Abstract
Ribosomal proteins are highly conserved components of basal cellular organelles, primarily involved in the translation of mRNA leading to protein synthesis. However, certain ribosomal proteins moonlight in the development and differentiation of organisms. In this study, the ribosomal protein L44 (RPL44), associated with salt resistance, was screened from the halophilic fungus Aspergillus glaucus (AgRPL44), and its activity was investigated in Saccharomyces cerevisiae and Nicotiana tabacum. Sequence alignment revealed that AgRPL44 is one of the proteins of the large ribosomal subunit 60S. Expression of AgRPL44 was upregulated via treatment with salt, sorbitol, or heavy metals to demonstrate its response to osmotic stress. A homologous sequence from the model fungus Magnaporthe oryzae, MoRPL44, was cloned and compared with AgRPL44 in a yeast expression system. The results indicated that yeast cells with overexpressed AgRPL44 were more resistant to salt, drought, and heavy metals than were yeast cells expressing MoRPL44 at a similar level of stress. When AgRPL44 was introduced into M. oryzae, the transformants displayed obviously enhanced tolerance to salt and drought, indicating the potential value of AgRPL44 for genetic applications. To verify the value of its application in plants, tobacco was transformed with AgRPL44, and the results were similar. Taken together, we conclude that AgRPL44 supports abiotic stress resistance and may have value for genetic application.
INTRODUCTION
Halotolerant and halophilic microorganisms are widely distributed in saline environments such as solar salterns, seawater, and concentrated brines (1–4). These microorganisms can adapt to extreme concentrations of sodium chloride (NaCl) and often to high concentrations of other ions, high UV irradiation, and in some cases, extremes of pH (3, 5). These characteristics make halotolerant and halophilic microorganisms valuable resources for genes that confer stress tolerance and the investigation of stress resistance mechanisms. Halophilic and halotolerant microbes are also of interest for their potential industrial applications, including as a source of transgenes for improving osmotolerance of industrially important yeasts. The gene source from these halophilic and halotolerant organisms might also provide aids to enhance the halotolerance of some plant species and help overcome the global problem of salinization of agricultural areas (5–7).
Previously, we isolated the halophilic fungus Aspergillus glaucus from air-dried wild vegetation at the surface periphery of a solar salt field. The salinity range of growth for this isolate, defined in vitro, was from 0 to 32% (saturation) NaCl, with a broad optimum from 5 to 15% NaCl (8). To elucidate the saline stress response mechanisms, a cDNA yeast library was constructed from mycelia exposed to NaCl for 24 h. By screening the cDNA library on potato dextrose agar supplemented with different concentrations of NaCl, we isolated some genes that have strong saline resistance. Among the identified sequences, a cDNA fragment representing an A. glaucus ribosomal protein (RP) L44 gene (designated AgRPL44) was found.
Ribosomal proteins make up the protein portion of the ribosome and with rRNA are essential for protein synthesis (9, 10). However, recent studies have shown that some ribosomal proteins may also participate in cellular events apart from protein biosynthesis, and this has inspired research interest in the genes that encode them. Such extraribosomal functions include DNA reparation, RNA chaperone activity, cell development, regulation of differentiation, pathogen resistance mechanisms, and tumor repression (10–14). RPS3e is involved in DNA repair and selective gene regulation via the NF-κB signaling pathway (15). Plastid-specific ribosomal protein 2 (PSRP2), with RNA chaperone activity, participates as a negative regulator in seed germination and seedling growth under abiotic stress conditions and thus has a role in the abiotic stress response (16). Overexpressed StoL13a in potato (Solanum tuberosum) was shown to enhance disease resistance against Verticillium dahliae infection in transgenic potato plants (17). An antimicrobial peptide purified from skin secretions and epithelial cells of rainbow trout (likely the 40S ribosomal protein S30) has a protective function against intracellular or extracellular pathogens (18). Research on ribosomal proteins RPL24, RPL31, and RPS23 of the giant panda (Ailuropoda melanoleuca) shows that they have anticancer activity (19–21). Overexpression of human ribosomal protein RPL36A may be associated with hepatocarcinogenesis and functionally relates to tumor cell proliferation; RPL36A may thus be a potential target gene for anticancer therapy in hepatocellular carcinoma (22). In summary, it is clear that the functions of ribosomal proteins extend beyond the ribosome, and such moonlighting may alert the cell to stresses or defects in ribosome protein synthesis itself. Indeed, there is increasing evidence that among ribosomal proteins extraribosomal moonlighting is widespread.
Ribosomal protein genes are regulated in response to environmental stresses (16, 23–25). However, the mechanism of their resistance to abiotic stresses has not been described. In the present study, to gain further insight into the function of the AgRPL44 gene, its expression patterns were measured in response to NaCl, sorbitol, and CuSO4 stresses. We used a yeast expression system to generate a transgenic Saccharomyces cerevisiae strain that overexpresses AgRPL44 or MoRPL44 (a homologous sequence from Magnaporthe oryzae), and then abiotic stress resistance of transgenic yeast was examined. To verify the value of its application in genetic engineering, we transformed AgRPL44 into the model fungus M. oryzae and model plant tobacco. Overexpressing AgRPL44 enhanced tolerance to salt and drought in transgenic M. oryzae and transgenic tobacco. The present study provides a candidate gene for genetic engineering of enhanced stress tolerance in crops and explores the possibility of a new screening marker for construction of an M. oryzae mutant library.
MATERIALS AND METHODS
Strains and culture conditions.
