Abstract
In a previous prospective multicenter study in Spain, we found that OXA-1 and inhibitor-resistant TEM (IRT) β-lactamases constitute the most common plasmid-borne mechanisms of genuine amoxicillin-clavulanate (AMC) resistance in Escherichia coli. In the present study, we investigated the population structure and virulence traits of clinical AMC-resistant E. coli strains expressing OXA-1 or IRT and compared these traits to those in a control group of clinical AMC-susceptible E. coli isolates. All OXA-1-producing (n = 67) and IRT-producing (n = 45) isolates were matched by geographical and temporal origin to the AMC-susceptible control set (n = 56). We performed multilocus sequence typing and phylogenetic group characterization for each isolate and then studied the isolates for the presence of 49 virulence factors (VFs) by PCR and sequencing. The most prevalent clone detected was distinct for each group: group C isolates of sequence type (ST) 88 (C/ST88) were the most common in OXA-1 producers, B2/ST131 isolates were the most common in IRT producers, and B2/ST73 isolates were the most common in AMC-susceptible isolates. The median numbers of isolates per ST were 3.72 in OXA-1 producers, 2.04 in IRT producers, and 1.69 in AMC-susceptible isolates; the proportions of STs represented by one unique isolate in each group were 19.4%, 31.1%, and 48.2%, respectively. The sum of all VFs detected, calculated as a virulence score, was significantly higher in AMC-susceptible isolates than OXA-1 and IRT producers (means, 12.5 versus 8.3 and 8.2, respectively). Our findings suggest that IRT- and OXA-1-producing E. coli isolates resistant to AMC have a different and less diverse population structure than AMC-susceptible clinical E. coli isolates. The AMC-susceptible population also contains more VFs than AMC-resistant isolates.
INTRODUCTION
Escherichia coli is an important etiologic agent that causes both nosocomial and community-acquired infections (1). Amoxicillin-clavulanate (AMC) is a widely used antibiotic in many countries and is often used to treat E. coli infections (2, 3). According to the European Antimicrobial Resistance Surveillance Network (EARS-Net), the percentage of E. coli blood isolates in Spain that are nonsusceptible to AMC increased from 9.3% in 2003 to 25.3% in 2012 (http://ecdc.europa.eu/en/activities/surveillance/EARS-Net/database/Pages/database.aspx). This increase in resistance coincides with a dramatic increase in the community consumption of AMC (4).
AMC resistance in E. coli results from a complex epidemiological background involving the clonal and nonclonal spread of several resistance mechanisms (5). The production of OXA-1 and inhibitor-resistant TEM (IRT) β-lactamases is the most common plasmid-borne mechanism of AMC resistance in E. coli and does not affect other broad-spectrum β-lactam antibiotics (5). Clinical data for patients infected by AMC-resistant E. coli have also been provided by our group (6). However, there is very little information available about the population structure and virulence-associated determinants of OXA-1- and IRT-producing E. coli isolates compared with the amount of information available for AMC-susceptible clinical isolates.
Most extraintestinal pathogenic E. coli (ExPEC) isolates belong to phylogenetic group B2 and, to a lesser extent, to group D. ExPEC strains possess high numbers of virulence factors (VFs), including toxins, adhesins, polysaccharide capsules, siderophores, and invasins, that may enable them to evade host defenses and invade host tissues. E. coli isolates of phylogroups A and B1 are mainly found as part of the intestinal commensal population and usually possess a lower number of VFs (7, 8). In E. coli, several studies have evaluated the linkage between virulence and resistance to antimicrobials, such as quinolones, trimethoprim-sulfamethoxazole, or cephalosporins. Most of these studies have shown that antibiotic-susceptible E. coli isolates are usually more virulent than resistant ones (9–11). However, in the last few years, E. coli group B2 clones possessing a high number of VFs and resistance to several antimicrobials have emerged (i.e., sequence type [ST] 131 [ST131]) (12–14). According to recent data (15), eight phylogroups are now recognized: seven (A, B1, B2, C, D, E, F) belong to E. coli sensu stricto, whereas the eighth is Escherichia cryptic clade I.
Our hypothesis was that AMC, the antibiotic most consumed by far in Spain and other countries (3, 4), could select for not only AMC resistance but also certain specific clones carrying AMC resistance (16). To clarify this subject, the population structure of both resistant and susceptible isolates was studied in parallel. The objective of this study was to determine the population structure and virulence traits of clinical AMC-resistant E. coli isolates due to OXA-1 or IRT production in comparison with those of a control group of clinical AMC-susceptible E. coli isolates.
MATERIALS AND METHODS
Study design and bacterial isolates.
As described previously (5), 257 nonduplicated, AMC-resistant E. coli isolates were collected from clinical samples at seven Spanish hospitals in six geographic regions between January and March of 2010. Of these, all 112 (43.6%) isolates producing either OXA-1 (n = 67) or IRT (n = 45) were included in this study. The IRT types were TEM-40 (n = 15), TEM-30 (n = 13), TEM-33 (n = 5), TEM-32 (n = 2), TEM-34 (n = 2), TEM-35 (n = 1), TEM-54 (n = 1), TEM-76 (n = 1), TEM-79 (n = 1), and TEM-185 (n = 4). Among the 67 OXA-1-producing isolates, 25 (37.3%) were also CTX-M-15 producers.
Additionally, 56 AMC-susceptible (MIC < 4/2 μg/ml) clinical isolates were simultaneously collected at the participant hospitals to constitute the AMC-susceptible control group. These AMC-susceptible isolates were matched by geographical and temporal origin and were susceptible to several other β-lactam antibiotics. Susceptibility to AMC and other antibiotics, including ampicillin, cephalosporins, carbapenems, quinolones, aminoglycosides, and co-trimoxazole, was confirmed at the central reference laboratory, as described previously (5).
In total, 168 E. coli isolates were included in this study: 67 OXA-1-producing isolates, 45 IRT-producing isolates, and 56 AMC-susceptible isolates. The origins of all the E. coli isolates included in this study are detailed in Table 1.
TABLE 1.
