Abstract
In combination with antibiotics, quinine is recommended as the second-line treatment for uncomplicated malaria, an alternative first-line treatment for severe malaria, and for treatment of malaria in the first trimester of pregnancy. Quinine has been shown to have frequent clinical failures, and yet the mechanisms of action and resistance have not been fully elucidated. However, resistance is linked to polymorphisms in multiple genes, including multidrug resistance 1 (Pfmdr1), the chloroquine resistance transporter (Pfcrt), and the sodium/hydrogen exchanger gene (Pfnhe1). Here, we investigated the association between in vitro quinine susceptibility and genetic polymorphisms in Pfmdr1codons 86 and 184, Pfcrt codon 76, and Pfnhe1 ms4760 in 88 field isolates from western Kenya. In vitro activity was assessed based on the drug concentration that inhibited 50% of parasite growth (the IC50), and parasite genetic polymorphisms were determined from DNA sequencing. Data revealed there were significant associations between polymorphism in Pfmdr1-86Y, Pfmdr1-184F, or Pfcrt-76T and quinine susceptibility (P < 0.0001 for all three associations). Eighty-two percent of parasites resistant to quinine carried mutant alleles at these codons (Pfmdr1-86Y, Pfmdr1-184F, and Pfcrt-76T), whereas 74% of parasites susceptible to quinine carried the wild-type allele (Pfmdr1-N86, Pfmdr1-Y184, and Pfcrt-K76, respectively). In addition, quinine IC50 values for parasites with Pfnhe1 ms4760 3 DNNND repeats were significantly higher than for those with 1 or 2 repeats (P = 0.033 and P = 0.0043, respectively). Clinical efficacy studies are now required to confirm the validity of these markers and the importance of parasite genetic background.
INTRODUCTION
Quinine (QN), a quinoline derivative, is used in many African countries as second-line treatment for uncomplicated malaria, as an alternate first-line treatment of severe malaria, and for treatment of malaria in the first trimester of pregnancy (1). World Health Organization (WHO) guidelines recommend a combination of QN plus doxycycline, tetracycline, or clindamycin for treatment of malaria (2). However, in most African countries, QN is used as a monotherapy (1, 2), probably due to the high cost of antibiotics (3). QN has been shown to have frequent clinical failures in Southeast Asia (4, 5), South America (5), and Africa (6–8). In a clinical trial conducted in Kenya concerning the treatment of severe Plasmodium falciparum malaria, QN was shown to have longer clearance times, longer fever clearance times, and higher recrudescence rates than malarone (9). The high rates of QN clinical failures can be explained by variations in pharmacokinetics, drug quality, and treatment compliance, but less so to resistance. Parasites with high-grade resistance to QN have not been documented (3). However, in vitro analysis has shown reduced sensitivity to QN in Asia (10) and South America (11), and much less so in Africa (12, 13). The high rates of reinfection and recrudescence can also be explained by the short half-life of QN (14).
Although the mechanisms of action and resistance to QN have not been fully elucidated, inhibition of heme detoxification in the parasite digestive vacuole has been implicated in QN antimalarial activity (15), and resistance is linked to polymorphisms in multiple genes, including the multidrug resistance 1 gene (Pfmdr1), chloroquine resistance transporter gene (Pfcrt), and the sodium/hydrogen exchanger gene (Pfnhe1) (16). In particular, mutations in Pfmdr1 at codons 86, 184, 1042, and 1246 and in Pfcrt at codon 76 have been associated with reduced QN sensitivity (17–19). Interestingly, however, in some studies polymorphism in Pfcrt-76 has been shown not to have any effect on QN activity (20, 21), and the Pfmdr1-86 mutation only resulted in a decreased QN activity that did not reach statistical significance (20). Other novel mutations in the Pfcrt gene that have recently been shown to alter QN sensitivity are Q352K/R and C350R (20, 22, 23).
Pfnhe1 is a 5,760-bp gene that encodes 1,920 amino acids, with a microsatellite polymorphism consisting of variable DNNND repeat units, designated ms4760. Sequence polymorphisms in the Pfnhe1 gene have been analyzed in laboratory strains and field isolates with varied susceptibilities to QN (16, 24–27). Several variants of ms4760 have been described in which ms4760–1, with 2 copies of the DNNND repeat unit, was significantly associated with reduced in vitro QN sensitivity in laboratory clones (16) and in field isolates (24). On the contrary, other studies have not found an association between polymorphisms in the Pfnhe1 gene and QN reduced susceptibility (26, 27). These conflicting results may indicate that the influence of Pfnhe1 on QN susceptibility is strain dependent and can vary depending on the geographic origin of the parasites (28). Polymorphisms in the Pfnhe1 ms4760 DDNHNDNHNND and DDNNNDNHNDD repeats have also been associated with varied in vitro QN susceptibilities (24–27, 29, 30).
Similar to most countries in Africa, QN is used as second-line treatment for uncomplicated malaria and treatment of severe malaria in Kenya (9, 12). A recent study analyzed the association of in vitro activities of QN with Pfmdr1 codon 86, Pfcrt codon 76, and Pfnhe1 polymorphism in isolates collected from Kilifi District, which is located on the Kenyan coast (24). These authors observed Pfmdr1-86 mutants, and 2 copies of DNNND repeat units of Pfnhe1were associated with a decrease in QN susceptibility. In this study, we investigated the association between in vitro QN susceptibility and genetic polymorphisms in Pfmdr1codons 86 and 184, Pfcrt codon 76, and Pfnhe1 ms4760 in field isolates from western Kenya, where malaria is holoendemic.
MATERIALS AND METHODS
Plasmodium falciparum parasites.
P. falciparum field isolates used in this study were collected from patients with uncomplicated malaria, ages 6 months and older, attending outpatient clinics in Kisumu, Kisii, and Kericho District hospitals in western Kenya between January 2010 and December 2011. Details of this protocol have been described elsewhere (31). The research protocol was approved by the Ethical Review Committee of the Kenya Medical Research Institute (KEMRI number 1330) and the Walter Reed Army Institute of Research institutional review board (WRAIR number 1384). P. falciparum reference strains3D7 (sensitive) and W2 (resistant) were used as controls. These clones were obtained from cryopreserved stocks and culture adapted for drug sensitivity assays.