Spores of Aspergillus glaucus were cultured (106 to 1010 spores/ml) in potato dextrose medium containing 10% NaCl at 30°C for about 6 days. The Escherichia coli strains employed in this study were DH5α and BL21. Standard procedures were used for manipulating bacterial cells and recombinant DNA (26, 27). The Saccharomyces cerevisiae strains used in the present study were His-, Leu-, Trp-, and Ura-AH109. Yeast cells were grown in yeast potato dextrose (YPD) containing 2% peptone, 1% yeast extract, and 2% glucose. Yeast strain maintenance was achieved by plating onto yeast nitrogen base-glucose medium supplemented with 2% (wt/vol) agar.
Isolation and sequence analysis of AgRPL44.
A yeast expression library containing full-length cDNAs of A. glaucus was constructed in our lab. Briefly, total RNAs of A. glaucus treated by different concentrations of NaCl (5%, 10%, and 20% [wt/vol]) were isolated and purified together. Then, full-length cDNAs were synthesized and cloned into pDONR222 and pYES2-DEST52 through the Gateway cloning system (Invitrogen, Shanghai, China). Lastly, the library was introduced into S. cerevisiae. The titer of the library was 1.28 × 106 CFU/ml, and the number of total colonies was 1.28 × 107 CFU. The rate of positive recombinants clones was 93%, and the size of the average insert cDNA was 1 kb. Salt-tolerant transformants were isolated by screening the yeast expression library. The inserted cDNA fragments were identified via PCR amplification using primers S1/S2. After purification, the PCR products were combined with the plasmid pMD-18T (TaKaRa, Dalian, China) and sequenced. Molecular weight (MW) and isoelectronic point (pI) predictions for the deduced AgRPL44 protein were performed using the ProtParam tool (http://www.expasy.org/tools/protparam.html). Multiple sequence alignment was performed with ClustalW and MEGA5.05.
AgRPL44 gene expression profiles in response to abiotic stresses.
Total RNA from A. glaucus was extracted before and after treatment with 20% NaCl, 2 M sorbitol, or 10 mM CuSO4 using TRIzol reagent (Invitrogen, USA) in accordance with the manufacturer's instructions. First-strand cDNAs were synthesized from 2 μg of total RNA (pretreated with DNase I) with avian myeloblastosis virus reverse transcriptase (TaKaRa). Semiquantitative reverse transcription (RT)-PCR was used to determine expression of the AgRPL44 gene. Primers R1/R2 (see Table S1 in the supplemental material) used in RT-PCR had high specificity, as determined by agarose gel electrophoresis, and were confirmed by sequencing. The RT-PCRs were performed using TaKaRa DNA polymerase (TaKaRa, Dalian, China). The cycle numbers of the PCRs were adjusted for each gene to obtain visible bands on agarose gels. The ACTB (β-actin) gene was used as the internal control.
Experiments for adverse tolerance analysis in yeast.
Ribosomal protein is highly conserved. We found that the M. oryzae ribosomal protein gene MoRPL44 is 85.85% homologous with AgRPL44 (see Fig. S1 in the supplemental material). To further investigate the salt tolerance of 60S ribosomal protein L44, we also cloned the MoRPL44 gene. The AgRPL44 open reading frame (ORF) was amplified from the A. glaucus cDNA clone via PCR using the primers Y1/Y2. The MoRPL44 ORF was amplified through RT-PCR using primers Y3/Y4. The PCR-amplified product was digested with BamHI-HindIII and directionally cloned into the yeast expression vector pURL129, which contains the URA3 selection marker, and was identified by PCR and sequencing. Sequence analysis confirmed that AgRPL44 or MoRPL44 was inserted into the pURL129 plasmid. The recombinant plasmid pURL129-AgRPL44 or pURL129-MoRPL44 was transformed into competent S. cerevisiae AH109 cells. Selection and growth of transformants were performed in synthetic medium (yeast nitrogen base, 2% glucose) lacking uracil.
For growth assays, yeast cells harboring the empty vector pURL129 (as a control) or recombinant plasmid were pregrown for 2 days in YPD and then resuspended in YPD to an optical density at 600 nm (OD600) of 0.5 and diluted in a 10-fold dilution series, of which 4 μl was spotted onto agar plates supplemented with 10%, 15%, or 20% NaCl, 1 M or 2 M sorbitol, or 10 mM CuSO4. Growth conditions were maintained for 2 to 5 days at 30°C.
Analysis of salt and drought tolerance in Magnaporthe oryzae.
M. oryzae and A. glaucus both are filamentous fungi. M. oryzae is often used as a model fungus, and the method for its genetic transformation has been well refined. We transformed the AgRPL44 gene into M. oryzae to verify its tolerance to salt and drought. The expression vector introduced into M. oryzae was constructed for expression of the AgRPL44 gene by cloning it (using primers M1/M2) at the XbaI site into the pGFPGUSPlus vector (28). The M. oryzae strain 70-15 was cultured on complete medium (CM), 5× yeast extract-glucose (YEG; 0.5% yeast extract and 2% glucose), and oatmeal agar as previously described (29). To enhance conidiation, culture plates were grown under constant fluorescent light at room temperature. Protoplasts were prepared and transformed by polyethylene glycol treatment as described previously (30). Transformants were selected on TB3 medium (0.3% yeast extract, 0.3% Casamino Acids, and 20% glucose) with 250 mg ml−1 hygromycin B (Calbiochem). Transformants overexpressing AgRPL44 were further confirmed by Western blot assay.
Mycelium blocks 10 mm in diameter from wild-type M. oryzae or transformed M. oryzae with an empty vector or AgRPL44 were obtained using a round punch, after which they were inoculated on 0%, 5%, 10%, or 15% NaCl and 5% sorbitol solid CM culture medium and cultured 7 to 10 days under normal conditions. Vegetative growth and mycelium growth diameters of wild-type and transformed M. oryzae were determined to investigate salt tolerance and drought resistance.
