Abstract
The influenza pH1N1 virus caused a global flu pandemic in 2009 and continues manifestation as a seasonal virus. Better understanding of the virus-host cell interaction could result in development of better prevention and treatment options. Here we show that the Akt inhibitor MK2206 blocks influenza pH1N1 virus infection in vitro. In particular, at noncytotoxic concentrations, MK2206 alters Akt signaling and inhibits endocytic uptake of the virus. Interestingly, MK2206 is unable to inhibit H3N2, H7N9, and H5N1 viruses, indicating that pH1N1 evolved specific requirements for efficient infection. Thus, Akt signaling could be exploited further for development of better therapeutics against pH1N1 virus.
INTRODUCTION
The influenza pH1N1 virus caused a global pandemic which started in March 2009 in Mexico and the United States (www.who.int). The virus was a reassortant derivative of several viruses circulating in swine, and the initial transmission to humans occurred several months before recognition of the outbreak (1). Since 2011 the virus has been detected mainly during influenza epidemic seasons. Infections with influenza pH1N1 viruses varies from asymptomatic to serious complicated illness, including exacerbation of other underlying conditions and severe viral pneumonia with multiorgan failure. WHO recommended oseltamivir for treatment of patients with severe or progressive illness, because pH1N1 viruses are resistant to amantadine. Zanamivir and peramivir were recommended as the treatment of choice for patients in whom oseltamivir resistance was demonstrated or highly suspected (www.who.int). However, these drugs are not available in many hospitals and are difficult to administer to patients who are mechanically ventilated, and moreover, the virus can also acquire resistance to them (2, 3). Therefore, there is a need for developing newer antiviral agents against pH1N1and influenza viruses in general (4).
Recent advances in understanding influenza virus-host interactions revealed a number of cellular targets for potential antiviral interventions (5). It was proposed that temporal inhibition of proviral cellular functions with small molecules is less likely to elicit viral drug resistance (6–8). There are dozens of commercially available small molecules that inhibit cellular functions. Importantly, some of these small molecules are approved or in clinical development for treatment of other diseases (www.clinicaltrials.gov). This could facilitate their repurposing and use for treatment of viral infections, since a great deal is already known about their pharmacokinetics and toxicity profiles.
Multiple cellular signaling pathways are involved in influenza virus replication (9, 10). In particular, Akt signaling is involved in influenza virus uptake as well as at the later stages of the virus replication cycle (11–14). Here, we tested MK2206, an Akt inhibitor, against different influenza virus infections and showed that MK2206 has anti-pH1N1 activity in vitro. In particular, it interrupts the virus-mediated Akt signaling cascade and blocks endocytic uptake of pH1N1 virus. Thus, MK2206 represents a novel research tool for studying pH1N1 influenza virus-host cell interaction, and we propose that Akt signaling is a valuable target for anti-pH1N1 interventions.
MATERIALS AND METHODS
Compounds, cells, and viruses.
MK2206 (catalog no. S1078), ABT-263 (catalog no. S1001), perifosine (catalog no. S1037), miltefosine (catalog no. S3056), and obatoclax (catalog no. S1057) were from Selleckchem, Akt inhibitor VIII and NH4Cl (catalog no. A6730 and 254134) were from Sigma-Aldrich, and GDC-0068 (catalog no. CT-G0068) was from ChemieTek. Saliphenylhalamide (SaliPhe) was synthetized by Jef De Brabander (15). All compounds were dissolved in 100% dimethyl sulfoxide (DMSO) (Sigma-Aldrich) to 10 mM stock solutions and stored at −80°C until use.
MDCK and A549 cells were maintained in Dulbecco modified Eagle medium (DMEM) (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS) (Gibco), 2 mM l-glutamine (Lonza), and 50 U/ml penicillin-streptomycin mix (PenStrep; Lonza). NCI-H1666, NCI-H1703, NCI-1437, Calu-1, NCI-460, and NCI-H2126 cells were cultured in RPMI 1640 medium (Lonza) supplemented with 10% FBS, 2 mM l-glutamine, and 50 U/ml PenStrep. All cells were grown at 37°C and 5% CO2. Influenza A/Helsinki/P14/2009(pH1N1), A/Helsinki/628/2013(pH1N1), A/Helsinki/Vi1/2009(pH1N1), A/Helsinki/668/2013(pH1N1), A/Helsinki/Vi2/2009(pH1N1), A/Helsinki/526/2013(pH1N1) (16), A/Brussels/BB/2009(pH1N1) (17), A/WSN/33(H1N1), A/Sydney/5/1997(H3N2), A/Chicken/Nigeria/BA211/2006 (H5N1) (18), and A/Anhui/01/2013(H7N9) (provided by the WHO Collaborating Centre for Reference and Research on Influenza at the National Institute for Medical Research, London, United Kingdom) virus strains were propagated, and titers were determined, in Madin-Darby canine kidney (MDCK) cells.
Compound efficacy testing.
The compound efficacy testing was performed in cells grown in 96-well plates. Typically, cells were seeded in 100 μl of appropriate cell growth medium and grown for 24 h to reach 95% confluence. The growth medium was changed to appropriate virus growth medium (VGM) containing 0.2% bovine serum albumin (BSA) (Sigma-Aldrich), 2 mM l-glutamine, and 1 μg/ml (MDCK) or 0.4 μg/ml (A549 and NCI-H1666) l-1-tosylamido-2-phenylethyl chloromethyl ketone-trypsin (TPCK) (Sigma-Aldrich) in DMEM (A549 and MDCK) or RPMI (NCI-H1666). The compounds were added to the medium, and DMSO was added to the control wells. The cells were infected with viruses at a multiplicity of infection (MOI) of 0.1 to 10 or were mock infected. Cell viability was measured using the Cell Titer Glo (CTG) assay (Promega) at 24, 48, or 72 h postinfection. Luminescence was read with a PHERAstar FS plate reader (BMG Labtech).
For each virus-compound pair, we calculated the activity and toxicity score (ATS) as described in reference 6. Briefly, ATS = (AM − AB) − TM − TB, where A is the maximal antiviral activity (with virus), T is the cytotoxicity (without virus), M is the concentration level of maximal activity (over the whole dose range), and B refers to the baseline concentration level (i.e., the lowest dose). If the maximal activity AM was >100%, we set AM = 100, and if M = B (i.e., the maximal activity was already reached at the lowest dose), we set ATS = 0. Using these constraints, ATS varies between −100 and +100, where negative values indicate excessive toxicity and the highest positive values indicate the most potent compounds.