In vitro drug sensitivity tests.
QN was obtained from the Walter Reed Army Institute of Research (WRAIR), Silver Spring, MD. Parasites were maintained in continuous culture by using a method described elsewhere (32). A previously described SYBR green I-based drug sensitivity assay was used for in vitro drug sensitivity testing (33–35). P. falciparum parasites in continuous culture attaining 3 to 8% parasitemia (field isolates or reference strains) were adjusted to 2% hematocrit and 1% parasitemia. QN was prepared in 5 ml of 70% ethanol to attain 5 mg/ml, which was lowered to 2,554 nM in tissue culture medium. QN was then diluted across 10 concentrations ranging from 2,554 nM to 5 nM and predosed on 96-well microtiter plates as previously described (36). The assay was initiated by addition of 100 μl reconstituted parasite components to each well of the drug plates and incubation at 37°C as previously described (35). The assay was terminated after 72 h, 100 μl of lysis buffer containing SYBR green I (1×, final concentration) was added directly to the plates, and the mixtures were kept at room temperature in the dark for 24 h. Parasite replication inhibition was quantified by measuring the per well relative fluorescence units (RFU) of SYBR green 1 dye, using the Tecan Genios Plus system with excitation and emission wavelengths of 485 nm and 535 nm. The 50% inhibition concentration (IC50) values for QN were calculated as described previously (31).
DNA extraction.
Total genomic DNA of each P. falciparum isolate was extracted from filter paper blots (FTA; Whatman Inc., Bound Brook, NJ) by using a QIAamp DNA blood minikit (Qiagen, Valencia, CA), according to the manufacturer's instructions.
Genotypic characterization of Pfmdr1 and Pfcrt genes.
The 2−ΔΔCT method (based on the threshold cycle [CT]) of relative quantification was used to estimate copy number variation in Pfmdr1, as previously published (37). Polymorphisms in Pfmdr1 codons 86 and 184 and Pfcrt codon 76 were determined by nested PCR and sequencing as previously described (31, 38).
Pfnhe1 microsatellite genotyping.
Microsatellite repeats in Pfnhe1 were characterized by nested PCR using 1.5 μl of each primer. Primary run primers were NHE-F (5′-AGTCGAAGGCGAATCAGATG-3′) and NHE-R (5′-GATACTTACGAACATGTTCATG-3′). Secondary run primers NHE-F (5′-ATCCCTGTTGATATATCGAATG-3′) and NHE-R (5′-TTGTCATTAGTACCCTTAGTTG-3′), as previously described (24). The 25-μl reaction volume setup consisted of 10× PCR buffer to a final concentration of 1×, 25 mM MgCl2 to a final concentration of 1.5 mM, 20 mM deoxynucleoside triphosphates mix to a final concentration of 2 mM, 10 μM each of the primers to a final concentration of 100 nM, 5 U/μl AmpliTaq DNA polymerase to a final concentration of 1 U/reaction tube. Amplification of the ms4760 region was done under the following amplification conditions for the first run: initial hybridization at 94°C for 5 min, a subsequent 35 cycles of denaturation at 94°C for 30 s, annealing at 57.0°C for 30 s, and extension at 72°C for 30 s, and a final extension at 72°C for 7 min. Secondary run conditions were the following: initial hybridization at 94°C for 5 min, a subsequent 35 cycles of denaturation at 94°C for 30 s, annealing at 59.0°C for 30 s, and extension at 72°C for 30 s, and a final extension at 72°C for 7 min. The PCR products were resolved by electrophoresis on a 2% agarose gel, stained with 0.5 μg/ml ethidium bromide, and visualized under UV light. Amplicons were purified and sequenced using ABI Prism BigDye Terminator v3.1 cycle sequencing ready reaction kits (Applied Biosystems, Foster City, CA), as directed by the manufacturer on a 3500XL genetic analyzer. Sequences were analyzed with the BioEdit sequence alignment editor (version 7.0.9.0). Sequences were analyzed for the number of DNNND, DDNHNDNHNND, and DDNNNDNHNDD repeats in the Pfnhe1 ms4760 microsatellite.
Statistical analysis.
Continuous data in the form of IC50 estimates were expressed as medians with interquartile ranges. Further, IC50 data for isolates with various genetic polymorphism in Pfmdr1, Pfcrt, and Pfnhe1 were compared by using nonparametric tests (one-way analysis of variance, Kruskal-Wallis test with Dunn's multiple comparison posttest, and the Mann-Whitney test). Associations between IC50 estimates and genetic markers were considered significant when P was <0.05. Analyses were conducted using the Prism program (version 5.0.2; GraphPad Software, Inc., San Diego, CA). For analyses, all isolates with mixed genotypes were considered mutants.
Nucleotide sequence accession numbers.
New Pfnhe1 ms4760 microsatellite profiles were deposited in GenBank under the following accession numbers: KF719182, KF719183, KF719184, KF719185, and KF719186.
RESULTS
QN chemosensitivity.
The QN IC50 values for the two reference strains, 3D7 and W2, were established and used as internal controls in subsequent experiments. The median IC50 values (in nM; interquartile ranges and n values in parentheses) were 27 (17–32; n = 4) for 3D7 and 362 (111–400; n = 4) for W2. A total of 88 culture-adapted field isolates were successfully analyzed, and QN IC50s were determined. The median IC50 (interquartile range) for the 88 isolates was 69.01 nM (19.05–336.0).
Pfmdr1 and Pfcrt mutations.
Genotype analyses revealed 67% and 52% of the isolates carried the wild-type genotype at codons N86 and Y184 of the Pfmdr1 gene, respectively, whereas 35% of the isolates carried the wild-type genotype at codon K76 of the Pfcrt gene. Notably, all the isolates had a single copy of the Pfmdr1 gene.
Associations of QN in vitro sensitivity with Pfmdr1 and Pfcrt polymorphisms.