Western blot assay.
The ORF of AgRPL44 was constructed into vector pET32a (Novagen, Billerica, MA, USA) to generate a fusion protein with a hexahistidine tag. The PCR product (using primers A1/A2) was digested with BamHI and HindIII. The identity of the recombinant pET32a-AgRPL44 construct was confirmed by DNA sequencing and then transformed into E. coli BL21 cells. The BL21 cells were induced by 1 mM isopropyl-beta-d-1-thiogalactopyranoside (IPTG) for 4 h at 37°C. Cells were then harvested by centrifugation and disrupted by physical fragmentation. Inclusion bodies were dissolved with 6 M urea on ice. The supernatant was filtered through a 0.45-μm membrane and purified by affinity chromatography using a nickel-nitrilotriacetic acid column (Ni-NTA agarose; Roche). The protein was renatured through step dialysis at 4°C for 12 h. The primary polyclonal antibody was produced via the immunization of a New Zealand rabbit with AgRPL44 protein (31).
The total proteins of M. oryzae overexpressing AgRPL44 were extracted and resolved via SDS-PAGE. After separation, proteins were transferred to a nitrocellulose membrane by electroblotting. The membrane was blocked overnight at 4°C in Tris-buffered saline with Tween 20 (TBST) buffer with 5% nonfat milk. The membrane was incubated with primary rabbit anti-AgRPL44 antibody (dilution, 1:10,000 in TBST) at room temperature for 2 h. The membrane was washed with TBST buffer three times and then incubated with 1:10,000 horseradish peroxidase-conjugated goat anti-rabbit IgG antibody for 1 h at 37°C (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Plant transformation and salt resistance assay, postgermination.
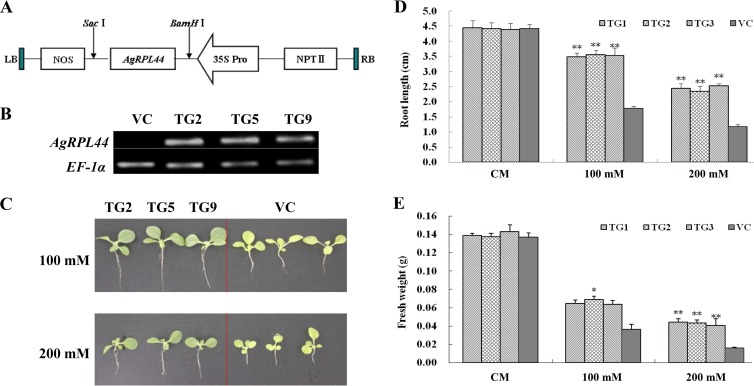
The role of the ribosomal protein RPL44 gene in salt stress tolerance was investigated by generating transgenic tobacco plants overexpressing AgRPL44. The transformation vector was constructed for expression of the ribosomal protein RPL44 gene by cloning it at the BamHI and SacI sites into plasmid pBI121 using primers P1/P2 and a plant expression vector carrying a cauliflower mosaic virus 35S (CaMV35S) promoter and nopaline synthase (NOS) terminator (see Fig. 5A). The construct was introduced into competent cells of Agrobacterium tumefaciens strain LBA4404 by the freeze-thaw method. Tobacco (Nicotiana tabacum L.) plants were transformed using the Agrobacterium-mediated leaf disc method. The regenerated seedlings were selected on Murashige and Skoog medium containing a final concentration of 40 mg/liter of kanamycin. The transgenic lines were also confirmed by RT-PCR using primers E1/E2. Three independent transgenic homozygous T2 line (TG) seedlings and the pBI121 vector control (VC) lines were used for subsequent experiments.
FIG 5.
Assay of early development of AgRPL44-transformed tobacco plants. (A) Schematic of the vector used for plant transformation. (B) RT-PCR analysis of transgenic lines with the pBI121::AgRPL44 construct. (C) Phenotype of wild-type and TG tobacco after 2 weeks of 100 mM or 200 mM NaCl treatment. (D) Root lengths of tobacco plants overexpressing the AgRPL44 gene. (E) Fresh weights of tobacco plants overexpressing the AgRPL44 gene. The data represent means ± SD of results from three experiments performed. Significant differences between the TG and control lines are indicated by asterisks (*, P < 0.05; **, P < 0.01).
For early growth assessments, well-grown tobacco plants (VC) and three transgenic line seedlings of 3 weeks were placed in Hoagland's nutrient solution containing NaCl at different concentrations (0 mM, 100 mM, or 200 mM). Tobacco plants were maintained vertically under normal conditions. The root length and fresh weight were monitored after the 15th day.
Nucleotide sequence accession number.
The sequence of the AgRPL44 gene determined in this study has been deposited in GenBank under accession number KJ679499.
RESULTS
Isolation and sequence analysis of the ribosomal protein AgRPL44 gene.
S. cerevisiae is an excellent single-cellular model for quickly identifying functional genes. Based on screening of the yeast expression library constructed in our lab, a yeast colony that was able to grow on YPD solid medium supplemented with 10%, 15%, or 20% NaCl was selected. The cDNA fragments inserted from the salt-tolerant transformants were identified by PCR amplification and then sequenced. Protein conservative domains were analyzed via NCBI BLAST alignment. We found that a protein encoded by AgRPL44 belongs to the 60S ribosomal protein family. The ribosomal protein RPL44 cDNA is 462 bp in length, including a complete ORF of 321 bp encoding a putative protein of 106 amino acids (predicted relative molecular mass, 12.09 kDa; theoretical pI, 10.43).
