Abstract
We report here the complete nucleotide sequence of two IncR replicons encoding multidrug resistance determinants, including β-lactam (blaDHA-1, blaSHV-12), aminoglycoside (aphA1, strA, strB), and fluoroquinolone (qnrB4, aac6′-1b-cr) resistance genes. The plasmids have backbones that are similar to each other, including the replication and stability systems, and contain a wide variety of transposable elements carrying known antibiotic resistance genes. This study confirms the increasing clinical importance of IncR replicons as resistance gene carriers.
TEXT
Since the first description of the novel IncR complex replicons in 2009 (1), multidrug resistance (MDR) plasmids containing IncR characteristic sequences and carrying multiple resistance genes, such as blaKPC-2, blaVIM-1, blaDHA-1, blaTEM-52, qnrS1, or armA, have been increasingly reported in Enterobacteriaceae isolates from various geographical regions (1–7). Moreover, IncR replicons have been found either as single replicons (7) or as part of multireplicon plasmids, i.e., plasmids harboring multiple replication initiation proteins (6, 8–10). However, the possible role of this emerging IncR complex in the spread of multidrug resistance is difficult to evaluate, since the plasmids belonging to this complex are known to be nontransferable. In order to further explore the role of these resistance plasmids, we analyzed the full nucleotide sequence of two IncR plasmids, pKPS30 and pKPS77, from Klebsiella pneumoniae strains isolated in France.
K. pneumoniae KPS30 (sequence type 11 [ST11]) was isolated from urine in 2008 at Tenon Hospital (Paris, France). KPS77 (ST15) was responsible for a urinary tract infection in a pregnant woman in 2008 at Saint Antoine Hospital (Paris, France). KPS30 exhibited an acquired ampC phenotype and was resistant to tobramycin, amikacin, fluoroquinolones, and cotrimoxazole. KPS77 exhibited an extended-spectrum β-lactamase (ESBL) phenotype and was susceptible to aminoglycosides but resistant to fluoroquinolones and cotrimoxazole. KPS30 carries the broad-spectrum β-lactamase genes blaSHV-11 and blaOXA-30 and the acquired ampC-type gene blaDHA-1, whereas KPS77 carries blaTEM-1 and the ESBL gene blaSHV-12. Resistance to β-lactams and cotrimoxazole was transferred by electroporation for the two parental strains, whereas resistance to aminoglycosides was transferred from only KPS30 to the E_KPS30 (electroporant KPS30) transformant (Table 1).
TABLE 1.
Susceptibility of parental and recipient strains
| Antibiotic | MICs (mg/liter) fora: |
|||
|---|---|---|---|---|
| KPS30 | E_KPS30 | KPS77 | E_KPS77 | |
| Piperacillin/tazobactam | 8 | 2 | 4 | 1 |
| Cefotaxime | >128 | 32 | 16 | 4 |
| Ceftazidime | >128 | 32 | >128 | 32 |
| Cefoxitin | >32 | 8 | 4 | 2 |
| Cefepime | 0.5 | 0.125 | 4 | 2 |
| Gentamicin | S | S | S | S |
| Tobramycin | R | I | S | S |
| Amikacin | R | I | S | S |
| Ofloxacin | R | I | R | S |
| Ciprofloxacin | R | S | R | S |
| Cotrimoxazole | R | R | R | R |
Resistance according to the Société Française de Microbiologie (see http://www.sfm-microbiologie.org/). S, susceptible; I, intermediate; R, resistant.
Plasmid-based replicon typing (PBRT) (1, 11) allowed the identification of an IncR replicon in the two parental strains, KPS30 and KPS77. An additional IncFIIk replicon was detected in KPS30. The plasmid contents of the parental strains and transformants were determined by the Kado method (12) and confirmed the presence of two distinct plasmids in KPS30, whereas only one plasmid was present in the transformant E_KPS30 (data not shown). IncR plasmids harboring blaDHA-1 and blaSHV-12 could be transferred into Escherichia coli DH10B (Invitrogen, Cergy-Pontoise, France) only by electroporation. In order to analyze the complete sequence of the IncR replicons of KPS30 and KPS77, plasmid DNA was extracted from transformants using the Qiagen (Courtaboeuf, France) large construct kit. Sequencing was performed using shotgun and 3-kb paired-end sequencing on a 454/Roche GS FLX analyzer (Roche, Basel, Switzerland). The sequences obtained were assembled to a unique scaffold, and the predicted gaps were closed using PCR followed by sequencing with a BigDye Terminator v3.0 cycle sequencing ready reaction kit (Applied Biosystems, Foster City, CA) in an ABI Prism 310 DNA sequencer (Applied Biosystems). Gene prediction and annotation were performed using the CAAT-Box tool (13).
