Abstract
The objective of this study is to describe the epidemiology of intestinal carriage with extended-spectrum-cephalosporin-resistant Enterobacteriaceae in children with index infections with these organisms. Patients with resistant Escherichia coli or Klebsiella bacteria isolated from the urine or a normally sterile site between January 2006 and December 2010 were included in this study. Available infection and stool isolates underwent phenotypic and molecular characterization. Clinical data relevant to the infections were collected and analyzed. Overall, 105 patients were identified with 106 extended-spectrum-cephalosporin-resistant E. coli (n = 92) or Klebsiella (n = 14) strains isolated from urine or a sterile site. Among the 27 patients who also had stool screening for resistant Enterobacteriaceae, 17 (63%) had intestinal carriage lasting a median of 199 days (range, 62 to 1,576). There were no significant differences in demographic, clinical, and microbiological variables between those with and those without intestinal carriage. Eighteen (17%) patients had 37 subsequent resistant Enterobacteriaceae infections identified: 31 urine and 6 blood. In a multivariable analysis, antibiotic intake in the 91 days prior to subsequent urine culture was significantly associated with subsequent urinary tract infection with a resistant organism (hazard ratio, 14.3; 95% confidence interval [CI], 1.6 to 130.6). Intestinal carriage and reinfection were most commonly due to bacterial strains of the same sequence type and with the same resistance determinants as the index extended-spectrum-cephalosporin-resistant Enterobacteriaceae, but carriage and reinfection with different resistant Enterobacteriaceae strains also occurred.
INTRODUCTION
Emerging antibiotic resistance has become a serious threat to global public health. Antibiotic resistance in Enterobacteriaceae specifically is a growing concern due to the continual increase in rates of resistance, the rapid emergence of new mechanisms of resistance, and the relative lack of effective antibacterial agents (1).
Available epidemiological data suggest that intestinal carriage with resistant Enterobacteriaceae may serve as an ongoing reservoir for infection, as well as a source for transmission of these bacteria between individuals. Studies, including mostly adults, have shown that intestinal carriage of extended-spectrum-cephalosporin-resistant (ESC-R) Enterobacteriaceae is common among patients with ESC-R index infections and can persist for months to years (2–4). Few pediatric studies include the in-depth clinical and molecular characterization required to understand the role that intestinal carriage plays in recurrent infection and to aid in the design of interventions focused on reducing intestinal carriage and limiting transmission.
The objective of this study is to describe the clinical and microbiological characteristics of intestinal carriage and subsequent infection due to ESC-R Escherichia coli and Klebsiella spp. in pediatric patients with prior infections with these organisms.
MATERIALS AND METHODS
Setting and institutional review.
Seattle Children's Hospital (SCH) is a 323-bed, tertiary-care pediatric hospital with over 350,000 patient visits annually, including admissions to the hospital and ambulatory visits. SCH's Institutional Review Board reviewed and approved this study.
Subjects.
SCH patients with ESC-R E. coli and Klebsiella isolates collected from the urine or a normally sterile site between January 2006 and December 2010 were eligible for inclusion in this study. For purposes of this study, an ESC-R isolate was defined as an E. coli or Klebsiella isolate nonsusceptible to ceftriaxone, cefotaxime, or ceftazidime. Patients were identified by searching a database maintained by the SCH Clinical Microbiology Laboratory of all resistant isolates identified at our institution. In total, 109 patients with 110 ESC-R index isolates were identified. Of these 110 isolates, 106 isolates from 105 patients were available. Four patients whose index isolates were not found were excluded from analyses. Follow-up isolates obtained through May 2011 were included in this study. Of 108 subsequent isolates from 28 subjects identified through the clinical record, 88 isolates from 26 patients were retrieved from archiving.
Clinical data.
Clinical data from January 2006 through May 2011 were collected from the medical record. These data included age, gender, comorbid conditions, characteristics of subsequent infections, and all antibiotics (inpatient and outpatient) received from 1 year prior to the index infection through each patient's last culture (positive or negative) during the study period. Clinical microbiology data on all ESC-R strains isolated from urine, a normally sterile site, or a stool surveillance screening culture designed to identify intestinal carriage of ESC-R Enterobacteriaceae were collected. Stool surveillance screening cultures were obtained under the direction of the clinicians involved in the care of the patients.
Definitions.
“Index infection” signifies an ESC-R E. coli or Klebsiella species strain isolated from urine or a normally sterile site (blood, peritoneal fluid, etc.) during the study period. “Subsequent infection” signifies an ESC-R E. coli or Klebsiella strain isolated from urine or a normally sterile site obtained ≥28 days after the index infection and during the study period. Our goal was to select a time point when patients would have completed their antibiotics for the index infection and after which any infection occurring would likely be a new infection rather than an unresolved previous infection. “Intestinal carriage” signifies an ESC-R E. coli or Klebsiella strain isolated from a stool sample obtained any time after the index infection and during the study period. “Clearance of intestinal carriage” signifies 3 consecutive negative surveillance stool cultures obtained after a positive stool culture in a patient identified as having intestinal carriage. “Hospital acquired” signifies a positive culture obtained >48 h after hospitalization from a patient without signs or symptoms of infection on hospital admission. “Health care associated” signifies a positive culture obtained in an outpatient setting, or ≤48 h after hospitalization, from a patient who had been hospitalized in the last year and/or had a chronic medical condition requiring frequent contact with health care facilities or prolonged/recurrent antibiotic courses. “Community acquired” signifies a positive culture obtained in an outpatient setting, or ≤48 h after hospitalization, from an otherwise healthy patient without hospitalization in the last year. “Immunocompromised” signifies a diagnosis of cancer requiring chemotherapy within the past year, medical conditions (such as lupus erythematosus, etc.) requiring immunosuppressants, or receipt of hematopoietic cell or solid organ transplantation.