Figure 1 shows profiles for in vitro QN sensitivity per genetic polymorphism in the Pfmdr1 and Pfcrt genes. The isolates revealed a significant association between the QN IC50 and mutations in Pfmdr1 codons 86 and 184 and Pfcrt codon 76 (P < 0.0001 for each association). The median IC50 for parasites for Pfmdr1 codons 86 and 184 were ∼15-fold higher in the mutant genotypes compared to the wild-type, whereas with Pfcrt codon 76, the median IC50 was ∼9-fold higher for the mutant genotype than for the wild type.
FIG 1.
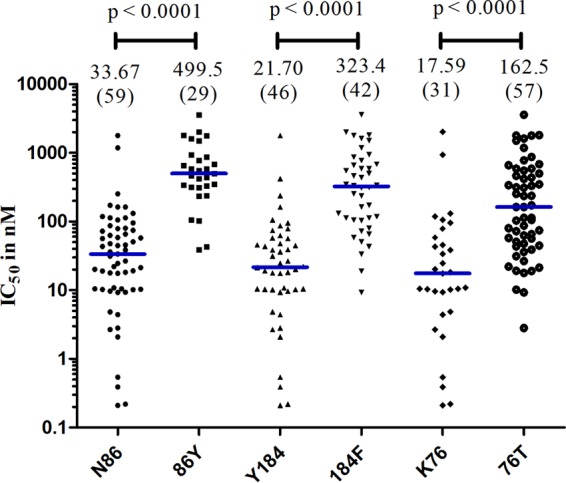
In vitro QN sensitivity for polymorphisms at each genetic marker in Pfmdr1-86, Pfmdr1-184, and Pfcrt-76 codons. The analysis comparing alleles at each codon was done using the Mann-Whitney test. The horizontal (blue) bars indicate medians. The numbers of isolates analyzed are shown in brackets. There was a significant association between the QN IC50 and mutation at each codon.
Pfnhe1 ms4760 polymorphism.
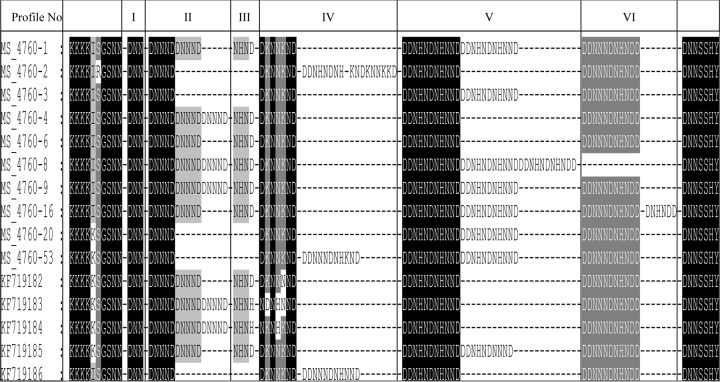
The genetic polymorphisms of ms4760 in Pfnhe1 gene was analyzed. Information on the QN IC50 values and the genetic profiles is summarized in Table S1 of the supplemental material. These isolates contained 15 different genetic polymorphisms of ms4760 in the Pfnhe1 gene. Five of the genetic profiles have not been previously described. The new profiles were deposited with GenBank and were assigned accession numbers KF719182, KF719183, KF719184, KF719185, and KF719186. Previously described profiles that were present in our isolates included the following: ms4760-1, ms4760-2, ms4760-3, ms4760-4, ms4760-6, ms4760-8, ms4760-9, ms4760-16, and ms4760-53. The most common genetic polymorphisms were the following: ms4760-1, ms4760-3, and ms4760-9, in 22, 16, and 12 of the isolates analyzed, respectively. Combined, they represented 60% of the total isolates analyzed. The least common genetic polymorphisms were ms4760-16, KF719184, and KF719186, each of which was present in only one isolate (Fig. 2). Figure 3 shows alignments of the 15 sequences of Pfnhe1 ms4760 identified in 88 western Kenya P. falciparum isolates.
FIG 2.
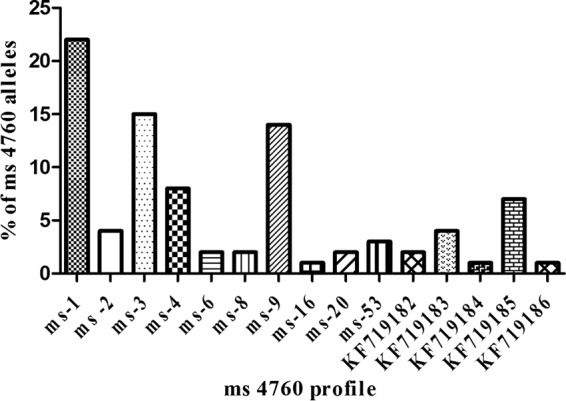
Prevalence rates of Pfnhe1 ms4760 profiles among the 88 western Kenya isolates. Profiles ms-1 to ms-53 have been described previously, while the sequences for KF719182 to KF719186 were first described in this study.
FIG 3.
Alignment of 15 sequences of Pfnhe1 ms4760 identified in 88 western Kenya P. falciparum isolates. Blocks I to VI have been described previously (29). The DNNND repeats are in block II, the DDNNNDNHNDD repeats are in block V, and the DDNNNDNHNDD repeats are in block VI. Profiles ms4760-1 to ms4760-53 have been described previously (25, 27, 32); sequences KF719182 to KF719186 are the described for the first time in this study. Other differences among sequences are highlighted.
Association between QN sensitivity and the number of DNNND repeats in Pfnhe1.
There was a significant association between the QN IC50 and the number of DNNND repeats (Fig. 4). The QN IC50 for parasites with 3 DNNND repeats was significantly higher than for those with 1 or 2 repeats (P = 0.033 and P = 0.0043, respectively). There was no statistical difference in QN IC50s between parasites with 1 or 2 repeats. Interestingly, there was no association between QN IC50 and DDNHNDNHNND repeats. The median IC50 (interquartile range; n) for parasites with 1 DDNHNDNHNND repeat was 80.72 nM (18.71–589.5; n = 28), for parasites with 2 repeats it was 57.79 nM (18.19–253.4; n = 59), and for those with 3 repeats it was 948.7 nM (118.4–1,779; n = 2).