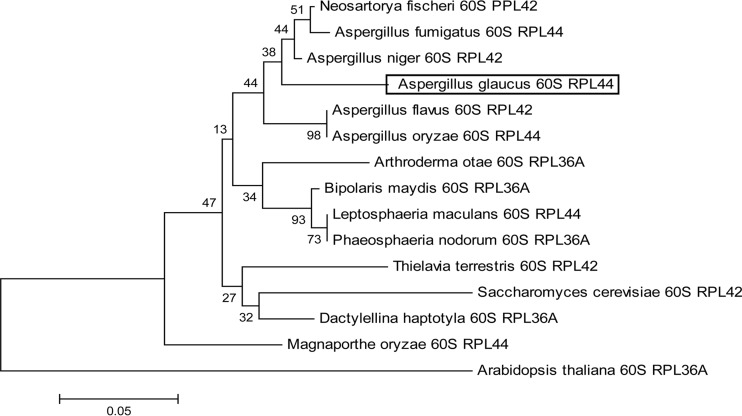
Analyzing the evolutionary relationships among the various species of RPL44 might provide clues to the evolution of their function. To further characterize the AgRPL44 protein, 15 RPL44 proteins in different species were aligned. In the phylogenetic tree (Fig. 1), AgRPL44 and other 60S ribosomal proteins of five Aspergillus species were on the same branch, indicating that AgRPL44 is closely related to the Aspergillus 60S ribosomal protein RPL44.
FIG 1.
Phylogenetic tree of AgRPL44 from various species. The multiple alignments generated by ClustalW and the phylogenetic tree were constructed with MEGA5.05 using a bootstrap test of phylogeny with the minimum evolution test and default parameters. GenBank accession numbers of RPL44 proteins used for drawing the phylogenetic tree are shown in Table S1 in the supplemental material.
Ribosomal protein-encoding AgRPL44 is more resistant to abiotic stresses than MoRPL44 in yeast.
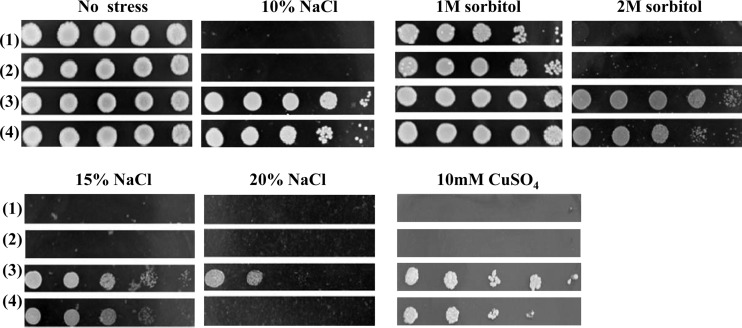
Wild-type yeast cells (AH109) and yeast transformants harboring the empty vector pURL129, pURL129-AgRPL44, or pURL129-MoRPL44 were spotted in a 1:10 dilution series on defined medium supplemented with NaCl or sorbitol as indicated and incubated for 3 to 5 days at 30°C. Each experiment was repeated at least three times, and a representative example is shown.
The data show that AgRPL44- or MoRPL44-transformed yeast could survive on 10% and 15% NaCl while the wild-type yeast could not. However, only AgRPL44-transformed yeast could survive on 20% NaCl YPD medium. At the same salt concentration, AgRPL44-transformed yeast had stronger vitality than MoRPL44-transformed yeast. In addition, AgRPL44-transformed yeast was more resistant to drought and heavy metals than MoRPL44-transformed yeast at a similar level of stress (Fig. 2).
FIG 2.
Expression of AgRPL44 or MoRPL44 in yeast enhances growth at high osmolarity. At high osmolarity, yeast cells (AH109) expressing AgRPL44 or MoRPL44 show increased growth compared with control cells transformed with an empty vector (pURL129). Transformants were spotted in 1:10 dilution series on YPD medium supplemented with NaCl, sorbitol, or CuSO4 as indicated and incubated for 3 to 5 days at 30°C. Rows: 1, wild-type yeast AH109; 2, transformants with empty vector (pURL129); 3, transformants with AgRPL44; 4, transformants with MoRPL44. Each experiment was replicated at least three times. A representative example is shown.
Expression of AgRPL44 in response to different abiotic stresses.
The expression of AgRPL44 in response to NaCl, sorbitol, or CuSO4 in A. glaucus was investigated using semiquantitative RT-PCR. AgRPL44 was induced at 1 h and reached a maximum at 8 h after treatment with NaCl (Fig. 3). During sorbitol or CuSO4 treatment, the expression of AgRPL44 peaked at 4 h. These results suggest that expression of the AgRPL44 gene is induced by NaCl, sorbitol, and CuSO4 treatments.
FIG 3.
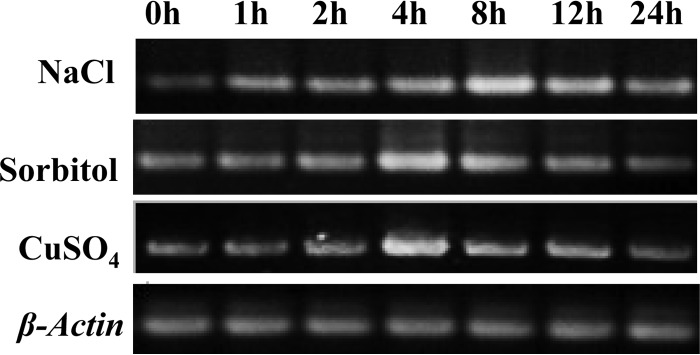
Semiquantitative RT-PCR analysis of AgRPL44 expression under different abiotic stresses. RT-PCR experiments were conducted with total RNA isolated from A. glaucus before and after stress treatments consisting of 20% NaCl, 2 M sorbitol, or 10 mM CuSO4 for the indicated times.
