Abstract
NDM-1 probably emerged in Acinetobacter species prior to its dissemination among Enterobacteriaceae, and NDM-1-like enzymes are increasingly reported in Acinetobacter species. Here, we report on the genetic context of blaNDM-1 in the earliest known NDM-1-producing organisms, clinical isolates of Acinetobacter from India in 2005. These strains harbor blaNDM-1 plasmids of different sizes. The gene is associated with the remnants of the Tn125 transposon normally associated with blaNDM-1 in Acinetobacter spp. The transposon has been disrupted by the IS26 insertion and subsequent movement events.
TEXT
Multidrug-resistant Enterobacteriaceae producing the New Delhi metallo-β-lactamase-1 (NDM-1) enzyme have been isolated around the world, with many cases linked to travel, especially to South Asia (1–3). Carbapenem resistance in the opportunistic pathogen, Acinetobacter baumannii, is mostly associated with OXA-type β-lactamases (4), but NDM-1-like enzymes have been increasingly reported (5–10). In all bacterial species, the blaNDM-1 gene is associated with at least one copy of a complete or partial ISAba125 gene, which provides blaNDM-1 with a strong promoter (11–13). ISAba125 almost certainly originates from Acinetobacter spp., and there is evidence that blaNDM-1 was probably formed by a fusion event between the aminoglycoside resistance gene, aphA6, and an ancestral carbapenemase in an Acinetobacter background (13), thus implying that blaNDM-1 spread to Enterobacteriaceae from Acinetobacter spp.
A small number of studies have reported a relatively high prevalence of NDM-1-producing A. baumannii, causing infections in intensive care patients in Indian hospitals (10, 14). There are also several case reports of colonization and infection with these organisms from European countries, many with epidemiological links with travel to North Africa or the Balkans (5, 6, 11), and from the Middle East (9). In China, blaNDM-1 has been frequently identified in Acinetobacter spp., including A. baumannii, both in clinical cases and from environmental sources (7, 15–17). The genetic context of blaNDM-1-like genes in Acinetobacter spp. shows less variation than that observed in other genera. In most strains, blaNDM-1 is bracketed by two copies of ISAba125 to form a Tn125 transposon. The content of Tn125 is usually conserved, with the occasional exceptions resulting from truncation by insertion of other IS elements. Tn125 is found inserted in several different gene locations, with direct repeats at either end indicating movement by transposition. All isolates from Europe and North Africa are chromosomally located, but in China, blaNDM-1 is reportedly found mostly on plasmids in Acinetobacter spp. other than A. baumannii (15–18).
We received 9 Acinetobacter isolates that were collected in 2005 from a hospital in Tamil Nadu, India. Most isolates were from patients receiving intensive care and were isolated from blood, pus, and respiratory secretions. The initial identification and sensitivity testing were performed using a BD Phoenix system (Becton, Dickinson, Franklin Lakes, NJ, USA), with supplementary sensitivity testing performed using the Etest (bioMérieux, LaPlane, France) or MIC test strip (Liofilchem, Roseto degli Abruzzi, Italy) method (Table 1). The confirmation of A. baumannii identification was performed by PCRs for blaOXA-51-like genes. All isolates were blaOXA-51-like positive by PCRs except CHI-40-1, and all were extensively drug resistant (19), with the A. baumannii isolates being resistant to all drugs tested except colistin and in some cases amikacin and tobramycin. Seven isolates were positive for blaNDM-1 by PCRs, with CHI-41 and CHI-44 being the only blaNDM-1-negative isolates. All of the A. baumannii isolates carried blaOXA-23-like genes, confirmed to be associated with ISAba1, and thus having a strong upstream promoter, by PCR. All PCR amplicons were confirmed by sequencing.
TABLE 1.