pKPS30 is a 61,228-bp plasmid with an average G+C content of 54.4%. It includes a 12,391-bp backbone (nucleotides [nt] 1 to 7,690 and 56,528 to 61,228), which showed maximum identity with regions of IncR replicons. Using the GenBank BLAST tool, the highest similarity scores (100% coverage, 99% identity) were obtained with the corresponding regions of the recently described plasmid pKP1780 (GenBank accession number JX424614) harboring the blaVIM-1 metallo-β-lactamase gene of a K. pneumoniae strain isolated in Greece (7). pKPS77 is a 45,867-bp plasmid with an average G+C content of 54%. Its IncR backbone (nt 1 to 7,690 and 41,711 to 45,867) is similar to that of pKPS30. It showed the highest similarity with the respective regions of plasmid p1 from K. pneumoniae JM45 (GenBank accession number CP006657) encoding the ESBLs VEB-3 and CTX-M-24. The linear map of the plasmids pKPS30 and pKPS77 and the direction of gene transcription are shown in Fig. 1.
FIG 1.
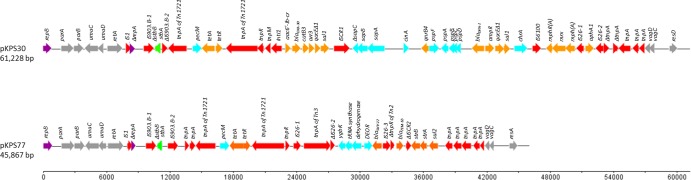
Linear maps of pKPS77 and pKPS30. Open reading frames (ORFs) are shown as arrows indicating the direction of transcription. Purple, replication initiation genes; gray, IncR backbone common to the two plasmids; red, transposon-related genes; orange, resistance genes; green, stbA and stbB genes; pale blue, other genes. ORFs encoding hypothetical proteins are not represented. The scale shows the number of base pairs.
A comparative search of GenBank revealed 11 similar IncR plasmid backbones (98% to 99% identity) in Enterobacteriaceae (Table 2).,Their backbone regions include a replication initiation gene, repB, 100% identical to repB of pKP1780, with a set of iterons suggested to be implicated in replication regulation. The upstream region specifies maintenance systems, including the vagCD operon encoding a toxin-antitoxin system and a resolvase probably contributing to multimer resolution (14). The downstream region encodes the ParA and ParB partition proteins. The scaffold also includes the umuDC operon, which is probably involved in SOS mutagenesis (15). As observed with all other fully sequenced IncR single replicon plasmids such as pKP1780 (GenBank accession number JX424614) (7), pKPC-LK30 (KC405622) (16), pKPN5 (CP000650), and pEFER (CU928144), no transfer-encoding region was identified. The remaining regions surrounding the IncR backbone in all 11 IncR plasmids whose sequences are stored in public databases contain various combinations of transposable elements and resistance genes. In pKPS30 and pKPS77, an IS1-like sequence and a truncated repA gene similar to the replication region of IncN plasmids were found at the boundary of the IncR region. It has an adjacent composite transposon containing stbA and truncated stbB genes (probably involved in plasmid stabilization [17]) flanked by two IS903.B insertion sequences (IS903.B-1 and IS903.B-2) (18, 19). A similar structure can be observed in pKP1780 (JX424614) (where Tn5393 had transposed within IS903.B-1 [7]), pEFER (CU928144), and p1 (CP006657). Apart from these related structures, pKPS30 and pKPS77 include various MDR determinants.
TABLE 2.
