
Expression of the putrescine synthesis enzymes in response to PmB.
To better understand the role of the different putrescine-synthesizing enzymes in response to oxidative stress and consequently to PmB, we investigated the expression profiles of their corresponding genes also using lux promoter fusions as before. BCAL2641::lux gene expression was stimulated by exposure to PmB (Fig. 5A); whereas neither BCAM1111::lux nor BCAM1112::lux fusions were responsive to PmB (Fig. 5B and C, respectively). This agrees with the behavior of the ΔBCAL2641 and ΔBCAM1111-1112 mutants in response to PmB in terms of antimicrobial resistance (Fig. 2C to E) and ROS production (Fig. 3). Moreover, this is consistent with our previous data showing increased transcription of BCAL2641, but not BCAM1111 or BCAM1112, in response to PmB (12). BCAL2641 also appears to regulate by an unknown mechanism the gene expression of the BCAM1111 and BCAM1112 putrescine synthesis enzymes, since the expression of both genes was significantly reduced in the ΔBCAL2641 background (Fig. 5B and C, respectively). This regulation is not mediated through the action of putrescine since 10 mM putrescine did not stimulate the gene expression of BCAM1111 or BCAM1112 (not shown). Other indirect regulatory pathways may be involved, which will require further investigation. On the other hand, the gene expression of BCAL2641 increased in the absence of BCAM1111 and BCAM1112 (Fig. 5A), which may explain the slight increase in survival of the ΔBCAM1111-1112 mutant when exposed to PmB shown in Fig. 2E. This might be due to compensation for the reduced synthesis of putrescine by these enzymes being normally stimulated by BCAL2641. Alternatively, BCAM1111 and BCAM1112 might provide feedback inhibition to BCAL2641; thus, their absence would lead to increased BCAL2641 gene expression. Notably, the expression of BCAM1112 (RLU/OD600, 0.2423) is much lower than that of the other 2 enzymes (RLU/OD600, 1.4829 and 1.5585 for BCAL2641 and BCAM1111, respectively). This suggests that B. cenocepacia does not preferentially utilize the arginine decarboxylase BCAM1112. This agrees with the fact that B. cepacia can degrade arginine only through the use of the succinyl transferase pathway, despite the possession of an arginine decarboxylase homologue (37, 38). Except for the ΔBCAL2641 mutant, which exhibited reduced growth in the presence of PmB, no differences in growth were observed in the other strains tested regardless of PmB exposure (see Fig. S3 in the supplemental material). Together, our findings expose BCAL2641 as a crucial contributor of putrescine synthesis in the response against antibiotics.
FIG 5.
Luciferase expression assay of the different putrescine-synthesizing enzymes in response to 500 μg/ml PmB at 3 h. Results are shown as percentage of relative light units (RLU)/OD600 relative to the control (untreated K56-2 background). The percentages of OD600 are shown in Fig. S3 in the supplemental material. (A) Expression of BCAL2641 in the wild-type (OME50) and ΔBCAM1111-1112 (OME51) backgrounds. n = 6 from 2 different clones. The mean RLU/OD600 of the control is 1.4829. (B) Expression of BCAM1111 in the wild-type (OME52) and ΔBCAL2641 (OME53) backgrounds. n = 6 from 2 different clones. The mean RLU/OD600 of the control is 1.5585. (C) Expression of BCAM1112 in the wild-type (OME54) and ΔBCAL2641 (OME55) backgrounds. n = 7 from 2 different clones. The mean RLU/OD600 of the control is 0.2423. ns, not significant. *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
ROS production in response to other bactericidal antibiotics.