Laboratory methods. (i) Isolation of bacterial isolates from urine and normally sterile sites.
Isolates from urine or normally sterile sites (e.g., blood) were identified using routine microbiology laboratory procedures according to current laboratory guidelines (5, 6).
(ii) Isolation of bacterial isolates from stool.
For the selective culture, stool specimens were inoculated into 2 vials of MacConkey broth (5 ml each), one with a cefpodoxime disc and one with an ertapenem disc (or imipenem if ertapenem was unavailable), resulting in antibiotic concentrations of 2 μg/ml. Specimens were incubated at 35°C in ambient air for 48 h. After 24 h, turbid broth was plated on blood and MacConkey agars.
(iii) Antibiotic susceptibility testing.
E. coli or Klebsiella strains isolated from urine or normally sterile sites or selective cultures of stool were screened for resistance using the Vitek GN30 panel (bioMérieux) prior to February 2010, or the standard disc diffusion method on Mueller-Hinton agar (Remel, USA). From February 2010 through the end of the study period, standard disc diffusion testing on Mueller-Hinton agar was the only susceptibility testing method used due to CLSI breakpoint changes associated with first- and third-generation cephalosporins and carbapenems (7). The antibiotic agents tested throughout the study period included ampicillin, amoxicillin-clavulanic acid (disc diffusion only), cefazolin, cefuroxime, cefoxitin, ceftazidime, ceftriaxone, cefepime, ciprofloxacin (for patients >1 year old), meropenem, piperacillin-tazobactam, gentamicin, nitrofurantoin (urine isolates only), and trimethoprim-sulfamethoxazole. Additional agents tested using the Vitek GN30 panel included amikacin, ampicillin-sulbactam, aztreonam, cefoxitin, imipenem, and tobramycin. Antimicrobial zones of inhibition were interpreted according to laboratory guidelines (7).
(iv) Phenotypic characterization of beta-lactamase-producing isolates.
Isolates demonstrating nonsusceptibility (either intermediate susceptibility or resistance) to ceftriaxone, ceftazidime, imipenem, or meropenem underwent further phenotypic resistance characterization as previously described (8). Briefly, discs with ceftazidime (30 μg), cefotaxime (30 μg), ceftazidime-clavulanic acid (30/10 μg), cefotaxime-clavulanic acid (30/10 μg), cefpodoxime (30 μg), and cefepime (30 μg) were used with Mueller-Hinton agar. The extended-spectrum beta-lactamase (ESBL) phenotype was defined as a ≥5-mm increase in zone of inhibition with the combination discs of ceftazidime or cefotaxime plus clavulanic acid compared to ceftazidime or cefotaxime alone (8). The AmpC phenotype was defined as nonsusceptibility to ceftriaxone, cefotaxime, or ceftazidime and nonsusceptibility to cefoxitin. In addition, the AmpC phenotype was further defined using a 2-sided Etest with one side containing cefotetan and the other containing cefotetan plus cloxacillin (cefotetan/cefotetan plus cloxacillin Etest; bioMérieux, SA, Lyon, France). For this Etest, a cefotetan/cefotetan plus cloxacillin mean inhibitory concentration ratio of ≥8 is indicative of an AmpC producer.
(v) Resistance genotyping.
All ESC-R isolates were tested by PCR for the presence of bla genes encoding the most common extended-spectrum cephalosporinases at our center, the (class A) CTX-M and the (class C) CMY enzymes (8), along with (class C) DHA enzymes, using previously published primers (see Table S1 in the supplemental material) (9, 10). To test for (class A) SHV enzymes, isolates were tested using universal primers annealing to conserved sequences inside the blaSHV coding region (11). Because in K. pneumoniae these primers could detect both plasmid-associated blaSHV associated with ESC resistance (e.g., SHV-12) and intrinsic, chromosomal ampicillinases (e.g., SHV-1 and SHV-11), we also tested the K. pneumoniae isolates using an SHV-12-associated, IS26-dependent primer (Table S1). Isolates exhibiting resistance to carbapenems were tested with primers for bla genes encoding the (class A) KPC and (class B) IMP and VIM carbapenemases (12). For all resistance genotyping, monoplex PCRs were carried out in a reaction mix of a total volume of 20 μl, containing bacterial lysis DNA template, JumpStart Taq polymerase Ready Mix (Sigma-Aldrich, Inc.), and primers. Thermocycling was carried out under the following conditions: initial denaturation (94°C for 2 min), followed by 30 cycles of denaturation (94°C for 30 s), annealing (temperature variable by primer set, per Table S1, for 15 s), and extension (72°C for 1 min). Assembly and alignment of nucleotide sequences were performed using BioNumerics software (version 6.5; Applied Maths, Inc., Sint-Martens-Latem, Belgium). Allele assignments for resistance determinants were accomplished using publicly available, reference-allele resources (http://www.lahey.org/Studies).
(vi) Sequence-based strain typing.
To characterize the clonal relatedness of ESC-R Enterobacteriaceae, PCR and sequencing were carried out as described above, using primers for fumC and fimH for E. coli or primers for tonB for K. pneumoniae (see Table S1 in the supplemental material) (13, 14). For sequence typing of E. coli, fumC alleles were identified using a publicly available reference-allele resource (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli), while fimH alleles were identified by comparison to a previously described reference collection (14), on the basis of which subsequent novel alleles have been assigned sequential identifiers. We identified four fumC/fimH types (40-30, 40-22, 40-41, and 40-27) that have previously been associated with strains of the globally disseminated, multidrug-resistant ST131 clone (15, 16) as “ST131-associated sequence types.” Isolates with a fumC type 40 (fumC40)/fimH0 profile (i.e., which did not amplify a fimH product) were characterized by sequencing of uxuA, a housekeeping gene immediately downstream of the fim cluster which is highly concordant with fimH phylogeny (S. J. Weissman, unpublished data). The fimH allele was then assigned by matching the uxuA sequence from the fimH-null strain to that of a fumC40/fimH-positive strain. For tonB typing of K. pneumoniae, alleles were identified using a publicly available reference-allele resource (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html).