The 50% cytotoxic concentration (CC50) of MK2206 treatment was determined in NCI-H1666 cells treated with different concentrations of MK2206 (0 to 30 μM) for 24 h by CTG assay. The antiviral effect (50% effective concentration [EC50]) of MK2206, i.e., the ability of MK2206 to reduce pH1N1 virus production to 50%, was measured by determining the titers of viruses grown in NCI-H1666 cells (initial MOI, 0.1) for 24 h in the presence of 0 to 30 μM MK2206. The selectivity index (SI) was defined as the CC50/EC50 ratio.
Virus titration.
Cells were nontreated or treated with MK2206 at noncytotoxic concentrations and infected with A/Helsinki/P14/2009 at an MOI of 0.1. Supernatants were collected at 48 h postinfection, 10-fold diluted in DMEM-based VGM, and added to MDCK cells on 6-well plates. After 1 h, the cells were overlaid with medium containing 1.2% Avicel (FMC Biopolymer), 0.2% BSA, 50 U/ml PenStrep, 2 mM l-glutamine, and 1 μg/ml TPCK-trypsin in minimal essential medium and incubated for 2 days. The cells were fixed with 4% formaldehyde in phosphate buffer solution (PBS) for 3 h and stained with 0.1% crystal violet in 1% methanol, 20% pure ethanol, and 3.6% formaldehyde. Virus titers were determined by counting the PFU (clear spots) for each sample and expressed as PFU/ml.
Quantitative real-time PCR.
Total cellular RNA was isolated from NCI-H1666 cells noninfected or infected with the A/Helsinki/p14/2009 strain using the RNeasy Plus minikit (Qiagen). The RNA was reverse transcribed using the high-capacity cDNA reverse transcription kit (Applied Biosystems) according to the manufacturer's instructions. Reverse transcription-quantitative PCR (RT-qPCR) was performed with an ABI Prism 7500 sequence detection system. The TaqMan primers and probes (18S, Hs99999901_s1; beta interferon [IFN-β], Hs01077958_s1; and interleukin-29 [IL-29], Hs00601677_g1) and the TaqMan Fast Advanced master mix from Applied Biosystems were used. SYBR green master mix (Applied Biosystems) was applied for the SYBR green runs. A mix of the following GAPDH (glyceraldehyde-3-phosphate dehydrogenase) oligomers (Oligomer Oy) was used: huGAPDH_r with the sequence 5′-TGACCTTGGCCAGGGGTGCT-3′ and huGAPDH_f with the sequence 5′-GGCTGGGGCTCATTTGCAGGG-3′. The sequences of the NS1 oligomers (Sigma-Aldrich) were 5′-AGAAAGCTCTTATCTCTTG (reverse) and 5′-GAAATGTCAMGAGACT (forward). The RT-qPCR data were processed and quantified as described before (19), and the results are represented as relative units (RU).
Fluorimetry.
The fluorescence spectra were recorded using a Carry Eclipse fluorescence spectrophotometer (Varian/Agilent). A Hitachi U-3900 spectrophotometer was used for the absorbance measurements. The absorbance and fluorescence spectra of MK2206 were measured in PBS. The pH titrations were done using a series of buffers based on the Hydrion Buffer Chemvelopes (Micro Essential Laboratory, USA).
Automated image acquisition and image analysis.
MDCK cells were grown in 96-well plates at 40,000 cells/well and treated with different concentrations of MK2206 alone or combined with saliphenylhalamide (SaliPhe) (0.4 μM), NH4Cl (20 mM), or obatoclax (0.4 μM). Nontreated cells were used as controls. Cells were fixed with 4% paraformaldehyde (in PBS) for 20 min at room temperature and imaged with a ScanR modular epifluorescence microscope (Olympus). Long-working-distance objectives (40× [numerical aperture {NA}, 0.6] and 20× [NA, 0.45]) were used for image acquisition. A standard filter set for DAPI (4′,6′-diamidino-2-phenylindole) (excitation, 377/50 nm) was used for visualizing the autofluorescence of MK2206 within the cells. Sample analysis was performed in duplicate, and for each well, 9 different fields of view were analyzed using the ScanR Analysis software (Olympus). An automated background correction was applied for the DAPI channel. The autofluorescent vesicles (size between 1 and 20 pixels) visible in the DAPI channel were defined as the main objects, using edge detection. These objects were counted, and their area, circularity, and mean intensity were measured. The images were manually controlled for quality and for potential errors. The average and standard deviation, for the number of objects as well as for the mean intensity of the objects, were calculated for each sample.
siRNA and transfections.
Human Akt1 small interfering RNA (siRNA) (catalog no. 633), Akt2 siRNA (catalog no. 103305), Akt3 siRNA (catalog no. 110901), and control siRNA (catalog no. AM4636) were from Applied Biosystems, CA, USA. A549 cells were transfected with siRNA (final concentration, 20 nM) in 96-well plates. The transfections were performed with the Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer's protocol. Infections with viruses or mock infections were performed 48 h after the transfection of siRNA.
Immunofluorescence.
NCI-H1666 cells were grown on cover glasses in 6-well plates. Cells were nontreated or MK2206 treated (10 μM) and mock or A/Helsinki/P14/2009 infected (MOI, 30) on ice for 1 h. Cells were then washed twice with ice-cold VGM, overlaid with prewarmed medium with or without compound, and incubated at 37°C. At 1, 2, and 4 h postinfection, cells were fixed with 4% paraformaldehyde (in PBS) for 20 min at room temperature. PBS with 1% BSA and 0.1% Triton X-100 was used for blocking and permeabilization of the fixed cells and for dilution of antibodies. Viral NP and M1 were detected with rabbit polyclonal antibodies (1:1,000; Ilkka Julkunen, Finland); the secondary antibody was Alexa Fluor 488-conjugated goat anti-rabbit IgG(H+L) (1:2,000; Invitrogen). Nuclei were counterstained with DAPI. Images were captured with a Nikon 90i microscope and processed with NIS Elements AR software.
Immunoblotting.