Study isolates, specimen types, and antimicrobial MICs for Acinetobacter isolates
| Isolate | Specimen type | MIC (μg/ml) fora: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Azt | Caz | Taz | Imp | Mem | Ami | Gent | Tob | Cip | Col | Rif | Tig | ||
| A. baumannii CHI-16 | Blood | >16 | ≥256 | >16/4 | ≥32 | ≥32 | ≤4 | >4 | 2 | ≥32 | ≤1 | 6 | 2 |
| A. baumannii CHI-18 | Blood | >16 | ≥256 | >16/4 | ≥32 | ≥32 | >16 | >4 | >4 | ≥32 | ≤1 | 4 | 3 |
| A. baumannii CHI-32 | Blood | >16 | ≥256 | >16/4 | ≥32 | ≥32 | ≤4 | >4 | 2 | ≥32 | ≤1 | 6 | 2 |
| A. baumannii CHI-34 | Sputum | >16 | ≥256 | >16/4 | ≥32 | ≥32 | ≤4 | >4 | >4 | ≥32 | ≤1 | 4 | 2 |
| Acinetobacter sp CHI-40-1 | Pus | >16 | ≥256 | >16/4 | ≥32 | ≥32 | >16 | >4 | 4 | ≥32 | ≤1 | ≥256 | 0.75 |
| A. baumannii CHI-40-2 | Pus | >16 | ≥256 | >16/4 | ≥32 | ≥32 | >16 | >4 | >4 | ≥32 | ≤1 | 6 | 2 |
| A. baumannii CHI-41 | Sputum | >16 | ≥256 | >16/4 | ≥32 | ≥32 | >16 | >4 | >4 | ≥32 | ≤1 | 6 | 1.5 |
| A. baumannii CHI-44 | Endotracheal aspirate | >16 | ≥256 | >16/4 | ≥32 | ≥32 | >16 | >4 | >4 | ≥32 | ≤1 | 6 | 2 |
| A. baumannii CHI-45-1 | Endotracheal aspirate | >16 | ≥256 | >16/4 | ≥32 | ≥32 | >16 | >4 | >4 | ≥32 | ≤1 | 3 | 2 |
MICs were determined by the BD Phoenix system except for ceftazidime, imipenem, meropenem, ciprofloxacin, rifampin, and tigecycline, which were determined by an Etest or a MIC test strip. Ami, amikacin; Azt, aztreonam; Caz, ceftazidime; Cip, ciprofloxacin; Col, colistin; Gent, gentamicin; Imp, imipenem; Mem, meropenem; Rif, rifampin; Taz, piperacillin-tazobactam; Tig, tigecycline; Tob, tobramycin.
A. baumannii isolates were then typed by the multilocus sequence typing (MLST) method described by Turton et al. (20) and by pulsed-field gel electrophoresis (PFGE) of ApaI-digested genomic DNA. All A. baumannii isolates producing NDM-1 by MLST were within group II, which corresponds with worldwide clone 1. The 2 blaNDM-1-negative isolates were within group I. ApaI profiles were similar but not identical for all group II A. baumannii isolates and differed substantially from those for group I isolates (Fig. 1a) and CHI-40-1.
FIG 1.
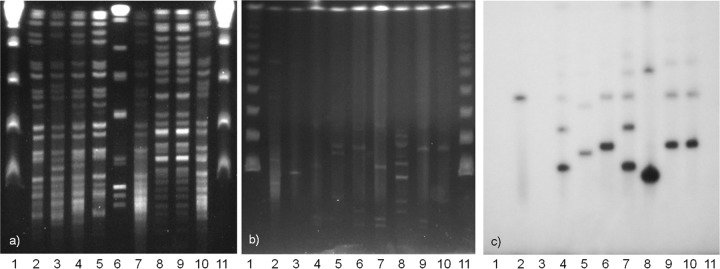
(a) PFGE of ApaI-digested genomic DNA. Lane 1, λ ladder (48.5-kb concatemers); lane 2, CHI-16; lane 3, CHI-18; lane 4, CHI-32; lane 5, CHI-34; lane 6, CHI-40-1; lane 7, CHI-40-2; lane 8, CHI-41; lane 9, CHI-44; lane 10, CHI-45-1; lane 11, λ ladder. (b) PFGE of S1 endonuclease-digested genomic DNA; (c) in-gel hybridization with 32P-labeled blaNDM-1 gene probe. Lanes 1, λ ladder; lanes 2, Klebsiella pneumoniae KP506 (NDM positive control); lanes 3, Escherichia coli UAB190 (NDM negative control); lanes 4, CHI-16; lanes 5, CHI-18; lanes 6, CHI-32; lanes 7, CHI-34; lanes 8, CHI-40-1; lanes 9, CHI-40-2; lanes 10, CHI-45-1; lanes 11, λ.
To investigate the genetic context of blaNDM-1, we performed sequencing by primer walking in CHI-32, CHI-34, and CHI-45-1 using primers designed against the context in A. baumannii 161/07 (21). Gene probing was performed by in-gel hybridization of S1 nuclease and NotI PFGE gels. The gene probes were made using a random priming method with blaNDM-1, ISAba125, IS26, and ISCR27 PCR products labeled with [32P]CTP. As the primer walking PCR results were consistent for all three isolates studied, products were only fully sequenced for CHI-45-1. The full Tn125 structure was present; however, ISCR27 contains an IS26 insertion (Fig. 2). The sequence analysis of PCR amplicons revealed that the fragment of ISCR27 downstream of the IS26 insertion is present both in its normal position and upstream of blaNDM-1. The results of probing NotI gels with blaNDM-1, IS26, and ISCR27 suggest that it is the latter context which is more common. S1 gels demonstrated multiple plasmids in all A. baumannii isolates. The probing showed that blaNDM-1 was on multiple bands, ranging in size from ∼45 kb to ∼300 kb. We believe most of the larger bands represent cointegrate formation rather than the presence of multiple plasmids carrying blaNDM-1, since the bands increase in size by intervals of approximately the size of the smallest band for each isolate (Fig. 1b). Mating experiments were performed on plates and in broth. A. baumannii CHI-32, CHI-34, and CHI-45-1 were used as donors, and Escherichia coli UAB190 and Acinetobacter pittii AG3528 were used as recipients (both rifampin resistant). No transconjugants were obtained after multiple mating experiments.