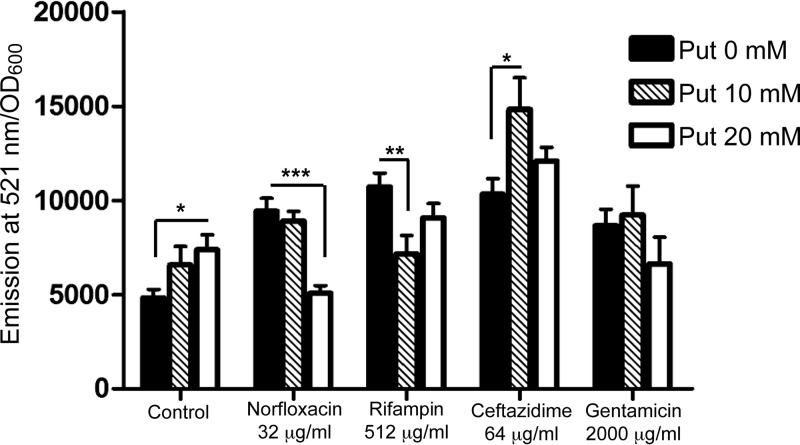
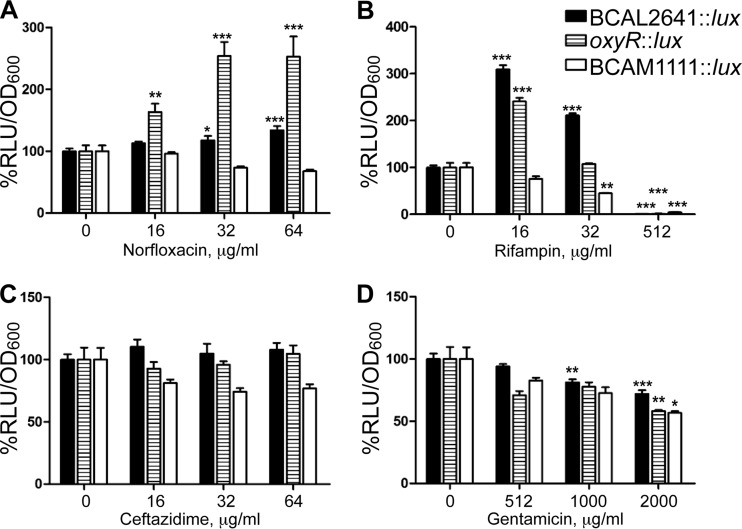
To evaluate whether the induction of oxidative stress and its amelioration by putrescine is a general phenomenon, we tested other bactericidal antibiotics. Exposure of B. cenocepacia to gentamicin, norfloxacin, ceftazidime, and rifampin led to increased ROS production, as detected by DCF (see Fig. S4 in the supplemental material) at sublethal concentrations (i.e., concentrations below but more specifically near the MIC of these antibiotics) (see Fig. S5 in the supplemental material). Putrescine reduced the antibiotic-induced elevation in ROS levels only for norfloxacin and rifampin (Fig. 6), and this correlated with induction of BCAL2641 gene expression (Fig. 7A and B, respectively). This agrees with the contribution of BCAL2641 in resistance to both antibiotics that we have previously reported (12). Moreover, oxyR transcription was also upregulated in response to both norfloxacin and rifampin (Fig. 7A and B, respectively), which was reflected in an increase in the catalase activity (Table 2), supporting the notion that both antibiotics lead to increased ROS production (Fig. 6; see Fig. S4). In contrast, neither antibiotic affected BCAM1111 gene expression (Fig. 7), indicating that this gene and its product are not directly involved in the response to antibiotic-mediated oxidative stress. It should be noted that higher rifampin concentrations resulted in a great reduction in the expression of BCAL2641, oxyR, and BCAM1111 (Fig. 7B), which might be attributed to nonspecific inhibition of transcription by rifampin, especially at 512 μg/ml, where expression from these genes was almost completely inhibited.
FIG 6.
Role of putrescine in the bactericidal antibiotic-mediated ROS accumulation in B. cenocepacia K56-2. n = 9 from 3 independent experiments. The 4 tested antibiotics alone significantly (P < 0.001) induced the accumulation of ROS compared to the effect in control cells. *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
FIG 7.