Different population markers indicating genetic and virulence variations between OXA-1-producing-, IRT-producing, and susceptible Escherichia coli isolatesa
| Population and virulence marker | Result by resistance mechanism |
P valueb |
|||||
|---|---|---|---|---|---|---|---|
| OXA-1 production (n = 67) | IRT production (n = 45) | AMC susceptible (n = 56) | OXA-1 vs IRT | OXA-1 vs AMC susceptible | IRT vs AMC susceptible | AMC resistant vs AMC susceptible | |
| No. (%) of isolates | |||||||
| From urine | 49 (73.1) | 35 (77.8) | 41 (73.2) | 0.65 | 1 | 0.65 | 0.85 |
| Invasive | 11 (16.4) | 1 (4.4) | 10 (17.9) | 0.07 | 1 | 0.07 | 0.22 |
| From females | 36 (53.7) | 30 (66.7) | 33 (58.9) | 0.23 | 0.70 | 0.54 | 1 |
| From patients >65 yr of age | 46 (68.7) | 20 (44.4) | 22 (39.3) | 0.01 | 0.002 | 0.69 | 0.01 |
| No. of STs | 18 | 22 | 33 | ||||
| Median (range) no. of isolates per ST | 3.72 (1–25) | 2.04 (1–8) | 1.69 (1–12) | ||||
| No. (%) of single isolates by ST | 13 (19.4) | 14 (31.1) | 27 (48.2) | 0.18 | 0.009 | 0.10 | 0.03 |
| No. (%) of isolates by phylogroup | |||||||
| B2 | 23 (34.3) | 24 (53.3) | 59 (69.6) | 0.53 | 0.0001 | 0.1 | 0.001 |
| B2/ST131 | 22 (32.8) | 8 (17.8) | 3 (5.36) | 0.08 | 0.0001 | 0.06 | 0.0008 |
| B2/ST73 | 1 (1.5) | 5 (11.1) | 12 (21.4) | 0.04 | 0.0005 | 0.19 | 0.003 |
| C | 36 (53.7) | 8 (17.8) | 5 (8.9) | 0.0002 | <0.0001 | 0.23 | <0.0001 |
| C/ST88 | 25 (37.3) | 0 | 0 | <0.0001 | <0.0001 | <0.0001 | |
| Virulence score (range) | 8.3 (1–13) | 8.2 (2–17) | 12.5 (1–19) | 0.2722 | <0.0001 | <0.0001 | <0.0001 |
IRT, inhibitor resistant TEM; AMC, amoxicillin-clavulanate; ST, sequence types.
Boldface data indicate statistically significant differences.
Molecular epidemiology, phylogenetic groups, and detection of serotypes O25b and O16.
The multilocus sequence types (MLSTs) and phylogenetic groups of all 168 E. coli isolates were determined. MLST was performed according to the University of Warwick (Warwick Medical School, Coventry, United Kingdom) scheme for E. coli developed by M. Achtman (http://mlst.warwick.ac.uk). The phylogenetic relationships among the different STs obtained were established according to the eBURST program, version 3.
In addition, serotypes O25b and O16 were identified by allele-specific PCRs as described previously (17, 18).
Phylogenetic groups were determined both by the former method of Clermont et al. (19) and by the recently updated method of Clermont et al. (15).
VFs.
In all 168 E. coli isolates, the presence of 49 virulence-associated genes, including 19 adhesins, 4 siderophores, 11 toxins, 6 capsule synthesis-associated genes, and 9 miscellaneous VF genes, was determined by multiplex PCRs using primers described previously (20–22). Virulence scores were calculated for each isolate as the sum of all VFs detected; pap, sfa-foc, and clbB-clbN were counted only once regardless of the number of elements or subunits identified.
Statistical analysis.
Differences in the prevalence of phylogroups and sequence types between the different groups were assessed by Fisher's exact test. Associations were determined by calculation of the odds ratio (OR) with 95% confidence intervals (CIs). The null hypothesis was rejected for data with P values of <0.05. Statistical analysis was performed using GraphPad Prism, version 3.02, software (GraphPad Software, Inc., San Diego, CA). Virulence scores were compared by use of the Mann-Whitney U test.
RESULTS
Phylogenetic groups.
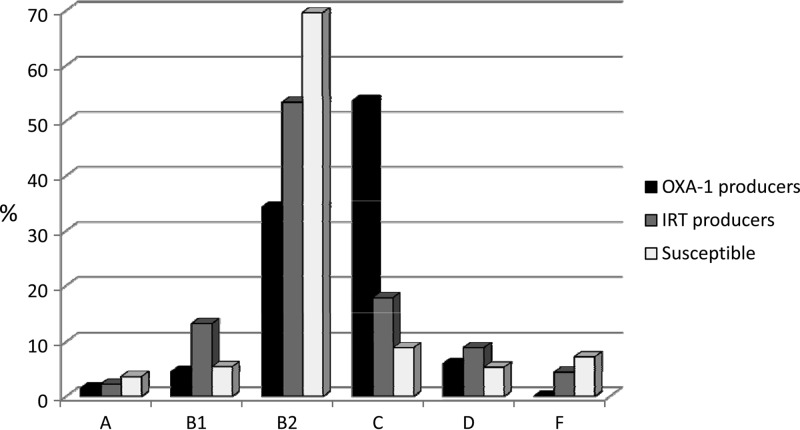
The distribution of the phylogenetic groups is depicted in Fig. 1. OXA-1 producers mostly belonged to phylogroups C and B2, most IRT producers belonged to phylogroup B2, and the vast majority of susceptible isolates were of the B2 phylogroup (Table 1).
FIG 1.
Distribution of phylogenetic groups among 67 OXA-1 producers, 45 IRT producers, and 56 susceptible Escherichia coli isolates. Phylogroup E isolates were not detected.
E. coli isolates producing OXA-1 but not CTX-M-15 extended-spectrum β-lactamases (ESBLs) belonged to phylogroup C, but the majority of isolates (78.3%) producing OXA-1 plus CTX-M-15 belonged to phylogroup B2.
Of the 168 E. coli isolates, 55 (32.7%) belonged to newly described phylogroup C (29.1%) or F (3.6%). No phylogroup E isolates were detected.
By the former method of Clermont et al. (19), 49 (29.2%) isolates were classified as phylogroup A; however, 45 of them (91.8%) were reclassified as phylogroup C.
MLST results.
Eighteen different STs were identified among the 67 OXA-1-producing isolates (mean number of isolates per ST, 3.72; range, 1 to 25). Twenty-two different STs were identified among the 45 IRT-producing isolates (mean of number of isolates per ST, 2.04; range, 1 to 8), and the 56 AMC-susceptible isolates were identified to be 33 different STs (mean number of isolates per ST, 1.69; range, 1 to 12) (Tables 1 and 2). The proportions of STs represented by one unique isolate were 19.4% for the OXA-1 group, 31.1% for the IRT group, and 48.2% for the AMC-susceptible group (Table 1).
TABLE 2.