FIG 2.
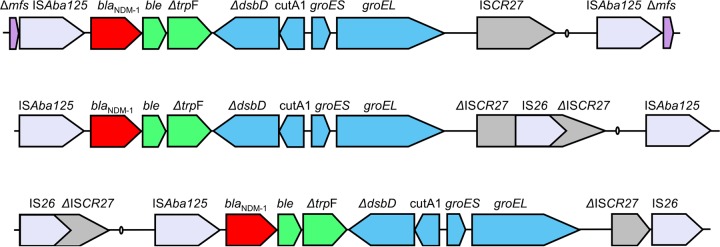
Gene maps of the genetic context of blaNDM-1 in Acinetobacter species. aphA6 codes for 3′ phosphotransferase VI aminoglycoside-modifying enzyme, ble for bleomycin resistance protein, trpF for phosphoribosylanthranilate isomerase, tat for twin-arginine translocation pathway signal sequence domain protein, cutA1 for periplasmic divalent cation tolerance protein, groES for cochaperonin, groEL for chaperonin, ISCR27 for insertion sequence common repeat 27 transposase, oriIS for origin of insertion of ISCR27, and Δmfs for major facilitator superfamily (MFS) metabolite/H+ symporter. Arrows indicate the direction of the transcription of genes. The genes tat, cutA1, groES, and groEL are shaded in the same color because they are believed to be from a common source, with similar genes found in synteny in both Xanthomonas and Pseudoxanthomonas spp.
The A. baumannii isolates analyzed in this study are among the earliest found to produce NDM-1, having been initially identified in 2005. This is the first time that the genetic context of A. baumannii isolates from the Indian subcontinent has been analyzed. The findings are compatible with the hypothesis that blaNDM-1 might have been disseminated from Acinetobacter to Enterobacteriaceae in South Asia. In these isolates, Tn125 has been disrupted by IS26, and subsequent rearrangement has resulted in blaNDM-1 being within an IS26 composite transposon, which might potentially mobilize blaNDM-1. Otherwise, the genes usually found on Tn125 are conserved, and so the genetic context is compatible with being the progenitor of the blaNDM-1 in many of the Enterobacteriaceae for which sequences are available. blaNDM-1 is located on plasmids in these isolates, which could facilitate mobilization of the gene to other bacterial species. The mating experiments suggest that the plasmids are nonconjugative, but they may be mobilizable with a helper plasmid. That these clinical isolates of A. baumannii producing NDM-1 were clonally related demonstrates the potential for blaNDM-1 establishing itself in successful strain backgrounds capable of being disseminated in the hospital environment and further compromising therapeutic options in the treatment of significant bacterial pathogens.
ACKNOWLEDGMENTS
The work was supported by British Society of Antimicrobial Chemotherapy grant GA2011-07P (to L.S.J.), Welsh Clinical Academic Track (WCAT) funding for project consumables grant AJ11212001, provided by Cardiff University from funds available from the Welsh Assembly Government (to L.S.J.), and Canadian Institutes of Health Research and Medical Research Council, Canada-UK Partnership on Antibiotic Resistance grant G1100135 (to M.A.T.).
We declare no conflict of interest.