For immunoblotting, proteins from total cell lysates were separated using SDS-PAGE and transferred onto an Immobilon-P membrane (Millipore, MA, USA). The membrane was blocked with 5% nonfat milk in PBS containing 0.05% Tween 20 or with 5% BSA in Tris-buffered saline (TBS) containing 0.05% Tween 20 and incubated with primary rabbit anti-NS1, rabbit anti-M1, rabbit anti-NP (1:5,000) (Ilkka Julkunen, Finland), rabbit anti-total Akt123 (1:1,000) (sc-8312; Santa Cruz Biotechnology), rabbit anti-phosphoSer473-Akt (1:1,000) (Cell Signaling), rabbit antiactin (1:500) (Santa Cruz Biotechnology), or mouse anti-GAPDH (1:1,000) (sc-47724; Santa Cruz Biotechnology) antibody overnight at 4°C. The membrane was washed three times for 10 min with TBS-Tween or PBS-Tween and incubated for 1 h at room temperature with the respective secondary antibodies conjugated to horseradish peroxidase (HRP) (DakoCytomation, Denmark). After three washes for 5 min with TBS-Tween or PBS-Tween buffer, the chemiluminescence of the labeled proteins was visualized with HRP substrate and captured with a photographic film.
Phosphoprotein, transcription, and cytokine profiling.
NCI-H1666 cells were nontreated or MK2206 treated (10 μM) and mock or virus infected with A/Helsinki/P14/2009 at an MOI of 3. For phosphoprotein profiling, the medium was discarded after 0.5, 4, and 12 h of infection, cells were lysed in lysis buffer from the human phosphokinase array kit (R&D Systems), and phosphorylation profiles of 43 kinases and 2 related substrates were analyzed using the human phosphokinase arrays according to the manufacturer's instructions. For cytokine profiling, the supernatants were collected after 24 h, and 26 different cytokines were assayed using the human cytokine array panel A according to the manufacturer's instructions (R&D Systems). The membranes were exposed to X-ray films, and the films were scanned. Each image was analyzed with ImageJ software (NIH, USA).
For the whole-genome gene expression analysis, cells were collected 8 h after infection and total RNA was isolated using RNeasy Plus minikit (Qiagen). The Illumina TotalPrep 96 RNA amplification kit was used to amplify and label the RNA. Human HT-12 V3 expression arrays were processed using the Limma and Beadarray packages from the Bioconductor suite. The raw data were exported from GenomeStudio. The data were log2 transformed prior to quintile normalization. We applied the IlluminaHumanv3.db annotation data freely available from Bioconductor in order to map Illumina probes to known genes and annotate the probes. To rank the genes in order of evidence for differential expression, we computed moderated t statistics and log odds of differential expression by empirical Bayes estimator.
Multipassage experiment.
NCI-H1666 cells were infected with A/Helsinki/P14/2009 at an MOI of 0.1 in the absence or presence of MK2206 (starting concentration, 0.1 μM). At 48 h postinfection, 20 μl of medium was used for the next passage in the presence or absence of the inhibitor. During passaging, we increased the MK2206 concentration from 0.1 to 10 µM. We performed 15 passages and determined the virus titers by plaque assay.
Ethics.
Virus experiments were carried out under biosafety level 2 (BSL2) conditions and in compliance with regulations of the University of Helsinki (permit no. 21/M/09) and under BSL3 conditions and in compliance with regulations of the Centre de Recherche Public de la Sante/Laboratoire National de Santé.
Microarray data accession number.
The whole-genome gene expression analysis data have been submitted to GEO (accession number GSE54293).
RESULTS
Agent.
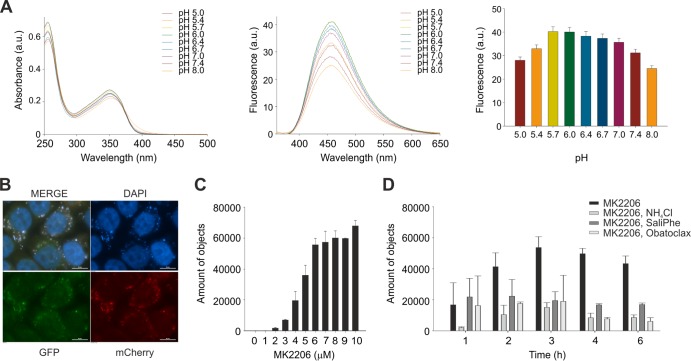
MK2206 is a highly selective allosteric inhibitor of Akt kinases which is in phase I/II clinical trials against cancer (20). We noticed that MK2206 has fluorescent properties, and therefore, we recorded and analyzed the fluorescence spectrum of MK2206. MK2206 has a single fluorescence emission peak at 456 nm (Fig. 1A). When we measured MK2206 fluorescence at different pHs, we observed maximal fluorescence at pH 5.7 to 6.0 (Fig. 1A). When we added MK2206 to Madine-Darby canine kidney (MDCK) cells, we observed fluorescent staining in vesicles in the cytoplasm (Fig. 1B). Moreover, we observed an increase in the number of fluorescent vesicles and their average intensities with increasing MK2206 concentration (Fig. 1C). Strikingly, the accumulation of MK2206 in the vesicles was abrogated by inhibitors of cellular v-ATPase and Mcl-1 (Fig. 1D) (6, 8, 20). These results suggest that endosome acidification and Mcl-1 function are required for MK2206 accumulation in the vesicles in cytoplasm.
FIG 1.
MK2206 is an autofluorescent small molecule whose accumulation in cellular cytoplasmic compartments is sensitive to endocytic pH. (A) Absorbance spectra, fluorescence emission spectra, and total fluorescence of MK2206 at different pHs. The absorbance peaks are at 259 nm and 354 nm; the shorter peak corresponds to the excitation of individual phenyl rings, while the longer peak possibly corresponds to the system of three conjugated rings. The emission has a rather broad spectrum with a half-width of about 100 nm, possibly reflecting the emission input from excitation of different rings of the compound. (B) MDCK cells were treated with 10 μM MK2206, fixed at 24 h, and imaged. Scale bars, 10 μm. (C) MDCK cells were treated with increasing concentrations of MK2206, fixed after 4 h, and imaged with ScanR. The autofluorescent vesicles visible in the DAPI channel were counted, and their area, circularity, and mean intensity were measured. (D) Cells were treated with 10 μM MK2206 or its combination with 20 mM NH4Cl, 0.4 μM SaliPhe, or 0.4 μM obatoclax (or remained nontreated), fixed at the indicated time points, and imaged with a ScanR. The points in panels A, C, and D are mean values, the number of observations used to derive the values is 2, and error bars represent the standard deviation (SD).