Effects of different antibiotics on the expression of BCAL2641 (in OME50), oxyR (in OME56), and BCAM1111 (in OME52), determined using a luciferase expression assay at 3 h. Results are shown as percentage of relative light units (RLU)/OD600 relative to the control (untreated K56-2 background). The percentages of OD600 are shown in Fig. S5 in the supplemental material. n = a minimum of 6 from at least 2 different clones. The mean RLU/OD600 values of the control are 1.0759 for BCAL2641, 0.1087 for oxyR, and 1.4723 for BCAM1111. *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Putrescine did not reduce ROS production generated in response to ceftazidime but rather further increased the generated ROS at a 10 mM but not 20 mM concentration of putrescine (Fig. 6). Ceftazidime did not affect the expression of BCAL2641, oxyR, or BCAM1111 (Fig. 7C) and did not alter the catalase enzyme activity (Table 2). However, in a previous study, we reported that BCAL2641 is involved in the response of B. cenocepacia to ceftazidime (12). This may suggest another role of BCAL2641 in the protective actions against ceftazidime not related to the oxidative stress.
Concerning the response to gentamicin, exogenous putrescine did not affect the level of gentamicin-induced superoxide anion (Fig. 6). Moreover, gentamicin did not alter the expression of BCAL2641 (Fig. 7D). This agrees with the previously reported lack of involvement of this enzyme in the response to gentamicin in B. cenocepacia (12). Furthermore, gentamicin did not affect the expression of oxyR (Fig. 7D). However, the highest tested concentrations of gentamicin did reduce the expression of both BCAL2641 and oxyR (Fig. 7D). Similarly, gentamicin reduced the catalase enzyme activity (Table 2). Such inhibition might be due to the mechanism of action of the aminoglycoside inhibiting translation and protein synthesis in general, since it also inhibited the expression of BCAM1111, which consequently might have led to increased ROS levels at high concentrations (Fig. 6).
Conclusions.
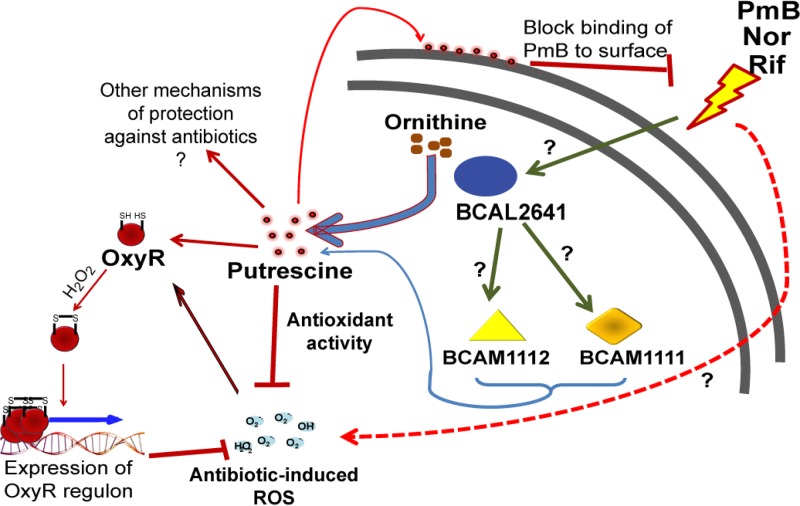
In this article, we have shown the following. (i) Sublethal concentrations of different bactericidal antibiotics (PmB at 1 mg/ml, rifampin at 256 and 512 μg/ml, and norfloxacin at 32 and 64 μg/ml) induce oxidative stress in B. cenocepacia that is manifested as induction of ROS formation, as detected by DCF, stimulation of expression of the transcription regulator OxyR involved in response to oxidative stress (at antibiotic concentrations similar to or even lower than those inducing ROS formation: PmB at 0.5 mg/ml, rifampin at 16 and 32 μg/ml, and norfloxacin at 16 to 64 μg/ml), and increased catalase enzyme activity (PmB at 0.5 mg/ml, rifampin at 16 μg/ml and norfloxacin at 8 μg/ml). (ii) This response does not apply to gentamicin and ceftazidime, which do not induce OxyR expression or increase catalase enzyme activity, suggesting that not all bactericidal antibiotics induce oxidative stress. (iii) Putrescine protects against oxidative stress induced by several bactericidal antibiotics (PmB, norfloxacin, and rifampin). (iv) Protection by putrescine correlates with increased BCAL2641 gene expression. (v) BCAL2641, in addition to synthesizing putrescine, regulates the other putrescine biosynthetic enzymes BCAM1111 and BCAM1112 by an unknown mechanism that does not directly involve putrescine. Together, these observations suggest a model (Fig. 8) by which B. cenocepacia responds to antibiotic stress by overproducing putrescine, and in turn, this polyamine protects bacterial cells by a surface effect blocking antibiotic binding (12) as well as by reducing oxidative damage.