FIG 4.
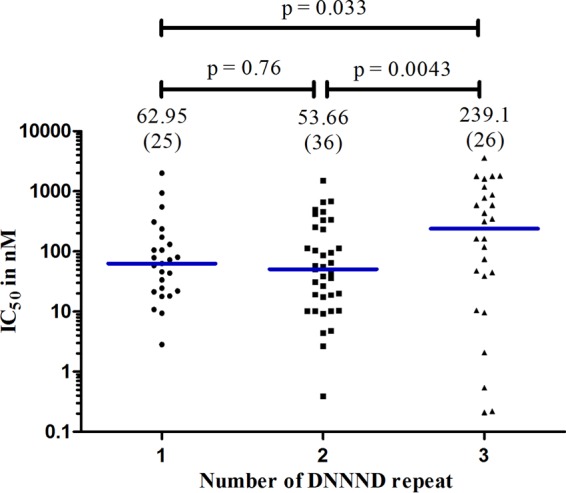
Association between the QN IC50 and the number of DNNND repeats. The analysis comparing polymorphisms in DNNND repeats was done using the Mann-Whitney test. The horizontal (blue) bars indicate the medians. The numbers of isolates analyzed are shown in brackets. P values comparing the repeats are indicated; 3 DNNND repeats had statistically higher QN IC50 values, compared to isolates with 1 or 2 repeats.
Associations of QN sensitivity and genotype combination for Pfmdr1, Pfcrt, and Pfnhe1.
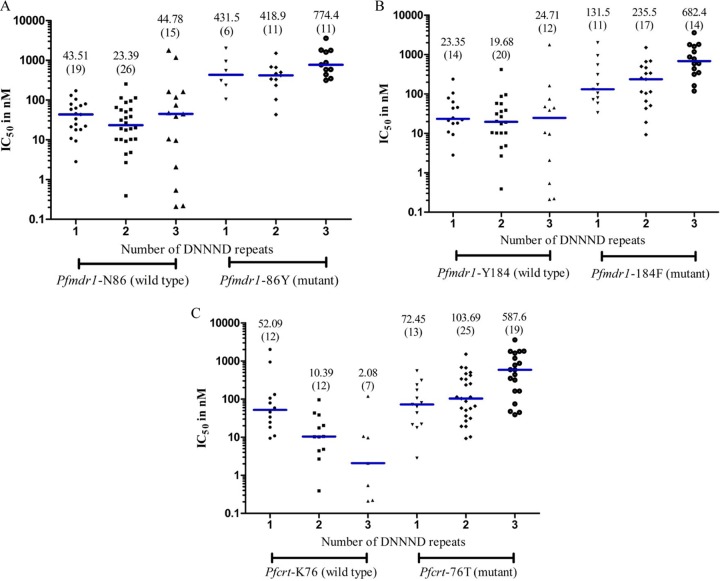
Our data showed a significant association of genetic polymorphisms in the Pfmdr1 gene at codons 86 and 184, the Pfcrt gene at codon 76, and the Pfnhe1 gene with DNNND repeats. To further understand this observation, we analyzed the relationship between the QN IC50 and the genotype combination of DNNND repeat number for the Pfmdr1 and Pfcrt genes. In Pfmdr1 codons 86Y (Fig. 5A) and 184F (Fig. 5B), data revealed parasites with 3 DNNND repeats had higher median IC50s than those with 1 or 2 repeats. For codon 86Y, there was a statistically significant difference between parasites with 3 DNNND repeats versus 2 repeats (P = 0.0302). For codon 184F, there was a statistically significant difference between parasites with 3 DNNND repeats versus 1 or 2 repeats (P = 0.0148 and P = 0.0031, respectively). For Pfcrt codon 76T (Fig. 5C), data revealed parasites with 3 DNNND repeats had median IC50s more than 5-fold higher than in parasites with 1 or 2 DNNND repeats, and there was a statistically significant difference between those with 3 DNNND repeats versus 1 or 2 repeats (P = 0.0019 and P = 0.0041, respectively).
FIG 5.
Relationship between the QN IC50 and the genotype combination of DNNND repeats for Pfmdr1-86 (A), Pfmdr1-184 (B), and Pfcrt-76 (C). The analysis comparing the number of DNNND repeats with each allele was done using the Mann-Whitney test. The horizontal bars (blue) indicate the medians. The numbers of isolates analyzed are shown in brackets. With one exception, 3 DNNND repeats had statistically significantly higher QN IC50 values than did isolates with 1 or 2 repeats for all mutant alleles analyzed. The association of Pfmdr1-86Y with 1 versus 3 DNNND repeats was not statistically significant (P = 0.21).
Genetic profiles of parasites with a QN IC50 outside the interquartile range.
To further investigate the role of genetic polymorphisms in the Pfmdr1, Pfcrt, and Pfnhe1 genes in determining parasite phenotypic characteristics in response to QN sensitivity, the genetic profiles of parasites with QN IC50s below the lower interquartile range or above the upper interquartile range were analyzed. There were 23 parasites with a QN IC50 of ≤19.05 and 22 parasites with a QN IC50 of ≥336. Table S2 in the supplemental material shows the genetic profiles of parasites with QN IC50s below and above the interquartile ranges. For the parasites with values below the lower interquartile range, the predominant genotype present in 74% of the parasites was Pfmdr1-N86 Pfmdr1-Y184 Pfcrt-K76 (all codons carrying the wild-type genotype), whereas above the upper interquartile range, the predominant genotype present in 82% of the parasites was Pfmdr1-86Y Pfmdr1-184F Pfcrt-76T (all codons carrying mutant genotypes). Two genotype profiles, Pfmdr1-N86 Pfmdr1-Y184 Pfcrt-76T and Pfmdr1-N86 Pfmdr1-184F Pfcrt-76T were present in parasites both in the lower and above the interquartile ranges. In the parasites with values below the lower interquartile range, 52% of the parasites had 2 DNNND repeats, whereas for parasites above the upper interquartile range, 55% had 3 DNNND repeats.