Cells.
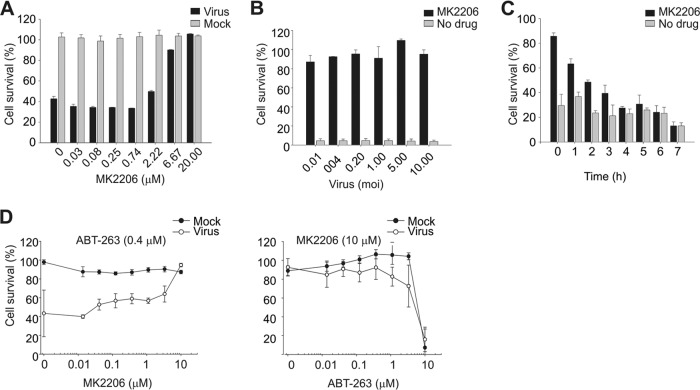
We next tested MK2206 for its ability to prevent influenza pH1N1 virus-mediated cell death at compound concentrations ranging from 0.002 to 20 μM. MK2206 rescued MDCK and human non-small-cell lung cancer NCI-H1666 cells from death mediated by A/Helsinki/P14/2009 virus at micromolar concentrations (Fig. 2A; see Fig. S1B in the supplemental material). Importantly, these concentrations are not toxic for the cells. Interestingly, the rescue of cells was independent of virus dose at selected compound concentrations (Fig. 2B; see Fig. S1C in the supplemental material).
FIG 2.
MK2206 prolongs survival of infected cells after lethal challenge with influenza pH1N1 virus when administered early in infection. (A) NCI-H1666 cells were treated with increasing concentrations of MK2206 and mock infected or A/Helsinki/P14/2009 infected (MOI, 3), and cell viability was measured by CTG assay after 48 h. The ATS was calculated. (B) NCI-H1666 cells were nontreated or MK2206 treated (8 μM) and infected with different MOIs of A/Helsinki/P14/2009, and cell viability was measured using the CTG assay after 48 h. (C) A/Helsinki/P14/2009-infected (MOI, 3) NCI-H1666 cells were treated with MK2206 (10 μM) at the indicated time points. Control cells remained nontreated. Cell viability was measured after 24 h using the CTG assay. (D) NCI-H1666 cells were treated with a constant concentration of ABT-263 (0.4 μM) and increasing concentrations of MK2206 (left panel) or with a constant concentration of MK2206 (3 μM) and increasing concentrations of ABT-263 (right panel). Cells were mock or A/Helsinki/P14/2009 infected (MOI, 3), and cell viability was measured using the CTG assay after 24 h. The data points are mean values, the number of observations used to derive the values is 3, and error bars represent the SD.
We next tested other Akt inhibitors for their ability to rescue MDCK cells from A/Helsinki/P14/2009 virus-mediated cell death. Perifosine, miltefosine, Akt inhibitor VIII, and GDC-0068 were unable to rescue the cells at the selected range of concentrations (see Fig. S2 in the supplemental material). We also tested the efficacy of MK2206 in cells infected with different pandemic influenza pH1N1 strains, as well as laboratory H1N1, seasonal H3N2, and potentially pandemic H7N9 and H5N1 strains. MK2206 rescued cells infected with influenza pH1N1 virus and to some extent the H1N1 WSN strain but not those infected with seasonal and potentially pandemic strains (see Fig. S3 in the supplemental material), indicating that MK2206 efficacy could be virus subtype specific.
Next, we performed an experiment on the time of compound addition in A/Helsinki/P14/2009-infected NCI-H1666 cells. When MK2206 was applied to the cells within the first 2 h of infection, cells were protected from virus-mediated death (Fig. 2C). We then analyzed whether MK2206 could protect cells from death by a combination of influenza virus infection and ABT-263 treatment: ABT-263 at submicromolar concentrations is known to induce premature apoptosis in response to viral RNA in infected but not noninfected cells (21). We found that MK2206 protects NCI-H1666 cells from simultaneous influenza virus infection and ABT-263 treatment (Fig. 2D). Thus, MK2206 protects cells during early steps of influenza pH1N1 virus infection.
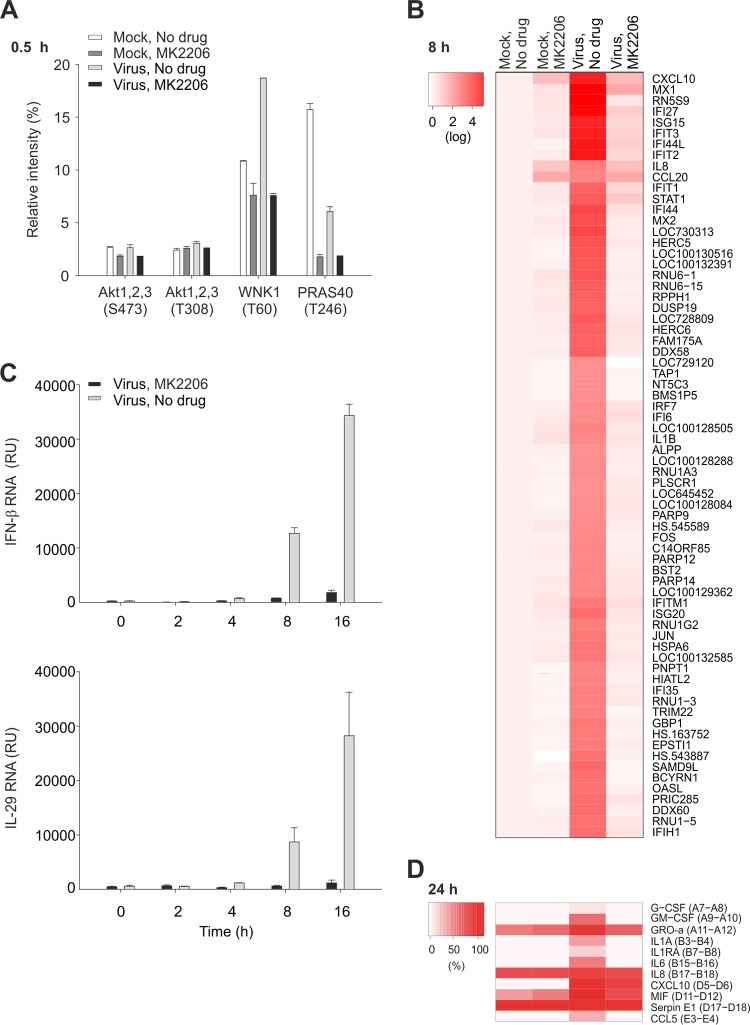
It was shown that Akt becomes phosphorylated during influenza virus entry (22). Therefore, we evaluated the effect of MK2206 on the phosphorylation status of Akt and several of its downstream targets, including AMP-activated protein kinase (AMPK), β-catenin, CREB-1, endothelial nitric oxide synthase (eNOS), extracellular signal-regulated kinase (ERK), focal adhesion kinase (FAK), glycogen synthase kinase 3 (GSK3), HSP27, P70S6, PRAS40, TOR, and WNK1 (23–33) in NCI-H1666 cells infected with A/Helsinki/P14/2009 for 0.5 h in the presence of absence of MK2206 and using mock-infected cells as controls. Phospho-proteome profiling revealed that MK2206 treatment did not affect the phosphorylation status of Akt but lowered the phosphorylation levels of its targets, PRAS40 and WNK1 (Fig. 3A; see Fig. S3 in the supplemental material). We also examined the effect of MK2206 on cellular signaling at 4 and 12 h postinfection. MK2206 abrogated virus-mediated modulation of phosphorylation of Akt, HSP27, p53, c-Jun, and HSP27 (see Fig. S3 in the supplemental material). This result suggests that MK2206 prevents Akt-mediated signaling and its feedback loops during pH1N1 virus infection.