FIG 8.
Model summarizing the role of putrescine in protecting B. cenocepacia from antibiotic-induced stress. Nor, norfloxacin; Rif, rifampin.
Putrescine was previously shown to communicate antibiotic resistance among different bacteria (12). Its increased production in B. cenocepacia occurs in response to a subset of bactericidal antibiotics (12) that induce oxidative stress in bacterial cells at nearly lethal concentration ranges. It is still controversial whether the generation of ROS is the cause of lethality of antimicrobial agents or a consequence of antibiotic stress (16, 20, 21). However, it is conceivable that the oxidative stress accompanying antibiotic treatment imposes a metabolic burden on the bacterial cells under near-death conditions. Thus, our results demonstrating a protective role for putrescine in the response to the oxidative stress generated in B. cenocepacia during antibiotic exposure represent another mechanism of protection from the antibacterial effects of bactericidal antibiotics. This agrees with previous reports on the antioxidant properties and protective effects of putrescine against antibiotic-induced ROS formation in E. coli (17).
While little is known about the physiological levels of putrescine, it seems that its level varies in different body sites. For example, the putrescine concentration was reported to be 3 mM in urine (11), whereas it was shown to be up to 0.2 mM in sputum samples from CF patients (39, 40). However, it is difficult to predict the local concentration of putrescine and other polyamines in the lung of CF patients, as infection alters the rheology of the mucus and the lung environment (41). Moreover, putrescine levels increase dramatically (by 10-fold or more) during exacerbations of bacterial infections in CF patients (39, 40). Hence, the concentrations used in this study could potentially resemble the physiological situation in certain body compartments. Furthermore, a direct relationship exists between increased putrescine concentration during infection and the proliferation of lung microbiota and specific pathogens, such as P. aeruginosa, in the lungs of CF patients (40). Also, putrescine and other polyamines in genital mucosal fluids increase the resistance of N. gonorrhoeae to antimicrobial peptides (PmB and LL-37), possibly enhancing its survival during infection by reducing bacterial susceptibility to host-derived antimicrobials (11). Interestingly, the expression of the ornithine decarboxylase BCAL2641 is induced in B. cenocepacia under CF conditions compared to soil environment-like conditions shown by comparative transcriptomics, underscoring the importance of putrescine, and this enzyme in particular, during infection (42).
This study also provides new information on the regulation of the putrescine synthesis enzymes. The ornithine decarboxylase BCAL2641 gene responds to the external antibiotic signals, while the other ornithine decarboxylase BCAM1111 and arginine decarboxylase BCAM1112 genes do not. Also, BCAL2641 regulates the expression of BCAM1111 and BCAM1112 since their expression depends on the presence of BCAL2641. This suggests that upon antibiotic stress, maximal production of putrescine is required, which arises from the upregulation of BCAL2641 and by maintaining the expression of the other two enzymes in a BCAL2641-dependent manner. The molecular mechanism of this regulation awaits further investigation.