DISCUSSION
In this study, we have clearly shown a significant association between polymorphisms in Pfmdr1 codons 86 and 184 and Pfcrt codon 76 and QN susceptibility in P. falciparum parasites from western Kenya. Furthermore, the diversity of microsatellite repeats in Pfnhe1 ms4760 within these isolates was underscored and also shown to be linked to QN susceptibility. Three repeats of the DNNND polymorphism in the Pfnhe1gene significantly reduced parasite susceptibility to QN. Most importantly, we have described a predominant parasite genotype, Pfmdr1-86Y Pfmdr1-184F Pfcrt-76T, in parasites with high QN IC50s.
The median IC50 (interquartile range) for isolates from our study was 69.01 nM (19.05–336.0). The chemosensitivity threshold for QN has not been clearly defined. However, the historical WHO IC50 against the W2 clone of P. falciparum, which is considered resistant, is 315 nM (39). Other different QN threshold cutoffs have been proposed: 300, 500, and 800 nM (40–42). In studies conducted in Senegal (43) and Kenya (24), only 1% and 7% (respectively) of the isolates tested against QN had IC50s of >500 nM. In the study conducted in Kilifi, Kenya, none of the isolates had an IC50 of >800 nM (24). In field isolates from the Republic of the Congo, 25.7% exceeded the 500 nM cutoff, whereas only 5.4% exceeded the 800 nM cutoff (29). In our study, 18.2% (16/88) of the isolates exceeded the 500 nM cuttoff and 11.4% (10/88) exceeded the 800 nM cutoff. Although 70.4% (62/88) of the isolates had IC50 estimates below 300 nM, and were therefore considered sensitive to QN, a relatively high number of samples, compared to those collected elsewhere, can be considered resistant to QN, with a threshold cutoff exceeding 800 nM. The in vitro data from our study were supported by the genotypic data. This was expected, since in western Kenya, the prevalence of the Pfmdr1-86Y Pfmdr1-184F Pfcrt-76T genotype, which is also responsible for chloroquine resistance, has remained persistently high (31). Also, in vitro data for field isolates from Uganda have shown a tight correlation between sensitivities to chloroquine and QN (44).
In a study that analyzed parasite isolates from Kilifi, Kenya, parasites carrying the Pfmdr1-86 mutation showed a trend toward decreased QN activity, but there was no significant association (24). Similarly, in a study that used field isolates from Uganda, polymorphisms in Pfmdr1- 86 and Pfmdr1-184 were implicated in decreased sensitivity to QN but did not reach significance (P = 0.22 and P = 0.34, respectively) (44). However, the Pfmdr1-1246 mutation was shown to be statistically significant for modulating QN activity (P = 0.029). Transfection studies have also shown that Pfmdr1-1034, Pfmdr1-1042, and Pfmdr1-1246 mutations modulate resistance to QN (17). Here, we have shown statistically significant associations of mutations in Pfmdr1 codons 86 and 184 with QN activity. The difference in these observations could be due to the genetic backgrounds of the parasites. QN resistance appears to be dependent on multiple genes, which indicates polymorphisms in Pfmdr1 contribute to the overall phenotype of the parasite but in the backdrop of the genetic background of the parasite (17). It will be important to go back and analyze Pfmdr1 codon 1246 in isolates from our study, as this mutation has been shown to be important in modulating QN activity both in transfection and among in vitro data of field isolates (16, 44).
Transfection studies have shown that K76T contributes to QN resistance, but the extent of its contribution differs between strains (16, 45). Similarly, our data clearly showed a significant association of K76T mutation with QN reduced sensitivity. This is contrary to what other studies have shown (24). However, similarly to the Pfmdr1gene, the genetic background seems to be important and must therefore ultimately determine parasite phenotype.
Studies of the association of polymorphisms in the Pfnhe1 ms4670 microsatellite with in vitro susceptibility to QN have had conflicting results (24, 27, 29, 46). In studies that showed a positive correlation, the number of repeats in block 2 (DNNND), block 5 (DDNHNDNHNNDD), and block 6 (DDNNNDNHNDD) have been associated with modulation of QN resistance (27, 46). In this study, we showed there was a significant association for parasites with 3 repeats of DNNND and QN susceptibility. Polymorphisms in DDNHNDNHNNDD or DDNNNDNHNDD repeats did not have an effect on QN susceptibility. Some studies have shown that an increase in DNNND repeats is associated with reduced susceptibility to QN (16, 46). Interestingly, the study that analyzed isolates from Kilifi, Kenya, showed 2 DNNND repeats, and not 3 DNNND repeats, were associated with reduced QN susceptibility (24). Further, that study showed that 3 DNNND repeats restored QN activity, contrary to our findings. Other studies did not find any association for polymorphisms in DNNND repeats with QN activity when evaluating isolates from African patient populations (27, 29). Interestingly, however, in one of the studies, there was a positive association for NHNDNHNNDDD repeats with increased an QN IC50 (P = 0.01) among isolates from an African population (27). This is in line with findings from studies conducted using isolates from Vietnam and the China-Myanmar border, where an increased number of DDNHNDNHNNDD repeats was associated with high in vitro QN susceptibility (30, 43). The seemingly conflicting association of polymorphisms in the Pfnhe1 ms4670 microsatellite with in vitro susceptibility to QN can also be explained by differences in parasite genetic backgrounds, which can be associated with geographic origins of the isolates.
Lastly, we established the contributions of Pfmdr1-86, Pfmdr1-184, and Pfcrt-76 in the context of DNNND polymorphisms to QN susceptibility. For all three genotypes (Pfmdr1-86Y, Pfmdr1-184F, and Pfcrt-76T), the cumulative effect was highest in the background of 3 DNNND repeats, reaching statistical significance in each one of the genotypes. When we analyzed the genotypes of samples with results above the upper interquartile range, comprising isolates with IC50 estimates above the WHO cutoff for resistance, the majority of these isolates (82%) carried mutant alleles at the three genetic markers. When analyzed in combination with DNNND repeats, 55% of these isolates carried 3 repeats, as opposed to 32% with 2 repeats, and the remainder with 1 repeat. On the contrary, the majority of the isolates (74%) with values below the lower interquartile range had wild-type genotypes for all three codons analyzed, with most (52%) carrying 2 DNNND repeats.