FIG 3.
MK2206 alters Akt signaling and does not allow development of cellular responses to pH1N1 infection. (A) NCI-H1666 cells were nontreated or MK2206 treated (10 μM) and mock or A/Helsinki/P14/2009 infected (MOI, 3), cells were collected after 0.5 h, and phosphorylation levels of kinases and their substrates were profiled using a human phosphokinase array. The relative intensities of spots were calculated in ImageJ and plotted. (B and C) NCI-H1666 cells were treated as for panel A, cells were collected, and total RNA was extracted at 8 h, labeled, and used for whole-genome gene expression analysis. A heat map of 70 significantly changed genes is shown. The heat map represents normalized expression data on the logarithmic scale (log fold change of >2 and <−2), where genes are ordered by means of hierarchical clustering. Total RNA was isolated at 0, 2, 4, 8, and 16 h and subjected to RT-qPCRs detecting IFN-β and IL-29 mRNA levels. The data points are mean values, the numbers of observations used to derive the values are 2 and 3, respectively, and error bars represent the SD. (D) NCI-H1666 cells were treated as for panel A, cell culture supernatants were collected at 24 h postinfection, and cytokine levels were determined using human cytokine array panel A. A heat map is also shown.
Next, we studied cellular responses to virus infection in MK2206-treated and nontreated NCI-H1666 cells at the transcription level. Whole-genome expression analysis at 8 h postinfection (fold change cutoff, >+2 and <−2) revealed that transcription of antiviral genes remained unchanged in response to pH1N1 infection in MK2206-treated cells (Fig. 3B). RT-qPCR analysis confirmed that IFN-β and IL-29 levels remained unchanged over time in MK2206-treated virus-infected cells, in contrast to the case for untreated virus-infected cells (Fig. 3C). Moreover, cytokine profiling also confirmed that secretion of antiviral and proinflammatory cytokines was strongly reduced in MK2206-treated virus-infected cells (Fig. 3D; see Fig. S4 in the supplemental material). Thus, MK2206 inhibits Akt signaling and prevents development of cellular antiviral responses at the transcriptional, translational, and posttranslational levels.
Virus.
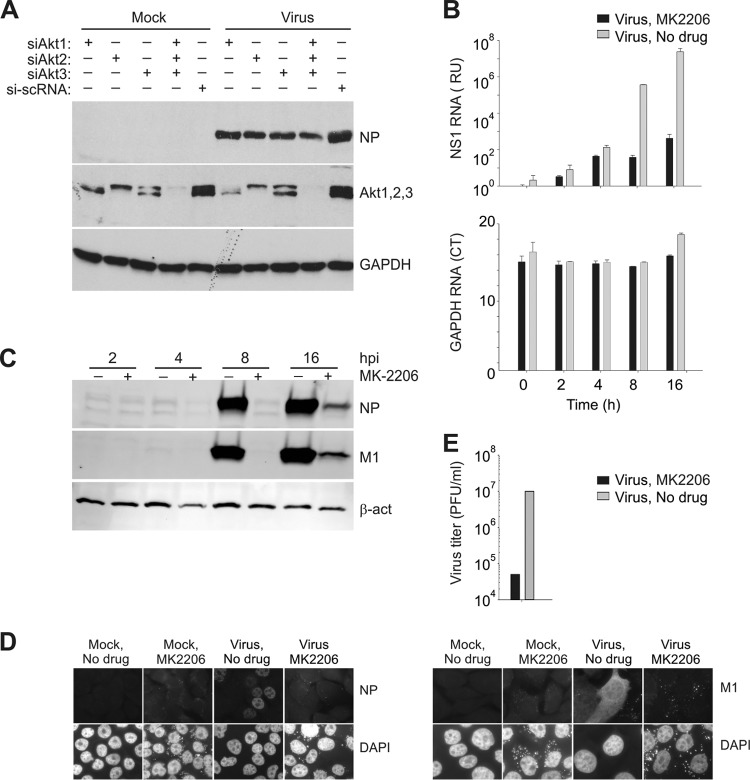
To understand whether the intracellular level of Akt is essential for efficient pH1N1 virus infection, we silenced different isoforms of Akt with isoform-specific siRNA (34). The effect of siRNA on expression of viral NP as well as cellular Akt and GAPDH in A549 cells was evaluated by Western blotting. Partial silencing of Akt1, Akt2, or Akt3, or all three Akt isoforms, resulted in a reduced NP expression (Fig. 4A). We concluded that intracellular steady-state concentrations of Akt are essential for virus protein synthesis.
FIG 4.