In conclusion, this study broadens our understanding of the mechanism of chemical communication of antibiotic resistance mediated by putrescine. In addition, it provides a clear target for the design of inhibitors targeting the ornithine decarboxylase BCAL2641, which is critically implicated in this phenomenon. Such inhibitors not only would reduce the resistance to antibiotics in B. cenocepacia but also would reduce its ability to communicate high-level resistance to other less-resistant bacteria.
Supplementary Material
ACKNOWLEDGMENTS
We thank past and present members of the Valvano laboratory for critical discussions of the experimental results in this article.
This work was funded by a grant from Cystic Fibrosis Canada and a Marie Curie Career Integration Grant (project 618095). O.M.E. was supported by an Ontario Graduate Scholarship.
Footnotes
Published ahead of print 12 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02649-14.
REFERENCES
- 1.Ejim L, Farha MA, Falconer SB, Wildenhain J, Coombes BK, Tyers M, Brown ED, Wright GD. 2011. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat. Chem. Biol. 7:348–350. 10.1038/nchembio.559 [DOI] [PubMed] [Google Scholar]
- 2.Waters V. 2012. New treatments for emerging cystic fibrosis pathogens other than Pseudomonas. Curr. Pharm. Des. 18:696–725. 10.2174/138161212799315939 [DOI] [PubMed] [Google Scholar]
- 3.Loutet SA, Valvano MA. 2010. A decade of Burkholderia cenocepacia virulence determinant research. Infect. Immun. 78:4088–4100. 10.1128/IAI.00212-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corey M, Farewell V. 1996. Determinants of mortality from cystic fibrosis in Canada, 1970–1989. Am. J. Epidemiol. 143:1007–1017. 10.1093/oxfordjournals.aje.a008664 [DOI] [PubMed] [Google Scholar]
- 5.Speert DP, Henry D, Vandamme P, Corey M, Mahenthiralingam E. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8:181–187. 10.3201/eid0802.010163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen JS, Witzmann KA, Spilker T, Fink RJ, LiPuma JJ. 2001. Endemicity and inter-city spread of Burkholderia cepacia genomovar III in cystic fibrosis. J. Pediatr. 139:643–649. 10.1067/mpd.2001.118430 [DOI] [PubMed] [Google Scholar]
- 7.De Soyza A, Ellis CD, Khan CM, Corris PA, Demarco de Hormaeche R. 2004. Burkholderia cenocepacia lipopolysaccharide, lipid A, and proinflammatory activity. Am. J. Respir. Crit. Care Med. 170:70–77. 10.1164/rccm.200304-592OC [DOI] [PubMed] [Google Scholar]
- 8.El-Halfawy OM, Valvano MA. 2012. Non-genetic mechanisms communicating antibiotic resistance: rethinking strategies for antimicrobial drug design. Expert Opin. Drug Discov. 7:923–933. 10.1517/17460441.2012.712512 [DOI] [PubMed] [Google Scholar]
- 9.Smith AL, Fiel SB, Mayer-Hamblett N, Ramsey B, Burns JL. 2003. Susceptibility testing of Pseudomonas aeruginosa isolates and clinical response to parenteral antibiotic administration: lack of association in cystic fibrosis. Chest 123:1495–1502. 10.1378/chest.123.5.1495 [DOI] [PubMed] [Google Scholar]
- 10.Peters BM, Jabra-Rizk MA, O'May GA, Costerton JW, Shirtliff ME. 2012. Polymicrobial interactions: impact on pathogenesis and human disease. Clin. Microbiol. Rev. 25:193–213. 10.1128/CMR.00013-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goytia M, Shafer WM. 2010. Polyamines can increase resistance of Neisseria gonorrhoeae to mediators of the innate human host defense. Infect. Immun. 78:3187–3195. 10.1128/IAI.01301-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Halfawy OM, Valvano MA. 2013. Chemical communication of antibiotic resistance by a highly resistant subpopulation of bacterial cells. PLoS One 8:e68874. 10.1371/journal.pone.0068874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tabor CW, Tabor H. 1984. Polyamines. Annu. Rev. Biochem. 53:749–790. 10.1146/annurev.bi.53.070184.003533 [DOI] [PubMed] [Google Scholar]