Conclusions.
In this study, we showed the associations of Pfmdr1-86, Pfmdr1-184, Pfcrt-76, and Pfnhe1 ms4670 polymorphisms with parasite responses to QN. Further, we showed that these genetic markers are only relevant in the context of the genetic background of the isolates. Andriantsoanirina et al. (27) strongly suggested polymorphisms in Pfnhe1 might not be valid molecular markers for in vitro susceptibility to QN in P. falciparum isolates from Africa. Those authors tested only 83 isolates from a few countries in Africa. Africa is a continent with extremely diverse geographical landscapes, different malaria ecologies, varied rates of malaria endemicity, and parasites with different genetic structures. The cumulative evidence is clearly showing that the genetic background of the parasite is critical in determining QN activity. This will have important implications, because parasites from each geographic region must therefore be analyzed to determine which markers confer reduced QN susceptibility. To further validate our observations and conclusions, it will be important that isolates from different malaria ecological zones and regions of malaria endemicity in Kenya are analyzed. Most importantly, more clinical efficacy studies must be conducted in Kenya and other African countries, given the importance of QN in the management of malaria.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Director of KEMRI, USAMRU-K, and the entire Malaria Drug Research team for their support during the study period. We also thank John Okombo of KEMRI—Wellcome Trust Kilifi for sharing the Pfnhe1 primers and guidance during data analyses.
This work was supported by the U.S. Department of Defense, Global Emerging Infections Surveillance and Response System (GEIS), a division at the Armed Forces Health Surveillance Center (AFHSC).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The opinions and assertions contained herein are the private opinions of the authors and are not to be construed as reflecting the views of the U.S. Army Medical Research Unit—Kenya or the U.S. Department of Defense.
We declare that there are no competing interests.
Footnotes
Published ahead of print 21 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02472-14.
REFERENCES
- 1.World Health Organization. 2009. Global antimalarial drug policies database—WHO African region, September ed. World Health Organization, Geneva, Switzerland [Google Scholar]
- 2.World Health Organization. 2010. Global report on antimalarial efficacy and drug resistance: 2000–2010. World Health Organization, Geneva, Switzerland [Google Scholar]
- 3.Achan J, Talisuna AO, Erhart A, Yeka A, Tibenderana JK, Baliraine FN, Rosenthal PJ, D'Alessandro U. 2011. Quinine, an old anti-malarial drug in a modern world: role in the treatment of malaria. Malar. J. 10:144. 10.1186/1475-2875-10-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pukrittayakamee S, Supanaranond W, Looareesuwan S, Vanijanonta S, White NJ. 1994. Quinine in severe falciparum malaria: evidence of declining efficacy in Thailand. Trans. R. Soc. Trop. Med. Hyg. 88:324–327. 10.1016/0035-9203(94)90102-3 [DOI] [PubMed] [Google Scholar]
- 5.Chongsuphajaisiddhi T, Sabchareon A, Attanath P. 1983. Treatment of quinine resistant falciparum malaria in Thai children. Southeast Asian J. Trop. Med. Public Health 14:357–362 [PubMed] [Google Scholar]
- 6.Kofoed PE, Mapaba E, Lopes F, Pussick F, Aaby P, Rombo L. 1997. Comparison of 3, 5 and 7 days' treatment with Quinimax for falciparum malaria in Guinea-Bissau. Trans. R. Soc. Trop. Med. Hyg. 91:462–464. 10.1016/S0035-9203(97)90286-8 [DOI] [PubMed] [Google Scholar]
- 7.Rogier C, Brau R, Tall A, Cisse B, Trape JF. 1996. Reducing the oral quinine-quinidine-cinchonin (Quinimax) treatment of uncomplicated malaria to three days does not increase the recurrence of attacks among children living in a highly endemic area of Senega. Trans. R. Soc. Trop. Med. Hyg. 90:175–178. 10.1016/S0035-9203(96)90128-5 [DOI] [PubMed] [Google Scholar]
- 8.Kremsner PG, Winkler S, Brandts C, Neifer S, Bienzle U, Graninger W. 1994. Clindamycin in combination with chloroquine or quinine is an effective therapy for uncomplicated Plasmodium falciparum malaria in children from Gabon. J. Infect. Dis. 169:467–470. 10.1093/infdis/169.2.467 [DOI] [PubMed] [Google Scholar]
- 9.Esamai F, Tenge CN, Ayuo PO, Ong'or WO, Obala A, Jakait B. 2005. A randomized open label clinical trial to compare the efficacy and safety of intravenous quinine followed by oral malarone vs. intravenous quinine followed by oral quinine in the treatment of severe malaria. J. Trop. Pediatr. 51:17–24. 10.1093/tropej/fmh069 [DOI] [PubMed] [Google Scholar]
- 10.Mayxay M, Barends M, Brockman A, Jaidee A, Nair S, Sudimack D, Pongvongsa T, Phompida S, Phetsouvanh R, Anderson T, White NJ, Newton PN. 2007. In vitro antimalarial drug susceptibility and Pfcrt mutation among fresh Plasmodium falciparum isolates from the Lao PDR (Laos). Am. J. Trop. Med. Hyg. 76:245–250 [PMC free article] [PubMed] [Google Scholar]
- 11.Legrand E, Volney B, Meynard JB, Mercereau-Puijalon O, Esterre P. 2008. In vitro monitoring of Plasmodium falciparum drug resistance in French Guiana: a synopsis of continuous assessment from 1994 to 2005. Antimicrob. Agents Chemother. 52:288–298. 10.1128/AAC.00263-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toure AO, Kone LP, Jambou R, Konan TD, Demba S, Beugre GE, Kone M. 2008. In vitro susceptibility of P. falciparum isolates from Abidjan (Cote d'Ivoire) to quinine, artesunate and chloroquine. Sante 18:43–47. 10.1684/san.2008.0103 [DOI] [PubMed] [Google Scholar]