Akt is essential for pH1N1 infection, and its inhibition by MK2206 attenuates pH1N1 replication. (A) A549 cells were transfected with siRNA (20 nM) or a mixture of siRNA (6.6 nM each Akt siRNA). After 48 h, cells were mock infected or infected with A/Helsinki/P14/2009 virus (MOI, 1). After 24 h, protein expression was evaluated by Western blotting. si-scRNA is a negative control. (B) NCI-H1666 cells were nontreated or MK2206 treated (10 μM) and mock or A/Helsinki/P14/2009 infected (MOI, 3), and total RNA was extracted at the indicated time points and subjected to RT-qPCRs detecting viral NS1 and cellular GAPDH RNAs. The data points are mean values, the number of observations used to derive the values is 3, and error bars represent the SD. (C) NCI-H1666 cells were nontreated or MK2206 treated (10 μM) and mock or A/Helsinki/P14/2009 infected (MOI, 3), and total cell lysates were obtained at the indicated times after infection. M1, NP, and β-actin (loading control) were analyzed by Western blotting. (D) NCI-H1666 cells were nontreated or MK2206 treated (10 μM) and mock or A/Helsinki/P14/2009 infected (MOI, 30), fixed at 4 h postinfection, and imaged for the viral NP and M1. DAPI stains the nucleus. Scale bars, 10 μm. (E) The titers in supernatants from NCI-H1666 cells that were nontreated or MK2206 treated (3 μM) and infected with A/Helsinki/P14/2009 (MOI, 0.1) were determined on MDCK cells.
We next examined the effect of MK2206 on the production of viral RNA and proteins. RT-qPCR analyses revealed that MK2206 treatment substantially lowered the transcription of viral but not cellular RNA (Fig. 4B). Immunoblotting and immunofluorescence microscopy analysis revealed that the production of viral M1 and NP was strongly attenuated in the presence of MK2206 (Fig. 4C and D). This inhibition of viral RNA and protein synthesis correlated with reduced infectious virus particle production (Fig. 4E). Thus, MK2206 has a clear anti-pH1N1 effect.
We next analyzed the efficacy of MK2206 against other pH1N1 strains isolated in 2009 or 2013 from Finnish and Belgian patients. We found that EC50s are in the micromolar range (Table 1). Thus, MK2206 efficiently inhibits infection of influenza pH1N1 viruses.
TABLE 1.
Anti-pH1N1 efficacy of MK2206a
| pH1N1 strain | EC50, μM (mean ± SD) | SI |
|---|---|---|
| A/Helsinki/P14/2009 | 0.79 ± 0.26 | 74 |
| A/Helsinki/Vi1/2009 | 0.99 ± 0.03 | 59 |
| A/Helsinki/Vi2/2009 | 4.55 ± 0.67 | 12 |
| A/Brussels/BB/2009 | 3.93 ± 1.45 | 15 |
| A/Helsinki/526/2013 | 3.36 ± 0.64 | 17 |
| A/Helsinki/628/2013 | 0.96 ± 0.01 | 61 |
| A/Helsinki/668/2013 | 1.11 ± 0.02 | 52 |
NCI-H1666 cells were treated with increasing concentrations of MK2206 (0 to 30 μM) and mock infected or infected with the indicated pH1N1virus (MOI, 0.1). The viability of mock-infected cells was measured by CTG assay after 24 h, and the CC50 was calculated (58.51 ± 5.6 μM). Cell culture supernatants from infected NCI-H166 cells were collected at 24 h postinfection, and the virus titers were determined in MDCK cells. The antiviral effect (EC50) of MK2206, i.e., the ability of MK2206 to reduce pH1N1 virus production to 50%, was calculated. The selectivity indexes (SI) was determined as the CC50/EC50 ratio. The number of independent experiments that were performed to derive the mean is 3.
We wondered whether the A/Helsinki/P14/2009 strain is able to acquire resistance to MK2206. We propagated the virus in cells in the presence of continuously increasing MK2206 concentrations for 15 passages and determined the titers of the resulting viruses (Fig. 5). The virus titers still dropped after passage 11 at 2 μM MK2206, indicating that the virus could not adapt to the selective pressure posed by MK2206 during 15 passages.
FIG 5.
pH1N1 virus remain sensitive after passaging in cells treated with increasing MK2206 concentrations. The influenza A/Helsinki/P14/2009 virus strain was propagated in NCI-H1666 cells for 15 passages (48 h per passage) with increasing MK2206 concentrations (from 0.1 to 10 μM). Cell culture supernatants were collected, and virus titers were determined in MDCK cells. The titers are plotted.
DISCUSSION
We showed here that Akt signaling plays an important role in influenza pH1N1 virus infection and that MK2206, a highly selective inhibitor of Akt, possesses anti-pH1N1 activity. Our results suggest that Akt signaling is essential for endocytic uptake of influenza pH1N1 virus, and that inhibition of Akt by MK2206 prevents release of viral RNA and triggering of cellular antiviral responses.
Importantly, MK2206 was unable to rescue cells from death associated with H5N1, H7N9, and H3N2 virus infections, indicating that MK2206 antiviral efficacy could be virus subtype specific. Given that viral hemagglutinin (HA) requires acid pH for the conformational switch which is needed for membrane fusion and release of viral genome from the endocytic compartment, we speculate that a more acidic pH is needed for fusogenic activation of HA of pH1N1, in contrast to HAs of H5N1, H7N9, and H3N2 viruses (35, 36). Perhaps this property was selected by pH1N1 during evolution in order to compete with other viruses for the host.
Akt was shown to regulate the endosome pH (37) and endosome/lysosome-associated mTORC1 activity (38). We showed that MK2206 treatment lowered the phosphorylation level of PRAS40, which is a substrate for Akt and a subunit of mTORC1. Our results suggest that the Akt/mTORC1 signaling axis could be essential for influenza pH1N1 virus infection. Future studies will undoubtedly provide further details on the role of Akt in pH1N1 virus entry and pH1N1 evolution.
Importantly, MK2206 is an orally bioavailable anticancer drug currently in phase I/II clinical trials. Anticancer and antiviral treatments with MK2206 could differ in dosage and duration, and therefore the anticancer side effects might not be relevant for treatment of pH1N1 infections in vivo. Thus, anticancer MK2206 and Akt signaling could be further exploited for treatment of severe pH1N1 infections.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Petri Jalovaara, Emmy Verschuren, Jenni Lahtela, Ashwini Nagaraj, Jhon Patrik Mpindi, Giuseppe Balistreri, Laura Kakkola, Maria Anastasina, Evgeni Kylesskiy, Ingrida Sauliene, and Triin Lakspere for fruitful scientific discussions and technical assistance.