AgRPL44 gene can promote adverse-stress resistance levels in M. oryzae.
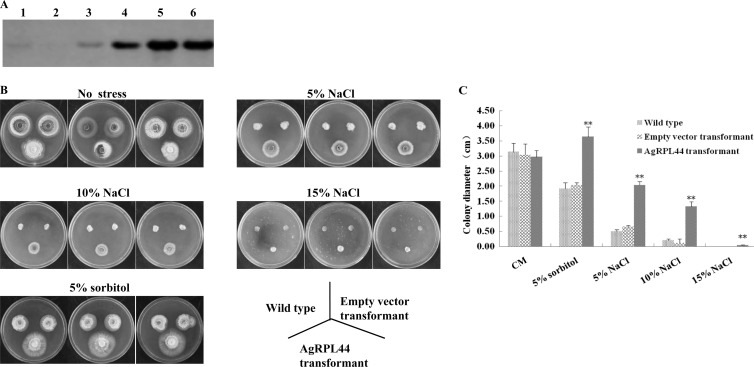
We constructed an expression recombinant plasmid, pGFPGUSPlus::AgRPL44, for M. oryzae and introduced it or an empty vector into M. oryzae. The AgRPL44 transformant showed high expression of the AgRPL44 protein, as confirmed by Western blotting (Fig. 4A).
FIG 4.
Analysis of stress resistance in Magnaporthe oryzae. (A) Western blot confirmation of AgRPL44 protein expression. Lane 1, wild-type M. oryzae; lane 2, M. oryzae transformed with empty vector; lane 3, halophilic A. glaucus; lanes 4 to 6, three M. oryzae strains transformed with AgRPL44. (B) The wild type and transformants were incubated on complete medium alone (CM) or CM supplemented with NaCl or sorbitol as indicated for 7 to 10 days at 30°C. (C) Mycelium growth diameter of wild-type and transformed M. oryzae. Each experiment was replicated at least three times. A representative example is shown. The data represent means ± standard deviations (SD) of results from three experiments performed. Significant differences between the AgRPL44 transformants and control groups are indicated with asterisks (**, P < 0.01).
We tested transgenic strains' responses to adverse stresses after introducing the AgRPL44 gene. The result showed that transformation with AgRPL44 was associated with greater salt and drought tolerance in transgenic M. oryzae. Under the same culture conditions (temperature, humidity, and time), on both the 5% and 10% NaCl solid CM, the wild-type M. oryzae and empty vector transformant remained alive, but the AgRPL44 transformant had greater salt tolerance and viability than either of the other two groups. On the 5% NaCl solid CM, the mycelium growth diameters of the wild type and empty vector transformant were 24.6% and 32% that of the AgRPL44 transformant, respectively. On the 10% NaCl solid CM, the mycelium growth diameters of the wild type and empty vector transformant were only 15% and 9% that of the AgRPL44 transformant, respectively (Fig. 4C). On the 15% NaCl solid CM, the AgRPL44 transformant could still grow, but the wild type and empty vector transformant could hardly stay alive (Fig. 4B).
We also inoculated the wild-type and transformed M. oryzae in 5% sorbitol solid CM under 28°C culture 7 to 10 days. Compared with that of the wild type, the mycelial growth area of the AgRPL44-transformed M. oryzae was large (Fig. 4B) and the mycelium growth diameters of the wild type and empty vector transformant were 25.3% and 55.9% that of the AgRPL44 transformant, respectively (Fig. 4C). These results show that AgRPL44 has a very important role in salt and drought tolerance. AgRPL44 in M. oryzae might provide a new, simple, and low-cost screening marker for M. oryzae mutant library screening.
Overexpression of AgRPL44 improves tolerance of transgenic tobacco plants to salt stress.
Tobacco leaf disks were transformed with the AgRPL44 gene. Transgenic lines were confirmed by RT-PCR (Fig. 5B). Three independent T2 lines (TG 2, TG 5, and TG 9) of the AgRPL44 transgenic plants were chosen for further analysis.
To evaluate the stress tolerance of the control (VC) and transgenic (TG) tobacco lines, seedlings were exposed to different concentrations of NaCl for salt tolerance analysis. The growths of the VC and TG lines under normal conditions were similar (data not shown). However, when treated with NaCl, the TG plants adapted much better to the stress than did the control lines (Fig. 5C). Under normal and 100 mM or 200 mM NaCl conditions, the root lengths and fresh weights of the VC and TG plants were comparable (Fig. 5D and E). As the NaCl concentration increased, the root lengths of the control lines were significantly arrested, whereas the roots of the TG plants continued to grow. At the NaCl concentrations of 100 and 200 mM, the root lengths of the control lines were 50.4% and 48.6% those of the TG lines, respectively, showing that the transgenic seedlings were significantly more salt resistant. It was easily observed that the health of the transgenic plants was superior to that of the control lines. The growth of the transgenic plants was robust, and their leaves were bright green, but the growth of the seedlings of the control lines was significantly inhibited and their leaves were chlorotic.
Transgenic plants also exhibited greater fresh weight than the VC. The fresh weight of the wild type was only 55.4% that of the transgenic lines, a significant difference between the two. At the NaCl concentration of 200 mM, the fresh weight of the wild type was 37.8% that of the transgenic lines (Fig. 5E). These results indicate that overexpression of AgRPL44 enhanced the salt stress tolerance of transgenic tobacco after germination.