- 13.Pradines B, Mabika Mamfoumbi M, Parzy D, Owono Medang M, Lebeau C, Mourou Mbina JR, Doury JC, Kombila M. 1998. In vitro susceptibility of Gabonese wild isolates of Plasmodium falciparum to artemether, and comparison with chloroquine, quinine, halofantrine and amodiaquine. Parasitology 117:541–545. 10.1017/S0031182098003400 [DOI] [PubMed] [Google Scholar]
- 14.Shann F, Stace J, Edstein M. 1985. Pharmacokinetics of quinine in children. J. Pediatr. 106:506–510 [DOI] [PubMed] [Google Scholar]
- 15.Hawley SR, Bray PG, Mungthin M, Atkinson JD, O'Neill PM, Ward SA. 1998. Relationship between antimalarial drug activity, accumulation, and inhibition of heme polymerization in Plasmodium falciparum in vitro. Antimicrob. Agents Chemother. 42:682–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferdig MT, Cooper RA, Mu J, Deng B, Joy DA, Su XZ, Wellems TE. 2004. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol. Microbiol. 52:985–997. 10.1111/j.1365-2958.2004.04035.x [DOI] [PubMed] [Google Scholar]
- 17.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. 2000. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403:906–909. 10.1038/35002615 [DOI] [PubMed] [Google Scholar]
- 18.Sidhu AB, Valderramos SG, Fidock DA. 2005. Pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol. Microbiol. 57:913–926. 10.1111/j.1365-2958.2005.04729.x [DOI] [PubMed] [Google Scholar]
- 19.Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. 2006. Decreasing Pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J. Infect. Dis. 194:528–535. 10.1086/507115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valderramos SG, Valderramos JC, Musset L, Purcell LA, Mercereau-Puijalon O, Legrand E, Fidock DA. 2010. Identification of a mutant Pfcrt-mediated chloroquine tolerance phenotype in Plasmodium falciparum. PLoS Pathog. 6:e1000887. 10.1371/journal.ppat.1000887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parola P, Pradines B, Simon F, Carlotti MP, Minodier P, Ranjeva MP, Badiaga S, Bertaux L, Delmont J, Morillon M, Silai R, Brouqui P, Parzy D. 2007. Antimalarial drug susceptibility and point mutations associated with drug resistance in 248 Plasmodium falciparum isolates imported from Comoros to Marseille, France in 2004 2006. Am. J. Trop. Med. Hyg. 77:431–437 [PubMed] [Google Scholar]
- 22.Cooper RA, Ferdig MT, Su XZ, Ursos L, Mu J, Nomura T, Fujioka H, Fidock DA, Roepe PD, Wellems TE. 2002. Alternative mutation at position 76 of the vaculolar transmembrane protein Pfcrt are associated with chloquine resistance and unique stereospecific quinine and quinidine response in Plasmodium falciparum. Mol. Pharmacol. 61:35–42. 10.1124/mol.61.1.35 [DOI] [PubMed] [Google Scholar]
- 23.Cooper RA, Lane KD, Deng B, Mu J, Patel JJ, Wellems TE, Su X, Ferdig MT. 2007. Mutations in transmembrane domains 1, 4 and 9 of the Plasmodium falciparum chloroquine resistance transporter alter susceptibility to chloroquine, quinine and quinidine. Mol. Microbiol. 63:270–282. 10.1111/j.1365-2958.2006.05511.x [DOI] [PubMed] [Google Scholar]
- 24.Okombo J, Kiara SM, Rono J, Mwai L, Pole L, Ohuma E, Borrmann S, Ochola LI, Nzila A. 2010. In vitro activities of quinine and other antimalarials and pfnhe polymorphisms in Plasmodium isolates from Kenya. Antimicrob. Agents Chemother. 54:3302–3307. 10.1128/AAC.00325-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baliraine FN, Nsobya SL, Achan J, Tibenderana JK, Talisuna AO, Greenhouse B, Rosenthal PJ. 2011. Limited ability of Plasmodium falciparum Pfcrt, Pfmdr1, and Pfnhe1 polymorphisms to predict quinine in vitro sensitivity or clinical effectiveness in Uganda. Antimicrob. Agents Chemother. 55:615–622. 10.1128/AAC.00954-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pelleau S, Bertaux L, Briolant S, Ferdig MT, Sinou V, Pradines B, Parzy D, Jambou R. 2011. Differential association of Plasmodium falciparum Na+/H+ exchanger polymorphism and quinine responses in field- and culture-adapted isolates of Plasmodium falciparum. Antimicrob. Agents Chemother. 55:5834–5841. 10.1128/AAC.00477-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andriantsoanirina V, Menard D, Rabearimanana S, Hubert V, Bouchier C, Tichit M, Bras JL, Durand R. 2010. Association of microsatellite variations of Plasmodium falciparum Na+/H+ exchanger (Pfnhe-1) gene with reduced in vitro susceptibility to quinine: lack of confirmation in clinical isolates from Africa. Am. J. Trop. Med. Hyg. 82:782–787. 10.4269/ajtmh.2010.09-0327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nkrumah LJ, Riegelhaupt PM, Moura P, Johnson DJ, Patel J, Hayton K, Ferdig MT, Wellems TE, Akabas MH, Fidock DA. 2009. Probing the multifactorial basis of Plasmodium falciparum quinine resistance: evidence for a strain-specific contribution of the sodium-proton exchanger PfNHE. Mol. Biochem. Parasitol. 165:122–131. 10.1016/j.molbiopara.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Briolant S, Pelleau S, Bogreau H, Hovette P, Zettor A, Castello J, Baret E, Amalvict R, Rogier C, Pradines B. 2011. In vitro susceptibility to quinine and microsatellite variations of the Plasmodium falciparum Na+/H+ exchanger (Pfnhe-1) gene: the absence of association in clinical isolates from the Republic of Congo. Malar. J. 10:37. 