- 14.Wortham BW, Patel CN, Oliveira MA. 2007. Polyamines in bacteria: pleiotropic effects yet specific mechanisms. Adv. Exp. Med. Biol. 603:106–115. 10.1007/978-0-387-72124-8_9 [DOI] [PubMed] [Google Scholar]
- 15.Johnson L, Mulcahy H, Kanevets U, Shi Y, Lewenza S. 2012. Surface-localized spermidine protects the Pseudomonas aeruginosa outer membrane from antibiotic treatment and oxidative stress. J. Bacteriol. 194:813–826. 10.1128/JB.05230-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. 10.1016/j.cell.2007.06.049 [DOI] [PubMed] [Google Scholar]
- 17.Tkachenko AG, Akhova AV, Shumkov MS, Nesterova LY. 2012. Polyamines reduce oxidative stress in Escherichia coli cells exposed to bactericidal antibiotics. Res. Microbiol. 163:83–91. 10.1016/j.resmic.2011.10.009 [DOI] [PubMed] [Google Scholar]
- 18.Kolodkin-Gal I, Sat B, Keshet A, Engelberg-Kulka H. 2008. The communication factor EDF and the toxin-antitoxin module mazEF determine the mode of action of antibiotics. PLoS Biol. 6:e319. 10.1371/journal.pbio.0060319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi H, Lee DG. 2012. Synergistic effect of antimicrobial peptide arenicin-1 in combination with antibiotics against pathogenic bacteria. Res. Microbiol. 163:479–486. 10.1016/j.resmic.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 20.Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. 2013. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339:1213–1216. 10.1126/science.1232688 [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Imlay JA. 2013. Cell death from antibiotics without the involvement of reactive oxygen species. Science 339:1210–1213. 10.1126/science.1232751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77:755–776. 10.1146/annurev.biochem.77.061606.161055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch EF, Maniatis T. 1990. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 24.Cohen SN, Chang AC, Hsu L. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. U. S. A. 69:2110–2114. 10.1073/pnas.69.8.2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Craig FF, Coote JG, Parton R, Freer JH, Gilmour NJ. 1989. A plasmid which can be transferred between Escherichia coli and Pasteurella haemolytica by electroporation and conjugation. J. Gen. Microbiol. 135:2885–2890 [DOI] [PubMed] [Google Scholar]
- 26.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652. 10.1073/pnas.76.4.1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tkachenko AG, Fedotova MV. 2007. Dependence of protective functions of Escherichia coli polyamines on strength of stress caused by superoxide radicals. Biochemistry (Mosc.) 72:109–116. 10.1134/S0006297907010130 [DOI] [PubMed] [Google Scholar]
- 28.Ortega XP, Cardona ST, Brown AR, Loutet SA, Flannagan RS, Campopiano DJ, Govan JR, Valvano MA. 2007. A putative gene cluster for aminoarabinose biosynthesis is essential for Burkholderia cenocepacia viability. J. Bacteriol. 189:3639–3644. 10.1128/JB.00153-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwase T, Tajima A, Sugimoto S, Okuda K, Hironaka I, Kamata Y, Takada K, Mizunoe Y. 2013. A simple assay for measuring catalase activity: a visual approach. Sci. Rep. 3:3081. 10.1038/srep03081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhee SG, Chang TS, Jeong W, Kang D. 2010. Methods for detection and measurement of hydrogen peroxide inside and outside of cells. Mol. Cells 29:539–549. 10.1007/s10059-010-0082-3 [DOI] [PubMed] [Google Scholar]