DISCUSSION
In the present study, a ribosomal protein gene (AgRPL44) was isolated by screening an A. glaucus cDNA library. Sequence alignment revealed that AgRPL44 belongs to the 60S ribosomal protein RPL44. AgRPL44 expression was strongly induced in A. glaucus by exposure to salinity (NaCl), drought (sorbitol), or heavy metal (CuSO4), suggesting that AgRPL44 has an important role in responses to these abiotic stressors. The yeast transformed with RPL44 exhibited significantly elevated tolerance to salt, drought, and heavy metal stresses. Interestingly, the tolerance to these conditions in the transformed AgRPL44 yeast strain was stronger than that of the MoRPL44 strain. This suggests that AgRPL44 expression confers tolerance to a variety of abiotic stresses. When AgRPL44 was introduced into M. oryzae, transformants displayed obviously enhanced tolerance to salt and drought, revealing a genetic application for AgRPL44. In addition, overexpression of AgRPL44 in tobacco resulted in enhanced salt stress tolerance.
Ribosomal protein (RP) gene expression can be dramatically altered in response to growth stimuli and environmental stresses. For example, plant ribosomal proteins S14, S15a, S16, L10, L13a, and L30 can be induced by diverse phytohormones, including auxin (32), cytokinin (32, 33), and abscisic acid (33), and by stress-inducing stimuli, including heat (32), cold (32, 34), and UVB stress (35). Recent analysis of the expression of RP genes and the differential accumulation of RPs in Arabidopsis roots upon phosphate or iron deficiency revealed that these 579 RP genes were associated mainly with abiotic stress responses. Some of the highly expressed RP genes were also involved in responses to cold, UVB, and salt stresses (36). It might thus be assumed that some ribosomal proteins have protective functions against multiple abiotic and nutrient stresses.
Although the ribosomal protein RPL44 is highly conserved in many fungal strains, under the same abiotic stresses AgRPL44 was associated with greater stress tolerance than MoRPL44, especially for salt; only the AgRPL44 transformant could survive on 20% NaCl YPD. Comparing the protein sequences of AgRPL44 and MoRPL44, it was found that they shared identical motifs: their amino acid residues 1 to 9, 24 to 33, 36 to 74, and 84 to 105 are nearly identical. Therefore, we could deduce that these regions were probably essential for the stress resistance function of RPL44. Although the two RP genes are highly conserved, there are some specific amino acid residues in the AgRPL44 sequence, for instance, lysines 12 and 80, histidine 17, glutamines 21 and 75, leucine 34, or serines 35 and 83, which may play a very key role in its remarkable abiotic-stress resistance. We will further proceed with site-specific mutagenesis to verify our deduction.
A. glaucus is halophilic and therefore grows more slowly under low-salt conditions than it does under high salt (8). However, higher salt no doubt affects the growth of most eukaryotic organisms. The expression patterns (Fig. 3) of AgRPL44 may in part explain the resistance to high salt in A. glaucus. Thus, we presume that when A. glaucus is under osmotic stresses, to eliminate saline damage, it enhances the expression levels of AgRPL44. Overexpression of AgRPL44 in M. oryzae markedly increased the transformants' ability to withstand salinity via exposure to NaCl. Moreover, mycelial growth, conidiation, and pathogenicity were evaluated, and there was no difference between the wild type and the transformed strains under normal conditions (data not shown). Thus, we could explore a new simple, practical, and low-cost selection marker for genetic transformation. Some ribosomal proteins have been utilized as selectable markers. For example, the small ribosomal protein S12P is used as an efficient positive-selection marker in allelic exchange mutation systems for Corynebacterium glutamicum (37, 38). A mutant protein of RPL44 from Aurantiochytrium sp. strain KRS101 affords cycloheximide resistance to strains, serving as a novel selection marker (39). The present M. oryzae transformants were selected with a high concentration of hygromycin B or phleomycin D1 (Zeocin) (40), both of which are toxic and expensive. In our research, we might use RPL44 to create a dominant selectable marker expressed with the aid of native transcriptional and translational machinery, and it is safe in biological transformation. Our results may be significant for M. oryzae mutation library screening. Despite this, further work is required. The optimization of transformation conditions and integration into various regions of ribosomal DNA (rDNA) may further increase conversion efficiency.
In summary, our work presented here has demonstrated ribosomal protein AgRPL44 as being involved in the response to abiotic stresses and revealed a previously unrecognized role of abiotic stress resistance in transgenic yeasts and transgenic plants. We might explore a new, simple, and safe screening marker for M. oryzae mutant library screening. However, more work will be needed to measure physiological indices involved in abiotic stresses of mature transgenic tobacco, analyze the expression of stress-related genes to verify whether these genes are directly regulated by AgRPL44, and explore its protecting mechanisms against abiotic stresses in transgenic plants.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alice Cheung (University of Massachusetts, Amherst, MA, USA) for her helpful suggestions during the course of these studies and Lam Han-Ming (CUHK, Hong Kong) for technical assistance.
This project was supported by the NSFC (31171794), the Ministry of Agriculture of China (20112X08009-001), and the Special Fund for Agro-Scientific Research in the Public Interest (201203014).
Footnotes
Published ahead of print 9 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00292-14.