10.1186/1475-2875-10-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinou V, Quang le H, Pelleau S, Huong VN, Huong NT, Tai le M, Bertaux L, Desbordes M, Latour C, Long LQ, Thanh NX, Parzy D. 2011. Polymorphism of Plasmodium falciparum Na+/H+ exchanger is indicative of a low in vitro quinine susceptibility in isolates from Viet Nam. Malar. J. 10:164. 10.1186/1475-2875-10-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eyase FL, Akala HM, Ingasia L, Cheruiyot A, Omondi A, Okudo C, Juma D, Yeda R, Andagalu B, Wanja E, Kamau E, Schnabel D, Bulimo W, Waters NC, Walsh DS, Johnson JD. 2013. The role of Pfmdr1 and Pfcrt in changing chloroquine, amodiaquine, mefloquine and lumefantrine susceptibility in western-Kenya P. falciparum samples during 2008–2011. PLoS One 8:e64299. 10.1371/journal.pone.0064299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trager W, Jensen JB. 1976. Human malaria parasites in continuous culture. Science 193:673–675. 10.1126/science.781840 [DOI] [PubMed] [Google Scholar]
- 33.Rason MA, Randriantsoa T, Andrianantenaina H, Ratsimbasoa A, Menard D. 2008. Performance and reliability of the SYBR Green I based assay for the routine monitoring of susceptibility of Plasmodium falciparum clinical isolates. Trans. R. Soc. Trop. Med. Hyg. 102:346–351. 10.1016/j.trstmh.2008.01.021 [DOI] [PubMed] [Google Scholar]
- 34.Bacon DJ, Latour C, Lucas C, Colina O, Ringwald P, Picot S. 2007. Comparison of a SYBR green I-based assay with a histidine-rich protein II enzyme-linked immunosorbent assay for in vitro antimalarial drug efficacy testing and application to clinical isolates. Antimicrob. Agents Chemother. 51:1172–1178. 10.1128/AAC.01313-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson JD, Dennull RA, Gerena L, Lopez-Sanchez M, Roncal NE, Waters NC. 2007. Assessment and continued validation of the malaria SYBR green I-based fluorescence assay for use in malaria drug screening. Antimicrob. Agents Chemother. 51:1926–1933. 10.1128/AAC.01607-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tyner SD, Lon C, Se Y, Bethell D, Socheat D, Noedl H, Sea D, Satimai W, Schaecher K, Rutvisuttinunt W, Fukuda MM, Chaorattanakawee S, Yingyuen K, Sundrakes S, Chaichana P, Saingam P, Buathong N, Sriwichai S, Chann S, Timmermans A, Saunders DL, Walsh DS. 2012. Ex vivo drug sensitivity profiles of Plasmodium falciparum field isolates from Cambodia and Thailand, 2005 to 2010, determined by a histidine-rich protein-2 assay. Malar. J. 11:198. 10.1186/1475-2875-11-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S. 2004. Mefloquine resistance in Plasmodium falciparum and increased Pfmdr1 gene copy number. Lancet 364:438–447. 10.1016/S0140-6736(04)16767-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vinayak S, Alam MT, Sem R, Shah NK, Susanti AI, Lim P, Muth S, Maguire JD, Rogers WO, Fandeur T, Barnwell JW, Escalante AA, Wongsrichanalai C, Ariey F, Meshnick SR, Udhayakumar V. 2010. Multiple genetic backgrounds of the amplified Plasmodium falciparum multidrug resistance (Pfmdr1) gene and selective sweep of 184F mutation in Cambodia. J. Infect. Dis. 201:1551–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.World Health Organization:. 2001. Mark III in vitro micro-test for the assessment of the response of Plasmodium falciparum to chloroquine, mefloquine, quinine, amodiaquine, sulfadoxine/pyrimethamine, and artemisinin. CTD/MAL/9720, revision 2. World Health Organization, Geneva, Switzerland [Google Scholar]
- 40.Ralaimazava P, Durand R, Godineau N, Jezic Z, Pradines B, Bouchaud O, Bras JL. 2002. Profile and evolution of the chemosusceptibility of falciparum malaria imported into France in 2000. Euro Surveill. 7(7):pii=355 http://eurosurveillance.org/ViewArticle.aspx?ArticleId=355 [DOI] [PubMed] [Google Scholar]
- 41.Brasseur P, Kouamouo J, Moyou-Somo R, Druilhe P. 1992. Multi-drug resistant falciparum malaria in Cameroon in 1987–1988. I. Stable figures of prevalence of chloroquine- and quinine-resistant isolates in the original foci. Am. J. Trop. Med. Hyg. 46:1–7 [DOI] [PubMed] [Google Scholar]
- 42.Pradines B, Rogier C, Fusai T, Tall A, Trape JF, Doury JC. 1996. In vitro sensitivity of 85 Plasmodium falciparum isolates in the Fatick region, Senegal. Med. Trop. (Mars.) 56:141–145 (In French.) [PubMed] [Google Scholar]
- 43.Meng H, Zhang R, Yang H, Fan Q, Su X, Miao J, Cui L, Yang Z. 2010. In vitro sensitivity of Plasmodium falciparum clinical isolates from the China-Myanmar border area to quinine and association with polymorphism in the Na+/H+ exchanger. Antimicrob. Agents Chemother. 54:4306–4313. 10.1128/AAC.00321-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nsobya SL, Kiggundu M, Nanyunja S, Joloba M, Greenhouse B, Rosenthal PJ. 2010. In vitro sensitivities of Plasmodium falciparum to different antimalarial drugs in Uganda. Antimicrob. Agents Chemother. 54:1200–1206. 10.1128/AAC.01412-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sidhu AB, Verdier-Pinard D, Fidock DA. 2002. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by Pfcrt mutations. Science 298:210–213. 10.1126/science.1074045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henry M, Briolant S, Zettor A, Pelleau S, Baragatti M, Baret E, Mosnier J, Amalvict R, Fusai T, Rogier C, Pradines B. 2009. Plasmodium falciparum Na+/H+ exchanger 1 transporter is involved in reduced susceptibility to quinine. Antimicrob. Agents Chemother. 53:1926–1930. 10.1128/AAC.01243-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.