Completely sequenced plasmids in GenBank containing IncR characteristic regions
| Plasmid name | Species | GenBank accession no. | No. of replication initiation proteins (Inc group)a | Transfer region | Reference or source (yr of submission) |
|---|---|---|---|---|---|
| pCROD1 | Citrobacter rodentium | FN543503.1 | 1 (IncR) | 27 (2010) | |
| pEFER | Escherichia fergusonii | CU928144 | 1 (IncR) | Direct submissionb (2012) | |
| pKP1780 | Klebsiella pneumoniae | JX424614 | 1 (IncR) | 7 (2013) | |
| pKPC-LK30 | K. pneumoniae | KC405622 | 1 (IncR) | 16 (2014) | |
| pKPN101-IT | K. pneumoniae | JX283456 | 2 (IncR, IncFIIk) | FIIk | 9 (2012) |
| pK245 | K. pneumoniae | DQ449578 | 3 (IncR, NT, NT) | 10 (2006) | |
| pKP048 | K. pneumoniae | FJ628167 | 2 (IncR, IncFIIk) | FIIk | 28 (2010) |
| pKPN5 | K. pneumoniae | CP000650 | 2 (IncR, NT) | Direct submission (2009) | |
| pKPHS2 | K. pneumoniae | CP003224 | 2 (IncR, IncFIIk) | FIIk | Direct submission (2011) |
| pTC2 | Providencia stuartii | JQ824049 | 2 (IncR, IncA/C) | A/C | 6 (2013) |
| p1 | K. pneumoniae | CP006657 | 2 (IncR, IncFIIk) | FIIk | Direct submission (2013) |
| pKPS30 | K. pneumoniae | KF793937 | 1 (IncR) | This study | |
| pKPS77 | K. pneumoniae | KF954150 | 1 (IncR) | This study |
Truncated replication initiation proteins were excluded.
Direct submission, plasmid was submitted directly to GenBank (no publications are associated with the submission).
In pKPS30, the MDR region consists of a 44,944-bp sequence in which we identified a mosaic structure, including 17 resistance genes, three complete and two truncated insertion sequences, a complete class 1 integron, and two composite transposons. The class 1 integron was part of an ISCR1-containing integron encoding blaDHA-1 and the sap and psp operons. The sap operon confers resistance to small cationic peptides (20), while the psp operon may play a role in maintaining cytoplasmic membrane integrity in response to various environmental stresses (21). These structures were previously described for the pRBDHA (GenBank accession number AJ971343) and pPMDHA (GenBank accession number AJ971344) plasmids isolated from K. pneumoniae in Paris, France (22). It has at its 3′ extremity (which includes IRt-IS6100) a macrolide resistance region [comprising the mph(R), mrx, and mph(A) genes], as is often observed with class 1 introns/transposons (In/Tn) (18). In addition to the IS903.B-containing element mentioned above, two other composite transposons were identified in pKPS30, (i) an aph(3′)-I gene as part of a Tn4352-like transposon (18) inserted downstream of the mph(A) region and (ii) a Tn1721-like transposon (18) carrying the tetracycline resistance tetA gene. This transposon was found not to be complete, as the class 1 In/Tn was inserted in its res site (TCAAG).
The remaining 28,051-bp sequence of pKPS77 includes genes conferring resistance to β-lactams (blaSHV-12, blaTEM-1), aminoglycosides (strA, strB), tetracycline (tetA), and sulfonamides (sul2). The blaSHV-12 gene is part of an IS26-flanked composite transposon, as previously described on other plasmids (18, 23), and tetA is part of a truncated Tn1721 transposon (18). The structure, including a truncated Tn2 element carrying blaTEM-1b followed by the ΔCR2-strB-strA-sul2 configuration (24), was similar to the one observed in the IncFIIk plasmid pKD01 (GenBank accession number JX424423) (25).
IncR plasmids that are able to carry various resistance genes are reported with increasing frequency in clinical strains (1–7). However, as they are nontransferable and nonmobilizable because they lack a transfer system and a relaxase (26), one might question their role in the spread of MDR elements in Enterobacteriaceae. In addition, IncR replicons are increasingly described as part of multireplicon plasmids (including associations with IncA/C, IncF, IncFIIk, or nontypeable backbones) (6, 8–10). These associations raise the issue of replication coordination and of the incompatibility phenomenon in multireplicon plasmids.
The present study included two IncR plasmids displaying a wide variety of transposable elements carrying resistance genes. It confirms the increasing clinical importance of this plasmid group. The pool of resistance genes carried by IncR replicons may spread to transmissible plasmids through transposition events or plasmid recombination leading to multireplicons, thus contributing to the high plasticity observed in bacterial plasmids.
Nucleotide sequence accession numbers.
The sequences of pKPS30 and pKPS77 have been submitted to GenBank under accession numbers KF793937 and KF954150, respectively.