(vii) PFGE.
DNA macrorestriction analysis was performed using pulsed-field gel electrophoresis (PFGE) according to the Centers for Disease Control and Prevention PulseNet protocol (17). Isolate DNA was digested using 50 U of XbaI and separated in a 1% agarose gel with a contour-clamped homogeneous electric field (CHEF) DR-III System (Bio-Rad, Hercules, CA) as described previously (18). Restriction patterns were analyzed using BioNumerics software (version 6.5; Applied Maths, Sint-Martens-Latem, Belgium). Dendrograms were constructed by the UPGMA (unweighted pair group method with arithmetic mean) clustering method and Dice similarity coefficients with an optimization setting of 0.5% and position tolerance of 1.25%.
Statistical analysis.
Descriptive statistics for resistance phenotypes, molecular resistance determinants, and sequence-based strain typing (E. coli and K. pneumoniae only) were tabulated and summarized.
Clinical and microbiological correlates of intestinal carriage with ESC-R Enterobacteriaceae were evaluated. Associations between dichotomous variables were evaluated using Fisher's exact test.
The frequency of subsequent infections due to any ESC-R strain was described. The duration of time between index and subsequent infection, as well as the characteristics of subsequent infections, was described.
We evaluated risk factors for subsequent urinary tract infection (UTI) due to ESC-R Enterobacteriaceae, with particular attention to exposure to antibiotics. The Cox proportional hazards model was used to explore the impact of antibiotic exposure on risk of first subsequent UTI due to an ESC-R organism occurring at least 28 days after an index UTI. We evaluated antibiotic exposure (whether prophylactic or treatment indicated) as a binary time-dependent variable, using the threshold of 14 days or more of exposure during the last 91 days. Prophylactic antibiotics that were administered 2 to 3 days per week were treated as daily antibiotics as long as the regimen continued. The analysis was stratified by immunocompromised status. Due to the relatively small numbers, we included only gender and presence of a urological abnormality as covariates, as both are known to be associated with risk of UTI.
Statistical analyses were performed using Stata (version 12.1; Stata Corp., College Station, TX) and R (version 3.0.0, R Core Team, 2013).
RESULTS
Subjects and their index infections.
Overall, 105 patients were identified with 106 ESC-R E. coli (n = 92) or Klebsiella species (n = 14) strains isolated from urine or a sterile site (Table 1). The median age of the patients was 5 years (interquartile range [IQR], 0.8, 14.5), 72 (69%) were female, and 70 (67%) had at least one underlying medical condition. Eighteen (17%) of the infections were classified as hospital acquired, 60 (57%) as health care associated, and 27 (26%) as community associated. Urine accounted for 88 (84%) of the index cultures, blood for 10 (10%), and other sterile sites for the remaining 7 (6%) cultures. One patient had both an ESC-R E. coli strain and an ESC-R Klebsiella pneumoniae strain isolated from an index urine culture. Resistance phenotypes included ESBL in 62 (58%) of the isolates, AmpC in 42 (40%), and carbapenem resistance in 2 isolates.
TABLE 1.
Demographic and clinical characteristics of patients with ESC-R Enterobacteriaceae infections and microbiological characteristics of the infections
| Characteristic | No. (%, unless otherwise noted) |
||||
|---|---|---|---|---|---|
| All patients (n = 105) | Subsequent infectionb |
Intestinal carriageb |
|||
| No (n = 42) | Yes (n = 18) | No (n = 10) | Yes (n = 17) | ||
| Median age in years (IQR)a | 5 (0.8, 14.5) | 4.8 (1.0, 13.5) | 11 (5.9, 18) | 2.6 (1.0, 9.3) | 13 (3.3, 16.2) |
| Female gender | 72 (69) | 30 (71) | 11 (61) | 6 (60) | 11 (65) |
| Any medical condition | 70 (67) | 36 (86) | 17 (94) | 8 (80) | 16 (94) |
| Urological condition | 37 (35) | 18 (38) | 8 (44) | 2 (20) | 4 (24) |
| Immunocompromised | 25 (24) | 14 (33) | 9 (50) | 4 (40) | 10 (59) |
| Hospitalized in year prior to index | 55 (52) | 27 (64) | 13 (72) | 7 (70) | 14 (83) |
| Median hospital days prior to index (IQR) | 17 (7, 57) | 28 (9,60) | 33 (15, 68) | 24 (7, 35) | 41 (16, 68) |
| ≥14 days of antibiotics in the last 91 days | 46 (44) | 23 (55) | 15 (83) | 6 (60) | 14 (82) |
| Acquisition of index infection | |||||
| Community acquired | 27 (26) | 6 (14) | 1 (5) | 2 (20) | |
| Health care associated | 60 (57) | 28 (67) | 12 (67) | 8 (80) | 9 (53) |
| Hospital acquired | 18 (17) | 8 (19) | 5 (28) | 8 (47) | |
| Primary site of index infection | |||||
| Urine | 88 (84) | 34 (81) | 13 (72) | 8 (80) | 11 (65) |
| Blood | 10 (10) | 6 (14) | 2 (11) | 1 (10) | 2 (12) |
| Otherc | 7 (6) | 2 (5) | 3 (17) | 1 (10) | 4 (23) |
| Species associated with index infectiond | |||||
| E. coli | 92 (88) | 37 (88) | 12 (67) | 8 (80) | 12 (71) |
| ST131-associated sequence type | 26 (25) | 9 (21) | 3 (17) | 2 (20) | 4 (24) |
| K. pneumoniae | 11 (10) | 4 (10) | 5 (28) | 2 (20) | 5 (29) |
| Klebsiella oxytoca | 3 (3) | 1 (2) | 1 (5) | ||
| Resistance phenotype of index isolate | |||||
| ESBL | 61 (58) | 24 (57) | 14 (78) | 7 (70) | 10 (59) |
| AmpC | 42 (40) | 17 (41) | 4 (22) | 3 (30) | 6 (35) |
| Carbapenem resistant | 2 (2) | 1 (2) | 1 (6) | ||
IQR, interquartile range.