Footnotes
Published ahead of print 21 April 2014
REFERENCES
- 1.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602. 10.1016/S1473-3099(10)70143-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nordmann P, Poirel L, Toleman MA, Walsh TR. 2011. Does broad-spectrum beta-lactam resistance due to NDM-1 herald the end of the antibiotic era for treatment of infections caused by Gram-negative bacteria? J. Antimicrob. Chemother. 66:689–692. 10.1093/jac/dkq520 [DOI] [PubMed] [Google Scholar]
- 4.Higgins PG, Dammhayn C, Hackel M, Seifert H. 2010. Global spread of carbapenem-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65:233–238. 10.1093/jac/dkp428 [DOI] [PubMed] [Google Scholar]
- 5.Bonnin RA, Cuzon G, Poirel L, Nordmann P. 2013. Multidrug-resistant Acinetobacter baumannii clone, France. Emerg. Infect. Dis. 19:822–823. 10.3201/eid1905.121618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnin RA, Poirel L, Naas T, Pirs M, Seme K, Schrenzel J, Nordmann P. 2012. Dissemination of New Delhi metallo-beta-lactamase-1-producing Acinetobacter baumannii in Europe. Clin. Microbiol. Infect. 18:E362−E365. 10.1111/j.1469-0691.2012.03928.x [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Zhou Z, Jiang Y, Yu Y. 2011. Emergence of NDM-1-producing Acinetobacter baumannii in China. J. Antimicrob. Chemother. 66:1255–1259. 10.1093/jac/dkr082 [DOI] [PubMed] [Google Scholar]
- 8.Espinal P, Fugazza G, Lopez Y, Kasma M, Lerman Y, Malhotra-Kumar S, Goossens H, Carmeli Y, Vila J. 2011. Dissemination of an NDM-2-producing Acinetobacter baumannii clone in an Israeli rehabilitation center. Antimicrob. Agents Chemother. 55:5396–5398. 10.1128/AAC.00679-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espinal P, Poirel L, Carmeli Y, Kaase M, Pal T, Nordmann P, Vila J. 2013. Spread of NDM-2-producing Acinetobacter baumannii in the Middle East. J. Antimicrob. Chemother. 68:1928−1930. 10.1093/jac/dkt109 [DOI] [PubMed] [Google Scholar]
- 10.Karthikeyan K, Thirunarayan MA, Krishnan P. 2010. Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J. Antimicrob. Chemother. 65:2253–2254. 10.1093/jac/dkq273 [DOI] [PubMed] [Google Scholar]
- 11.Poirel L, Bonnin RA, Boulanger A, Schrenzel J, Kaase M, Nordmann P. 2012. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56:1087–1089. 10.1128/AAC.05620-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poirel L, Dortet L, Bernabeu S, Nordmann P. 2011. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob. Agents Chemother. 55:5403–5407. 10.1128/AAC.00585-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toleman MA, Spencer J, Jones L, Walsh TR. 2012. blaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56:2773–2776. 10.1128/AAC.06297-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bharadwaj R, Joshi S, Dohe V, Gaikwad V, Kulkarni G, Shouche Y. 2012. Prevalence of New Delhi metallo-beta-lactamase (NDM-1)-positive bacteria in a tertiary care centre in Pune, India. Int. J. Antimicrob. Agents 39:265–266. 10.1016/j.ijantimicag.2011.09.027 [DOI] [PubMed] [Google Scholar]
- 15.Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang X, Hao Q, Yang X, Xiao X, Luan C, Yang Y, Cui Y, Yang R, Gao GF, Song Y, Zhu B. 2012. Novel plasmid and its variant harboring both a bla(NDM-1) gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob. Agents Chemother. 56:1698–1702. 10.1128/AAC.06199-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Wu C, Zhang Q, Qi J, Liu H, He T, Ma L, Lai J, Shen Z, Liu Y, Shen J. 2012. Identification of New Delhi metallo-beta-lactamase 1 in Acinetobacter lwoffii of food animal origin. PLoS One 7:e37152. 10.1371/journal.pone.0037152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang WJ, Lu Z, Schwarz S, Zhang RM, Wang XM, Si W, Yu S, Chen L, Liu S. 2013. Complete sequence of the blaNDM-1-carrying plasmid pNDM-AB from Acinetobacter baumannii of food animal origin. J. Antimicrob. Chemother. 68:1681−1682. 10.1093/jac/dkt066 [DOI] [PubMed] [Google Scholar]
- 18.Zhou Z, Guan R, Yang Y, Chen L, Fu J, Deng Q, Xie Y, Huang Y, Wang J, Wang D, Liao C, Gong S, Xia H. 2012. Identification of New Delhi metallo-beta-lactamase gene (NDM-1) from a clinical isolate of Acinetobacter junii in China. Can. J. Microbiol. 58:112–115. 10.1139/w11-112 [DOI] [PubMed] [Google Scholar]
- 19.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18:268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 20.Turton JF, Gabriel SN, Valderrey C, Kaufmann ME, Pitt TL. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:807–815. 10.1111/j.1469-0691.2007.01759.x [DOI] [PubMed] [Google Scholar]
- 21.Pfeifer Y, Wilharm G, Zander E, Wichelhaus TA, Gottig S, Hunfeld KP, Seifert H, Witte W, Higgins PG. 2011. Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J. Antimicrob. Chemother. 66:1998–2001. 10.1093/jac/dkr256 [DOI] [PubMed] [Google Scholar]