Distribution of STs and phylogenetic groups among IRT-producing, OXA-1-producing, and susceptible isolatesa
| Resistance mechanism (no. of isolates) | ST complex (no. of isolates) | ST(s) (no. of isolates, phylogenetic group) |
|---|---|---|
| IRT | ||
| TEM-40 (15) | ST12 (1) | 12 (1, B2) |
| ST38 (1) | 38 (1, D) | |
| ST73 (2) | 73 (2, B2) | |
| Singletons (11) | 131 (3, B2), 117 (2, F), 372 (2, B2), 491 (2, B2), 1193 (1, B2), 3312 (1, B2) | |
| TEM-30 (13) | ST12 (2) | 12 (2, B2) |
| ST23 (2) | 23 (1, C), 410 (1, C) | |
| ST38 (1) | 38 (1, D) | |
| ST73 (2) | 73 (2, B2) | |
| ST155 (1) | 155 (1, B1) | |
| Singletons (5) | 127 (1, B2), 131 (3, B2), 767 (1, B1) | |
| Other IRT (17)b | ST10 (2) | 10 (1, C), 167 (1, C) |
| ST38 (2) | 38 (2, D) | |
| ST73 (2) | 73 (1, B2), 156 (1, B1) | |
| ST23 (4) | 23 (4, C) | |
| ST95 (1) | 95 (1, B2) | |
| Singletons (6) | 131 (2, B2), 500 (1, B1), 949 (1, B1), 1196 (1, B1), 2817 (1, A) | |
| OXA-1 | ST23 (30) | 88 (25, C), 23 (2, C), 90 (1, C), 410 (2, C) |
| ST10 (5) | 10 (3, C), 167 (1, C), 617 (1, C) | |
| ST38 (1) | 38 (1, D) | |
| ST73 (1) | 73 (1, B2) | |
| ST155 (1) | 58 (1, B1) | |
| ST156 (1) | 156 (1, B1) | |
| ST448 (1) | 448 (1, B1) | |
| Singletons (27) | 131 (22, B2), 224 (1, A), 648 (1, D), 1412 (1, D), 2815 (1, C), 2816 (1, D) | |
| Susceptible | ST73 (12) | 73 (12, B2) |
| ST95 (5) | 95 (5, B2) | |
| ST10 (4) | 10 (4, C) | |
| ST12 (2) | 12 (2, B2) | |
| ST14 (1) | 14 (1, B2) | |
| ST38 (1) | 38 (1, D) | |
| ST59 (1) | 59 (1, F) | |
| ST69 (1) | 69 (1, F) | |
| ST101 (1) | 101 (1, B1) | |
| ST168 (1) | 93 (1, A) | |
| Singletons (27) | 62 (1, F), 91 (1, B2), 127 (1, B2), 131 (3, B2), 141 (3, B2), 224 (1, B2), 372 (1, B2), 420 (1, F), 681 (1, B2), 747 (1, B2), 971 (1, B1), 978 (1, B2), 1057 (1, B2), 1304 (1, B1), 1571 (1, A), 1829 (1, D), 2013 (1, B2), 2230 (1, C), 2346 (1, B2), 3018 (1, D), 3292 (1, B2), 3312 (1, B2), 3361 (1, B2) |
IRT, inhibitor-resistant TEM; ST, sequence type.
Five TEM-33 isolates, four TEM-185 isolates, two each isolates of TEM-32 and TEM-34, and one isolate each of TEM-35, TEM-54, TEM-76, and TEM-79.
The most prevalent ST was different for each group. In the OXA-1-producing isolates, ST88 (25 isolates, 37.3%) and ST131 (22 isolates, 32.8%) were the most common. In contrast, the IRT-producing isolates were most commonly ST131 (8 isolates, 17.8%), ST73 (5 isolates, 11.1%), and ST23 (5 isolates, 11.1%). Finally, ST73 (12 isolates, 21.4%) and ST95 (5 isolates, 8.9%) were the most common among the AMC-susceptible isolates (Table 2). Four novel STs were identified: STs 3292, 3312, and 3361 in the AMC-susceptible group and STs 2817 and 3312 in the IRT-producing group.
To determine whether a specific sequence type was significantly correlated with an AMC resistance mechanism, we conducted further statistical analysis. We found that a number of sequence types were significantly more prevalent in one specific group. ST131 was more prevalent in the OXA-1-producing group (P = 0.0001) and in the IRT-producing group (P = 0.06) than in the AMC-susceptible group (Table 1). ST88 was significantly more prevalent in the OXA-1-producing group than in the IRT-producing and AMC-susceptible groups (P < 0.0001) (Table 1). ST73 was significantly more prevalent in the AMC-susceptible group than in the OXA-1-producing group (P = 0.0005) (Table 1). ST131 and ST73 isolates were detected in all seven participating hospitals, and ST88 was detected in five of them.
Isolates of ST131, ST73, and ST95 belonged to phylogroup B2, and isolates of ST88, ST23, and ST10 belonged to phylogroup C.
Most OXA-1- and CTX-M-15-producing isolates belonged to ST131 (18/25 isolates, 72%), while isolates producing only OXA-1 and not CTX-M-15 mainly belonged to ST88 (25/48 isolates, 52.1%).
All except four isolates of group B2 and ST131 (B2/ST131) belonged to serotype O25b, three of the non-O25b isolates (two IRT producers and one AMC-susceptible isolate) were serotype O16, and the other one was serotype non-O25b non-O16.
Virulence factors.
To determine whether there was a relationship between virulence factors and resistance mechanisms, we thoroughly screened the isolates for 49 different VFs and analyzed their virulence relative to the AMC resistance mechanisms present. Data showing the virulence gene content among OXA-1-producing, IRT-producing, and AMC-susceptible isolates are summarized in Table 3. Most of the 49 VFs studied were more frequently detected in the AMC-susceptible isolates than in the OXA-1- or IRT-producing isolates. Nineteen VFs were significantly associated with the AMC-susceptible group, whereas only six were associated with the OXA-1 group (P = 0.005; Table 3). Overall, the AMC-susceptible group exhibited a significantly higher virulence score than the OXA-1 group (mean virulence score, 12.5 versus 8.3; P < 0.0001). In relation to the AMC-susceptible group and the group producing IRTs, 12 VFs were significantly associated with the AMC-susceptible group, whereas only 1 was associated with the IRT group (P < 0.001; Table 3); the AMC-susceptible group also exhibited a significantly higher virulence score than the IRT group (mean score, 12.5 versus 8.2; P < 0.0001). Overall, these data suggest that AMC-susceptible isolates may have a high potential for virulence.
TABLE 3.