These categories are not mutually exclusive.
Other sites of index infection included peritoneal fluid, bronchoalveolar lavage fluid, and wounds.
Values for this variable will add up to >100% as one patient had >1 ESC-R species identified from the index isolate.
ESC-R Enterobacteriaceae intestinal carriage.
Follow-up stool screening cultures were obtained in 27 (26%) of the 105 patients. A median of 3 (range, 1 to 15) stool screenings was performed per patient. The first stool culture was obtained a median 67 days after the index infection (range, 14 to 1,418). The median days to first culture did not differ between patients who ultimately had intestinal carriage documented and those who did not (P = 0.9). The last stool sample was obtained a median 221 days (range, 31 to 1,576 days) after the index infection. Among the 27 patients with stool screening, 17 (63%) had ESC-R Enterobacteriaceae intestinal carriage. The median duration of carriage was 199 days (range, 62 to 1,576). Five (29%) of the 17 patients exhibited carriage that persisted for over 300 days. Clearance of intestinal carriage with ESC-R Enterobacteriaceae was documented in only 2 patients; the last positive stool cultures in these patients occurred 239 and 305 days after the index infection, respectively. Only 2 other patients with documented intestinal carriage had any negative stool cultures; each had 2 consecutive negative stool cultures; in one there was no other subsequent testing, and the other was followed by additional positive cultures.
Subsequent infections.
Sixty of the 105 subjects had subsequent urine or sterile site cultures obtained at the study hospital at least 28 days after the index infection. Eighteen (30%) of the 60 patients had 37 subsequent ESC-R Enterobacteriaceae infections identified: 31 from urine and 6 from blood. The last subsequent ESC-R infection was identified a median of 284 days after the index infection (range, 29 to 1,479 days).
Sequence-based typing and resistance genotype of bacterial isolates.
Among 142 index, stool, and subsequent infection E. coli isolates from 94 patients, we identified 53 sequence types (Table 2), with 3 types (fumC/fimH types 40-30, 37-27, and 40-22) accounting for 47 (33%) isolates from 28 patients. ST131-associated sequence types (40-30, 40-22, 40-41, and 40-27) included 2 of the most common sequence types and accounted for 47 isolates from 27 patients. Among the 28 K. pneumoniae isolates from 11 patients, we identified 9 sequence types, with 2 types (tonB26 and tonB4) accounting for 13 isolates from 5 patients.
TABLE 2.
Distribution of ESC resistance determinants by species and sequence type among infection and stool isolates of ESC-R Enterobacteriaceae
| Sequence type | No. of patients | No. of isolates |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Index | Total | CTX-M-15 | CTX-M non-15a | SHV-12 | SHV non-12a | CMY-2 | DHA | None | ||
| E. coli fumC/fimH sequence types | ||||||||||
| ST131-associated sequence types | ||||||||||
| 40-30 | 19 | 19 | 28 | 26 | 1M-14 | 0 | 0 | 1 | 0 | 0 |
| 40-22 | 4 | 3 | 14 | 0 | 1M-22 | 0 | 0 | 13 | 0 | 0 |
| 40-41 | 3 | 3 | 4 | 0 | 3M-14 | 0 | 0 | 0 | 0 | 1 |
| 40-27 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Other frequent types | ||||||||||
| 37-27 | 5 | 5 | 5 | 1 | 2M-14, 1M-27 | 0 | 0 | 1 | 0 | 0 |
| 26-26 | 3 | 3 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| 4-24 | 3 | 3 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 2 |
| 45-97 | 3 | 3 | 5 | 0 | 0 | 0 | 0 | 5 | 0 | 0 |
| 4-27 | 3 | 2 | 3 | 1 | 0 | 0 | 0 | 1 | 0 | 1 |
| 11-54 | 3 | 3 | 3 | 2 | 0 | 0 | 0 | 2 | 0 | 0 |
| 11-0 | 3 | 3 | 7 | 6 | 0 | 0 | 0 | 4 | 0 | 0 |
| 100-96 | 3 | 3 | 4 | 1 | 0 | 0 | 0 | 3 | 1 | 0 |
| Other typesb | 50 | 41 | 62 | 12 | 7M-14, 2M-1, 1M-22 | 1 | 1−110 | 36 | 2 | 6 |
| E. coli total | 103 | 92 | 142 | 51 | 18 | 1 | 1 | 69 | 3 | 10 |
| K. pneumoniae tonB sequence types | ||||||||||
| 26 | 3 | 3 | 7 | 0 | 0 | 0 | 2−2, 2−5 | 1 | 0 | 3 |
| 4 | 2 | 2 | 6 | 5 | 1M-22 | 0 | 0 | 0 | 0 | 0 |
| Otherc | 6 | 6 | 15 | 10 | 2M-14 | 3 | 1−27 | 0 | 0 | 2 |
| K. pneumoniae total | 11 | 11 | 28 | 15 | 3 | 3 | 5 | 0 | 0 | 5 |
| Overall total | 114 | 103 | 170 | 66 | 21 | 4 | 6 | 69 | 3 | 15 |
CTX-M variants other than CTX-M-15 and SHV variants other than SHV-12 are indicated by superscripts (e.g., 1M-14 indicates 1 isolate with CTX-M-14).