ACKNOWLEDGMENTS
This work was funded in part by the Université Pierre et Marie Curie (Paris, France) and by a grant from the Assistance Publique-Hôpitaux de Paris (contract CRC 08007 [KPath]).
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 21 April 2014
REFERENCES
- 1.García-Fernández A, Fortini D, Veldman K, Mevius D, Carattoli A. 2009. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J. Antimicrob. Chemother. 63:274–281. 10.1093/jac/dkn470 [DOI] [PubMed] [Google Scholar]
- 2.Hidalgo L, Gutierrez B, Ovejero CM, Carrilero L, Matrat S, Saba CK, Santos-Lopez A, Thomas-Lopez D, Hoefer A, Santurde G, Martin-Espada C, Gonzalez-Zorn B. 2013. Klebsiella pneumoniae ST11 from companion animals bearing ArmA methyltransferase, DHA-1 β-lactamase and QnrB4. Antimicrob. Agents Chemother. 57:4532–4534. 10.1128/AAC.00491-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baraniak A, Grabowska A, Izdebski R, Fiett J, Herda M, Bojarska K, Żabicka D, Kania-Pudło M, Młynarczyk G, Żak-Puławska Z, Hryniewicz W, Gniadkowski M, KPC-PL Study Group 2011. Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008-2009. Antimicrob. Agents Chemother. 55:5493–5499. 10.1128/AAC.05118-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bielak E, Bergenholtz RD, Jørgensen Sørensen MSSJ, Hansen LH, Hasman H. 2011. Investigation of diversity of plasmids carrying the blaTEM-52 gene. J. Antimicrob. Chemother. 66:2465–2474. 10.1093/jac/dkr331 [DOI] [PubMed] [Google Scholar]
- 5.Mataseje LF, Boyd DA, Willey BM, Prayitno N, Kreiswirth N, Gelosia A, Poutanen SM, Low DE, Jenkins SG, Katz K, Mulvey MR. 2011. Plasmid comparison and molecular analysis of Klebsiella pneumoniae harbouring bla(KPC) from New York City and Toronto. J. Antimicrob. Chemother. 66:1273–1277. 10.1093/jac/dkr092 [DOI] [PubMed] [Google Scholar]
- 6.Drieux L, Decré D, Frangeul L, Arlet G, Jarlier V, Sougakoff W. 2013. Complete nucleotide sequence of the large conjugative pTC2 multireplicon plasmid encoding the VIM-1 metallo-β-lactamase. J. Antimicrob. Chemother. 68:97–100. 10.1093/jac/dks367 [DOI] [PubMed] [Google Scholar]
- 7.Papagiannitsis CC, Miriagou V, Giakkoupi P, Tzouvelekis LS, Vatopoulos AC. 2013. Characterization of pKP1780, a novel IncR plasmid from the emerging Klebsiella pneumoniae ST147, encoding the VIM-1 metallo-β-lactamase. J. Antimicrob. Chemother. 68:2259–2262. 10.1093/jac/dkt196 [DOI] [PubMed] [Google Scholar]
- 8.Shen P, Wei Z, Jiang Y, Du X, Ji S, Yu Y, Li L. 2009. Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob. Agents Chemother. 53:4333–4338. 10.1128/AAC.00260-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frasson I, Lavezzo E, Franchin E, Toppo S, Barzon L, Cavallaro A, Richter SN, Palù G. 2012. Antimicrobial treatment and containment measures for an extremely drug-resistant Klebsiella pneumoniae ST101 isolate carrying pKPN101-IT, a novel fully sequenced bla(KPC-2) plasmid. J. Clin. Microbiol. 50:3768–3772. 10.1128/JCM.01892-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen YT, Shu HY, Li LH, Liao TL, Wu KM, Shiau YR, Yan JJ, Su IJ, Tsai SF, Lauderdale TL. 2006. Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-beta-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob. Agents Chemother. 50:3861–3866. 10.1128/AAC.00456-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villa L, García-Fernández A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65:2518–2529. 10.1093/jac/dkq347 [DOI] [PubMed] [Google Scholar]
- 12.Kado CI, Liu ST. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frangeul L, Glaser P, Rusniok C, Buchrieser C, Duchaud E, Dehoux P, Kunst F. 2004. CAAT-Box, contigs-assembly and annotation tool-box for genome sequencing projects. Bioinformatics 20:790–797. 10.1093/bioinformatics/btg490 [DOI] [PubMed] [Google Scholar]