Distribution of virulence determinants among IRT-producing, OXA-1-producing, and AMC-susceptible isolates
| Virulence determinanta | No. (%) of the following isolates with virulence determinant: |
P value |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Susceptible (n = 56) | OXA-1 (n = 67) | IRT (n = 45) | B2/ST131 (n = 33) | B2/ST73 (n = 18) | C/ST88 (n = 25) | B2/non-ST131 (n = 53) | B2/non-ST73 (n = 68) | C/non-ST88 (n = 23) | Susceptible vs OXA-1 | Susceptible vs IRT | B2/ST131 vs B2/non-ST131 | B2/ST73 vs B2/non-ST73 | B2/ST131 vs B2/ST73 | C/ST88 vs C/non-ST88 | |
| Adhesins | |||||||||||||||
| papA | 20 (35.7) | 15 (22.4) | 12 (26.7) | 7 (21.2) | 6 (33.3) | 10 (40) | 22 (41.5) | 23 (33.8) | 3 (13) | ||||||
| papC | 22 (39.3) | 31 (46.3) | 16 (35.6) | 9 (27.3) | 7 (38.9) | 20 (80) | 24 (45.3) | 26 (38.2) | 2 (8.7) | <0.001 | |||||
| papEF | 19 (33.9) | 5 (7.5) | 7 (15.6) | 5 (15.2) | 6 (33.3) | 1 (4) | 21 (39.6) | 20 (29.4) | 2 (8.7) | 0.001 | 0.041 | 0.018 | |||
| papG | 20 (35.7) | 11 (16.4) | 11 (24.4) | 7 (21.2) | 8 (44.4) | 4 (16) | 22 (41.5) | 21 (30.9) | 1 (4.3) | <0.001 | |||||
| papG I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ||||||
| papG I′ | 1 (1.8) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1.9) | 1 (1.5) | 0 (0) | ||||||
| papG II | 5 (8.9) | 6 (9) | 10 (22.2) | 5 (15.2) | 4 (22.2) | 1 (4) | 12 (22.6) | 13 (19.1) | 0 (0) | ||||||
| papG III | 14 (25) | 6 (9) | 1 (2.2) | 2 (6.1) | 4 (22.2) | 4 (16) | 9 (17) | 7 (10.3) | 1 (4.3) | 0.012 | <0.001 | ||||
| fimH | 54 (96.4) | 63 (94) | 44 (97.8) | 33 (100) | 18 (100) | 25 (100) | 53 (100) | 68 (100) | 18 (78.3) | 0.020 | |||||
| afa | 0 (0) | 11 (16.4) | 2 (4.4) | 9 (27.3) | 0 (0) | 1 (4) | 0 (0) | 9 (13.2) | 2 (8.7) | 0.002 | <0.001 | 0.019 | |||
| afaE8 | 0 (0) | 22 (32.8) | 1 (2.2) | 2 (6.1) | 0 (0) | 19 (76) | 0 (0) | 2 (2.9) | 0 (0) | <0.001 | <0.001 | ||||
| sfa | 27 (48.2) | 1 (1.5) | 4 (8.9) | 1 (3) | 6 (33.3) | 0 (0) | 20 (37.7) | 15 (22.1) | 4 (17.4) | <0.001 | <0.001 | <0.001 | 0.046 | ||
| focG | 10 (17.9) | 15 (22.4) | 4 (28.9) | 8 (24.2) | 10 (55.6) | 4 (16) | 13 (24.5) | 11 (16.2) | 7 (30.4) | 0.001 | 0.035 | ||||
| f17 | 0 (0) | 0 (0) | 0 (0) | 1 (3) | 1 (5.6) | 0 (0) | 3 (5.7) | 3 (4.4) | 0 (0) | ||||||
| clpG | 5 (8.9) | 0 (0) | 0 (0) | 16 (48.5) | 3 (16.7) | 2 (8) | 9 (17) | 22 (32.4) | 0 (0) | 0.013 | 0.003 | ||||
| iha | 5 (8.9) | 16 (23.9) | 11 (24.4) | 5 (15.2) | 6 (33.3) | 16 (64) | 19 (35.8) | 18 (26.5) | 2 (8.7) | 0.049 | 0.035 | <0.001 | |||
| hra | 27 (48.2) | 26 (38.8) | 10 (22.2) | 2 (6.1) | 4 (22.2) | 11 (44) | 16 (30.2) | 14 (20.6) | 7 (30.4) | 0.007 | 0.0071 | ||||
| bmaE | 25 (44.6) | 16 (23.9) | 2 (6.7) | 0 (0) | 1 (5.6) | 1 (4) | 1 (1.9) | 0 (0) | 7 (30.4) | 0.020 | <0.001 | 0.020 | |||
| gafD | 0 (0) | 2 (3) | 1 (4.4) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ||||||
| Siderophores | |||||||||||||||
| fyuA | 27 (48.2) | 27 (40.3) | 12 (26.7) | 19 (57.6) | 6 (33.3) | 7 (28) | 20 (37.7) | 33 (48.5) | 11 (47.8) | 0.039 | 0.049 | ||||
| iroN | 28 (50) | 21 (31.3) | 10 (22.2) | 5 (15.2) | 8 (44.4) | 12 (48) | 23 (43.4) | 20 (29.4) | 7 (30.4) | 0.042 | 0.006 | 0.009 | 0.041 | ||
| ireA | 9 (16.1) | 1 (1.5) | 3 (6.7) | 0 (0) | 5 (27.8) | 0 (0) | 11 (20.8) | 6 (8.8) | 0 (0) | 0.002 | 0.006 | 0.047 | 0.004 | ||
| iutA | 19 (33.9) | 43 (64.2) | 20 (44.4) | 19 (57.6) | 6 (33.3) | 18 (72) | 16 (30.2) | 29 (42.6) | 8 (34.8) | 0.001 | 0.014 | 0.019 | |||
| Toxins | |||||||||||||||
| hlyD | 16 (28.6) | 6 (9) | 11 (24.4) | 6 (18.2) | 8 (44.4) | 1 (4) | 20 (37.7) | 18 (26.5) | 1 (4.3) | 0.003 | |||||
| hlyF | 5 (8.9) | 22 (32.8) | 14 (31.1) | 7 (21.2) | 0 (0) | 12 (48) | 5 (9.4) | 12 (17.6) | 10 (43.5) | 0.003 | 0.010 | 0.043 | |||
| cnf1 | 4 (7.1) | 3 (4.5) | 5 (11.1) | 2 (6.1) | 4 (22.2) | 2 (8) | 5 (9.4) | 3 (4.4) | 1 (4.3) | 0.032 | |||||
| cdtB | 4 (7.1) | 1 (1.5) | 3 (6.7) | 0 (0) | 6 (33.3) | 0 (0) | 8 (15.1) | 2 (2.9) | 0 (0) | 0.021 | 0.001 | 0.001 | |||
| clbB | 17 (30.4) | 1 (1.5) | 8 (17.8) | 1 (3) | 8 (44.4) | 0 (0) | 20 (37.7) | 13 (19.1) | 2 (8.7) | <0.001 | <0.001 | 0.035 | <0.001 | ||
| clbN | 31 (55.4) | 2 (3) | 12 (26.7) | 2 (6.1) | 17 (94.4) | 0 (0) | 39 (73.6) | 24 (35.3) | 1 (4.3) | <0.001 | 0.004 | <0.001 | 0.026 | <0.001 | |