One of two index isolates was positive for metallo-beta-lactamase IMP-4, as well as CMY-2.
One isolate was also positive for carbapenemase KPC-3 and represented the ST258 clone.
ESC resistance determinants were identified in all but 10 E. coli isolates and 5 K. pneumoniae isolates. These determinants were variably distributed among species and sequence types (Table 2), but striking associations were noted among the ST131-associated fumC/fimH types: CTX-M-15 was detected in 26/28 isolates of fumC/fimH 40-30 and CMY-2 in 13/14 isolates of fumC/fimH 40-22.
Comparison of index, stool, and subsequent infection isolates.
Sixteen patients had a total of 42 ESC-R stool isolates available for sequence typing and comparison to the index isolate. In 14 of the 16 patients, at least one ESC-R stool isolate shared the same sequence type and resistance determinant as the index infection isolate; 4 of the 14 patients had 5 additional ESC-R stool isolates that differed from their respective index isolate by sequence type and resistance determinant. Fifteen patients had 29 subsequent infection isolates available for comparison to their index isolates. In 10 of these 15 patients, at least one subsequent isolate shared both the sequence type and resistance determinant with the index infection isolate; one of the 10 patients had 2 additional subsequent infections with same-species isolates that differed by sequence type but shared a resistance determinant with the index isolate. Nine patients had subsequent infection isolates (n = 17) and stool isolates (n = 33) available for comparison. In 4 of these 9, at least one infection isolate and stool isolate shared both the same sequence type and resistance determinant as the index infection isolate. Patients with index, subsequent infection, and stool isolate pairs that differed included instances of different sequence types with shared resistance determinants as well as instances where both sequence type and resistance determinant differed.
PFGE analysis of cases with multiple isolates.
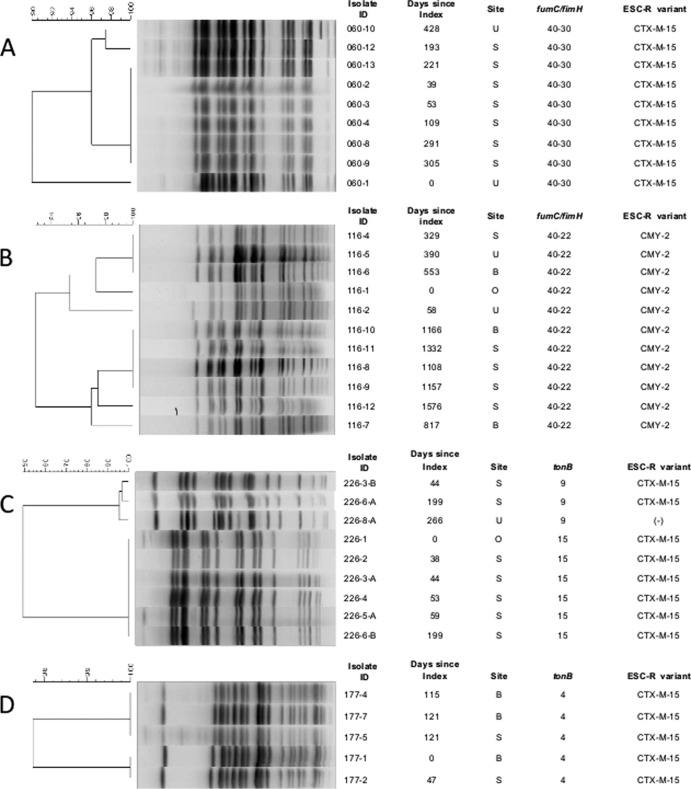
The two patients with the greatest numbers of same-type E. coli isolates recovered from sterile site and stool samples exhibited ST131-related sequence types (fumC/fimH types 40-30 and 40-22). Pulsed-field gel electrophoresis (PFGE) of each patient's isolates indicated genetic relatedness, even with restriction profile diversity levels up to 10% and 7% (Fig. 1A and B, respectively). In addition, uniform carriage of resistance determinants was demonstrated. Similar patterns were seen in patients with multiple K. pneumoniae isolates. One patient with 2 tonB sequence types encompassing 9 isolates fell, as expected, into 2 unrelated (<60% similarity) pulsotypes (Fig. 1C). Isolates within one pulsotype were indistinguishable from each other, while up to 5% diversity was seen within the second pulsotype. The two pulsotypes shared the CTX-M-15 resistance determinant. A second patient had 5 isolates of tonB4 encompassing 2 restriction fingerprints that were minimally distinct, at 3% dissimilarity (Fig. 1D); all 5 isolates displayed CTX-M-15 profiles.
FIG 1.
Molecular analysis of patients with multiple E. coli and K. pneumoniae isolates by pulsed-field gel electrophoresis. (A and B) E. coli isolates from two patients, respectively. (C and D) K. pneumoniae strains from two additional patients. The dendrogram, indicating genetic similarity of isolates, is shown on the left. Isolates with restriction patterns ≥90% similar were considered to belong to the same pulsotype. B, blood; U, urine; S, stool; O, other sterile site.
Risk factor analyses for intestinal carriage and subsequent ESC-R UTI.
Patients with intestinal carriage were compared to those without intestinal carriage. Demographic, clinical, and microbiological variables were not significantly different between the two groups (Table 3).
TABLE 3.