- 14.Lane D, de Feyter R, Kennedy M, Phua SH, Semon D. 1986. D protein of miniF plasmid acts as a repressor of transcription and as a site-specific resolvase. Nucleic Acids Res. 14:9713–9728 [PMC free article] [PubMed] [Google Scholar]
- 15.Smith CM, Eisenstadt E. 1989. Identification of a umuDC locus in Salmonella Typhimurium LT2. J. Bacteriol. 171:3860–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen YT, Lin JC, Fung CP, Lu PL, Chuang YC, Wu TL, Siu LK. 2014. KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J. Antimicrob. Chemother. 69:628–631. 10.1093/jac/dkt409 [DOI] [PubMed] [Google Scholar]
- 17.Guynet C, Cuevas A, Moncalián G, de la Cruz F. 2011. The stb operon balances the requirements for vegetative stability and conjugative transfer of plasmid R388. PLoS Genet. 7:e1002073. 10.1371/journal.pgen.1002073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Partridge SR. 2011. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol. Rev. 35:820–855. 10.1111/j.1574-6976.2011.00277.x [DOI] [PubMed] [Google Scholar]
- 19.Mollet B, Clerget M, Meyer J, Iida S. 1985. Organization of the Tn6-related kanamycin resistance transposon Tn2680 carrying two copies of IS26 and an IS903 variant, IS903.B. J. Bacteriol. 163:55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groisman EA, Parra-Lopez C, Salcedo M, Lipps CJ, Heffron F. 1992. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc. Natl. Acad. Sci. U. S. A. 89:11939–11943. 10.1073/pnas.89.24.11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darwin AJ. 2005. The phage-shock-protein response. Mol. Microbiol. 57:621–628. 10.1111/j.1365-2958.2005.04694.x [DOI] [PubMed] [Google Scholar]
- 22.Verdet C, Benzerara Y, Gautier V, Adam O, Ould-Hocine Z, Arlet G. 2006. Emergence of DHA-1-producing Klebsiella spp. in the Parisian region: genetic organization of the ampC and ampR genes originating from Morganella morganii. Antimicrob. Agents Chemother. 50:607–617. 10.1128/AAC.50.2.607-617.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ford PJ, Avison MB. 2004. Evolutionary mapping of the SHV beta-lactamase and evidence for two separate IS26-dependent blaSHV mobilization events from the Klebsiella pneumoniae chromosome. J. Antimicrob. Chemother. 54:69–75. 10.1093/jac/dkh251 [DOI] [PubMed] [Google Scholar]
- 24.Anantham S, Hall RM. 2012. pCERC1, a small, globally disseminated plasmid carrying the dfrA14 cassette in the strA gene of the sul2-strA-strB gene cluster. Microb. Drug Resist. 18:364–371. 10.1089/mdr.2012.0008 [DOI] [PubMed] [Google Scholar]
- 25.Dolejska M, Villa L, Dobiasova H, Fortini D, Feudi C, Carattoli A. 2013. Plasmid content of a clinically relevant Klebsiella pneumoniae clone from the Czech Republic producing CTX-M-15 and QnrB1. Antimicrob. Agents Chemother. 57:1073–1076. 10.1128/AAC.01886-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alvarado A, Garcillán-Barcia MP, de la Cruz F. 2012. A degenerate primer MOB typing (DPMT) method to classify gamma-proteobacterial plasmids in clinical and environmental settings. PLoS One 7:e40438. 10.1371/journal.pone.0040438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petty NK, Bulgin R, Crepin VF, Cerdeño-Tárraga AM, Schroeder GN, Quail MA, Lennard N, Corton C, Barron A, Clark L, Toribio AL, Parkhill J, Dougan G, Frankel G, Thomson NR. 2010. The Citrobacter rodentium genome sequence reveals convergent evolution with human pathogenic Escherichia coli. J. Bacteriol. 192:525–538. 10.1128/JB.01144-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Y, Yu D, Wei Z, Shen P, Zhou Z, Yu Y. 2010. Complete nucleotide sequence of Klebsiella pneumoniae multidrug resistance plasmid pKP048, carrying blaKPC-2, blaDHA-1, qnrB4, and armA. Antimicrob. Agents Chemother. 54:3967–3969. 10.1128/AAC.00137-10 [DOI] [PMC free article] [PubMed] [Google Scholar]