| astA | 6 (10.7) | 1 (1.5) | 7 (15.6) | 0 (0) | 2 (11.1) | 0 (0) | 6 (11.3) | 4 (5.9) | 4 (17.4) | 0.042 | 0.046 | ||||
| vat | 11 (19.6) | 11 (16.4) | 11 (24.4) | 7 (21.2) | 6 (33.3) | 3 (12) | 16 (30.2) | 17 (25) | 1 (4.3) | ||||||
| tsh | 4 (7.1) | 0 (0) | 3 (6.7) | 0 (0) | 0 (0) | 0 (0) | 3 (5.7) | 3 (4.4) | 3 (13) | 0.032 | |||||
| sat | 10 (17.9) | 22 (32.8) | 11 (24.4) | 23 (69.7) | 8 (44.4) | 3 (12) | 12 (22.6) | 27 (39.7) | 1 (4.3) | <0.001 | |||||
| pic | 14 (25) | 0 (0) | 0 (0) | 0 (0) | 6 (33.3) | 0 (0) | 10 (18.9) | 4 (5.9) | 2 (8.7) | <0.001 | <0.001 | 0.012 | 0.005 | 0.001 | |
| Capsules | |||||||||||||||
| kpsM II | 36 (64.3) | 19 (28.4) | 21 (46.7) | 15 (45.5) | 15 (83.3) | 4 (16) | 41 (77.4) | 36 (52.9) | 5 (21.7) | <0.001 | 0.005 | 0.029 | 0.016 | ||
| kpsM II K1 | 14 (25) | 1 (1.5) | 6 (13.3) | 2 (6.1) | 0 (0) | 1 (4) | 13 (24.5) | 15 (22.1) | 1 (4.3) | <0.001 | 0.040 | 0.033 | |||
| kpsM II K2 | 8 (14.3) | 14 (20.9) | 3 (6.7) | 9 (27.3) | 4 (22.2) | 3 (12) | 9 (17) | 14 (20.6) | 3 (13) | ||||||
| kpsM II K5 | 2 (3.6) | 1 (1.5) | 4 (8.9) | 0 (0) | 2 (11.1) | 1 (4) | 4 (7.5) | 2 (2.9) | 1 (4.3) | ||||||
| kpsM III | 1 (1.8) | 1 (1.5) | 1 (2.2) | 1 (3) | 0 (0) | 0 (0) | 1 (1.9) | 2 (2.9) | 0 (0) | ||||||
| kpsM K15 | 1 (1.8) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1.9) | 1 (1.5) | 0 (0) | ||||||
| Miscellaneous | |||||||||||||||
| iss | 8 (14.3) | 30 (44.8) | 11 (24.4) | 4 (12.1) | 1 (5.6) | 18 (72) | 9 (17) | 12 (17.6) | 8 (34.8) | 0.001 | 0.019 | ||||
| cvaC | 1 (1.8) | 15 (22.4) | 1 (2.2) | 2 (6.1) | 0 (0) | 10 (40) | 1 (1.9) | 3 (4.4) | 2 (8.7) | 0.019 | |||||
| traT | 16 (28.6) | 33 (49.3) | 18 (40) | 16 (48.5) | 6 (33.3) | 14 (56) | 21 (39.6) | 31 (45.6) | 3 (13) | 0.026 | 0.003 | ||||
| rfc | 4 (7.1) | 1 (1.5) | 0 (0) | 0 (0) | 1 (5.6) | 1 (4) | 4 (7.5) | 3 (4.4) | 0 (0) | ||||||
| ompT | 31 (55.4) | 9 (13.4) | 9 (20) | 9 (27.3) | 15 (83.3) | 2 (8) | 26 (49.1) | 20 (29.4) | 2 (8.7) | <0.001 | <0.001 | <0.001 | <0.001 | ||
| fliCH7 | 6 (10.7) | 0 (0) | 0 (0) | 0 (0) | 1 (5.6) | 0 (0) | 5 (9.4) | 4 (5.9) | 0 (0) | ||||||
| ibeA | 45 (80.4) | 2 (3) | 5 (11.1) | 4 (12.1) | 10 (55.6) | 0 (0) | 32 (60.4) | 26 (38.2) | 6 (26.1) | <0.001 | <0.001 | <0.001 | 0.002 | 0.008 | |
| malX | 38 (67.9) | 30 (44.8) | 26 (57.89) | 29 (87.9) | 13 (72.2) | 4 (16) | 41 (77.4) | 57 (83.8) | 6 (26.1) | ||||||
| usp | 38 (67.9) | 17 (25.4) | 13 (28.9) | 21 (63.6) | 14 (77.8) | 3 (12) | 37 (69.8) | 44 (64.7) | 1 (4.3) | <0.001 | <0.001 | ||||
| Mean (range) virulence scoreb | 12.5 (1–19) | 8.3 (1–13) | 8.2 (2–17) | 9.1 (5–13) | 13.2 (8–18) | 9.2 (5–13) | 12.5 (4–19) | 10.7 (4–19) | 6.6 (1–15) | <0.001 | < 0.001 | <0.001 | 0.003 | <0.001 | 0.004 |
Adhesins fimH (mannose-specific adhesin of type 1 fimbriae), papA, papC, papEF, and papG alleles I, I′, II, and III (P fimbria subunits), sfaS (S fimbrial adhesin), focG (putative F1C fimbrial adhesin), afa (Dr antigen-specific adhesin), afaE8 (afimbrial adhesin VIII), iha (nonhemagglutinin adhesin), bmaE (blood group M-specific adhesin), gafD (glucosamine-specific G fimbriae), f17 (F17c fimbriae), clpG (CS31A adhesin), and hra (heat-resistant agglutinin); toxins hlyD (α-hemolysin), hlyF (hemolysin F), cnf1 (cytotoxic necrotizing factor 1), cdtB (cytolethal distending toxin), clbB and clbN (colibactin), astA (enteroaggregative E. coli heat-stable toxin), vat (serine protease), tsh (temperature-sensitive hemagglutinin-serine protease), sat (secreted autotransporter toxin), and pic (serine protease); siderophores fyuA (yersiniabactin), iutA (aerobactin), and iroN and ireA; capsule synthesis-associated genes kpsM (groups II and III) specifically targeting the K1, K2, and K5 genes of group II capsules, as well as K15; and the miscellaneous VF genes iss (surface exclusion serum survival protein), cvaC (colicin V from serum resistance-associated plasmids), traT (serum resistance), rfc (O4 lipopolysaccharide synthesis), ompT (protease), fliCH7 (H7 flagellin variant), ibeA (invasion of brain endothelium), usp (uropathogenic-specific protein), and malX (pathogenicity island marker from the archetypal ExPEC strain CFT073).