Demographic, clinical, and microbiological characteristics and their relationships with intestinal carriage of ESC-R Enterobacteriaceae
| Characteristic | No. (%) |
P valuea | |
|---|---|---|---|
| No intestinal carriage (n = 10) | Intestinal carriage (n = 17) | ||
| Age >6 yr at index infection | 3 (30) | 10 (59) | 0.24 |
| Female gender | 6 (60) | 11 (65) | 0.99 |
| Any medical condition | 8 (80) | 16 (94) | 0.54 |
| Urological condition | 2 (20) | 4 (24) | 0.99 |
| Immunocompromised | 4 (40) | 10 (59) | 0.44 |
| Hospitalized in yr prior to index | 7 (70) | 14 (82) | 0.64 |
| ≥14 days of antibiotics in the last 91 days | 6 (60) | 14 (82) | 0.10 |
| Nonurine index isolate | 2 (20) | 6 (35) | 0.67 |
| ESBL index phenotype | 7 (70) | 10 (59) | 0.69 |
| ST131-associated sequence type | 2 (20) | 4 (24) | 0.99 |
Value derived using Fisher's exact test.
Patients with ESC-R intestinal carriage were significantly more likely than those without carriage to have a subsequent ESC-R urine or sterile site infection (10 or 58% of 17 patients versus 1 or 13% of 8 patients without intestinal carriage, respectively; P = 0.04). The limited number of patients with screening for intestinal carriage precluded multivariable analysis. In addition, given differences in underlying disease in patients who had blood cultures obtained (predominantly oncology patients), a multivariable analysis of risk factors for recurrent infection was restricted to patients with an index ESC-R UTI (n = 46) and intestinal carriage was not evaluated. Twelve of the 46 patients with an index ESC-R UTI developed a subsequent ESC-R UTI. Eleven (92%) of these 12 patients received at least 14 days of antibiotics in the 91 days prior to urine culture, compared to 17 (50%) of 34 patients without a subsequent ESC-R UTI. None of the 12 patients with a subsequent ESC-R UTI had an intercurrent UTI due to susceptible organisms of any species. In the 11 receiving antibiotics, 10 were receiving long-term prophylactic regimens to prevent UTI or Pneumocystis pneumonia and one patient received two 14-day courses of clindamycin for cellulitis. In a multivariable model, antibiotic intake in the 91 days prior to subsequent culture was significantly associated with subsequent UTI with an ESC-R organism (hazard ratio, 14.3; 95% confidence interval [CI], 1.6 to 130.6) (Fig. 2). The model was adjusted for gender and underlying urological condition, neither of which was statistically significant.
FIG 2.
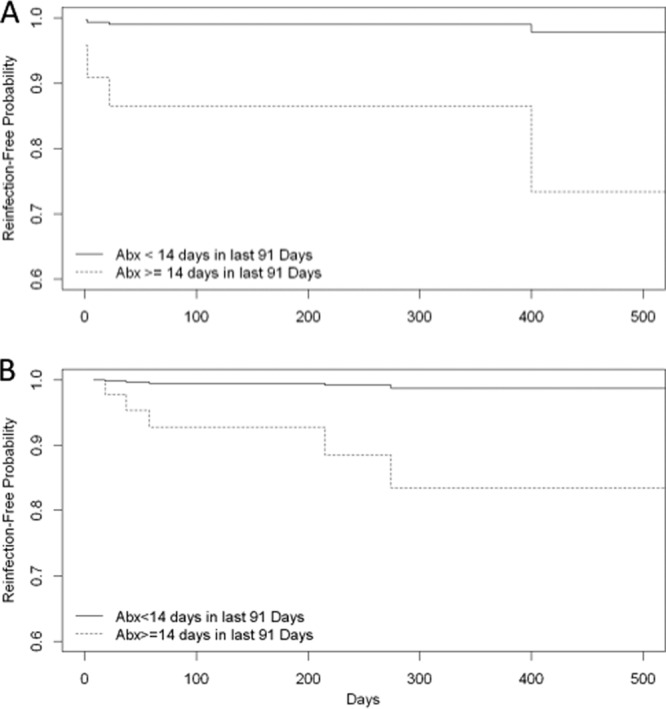
Kaplan-Meier curves modeling effect of antibiotic consumption on predicted time to first subsequent urinary tract infection due to ESC-R Enterobacteriaceae in patients with an index urinary tract infection due to ESC-R Enterobacteriaceae. The curves depict a model patient who is female without a urological abnormality. (A) The patient is immunocompromised; (B) the patient is not immunocompromised.
DISCUSSION
We found that intestinal carriage of ESC-R Enterobacteriaceae was common in pediatric patients with index infections due to these organisms and that carriage persisted for up to 4 years in some children. We also found that reinfection due to ESC-R Enterobacteriaceae occurred in a large proportion of children with an index infection and that subsequent ESC-R UTI was associated with previous antibiotic exposure. Finally, we found that intestinal carriage and reinfection were most commonly due to bacterial strains of the same sequence type and with the same resistance determinants as the index ESC-R Enterobacteriaceae, but that carriage and reinfection with ESC-R Enterobacteriaceae strains different from the index isolate also occurred.
Approximately 60% of children with index ESC-R infections who had stool cultures obtained had intestinal carriage with ESC-R Enterobacteriaceae, and it was persistent in most patients. Similar frequencies and durations of intestinal carriage of ESC-R Enterobacteriaceae have been documented in adults with index infections due to these organisms (2–4), but pediatric patient-specific longitudinal data have been relatively lacking (19). Transmission of an E. coli strain carrying the New Delhi metallo-beta-lactamase (NDM) carbapenemase from an adult with persistent intestinal carriage to 3 other patients has been reported (20). In addition, intra- and interhousehold transmission of ESC-R Enterobacteriaceae has been observed in studies from Spain and China, where high levels of intestinal carriage were documented (3, 21). In our study, most stool and subsequent infection isolates shared sequence types and resistance determinants with their respective index isolates. These data provide evidence that intestinal carriage of resistant organisms serves as a reservoir for infection, as well as a source for transmission of these bacteria between individuals. If true, this issue may be especially relevant in settings such as day care facilities and schools, where fecal-oral transmission of infectious organisms between children is common.