REFERENCES
- 1.Norkrans B. 1966. Studies on marine occurring yeasts: growth related to pH, NaCl concentration and temperature. Arch. Mikrobiol. 54:374–392. 10.1007/BF00406719 [DOI] [Google Scholar]
- 2.Onishi H. 1963. Osmophilic yeasts. Adv. Food Res. 12:53–94. 10.1016/S0065-2628(08)60006-3 [DOI] [PubMed] [Google Scholar]
- 3.Anton J, Rossello-Mora R, Rodriguez-Valera F, Amann R. 2000. Extremely halophilic bacteria in crystallizer ponds from solar salterns. Appl. Environ. Microbiol. 66:3052–3057. 10.1128/AEM.66.7.3052-3057.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantrell SA, Casillas-Martinez L, Molina M. 2006. Characterization of fungi from hypersaline environments of solar salterns using morphological and molecular techniques. Mycol. Res. 110:962–970. 10.1016/j.mycres.2006.06.005 [DOI] [PubMed] [Google Scholar]
- 5.Gunde-Cimerman N, Ramos J, Plemenitaš A. 2009. Halotolerant and halophilic fungi. Mycol. Res. 113:1231–1241. 10.1016/j.mycres.2009.09.002 [DOI] [PubMed] [Google Scholar]
- 6.Oren A. 2002. Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. J. Ind. Microbiol. Biotechnol. 28:56–63. 10.1038/sj.jim.7000176 [DOI] [PubMed] [Google Scholar]
- 7.Tehei M, Franzetti B, Maurel MC, Vergne J, Hountondji C, Zaccai G. 2002. The search for traces of life: the protective effect of salt on biological macromolecules. Extremophiles 6:427–430. 10.1007/s00792-002-0275-6 [DOI] [PubMed] [Google Scholar]
- 8.Liu XD, Liu JL, Wei Y, Tian YP, Fan FF, Pan HY, Zhang SH. 2011. Isolation, identification and biologic characteristics of an extreme halotolerant Aspergillus sp. J. Jilin Univ. 49:548–552 (In Chinese; abstract in English.) http://www.cnki.com.cn/Article/CJFDTotal-JLDX201103039.htm. [Google Scholar]
- 9.Ramakrishnan V, White SW. 1998. Ribosomal protein structures: insights into the architecture, machinery and evolution of the ribosome. Trends Biochem. Sci. 23:208–212. 10.1016/S0968-0004(98)01214-6 [DOI] [PubMed] [Google Scholar]
- 10.Cherepneva G, Schmidt KH, Kulaeva O, Oelmüller R, Kusnetsov V. 2003. Expression of the ribosomal proteins S14, S16, L13a and L30 is regulated by cytokinin and abscisic acid: implication of the involvement of phytohormones in translational processes. Plant Sci. 165:925–932. 10.1016/S0168-9452(03)00204-8 [DOI] [Google Scholar]
- 11.Bouakaz L, Bouakaz E, Murgola EJ, Ehrenberg M, Sanyal S. 2006. The role of ribosomal protein L11 in class I release factor mediated translation termination and translational accuracy. J. Biol. Chem. 281:4548–4556. 10.1074/jbc.M510433200 [DOI] [PubMed] [Google Scholar]
- 12.Martínez-Guitarte J, Planelló R, Morcillo G. 2007. Characterization and expression during development and under environmental stress of the genes encoding ribosomal proteins L11 and L13 in Chironomus riparius. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 147:590–596. 10.1016/j.cbpb.2007.03.015 [DOI] [PubMed] [Google Scholar]
- 13.Mazumder B, Sampath P, Seshadri V, Maitra RK, Corleto PE, Fox PL. 2003. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 115:187–198. 10.1016/S0092-8674(03)00773-6 [DOI] [PubMed] [Google Scholar]
- 14.Warner JR, McIntosh KB. 2009. How common are extra-ribosomal functions of ribosomal proteins? Mol. Cell 34:3–11. 10.1016/j.molcel.2009.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graifer D, Malygin A, Zharkov DO, Karpova G. 2014. Eukaryotic ribosomal protein S3: a constituent of translational machinery and an extraribosomal player in various cellular processes. Biochimie 99:8–18. 10.1016/j.biochi.2013.11.001 [DOI] [PubMed] [Google Scholar]
- 16.Xu T, Lee K, Gu L, Kim JI, Kang H. 2013. Functional characterization of a plastid-specific ribosomal protein PSRP2 in Arabidopsis thaliana under abiotic stress conditions. Plant Physiol. Biochem. 73:405–411. 10.1016/j.plaphy.2013.10.027 [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Chao X, Wang L, Zhang RJ, Jue DW, Qing Y. 10 August 2013. Expression of a wild eggplant ribosomal protein L13a in potato enhances resistance to Verticillium dahliae. Plant Cell Tissue Organ Cult. 10.1007/s11240-013-0365-4 [DOI] [Google Scholar]
- 18.Fernandes JM, Smith VJ. 2002. A novel antimicrobial function for a ribosomal peptide from rainbow trout skin. Biochem. Biophys. Res. Commun. 296:167–171. 10.1016/S0006-291X(02)00837-9 [DOI] [PubMed] [Google Scholar]
- 19.Hou YL, Ding X, Hou W, Song B, Wang T, Wang F, Li J, Zhong J, Xu T, Ma BX, Zhu HQ, Li JH, Zhong JC. 2013. Overexpression, purification, and pharmacologic evaluation of anticancer activity of ribosomal protein L24 from the giant panda (Ailuropoda melanoleuca). Genet. Mol. Res. 12:4735–4750. 10.4238/2013.October.18.11 [DOI] [PubMed] [Google Scholar]
- 20.Su XL, Hou YL, Yan XH, Ding X, Hou WR, Sun B, Zhang SN. 2012. Expression, purification, and evaluation for anticancer activity of ribosomal protein L31 gene (RPL31) from the giant panda (Ailuropoda melanoleuca). Mol. Biol. Rep. 39:8945–8954. 10.1007/s11033-012-1763-0 [DOI] [PubMed] [Google Scholar]