The virulence score was the number of virulence genes detected, adjusted for multiple detection of the pap, sfa and foc, clbB and clbN, and kpsM II operons. Virulence scores were compared by use of the Mann-Whitney U test.
Virulence factors in B2 and non-B2 isolates.
In order to determine whether there was a relationship between phylogroups and virulence gene content, we next analyzed these data relative to each phylogroup. All isolates, whether they were AMC susceptible or AMC resistant, that belonged to phylogroup B2 exhibited a higher number of VFs and, consequently, a higher virulence score than non-B2 strains (see Table S1 in the supplemental material). The highest virulence scores were observed in the group B2 AMC-susceptible isolates, which possessed a mean VF score of 13.5, whereas the non-B2 AMC-susceptible isolates had a mean VF score of 10.2 (P = 0.021). The lowest virulence scores were observed in the non-B2 AMC-resistant isolates, and within this group, the OXA-1 and IRT producers possessed mean virulence scores of 7.7 and 7.3, respectively. The virulence gene content of the isolates belonging to phylogenetic groups B2 (53.4%) and non-B2 (46.6%) in relation to the AMC resistance mechanism is summarized in Table S1 in the supplemental material. Overall, we found that, as expected, phylogroup B2 contained the largest number of virulence factors and the highest virulence score.
Virulence factors in isolates of the most prevalent sequence types.
To determine whether there was a relationship between the most prevalent clones detected and the virulence traits, we next performed a statistical analysis of these parameters, as shown in Table 3. ST131 isolates exhibited virulence scores and a range of VFs (mean, 9.1; range, 5 to 13) similar to those of isolates belonging to ST88 (mean, 9.2; range, 5 to 13). ST73 isolates possessed the highest virulence score (mean, 13.2; range, 8 to 18). There were more virulence factors statistically associated with B2/non-ST131 isolates than with B2/ST131 isolates (13 versus 6), and the virulence score was significantly higher for the former isolates (means, 12.5 and 9.1, respectively; P < 0.001). In contrast, the analysis comparing B2/ST73 and B2/non-ST73 isolates revealed that the former had a significantly higher virulence score than the latter (means, 13.2 and 10.7, respectively; P = 0.003) (Table 3). In relation to the ST88 clone, C/ST88 isolates possessed a higher virulence score than C/non-ST88 isolates (means, 9.2 and 6.6, respectively; P = 0.004) (Table 3).
Among ST131 isolates, those of serotypes O16 and O25b showed similar virulence scores and ranges of VFs (mean scores, 9.3 [range, 7 to 12] and 9.2 [range 5 to 13], respectively).
DISCUSSION
To our knowledge, this is the first study comparing the population structure and virulence-associated genes of AMC-resistant E. coli isolates producing either IRTs or OXA-1 with those of AMC-susceptible isolates. Historically, the population of E. coli, both environmental and human associated, has been genetically very diverse (23); however, the emergence and dissemination of multiresistant and virulent clones of ExPEC have recently been described (24, 25) and have mainly been associated with the successful B2/ST131 clone (25, 26). Our data suggest that AMC-resistant E. coli isolates have a different and less diverse population structure than AMC-susceptible E. coli isolates, mainly due to the OXA-1-producing isolates. The mechanism by which antibiotic consumption leads to the selection of certain clones is poorly understood; a recent study suggests that different clones of E. coli vary markedly in their response to antibiotics, despite comparable MICs; these results seem to support the ability of antibiotics to select certain successful clones (27).
The association between antimicrobial resistance and virulence in E. coli is a controversial topic (9–11, 25, 26). We observed an inverse relationship between resistance to AMC due to OXA-1 and IRT production and virulence potential. These results are in agreement with those of previous studies concluding that E. coli resistance to nonfluorinated quinolones, fluoroquinolones, or trimethoprim-sulfamethoxazole is associated with reductions in the virulence traits of the isolates (9–11). In contrast, E. coli multiresistant and virulent clones have been described in past years (24–26).
The pandemic clone B2/ST131, previously associated with multiple mechanisms of antibiotic resistance (25), was predominant in the isolates producing both OXA-1 and IRTs, but it was uncommon in AMC-susceptible isolates. The virulence profile of B2/ST131 isolates observed in this study was similar to that of other B2/ST131 isolates in Spain producing other resistance mechanisms, such as an extended-spectrum β-lactamase (12, 13). However, the virulence score of B2/ST131 isolates was lower than that of other B2 isolates not belonging to ST131 (means, 9.10 and 12.5, respectively), which were mainly found in the AMC-susceptible group. This finding is in agreement with that of a previous study suggesting that ST131 isolates could be less virulent than was previously supposed and less virulent than other B2/non-ST131 clones (14).
Although the genetic diversity detected in AMC-susceptible isolates was great, the high prevalence of isolates belonging to the B2/ST73 clone in this AMC-susceptible group (21.4% of all susceptible isolates) is remarkable. The ST73 lineage has recently been found to be one of the most prevalent STs in uropathogenic isolates in England (16.6% of 300 isolates) (28) and in isolates causing spontaneous bacterial peritonitis and bacteremia in patients with cirrhosis in France (8% of 110 isolates) (29). Most ST73 isolates were antibiotic susceptible, in accordance with the findings of previous studies (28, 29); however, ST73 has also been associated with the production of ESBLs of the CTX-M type in Egypt and Japan (30, 31). In addition, our study showed that isolates belonging to ST73 exhibited the highest virulence score (mean, 13.2; Table 3), in agreement with the findings of another study showing that ST73 was one of the most virulent clones detected in the United Kingdom (28). Although several authors have demonstrated that the overall virulence score of an E. coli isolate is directly related to its ability to cause invasive infections and lethality (32, 33), a single, specific VF may enhance the virulence potential of a defined strain (34, 35) beyond the virulence score.
As we described previously (5), 37.3% of the OXA-1-producing isolates belonged to ST88. ST88 has also been described in association with chromosome-mediated AmpC overproduction in a French hospital (36), but so far its virulence profile had not been reported. Our ST88 isolates belonged to the recently proposed phylogroup C, and they possessed a high virulence score (mean, 9.2). Most of the virulence-associated traits of isolates belonging to ST88 were adhesins, protectins, and siderophores that may facilitate persistence and survival in adverse circumstances. Phylogroup C has previously been identified in a virulent strain causing an outbreak in a neonatal ward (37). Interestingly, in this study, 30% of E. coli isolates belonged to phylogroup C, mainly due to the reclassification of prevalent clonal complexes ST10 and ST23 (Table 2) previously classified as phylogroup A by the former method of Clermont et al. (19). In this study, carried out in clinical isolates, phylogroup A was very uncommon (2.4%), in contrast to the 18 to 28% prevalence described previously in two different collections of human fecal isolates (15).
Concluding remarks.