While most intestinal carriage and subsequent infection isolates matched the respective index isolates in this study, we also observed a number of pairings where resistance determinants were shared but species or sequence types differed, suggesting the transmission of resistance plasmids between organisms of the same or different species within a given host, presumably under ongoing antibiotic selection pressure. We also observed patients with nonmatching sequence types and resistance determinants. Although sampling and colony selection bias cannot be completely ruled out, this observation may be evidence of the susceptibility of the host to new acquisition of resistant organisms, perhaps due to ongoing dysbiosis resulting from prolonged or repeated exposure to systemic antibiotics.
Sequence typing indicated that E. coli ST131-associated sequence types were prevalent among E. coli sterile site and stool isolates and accounted for the most prolonged carriage/infection episodes in the study cohort. Given the known epidemiologic significance of this recently disseminated clone as the major contributor to ESC resistance among Enterobacteriaceae worldwide (16, 22), the prevalence of these sequence types throughout the study period is not surprising; however, their persistence as colonizers and pathogens in individual patients appears to reflect the remarkable fitness that has driven the successful spread of ST131 through human populations on a global level. Also noteworthy in this study is the distinctive partitioning of ESC resistance determinants between ST131-associated sequence types. The striking association between CTX-M-15 and ST131 (15, 16) has now been attributed to dissemination of a virulent subclone of 40-30, known as H30-Rx (23), consistent with our findings.
Restriction fingerprints and resistance genotypes were generally highly related among same-type isolates from individual patients, but diversity up to 10% was observed within some patient series. Without whole-genome sequencing, we cannot determine whether within-patient variability in restriction fingerprints reflects genomic rearrangements in functionally neutral regions or in loci that may be related to fitness, virulence, or resistance. However, the findings do appear to validate the study of serial infection and stool isolates in patients with significant comorbidities in order to clarify the relationship between host factors, antibiotic exposure, and bacterial adaptation. It would be of value to determine, for example, whether qualitative (antimicrobial spectrum) or quantitative (dosage and duration) differences in antibiotic exposure produce measurable differences in markers of bacterial adaptation, such as loss or acquisition of resistance plasmids or virulence determinants. Such insights may inform antibiotic selection and dosing strategies for patients with persistent colonization or recurrent infection, as well as provide a rationale for further study of emerging strategies, including fecal microbiota transplant.
Subsequent infections due to ESC-R Enterobacteriaceae occurred in 17% of our overall cohort and 30% of patients who had follow-up testing. In univariate analysis, intestinal carriage with ESC-R Enterobacteriaceae was associated with subsequent infections with these organisms. Unfortunately, we were unable to assess the role of intestinal carriage on reinfection in a multivariable model due to the small number of patients who had stool screening performed. In multivariable time-dependent analyses of subsequent ESC-R UTI, we found that receipt of antibiotics was associated with subsequent UTI. It is possible that receipt of antibiotics impacts the intestinal microbiota, leaving the ESC-R organism as the predominant population, which may lead to reinfection. Further study of this issue is warranted.
This study is limited by the small number of patients who were identified from a single hospital. Thus, the results may not be representative of patients in the community or in other urban regions of the country. Also, given the retrospective design, stool screening was not performed on all patients. This may have introduced biases, and the resulting number of subjects identified with versus without intestinal carriage of ESC-R organisms was very small. As a result, the study was not powered to show potentially important differences between these groups. In addition, our capture of outpatient antibiotic use data was likely incomplete. Finally, some variability in resistance genotype profiles, specifically the occurrence of isolates negative for specific resistance genes within the context of patient isolate series that otherwise exhibited consensus or majority profiles, may have been due to plasmid loss during the freeze/thaw cycles of archive storage. Despite these limitations, we feel that this census study adds significantly to what is known about the epidemiology of intestinal carriage of ESC-R Enterobacteriaceae in pediatric patients.
We found that intestinal carriage with ESC-R Enterobacteriaceae was common and persistent in pediatric patients who had infections due these organisms. In addition, recurrent infection due to ESC-R Enterobacteriaceae occurred in a large proportion of patients and was associated with previous receipt of antibiotics. Research focusing on efforts to prevent reinfection due to ESC-R Enterobacteriaceae is needed, including such strategies as intestinal decontamination and restoration of healthy intestinal flora through fecal microbiota transplantation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Seattle Children's Hospital Research Institute's Center for Clinical and Translational Research Translational Research Ignition Projects Program.
Research reported in this publication was partially supported by the NIH: the NIAID under award number R01AI083413 and the NHLBI under K02HL105543. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published ahead of print 5 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02558-14.