- 21.Wang T, Hou Y, Ding X, Song B, Wang F, Hou W. 2013. Overexpression, purification, molecular characterization and pharmacological evaluation for anticancer activity of ribosomal protein S23 from the giant panda (Ailuropoda melanoleuca). Mol. Med. Rep. 7:1875–1882. 10.3892/mmr.2013.1430 [DOI] [PubMed] [Google Scholar]
- 22.Kim JH, You KR, Kim IH, Cho BH, Kim CY, Kim DG. 2004. Over-expression of the ribosomal protein L36a gene is associated with cellular proliferation in hepatocellular carcinoma. Hepatology 39:129–138. 10.1002/hep.20017 [DOI] [PubMed] [Google Scholar]
- 23.Seshadri T, Uzman JA, Oshima J, Campisi J. 1993. Identification of a transcript that is downregulated in senescent human fibroblasts. Cloning, sequence analysis, and regulation of the human L7 ribosomal protein gene. J. Biol. Chem. 268:18474–18480 [PubMed] [Google Scholar]
- 24.Pierandrei-Amaldi P, Amaldi F. 1994. Aspects of regulation of ribosomal protein synthesis in Xenopus laevis. Genetica 94:181–193. 10.1007/BF01443432 [DOI] [PubMed] [Google Scholar]
- 25.Warner JR. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437–440. 10.1016/S0968-0004(99)01460-7 [DOI] [PubMed] [Google Scholar]
- 26.Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1999. Short protocols in molecular biology, 4th ed. Wiley, Hoboken, NJ [Google Scholar]
- 27.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 28.Vickers CE, Schenk PM, Li D, Mullineaux PM, Gresshoff PM. 2007. pGFPGUSPlus, a new binary vector for gene expression studies and optimising transformation systems in plants. Biotechnol. Lett. 29:1793–1796. 10.1007/s10529-007-9467-6 [DOI] [PubMed] [Google Scholar]
- 29.Li L, Xue CY, Bruno K, Nishimura M, Xu JR. 2004. Two PAK kinase genes, CHM1 and MST20, have distinct functions in Magnaporthe grisea. Mol. Plant Microbe Interact. 17:547–556. 10.1094/MPMI.2004.17.5.547 [DOI] [PubMed] [Google Scholar]
- 30.Sweigard JA, Chumley FG, Valent B. 1992. Disruption of a Magnaporthe grisea cutinase gene. Mol. Gen. Genet. 232:183–190 [PubMed] [Google Scholar]
- 31.Chen P, Wang CD, Li KX, Chang JL, Wang YS, Yang GX, Shewry PR, He GY. 2008. Cloning, expression and characterization of novel avenin-like genes in wheat and related species. J. Cereal Sci. 48:734–740. 10.1016/j.jcs.2008.04.002 [DOI] [Google Scholar]
- 32.Hulm JL, McIntosh KB, Bonham-Smith PC. 2005. Variation in transcript abundance among the four members of the Arabidopsis thaliana ribosomal protein S15a gene family. Plant Sci. 169:267–278. 10.1016/j.plantsci.2005.04.001 [DOI] [Google Scholar]
- 33.Cherepneva GN, Schmidt KH, Kulaeva ON, Oelmüller R, Kusnetsov VV. 2003. Expression of the ribosomal proteins S14, S16, L13a and L30 is regulated by cytokinin and abscisic acid: implication of the involvement of phytohormones in translational processes. Plant Sci. 165:925–932. 10.1016/S0168-9452(03)00204-8 [DOI] [Google Scholar]
- 34.Kim KY, Park SW, Chung YS, Chung CH, Kim JI, Lee JH. 2004. Molecular cloning of low temperature-inducible ribosomal proteins from soybean. J. Exp. Bot. 55:1153–1155. 10.1093/jxb/erh125 [DOI] [PubMed] [Google Scholar]
- 35.Falcone Ferreyra ML, Pezza A, Biarc J, Burlingame AL, Casati P. 2010. Plant L10 ribosomal proteins have different roles during development and translation under ultraviolet-B stress. Plant Physiol. 153:1878–1894. 10.1104/pp.110.157057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang JY, Lan P, Gao HM, Zheng L, Li WF, Schmidt W. 2013. Expression changes of ribosomal proteins in phosphate- and iron-deficient Arabidopsis roots predict stress-specific alterations in ribosome composition. BMC Genomics 14:783–796. 10.1186/1471-2164-14-783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersson DI. 2003. Persistence of antibiotic resistant bacteria. Curr. Opin. Microbiol. 6:452–456. 10.1016/j.mib.2003.09.001 [DOI] [PubMed] [Google Scholar]
- 38.Kim IK, Jeong WK, Lim SH, Hwang IK, Kim YH. 2011. The small ribosomal protein S12P gene rpsL as an efficient positive selection marker in allelic exchange mutation systems for Corynebacterium glutamicum. J. Microbiol. Methods 84:128–130. 10.1016/j.mimet.2010.10.007 [DOI] [PubMed] [Google Scholar]
- 39.Hong WK, Heo SY, Oh BR, Kim CH, Sohn JH, Yang JW, Kondo A, Seo JW. 2013. A transgene expression system for the marine microalgae Aurantiochytrium sp. KRS101 using a mutant allele of the gene encoding ribosomal protein L44 as a selectable transformation marker for cycloheximide resistance. Bioprocess. Biosyst. Eng. 36:1191–1197 [DOI] [PubMed] [Google Scholar]
- 40.Zhou X, Liu W, Wang C, Xu Q, Wang Y, Ding S, Xu JR. 2011. A MADS-box transcription factor MoMcm1 is required for male fertility, microconidium production and virulence in Magnaporthe oryzae. Mol. Microbiol. 80:33–53. 10.1111/j.1365-2958.2011.07556.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.