This work was supported by the Jane and Aatos Erkko Foundation, Academy of Finland (255852), European Union Structural Funds (VP1-3.1-ŠMM-07-K-03-069), and the Erasmus program.
Footnotes
Published ahead of print 21 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02798-13.
REFERENCES
- 1.Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125. 10.1038/nature08182 [DOI] [PubMed] [Google Scholar]
- 2.Hurt AC, Holien JK, Parker M, Kelso A, Barr IG. 2009. Zanamivir-resistant influenza viruses with a novel neuraminidase mutation. J. Virol. 83:10366–10373. 10.1128/JVI.01200-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samson M, Pizzorno A, Abed Y, Boivin G. 2013. Influenza virus resistance to neuraminidase inhibitors. Antiviral Res. 98:174–185. 10.1016/j.antiviral.2013.03.014 [DOI] [PubMed] [Google Scholar]
- 4.Hayden FG. 2013. Newer influenza antivirals, biotherapeutics and combinations. Influenza and other respiratory viruses. 7(Suppl 1):63–75. 10.1111/irv.12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller KH, Kakkola L, Nagaraj AS, Cheltsov AV, Anastasina M, Kainov DE. 2012. Emerging cellular targets for influenza antiviral agents. Trends Pharmacol. Sci. 33:89–99. 10.1016/j.tips.2011.10.004 [DOI] [PubMed] [Google Scholar]
- 6.Denisova OV, Kakkola L, Feng L, Stenman J, Nagaraj A, Lampe J, Yadav B, Aittokallio T, Kaukinen P, Ahola T, Kuivanen S, Vapalahti O, Kantele A, Tynell J, Julkunen I, Kallio-Kokko H, Paavilainen H, Hukkanen V, Elliott RM, De Brabander JK, Saelens X, Kainov DE. 2012. Obatoclax, saliphenylhalamide, and gemcitabine inhibit influenza a virus infection. J. Biol. Chem. 287:35324–35332. 10.1074/jbc.M112.392142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Theisen LL, Muller CP. 2012. EPs 7630 (Umckaloabo), an extract from Pelargonium sidoides roots, exerts anti-influenza virus activity in vitro and in vivo. Antiviral Res. 94:147–156. 10.1016/j.antiviral.2012.03.006 [DOI] [PubMed] [Google Scholar]
- 8.Muller KH, Kainov DE, El Bakkouri K, Saelens X, De Brabander JK, Kittel C, Samm E, Muller CP. 2011. The proton translocation domain of cellular vacuolar ATPase provides a target for the treatment of influenza A virus infections. Br. J. Pharmacol. 164:344–357. 10.1111/j.1476-5381.2011.01346.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Planz O. 2013. Development of cellular signaling pathway inhibitors as new antivirals against influenza. Antiviral Res. 98:457–468. 10.1016/j.antiviral.2013.04.008 [DOI] [PubMed] [Google Scholar]
- 10.Ludwig S. 2009. Targeting cell signalling pathways to fight the flu: towards a paradigm change in anti-influenza therapy. J. Antimicrob. Chemother. 64:1–4. 10.1093/jac/dkp161 [DOI] [PubMed] [Google Scholar]
- 11.Ehrhardt C, Marjuki H, Wolff T, Nurnberg B, Planz O, Pleschka S, Ludwig S. 2006. Bivalent role of the phosphatidylinositol-3-kinase (PI3K) during influenza virus infection and host cell defence. Cell. Microbiol. 8:1336–1348. 10.1111/j.1462-5822.2006.00713.x [DOI] [PubMed] [Google Scholar]
- 12.Shin YK, Liu Q, Tikoo SK, Babiuk LA, Zhou Y. 2007. Effect of the phosphatidylinositol 3-kinase/Akt pathway on influenza A virus propagation. J. Gen. Virol. 88:942–950. 10.1099/vir.0.82483-0 [DOI] [PubMed] [Google Scholar]
- 13.Murray JL, McDonald NJ, Sheng J, Shaw MW, Hodge TW, Rubin DH, O'Brien WA, Smee DF. 2012. Inhibition of influenza A virus replication by antagonism of a PI3K-AKT-mTOR pathway member identified by gene-trap insertional mutagenesis. Antiviral Chem. Chemother. 22:205–215. 10.3851/IMP2080 [DOI] [PubMed] [Google Scholar]
- 14.Zhirnov OP, Klenk HD. 2007. Control of apoptosis in influenza virus-infected cells by up-regulation of Akt and p53 signaling. Apoptosis 12:1419–1432. 10.1007/s10495-007-0071-y [DOI] [PubMed] [Google Scholar]
- 15.Lebreton S, Jaunbergs J, Roth MG, Ferguson DA, De Brabander JK. 2008. Evaluating the potential of vacuolar ATPase inhibitors as anticancer agents and multigram synthesis of the potent salicylihalamide analog saliphenylhalamide. Bioorg. Med. Chem. Lett. 18:5879–5883. 10.1016/j.bmcl.2008.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lakspere T, Tynell J, Kaloinen M, Vanlede M, Parsons A, Ikonen N, Kallio-Kokko H, Kantele A, Mattila P, Almusa H, Julkunen I, Kainov D, Kakkola L. 2014. Full-genome sequences of influenza A(H1N1)pdm09 viruses isolated from Finnish patients from 2009 to 2013. Genome Announc. 2(1):e01004–13. 10.1128/genomeA.01004-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schotsaert M, Ysenbaert T, Neyt K, Ibanez LI, Bogaert P, Schepens B, Lambrecht BN, Fiers W, Saelens X. 2013. Natural and long-lasting cellular immune responses against influenza in the M2e-immune host. Mucosal Immunol. 6:276–287. 10.1038/mi.2012.69 [DOI] [PubMed] [Google Scholar]
- 18.Ducatez MF, Olinger CM, Owoade AA, De Landtsheer S, Ammerlaan W, Niesters HG, Osterhaus AD, Fouchier RA, Muller CP. 2006. Avian flu: multiple introductions of H5N1 in Nigeria. Nature 442:37. 10.1038/442037a [DOI] [PubMed] [Google Scholar]
- 19.Ohman T, Lietzen N, Valimaki E, Melchjorsen J, Matikainen S, Nyman TA. 2010. Cytosolic RNA recognition pathway activates 14-3-3 protein mediated signaling and caspase-dependent disruption of cytokeratin network in human keratinocytes. J. Proteome Res. 9:1549–1564. 10.1021/pr901040u [DOI] [PubMed] [Google Scholar]