REFERENCES
- 1.Kollef MH, Golan Y, Micek ST, Shorr AF, Restrepo MI. 2011. Appraising contemporary strategies to combat multidrug resistant gram-negative bacterial infections—proceedings and data from the Gram-Negative Resistance Summit. Clin. Infect. Dis. 53(Suppl 2):S33–S55. 10.1093/cid/cir475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alsterlund R, Axelsson C, Olsson-Liljequist B. 2012. Long-term carriage of extended-spectrum beta-lactamase-producing Escherichia coli. Scand. J. Infect. Dis. 44:51–54. 10.3109/00365548.2011.592987 [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez-Bano J, Lopez-Cerero L, Navarro MD, Diaz de Alba P, Pascual A. 2008. Faecal carriage of extended-spectrum beta-lactamase-producing Escherichia coli: prevalence, risk factors and molecular epidemiology. J. Antimicrob. Chemother. 62:1142–1149. 10.1093/jac/dkn293 [DOI] [PubMed] [Google Scholar]
- 4.Zahar JR, Lanternier F, Mechai F, Filley F, Taieb F, Mainot EL, Descamps P, Corriol O, Ferroni A, Bille E, Nassif X, Lortholary O. 2010. Duration of colonisation by Enterobacteriaceae producing extended-spectrum beta-lactamase and risk factors for persistent faecal carriage. J. Hosp. Infect. 75:76–78. 10.1016/j.jhin.2009.11.010 [DOI] [PubMed] [Google Scholar]
- 5.Isenberg HD. (ed). 2007. Clinical microbiology procedures handbook, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 6.Versalovic J, Carroll KC, Funke G, Jorgensen JH, Landry ML, Warnock DW. (ed). 2011. Manual of clinical microbiology, 10th ed. ASM Press, Washington, DC [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 1999 to 2013 Performance standards for antimicrobial susceptibility testing. M100-S9 to M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8.Qin X, Zerr DM, Weissman SJ, Englund JA, Denno DM, Klein EJ, Tarr PI, Kwong J, Stapp JR, Tulloch LG, Galanakis E. 2008. Prevalence and mechanisms of broad-spectrum beta-lactam resistance in Enterobacteriaceae: a children's hospital experience. Antimicrob. Agents Chemother. 52:3909–3914. 10.1128/AAC.00622-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batchelor M, Hopkins K, Threlfall EJ, Clifton-Hadley FA, Stallwood AD, Davies RH, Liebana E. 2005. bla(CTX-M) genes in clinical Salmonella isolates recovered from humans in England and Wales from 1992 to 2003. Antimicrob. Agents Chemother. 49:1319–1322. 10.1128/AAC.49.4.1319-1322.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez-Perez FJ, Hanson ND. 2002. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153–2162. 10.1128/JCM.40.6.2153-2162.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan JJ, Wu SM, Tsai SH, Wu JJ, Su IJ. 2000. Prevalence of SHV-12 among clinical isolates of Klebsiella pneumoniae producing extended-spectrum beta-lactamases and identification of a novel AmpC enzyme (CMY-8) in Southern Taiwan. Antimicrob. Agents Chemother. 44:1438–1442. 10.1128/AAC.44.6.1438-1442.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17:1791–1798. 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178–4182. 10.1128/JCM.43.8.4178-4182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weissman SJ, Johnson JR, Tchesnokova V, Billig M, Dykhuizen D, Riddell K, Rogers P, Qin X, Butler-Wu S, Cookson BT, Fang FC, Scholes D, Chattopadhyay S, Sokurenko E. 2012. High-resolution two-locus clonal typing of extraintestinal pathogenic Escherichia coli. Appl. Environ. Microbiol. 78:1353–1360. 10.1128/AEM.06663-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weissman SJ, Adler A, Qin X, Zerr DM. 2013. Emergence of extended-spectrum beta-lactam resistance among Escherichia coli at a US academic children's hospital is clonal at the sequence type level for CTX-M-15, but not for CMY-2. Int. J. Antimicrob. Agents 41:414–420. 10.1016/j.ijantimicag.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson JR, Tchesnokova V, Johnston B, Clabots C, Roberts PL, Billig M, Riddell K, Rogers P, Qin X, Butler-Wu S, Price LB, Aziz M, Nicolas-Chanoine MH, Debroy C, Robicsek A, Hansen G, Urban C, Platell J, Trott DJ, Zhanel G, Weissman SJ, Cookson BT, Fang FC, Limaye AP, Scholes D, Chattopadhyay S, Hooper DC, Sokurenko EV. 2013. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J. Infect. Dis. 207:919–928. 10.1093/infdis/jis933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67. 10.1089/fpd.2006.3.59 [DOI] [PubMed] [Google Scholar]
- 18.Arlet G, Rouveau M, Casin I, Bouvet PJ, Lagrange PH, Philippon A. 1994. Molecular epidemiology of Klebsiella pneumoniae strains that produce SHV-4 beta-lactamase and which were isolated in 14 French hospitals. J. Clin. Microbiol. 32:2553–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lohr IH, Rettedal S, Natas OB, Naseer U, Oymar K, Sundsfjord A. 2013. Long-term faecal carriage in infants and intra-household transmission of CTX-M-15-producing Klebsiella pneumoniae following a nosocomial outbreak. J. Antimicrob. Chemother. 68:1043–1048. 10.1093/jac/dks502 [DOI] [PubMed] [Google Scholar]
- 20.D'Andrea MM, Venturelli C, Giani T, Arena F, Conte V, Bresciani P, Rumpianesi F, Pantosti A, Narni F, Rossolini GM. 2011. Persistent carriage and infection by multidrug-resistant Escherichia coli ST405 producing NDM-1 carbapenemase: report on the first Italian cases. J. Clin. Microbiol. 49:2755–2758. 10.1128/JCM.00016-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo WU, Ho PL, Chow KH, Lai EL, Yeung F, Chiu SS. 2010. Fecal carriage of CTXM type extended-spectrum beta-lactamase-producing organisms by children and their household contacts. J. Infect. 60:286–292. 10.1016/j.jinf.2010.02.002 [DOI] [PubMed] [Google Scholar]
- 22.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Canica MM, Park YJ, Lavigne JP, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61:273–281. 10.1093/jac/dkm464 [DOI] [PubMed] [Google Scholar]
- 23.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of extended-spectrum-beta-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 4(6):e00377-13. 10.1128/mBio.00377-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.