Our findings suggest that IRT- and OXA-1-producing E. coli isolates resistant to AMC have a population structure different from and less diverse than that of AMC-susceptible clinical E. coli isolates, mainly due to OXA-1 producers. AMC-susceptible isolates had more VFs than AMC-resistant isolates. We also provide information about the higher numbers of virulence traits in the B2/ST73 clone than the B2/ST131 and C/ST88 clones.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Plan Nacional de I+D+i 2008-2011 and the Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD12/0015). The study was cofinanced by the European Development Regional Fund (ERDF; A way to achieve Europe) and the Fondo de Investigación Sanitaria (grant PI09/0917).
We thank the Genomics Unit of the Centro Nacional de Microbiología for support with DNA sequencing and James R. Johnson and Brian D. Johnston (Department of Veterans Affairs Medical Center, University of Minnesota, Minneapolis, MN) for providing us with advice for the detection of the virulence factors and some of the positive-control strains used in this study.
Footnotes
Published ahead of print 28 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02738-13.
REFERENCES
- 1.Oteo J, Lázaro E, de Abajo FJ, Baquero F, Campos J, and Spanish Members of EARSS 2005. Antimicrobial-resistant invasive Escherichia coli, Spain. Emerg. Infect. Dis. 1:546–553. 10.3201/eid1104.040699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campos J, Ferech M, Lázaro E, de Abajo F, Oteo J, Stephens P, Goossens H. 2007. Surveillance of outpatient antibiotic consumption in Spain according to sales data and reimbursement data. J. Antimicrob. Chemother. 60:698–701. 10.1093/jac/dkm248 [DOI] [PubMed] [Google Scholar]
- 3.Mimica Matanovic S, Bergman U, Vukovic D, Wettermark B, Vlahovic-Palcevski V. 2010. Impact of restricted amoxicillin/clavulanic acid use on Escherichia coli resistance-antibiotic DU90% profiles with bacterial resistance rates: a visual presentation. Int. J. Antimicrob. Agents 36:369–373. 10.1016/j.ijantimicag.2010.05.019 [DOI] [PubMed] [Google Scholar]
- 4.Oteo J, Campos J, Lázaro E, Cuevas O, García-Cobos S, Pérez-Vázquez M, de bajo FJ, Spanish Members of EARSS 2008. Increased amoxicillin-clavulanic acid resistance in Escherichia coli blood isolates, Spain. Emerg. Infect. Dis. 14:1259–1262. 10.3201/eid1408.071059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ortega A, Oteo J, Aranzamendi-Zaldumbide M, Bartolomé RM, Bou G, Cercenado E, Conejo MC, González-López JJ, Marín M, Martínez-Martínez L, Merino M, Navarro F, Oliver A, Pascual A, Rivera A, Rodríguez-Baño J, Weber I, Aracil B, Campos J. 2012. Spanish multicenter study of the epidemiology and mechanisms of amoxicillin-clavulanate resistance in Escherichia coli. Antimicrob. Agents Chemother. 12:576–581. 10.1128/AAC.06393-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodríguez-Baño J, Oteo J, Ortega A, Villar M, Conejo MC, Bou G, Aranzamendi-Zaldumbide M, Cercenado E, Gurguí M, Martínez-Martínez L, Merino M, Rivera A, Oliver A, Weber I, Pascual A, Bartolomé RM, Gónzalez-López JJ, Campos J. 2013. Epidemiological and clinical complexity of amoxicillin-clavulanate-resistant Escherichia coli. J. Clin. Microbiol. 51:2414–2417. 10.1128/JCM.00999-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moreno E, Prats G, Planells I, Planes AM, Perez T, Andreu A. 2006. Characterization of Escherichia coli isolates derived from phylogenetic groups A and B1 causing extraintestinal infection. Enferm. Infecc. Microbiol. Clin. 24:483–489 (In Spanish.) 10.1157/13092463 [DOI] [PubMed] [Google Scholar]
- 8.Duriez P, Clermont O, Bonacorsi S, Bingen E, Chaventre A, Elion J, Picard B, Denamur E. 2001. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 147:1671–1676 [DOI] [PubMed] [Google Scholar]
- 9.Moreno E, Prats G, Sabaté M, Pérez T, Johnson JR, Andreu A. 2006. Quinolone, fluoroquinolone and trimethoprim/sulfamethoxazole resistance in relation to virulence determinants and phylogenetic background among uropathogenic Escherichia coli. J. Antimicrob. Chemother. 57:204–211. 10.1093/jac/dki468 [DOI] [PubMed] [Google Scholar]
- 10.Horcajada JP, Soto S, Gajewski A, Smithson A, Jiménez de Anta MT, Mensa J, Vila J, Johnson JR. 2005. Quinolone-resistant uropathogenic Escherichia coli strains from phylogenetic group B2 have fewer virulence factors than their susceptible counterparts. J. Clin. Microbiol. 43:2962–2964. 10.1128/JCM.43.6.2962-2964.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vila J, Simon K, Ruiz J, Horcajada JP, Velasco M, Barranco M, Moreno A, Mensa J. 2002. Are quinolone-resistant uropathogenic Escherichia coli less virulent? J. Infect. Dis. 186:1039–1042. 10.1086/342955 [DOI] [PubMed] [Google Scholar]
- 12.Blanco J, Mora A, Mamani R, López C, Blanco M, Dahbi G, Herrera A, Blanco JE, Alonso MP, García-Garrote F, Chaves F, Orellana MÁ, Martínez-Martínez L, Calvo J, Prats G, Larrosa MN, González-López JJ, López-Cerero L, Rodríguez-Baño J, Pascual A. 2011. National survey of Escherichia coli causing extraintestinal infections reveals the spread of drug-resistant clonal groups O25b:H4-B2-ST131, O15:H1-D-ST393 and CGA-D-ST69 with high virulence gene content in Spain. J. Antimicrob. Chemother. 66:2011–2121. 10.1093/jac/dkr235 [DOI] [PubMed] [Google Scholar]
- 13.Coelho A, Mora A, Mamani R, López C, González-López JJ, Larrosa MN, Quintero-Zarate JN, Dahbi G, Herrera A, Blanco JE, Blanco M, Alonso MP, Prats G, Blanco J. 2011. Spread of Escherichia coli O25b:H4-B2-ST131 producing CTX-M-15 and SHV-12 with high virulence gene content in Barcelona (Spain). J. Antimicrob. Chemother. 66:517–526. 10.1093/jac/dkq491 [DOI] [PubMed] [Google Scholar]
- 14.Lavigne JP, Vergunst AC, Goret L, Sotto A, Combescure C, Blanco J, O'Callaghan D, Nicolas-Chanoine MH. 2012. Virulence potential and genomic mapping of the worldwide clone Escherichia coli ST131. PLoS One 7:e34294. 10.1371/journal.pone.0034294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5:58–65. 10.1111/1758-2229.12019 [DOI] [PubMed] [Google Scholar]