- 20.Bimbo LM, Denisova OV, Makila E, Kaasalainen M, De Brabander JK, Hirvonen J, Salonen J, Kakkola L, Kainov D, Santos HA. 2013. Inhibition of influenza a virus infection in vitro by saliphenylhalamide-loaded porous silicon nanoparticles. ACS Nano 7:6884–6893. 10.1021/nn402062f [DOI] [PubMed] [Google Scholar]
- 21.Kakkola L, Denisova OV, Tynell J, Viiliainen J, Ysenbaert T, Matos RC, Nagaraj A, Ohman T, Kuivanen S, Paavilainen H, Feng L, Yadav B, Julkunen I, Vapalahti O, Hukkanen V, Stenman J, Aittokallio T, Verschuren EW, Ojala PM, Nyman T, Saelens X, Dzeyk K, Kainov DE. 2013. Anticancer compound ABT-263 accelerates apoptosis in virus-infected cells and imbalances cytokine production and lowers survival rates of infected mice. Cell Death Dis. 4:e742. 10.1038/cddis.2013.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, Nikrad MP, Phang T, Gao B, Alford T, Ito Y, Edeen K, Travanty EA, Kosmider B, Hartshorn K, Mason RJ. 2011. Innate immune response to influenza A virus in differentiated human alveolar type II cells. Am. J. Respir. Cell Mol. Biol. 45:582–591. 10.1165/rcmb.2010-0108OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Socodato R, Brito R, Portugal CC, de Oliveira NA, Calaza KC, Paes-de-Carvalho R. 14 February 2014. The nitric oxide-cGKII system relays death and survival signals during embryonic retinal development via AKT-induced CREB1 activation. Cell Death Differ. 10.1038/cdd.2014.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dent P. 2014. Crosstalk between ERK, AKT, and cell survival. Cancer Biol. Ther. 15:245–246. 10.4161/cbt.27541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saraswati S, Kumar S, Alhaider AA. 2013. Alpha-santalol inhibits the angiogenesis and growth of human prostate tumor growth by targeting vascular endothelial growth factor receptor 2-mediated AKT/mTOR/P70S6K signaling pathway. Mol. Cancer 12:147. 10.1186/1476-4598-12-147 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Zhang J, Shemezis JR, McQuinn ER, Wang J, Sverdlov M, Chenn A. 2013. AKT activation by N-cadherin regulates beta-catenin signaling and neuronal differentiation during cortical development. Neural Dev. 8:7. 10.1186/1749-8104-8-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitagishi Y, Kobayashi M, Kikuta K, Matsuda S. 2012. Roles of PI3K/AKT/GSK3/mTOR pathway in cell signaling of mental illnesses. Depress. Res. Treat. 2012:752563. 10.1155/2012/752563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higuchi M, Kihara R, Okazaki T, Aoki I, Suetsugu S, Gotoh Y. 2013. Akt1 promotes focal adhesion disassembly and cell motility through phosphorylation of FAK in growth factor-stimulated cells. J. Cell Sci. 126:745–755. 10.1242/jcs.112722 [DOI] [PubMed] [Google Scholar]
- 29.Wiza C, Nascimento EB, Ouwens DM. 2012. Role of PRAS40 in Akt and mTOR signaling in health and disease. Am. J. Physiol. Endocrinol. Metab. 302:E1453–E1460. 10.1152/ajpendo.00660.2011 [DOI] [PubMed] [Google Scholar]
- 30.Hayashi N, Peacock JW, Beraldi E, Zoubeidi A, Gleave ME, Ong CJ. 2012. Hsp27 silencing coordinately inhibits proliferation and promotes Fas-induced apoptosis by regulating the PEA-15 molecular switch. Cell Death Differ. 19:990–1002. 10.1038/cdd.2011.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ning J, Xi G, Clemmons DR. 2011. Suppression of AMPK activation via S485 phosphorylation by IGF-I during hyperglycemia is mediated by AKT activation in vascular smooth muscle cells. Endocrinology 152:3143–3154. 10.1210/en.2011-0155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu S, Wong SL, Lau CW, Huang Y, Yu CM. 2011. Oxidized LDL at low concentration promotes in-vitro angiogenesis and activates nitric oxide synthase through PI3K/Akt/eNOS pathway in human coronary artery endothelial cells. Biochem. Biophys. Res. Commun. 407:44–48. 10.1016/j.bbrc.2011.02.096 [DOI] [PubMed] [Google Scholar]
- 33.Jiang ZY, Zhou QL, Holik J, Patel S, Leszyk J, Coleman K, Chouinard M, Czech MP. 2005. Identification of WNK1 as a substrate of Akt/protein kinase B and a negative regulator of insulin-stimulated mitogenesis in 3T3-L1 cells. J. Biol. Chem. 280:21622–21628. 10.1074/jbc.M414464200 [DOI] [PubMed] [Google Scholar]
- 34.Cheshenko N, Trepanier JB, Stefanidou M, Buckley N, Gonzalez P, Jacobs W, Herold BC. 2013. HSV activates Akt to trigger calcium release and promote viral entry: novel candidate target for treatment and suppression. FASEB J. 27:2584–2599. 10.1096/fj.12-220285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cotter CR, Jin H, Chen Z. 2014. A single amino acid in the stalk region of the H1N1pdm influenza virus HA protein affects viral fusion, stability and infectivity. PLoS Pathog. 10:e1003831. 10.1371/journal.ppat.1003831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lozach PY, Huotari J, Helenius A. 2011. Late-penetrating viruses. Curr. Opin. Virol. 1:35–43. 10.1016/j.coviro.2011.05.004 [DOI] [PubMed] [Google Scholar]
- 37.Diering GH, Numata Y, Fan S, Church J, Numata M. 2013. Endosomal acidification by Na+/H+ exchanger NHE5 regulates TrkA cell-surface targeting and NGF-induced PI3K signaling. Mol. Biol. Cell 24:3435–3448. 10.1091/mbc.E12-06-0445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. 2014. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156:771–785. 10.1016/j.cell.2013.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.