- 31.Chou HT, Kwon DH, Hegazy M, Lu CD. 2008. Transcriptome analysis of agmatine and putrescine catabolism in Pseudomonas aeruginosa PAO1. J. Bacteriol. 190:1966–1975. 10.1128/JB.01804-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renggli S, Keck W, Jenal U, Ritz D. 2013. The role of autofluorescence in flow-cytometric analysis of Escherichia coli treated with bactericidal antibiotics. J. Bacteriol. 195:4067–4073. 10.1128/JB.00393-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kooy NW, Royall JA, Ischiropoulos H. 1997. Oxidation of 2′,7′-dichlorofluorescein by peroxynitrite. Free Radic. Res. 27:245–254. 10.3109/10715769709065763 [DOI] [PubMed] [Google Scholar]
- 34.Aslund F, Zheng M, Beckwith J, Storz G. 1999. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl. Acad. Sci. U. S. A. 96:6161–6165. 10.1073/pnas.96.11.6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tkachenko AG, Nesterova LY. 2003. Polyamines as modulators of gene expression under oxidative stress in Escherichia coli. Biochemistry (Mosc.) 68:850–856. 10.1023/A:1025790729797 [DOI] [PubMed] [Google Scholar]
- 36.Reen FJ, Haynes JM, Mooij MJ, O'Gara F. 2013. A non-classical LysR-type transcriptional regulator PA2206 is required for an effective oxidative stress response in Pseudomonas aeruginosa. PLoS One 8:e54479. 10.1371/journal.pone.0054479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vander Wauven C, Stalon V. 1985. Occurrence of succinyl derivatives in the catabolism of arginine in Pseudomonas cepacia. J. Bacteriol. 164:882–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stalon V, Mercenier A. 1984. l-Arginine utilization by Pseudomonas species. J. Gen. Microbiol. 130:69–76 [DOI] [PubMed] [Google Scholar]
- 39.Grasemann H, Shehnaz D, Enomoto M, Leadley M, Belik J, Ratjen F. 2012. l-Ornithine derived polyamines in cystic fibrosis airways. PLoS One 7:e46618. 10.1371/journal.pone.0046618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Twomey KB, Alston M, An SQ, O'Connell OJ, McCarthy Y, Swarbreck D, Febrer M, Dow JM, Plant BJ, Ryan RP. 2013. Microbiota and metabolite profiling reveal specific alterations in bacterial community structure and environment in the cystic fibrosis airway during exacerbation. PLoS One 8:e82432. 10.1371/journal.pone.0082432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laube BL, Sharpless G, Benson J, Carson KA, Mogayzel PJ., Jr 2014. Mucus removal is impaired in children with cystic fibrosis who have been infected by Pseudomonas aeruginosa. J. Pediatr. 164:839–845. 10.1016/j.jpeds.2013.11.031 [DOI] [PubMed] [Google Scholar]
- 42.Yoder-Himes DR, Konstantinidis KT, Tiedje JM. 2010. Identification of potential therapeutic targets for Burkholderia cenocepacia by comparative transcriptomics. PLoS One 5:e8724. 10.1371/journal.pone.0008724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahenthiralingam E, Coenye T, Chung JW, Speert DP, Govan JR, Taylor P, Vandamme P. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore RA, Reckseidler-Zenteno S, Kim H, Nierman W, Yu Y, Tuanyok A, Warawa J, DeShazer D, Woods DE. 2004. Contribution of gene loss to the pathogenic evolution of Burkholderia pseudomallei and Burkholderia mallei. Infect. Immun. 72:4172–4187. 10.1128/IAI.72.7.4172-4187.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernier SP, Nguyen DT, Sokol PA. 2008. A LysR-type transcriptional regulator in Burkholderia cenocepacia influences colony morphology and virulence. Infect. Immun. 76:38–47. 10.1128/IAI.00874-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.