- 16.Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35:736–755. 10.1111/j.1574-6976.2011.00268.x [DOI] [PubMed] [Google Scholar]
- 17.Clermont O, Lavollay M, Vimont S, Deschamps C, Forestier C, Branger C, Denamur E, Arlet G. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J. Antimicrob. Chemother. 61:1024–1028. 10.1093/jac/dkn084 [DOI] [PubMed] [Google Scholar]
- 18.Clermont O, Johnson JR, Menard M, Denamur E. 2007. Determination of Escherichia coli O types by allele-specific polymerase chain reaction: application to the O types involved in human septicemia. Diagn. Microbiol. Infect. Dis. 57:129–136. 10.1016/j.diagmicrobio.2006.08.007 [DOI] [PubMed] [Google Scholar]
- 19.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558. 10.1128/AEM.66.10.4555-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sannes MR, Kuskowski MA, Owens K, Gajewski A, Johnson JR. 2004. Virulence factor profiles and phylogenetic background of Escherichia coli isolates from veterans with bacteremia and uninfected control subjects. J. Infect. Dis. 190:2121–2128. 10.1086/425984 [DOI] [PubMed] [Google Scholar]
- 21.Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261–272. 10.1086/315217 [DOI] [PubMed] [Google Scholar]
- 22.Johnson JR, Johnston B, Kuskowski MA, Nougayrede JP, Oswald E. 2008. Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. J. Clin. Microbiol. 46:3906–3911. 10.1128/JCM.00949-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.González-González A, Sánchez-Reyes LL, Delgado Sapien G, Eguiarte LE, Souza V. 2013. Hierarchical clustering of genetic diversity associated to different levels of mutation and recombination in Escherichia coli: a study based on Mexican isolates. Infect. Genet. Evol. 13:187–197. 10.1016/j.meegid.2012.09.003 [DOI] [PubMed] [Google Scholar]
- 24.Mihaila L, Wyplosz B, Clermont O, Garry L, Hipeaux MC, Vittecoq D, Dussaix E, Denamur E, Branger C. 2010. Probable intrafamily transmission of a highly virulent CTX-M-3-producing Escherichia coli belonging to the emerging phylogenetic subgroup D2 O102-ST405 clone. J. Antimicrob. Chemother. 65:1537–1539. 10.1093/jac/dkq155 [DOI] [PubMed] [Google Scholar]
- 25.Oteo J, Pérez-Vázquez M, Campos J. 2010. Extended-spectrum β-lactamase producing Escherichia coli: changing epidemiology and clinical impact. Curr. Opin. Infect. Dis. 23:320–326. 10.1097/QCO.0b013e3283398dc1 [DOI] [PubMed] [Google Scholar]
- 26.Clermont O, Lavollay M, Vimont S, Deschamps C, Forestier C, Branger C, Denamur E, Arlet G. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J. Antimicrob. Chemother. 61:1024–1028. 10.1093/jac/dkn084 [DOI] [PubMed] [Google Scholar]
- 27.Stewart B, Rozen DE. 2012. Genetic variation for antibiotic persistence in Escherichia coli. Evolution 66:933–939. 10.1111/j.1558-5646.2011.01467.x [DOI] [PubMed] [Google Scholar]
- 28.Gibreel TM, Dodgson AR, Cheesbrough J, Fox AJ, Bolton FJ, Upton M. 2012. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from northwest England. J. Antimicrob. Chemother. 67:346–356. 10.1093/jac/dkr451 [DOI] [PubMed] [Google Scholar]
- 29.Bert F, Johnson JR, Ouattara B, Leflon-Guibout V, Johnston B, Marcon E, Valla D, Moreau R, Nicolas-Chanoine MH. 2010. Genetic diversity and virulence profiles of Escherichia coli isolates causing spontaneous bacterial peritonitis and bacteremia in patients with cirrhosis. J. Clin. Microbiol. 48:2709–2714. 10.1128/JCM.00516-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fam N, Leflon-Guibout V, Fouad S, Aboul-Fadl L, Marcon E, Desouky D, El-Defrawy I, Abou-Aitta A, Klena J, Nicolas-Chanoine MH. 2011. CTX-M-15-producing Escherichia coli clinical isolates in Cairo (Egypt), including isolates of clonal complex ST10 and clones ST131, ST73, and ST405 in both community and hospital settings. Microb. Drug Resist. 17:67–73. 10.1089/mdr.2010.0063 [DOI] [PubMed] [Google Scholar]
- 31.Suzuki S, Shibata N, Yamane K, Wachino J, Ito K, Arakawa Y. 2009. Change in the prevalence of extended-spectrum-beta-lactamase-producing Escherichia coli in Japan by clonal spread. J. Antimicrob. Chemother. 63:72–79. 10.1093/jac/dkn463 [DOI] [PubMed] [Google Scholar]
- 32.Kudinha T, Johnson JR, Andrew SD, Kong F, Anderson P, Gilbert GL. 2013. Distribution of phylogenetic groups, sequence type ST131, and virulence-associated traits among Escherichia coli isolates from men with pyelonephritis or cystitis and healthy controls. Clin. Microbiol. Infect. 19:E173–E180. 10.1111/1469-0691.12123 [DOI] [PubMed] [Google Scholar]
- 33.Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson JR, Porter SB, Zhanel G, Kuskowski MA, Denamur E. 2012. Virulence of Escherichia coli clinical isolates in a murine sepsis model in relation to sequence type ST131 status, fluoroquinolone resistance, and virulence genotype. Infect. Immun. 80:1554–1562. 10.1128/IAI.06388-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson JR, Clermont O, Menard M, Kuskowski MA, Picard B, Denamur E. 2006. Experimental mouse lethality of Escherichia coli isolates, in relation to accessory traits, phylogenetic group, and ecological source. J. Infect. Dis. 194:1141–1150. 10.1086/507305 [DOI] [PubMed] [Google Scholar]
- 36.Crémet L, Caroff N, Giraudeau C, Dauvergne S, Lepelletier D, Reynaud A, Corvec S. 2010. Occurrence of ST23 complex phylogroup A Escherichia coli isolates producing extended-spectrum AmpC beta-lactamase in a French hospital. Antimicrob. Agents Chemother. 54:2216–2218. 10.1128/AAC.01580-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moissenet D, Salauze B, Clermont O, Bingen E, Arlet G, Denamur E, Mérens A, Mitanchez D, Vu-Thien H. 2010. Meningitis caused by Escherichia coli producing TEM-52 extended-spectrum β-lactamase within an extensive outbreak in a neonatal ward: epidemiological investigation and characterization of the strain. J. Clin. Microbiol. 48:2459–2463. 10.1128/JCM.00529-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.