Abstract
Hepatitis E virus (HEV), an enteric pathogen of both humans and animals, is excreted by infected individuals and is therefore present in wastewaters and coastal waters. As bivalve molluscan shellfish are known to concentrate viral particles during the process of filter feeding, they may accumulate this virus. The bioaccumulation efficiencies of oysters (Crassostrea gigas), flat oysters (Ostrea edulis), mussels (Mytilus edulis), and clams (Ruditapes philippinarum) were compared at different time points during the year. Tissue distribution analysis showed that most of the viruses were concentrated in the digestive tissues of the four species. Mussels and clams were found to be more sensitive to sporadic contamination events, as demonstrated by rapid bioaccumulation in less than 1 h compared to species of oysters. For oysters, concentrations increased during the 24-h bioaccumulation period. Additionally, to evaluate environmental occurrence of HEV in shellfish, an environmental investigation was undertaken at sites potentially impacted by pigs, wild boars, and human waste. Of the 286 samples collected, none were contaminated with hepatitis E virus, despite evidence that this virus is circulating in some French areas. It is possible that the number of hepatitis E viral particles discharged into the environment is too low to detect or that the virus may have a very short period of persistence in pig manure and human waste.
INTRODUCTION
Hepatitis E virus (HEV) is a small, nonenveloped single-stranded positive-sense RNA virus that is an enteric pathogen of humans and animals and the sole member of the genus Hepevirus in the family Hepeviridae (1). Four major genotypes have been defined, and two additional genotypes infecting wild boars have been suggested (2, 3). Genotypes 1 and 2, which mainly infect humans, have been described in Asia, Africa, or Mexico, whereas genotypes 3 and 4 have been described worldwide. Genotypes 3 and 4 have been described in a variety of animal species and present a potential zoonotic risk (4–6). As with other enteric viruses, HEV may be detected in high concentrations in feces from infected individuals and thus can be detected in raw or treated sewage (5, 7). The fecal-oral transmission route within an animal species and from contaminated food or waters to humans has been documented (4, 6, 8).
Bivalve molluscan shellfish are known to concentrate viral particles during filtration undertaken as part of their feeding process. A large variety of human enteric viruses have been detected in different shellfish species during either environmental studies or outbreak investigations (9, 10). HEV has been reported in shellfish collected in different European and Asian countries (11–14). Few outbreaks have been reported until now. One reason may be the lack of detection methods for HEV, as reported for the outbreak in New Delhi, India, between December 1955 and January 1956 (15). Additionally, the long incubation period often means that leftover food is not available for analysis. Thus, often the food responsible for the outbreak cannot be identified through testing and is implicated from epidemiological data linking human cases to shellfish consumption. For example, in Spain, a 10-year-old girl developed a hepatitis E infection following oyster and mussel consumption (16). More recently, it was reported that a Japanese man acquired an HEV infection after ingesting uncooked shellfish while traveling in Vietnam (17). Similarly, in 2008, English tourists returning from a cruise ship developed hepatitis E, and the main risk factors were gender, age, and shellfish consumption while on board (18). More generally, in countries where seafood dishes are common, as in China, shellfish consumption is considered a key risk factor for developing hepatitis E (19). In France, as in most European countries, genotype 3 is predominant and evidence of virus circulation among the French population has been reported (20–22). Until now, pork meat has been the food type most frequently implicated in outbreaks (8, 20, 23, 24). However, as shellfish is a common food in France, this transmission route cannot be excluded from consideration, although shellfish have not yet been implicated in a hepatitis E outbreak in France.
The aim of the present study was to obtain quantitative data on HEV bioaccumulation in different shellfish species using real-time reverse transcription-PCR (rRT-PCR) quantification following in vitro virus bioaccumulation experiments performed at several time points during the year. Between-species variations in bioaccumulation efficiency, tissue distribution, and contamination kinetics were evaluated by rRT-PCR based on triplicate analysis and triplicate extractions. In order to get a more complete picture of the potential contamination of shellfish, an environmental investigation was also performed.
MATERIALS AND METHODS
Virus strains.
Stool obtained from a pig experimentally infected with HEV genotype 3e (kindly provided by N. Pavio, Anses, Paris, France) was used for the laboratory experiments. The stool suspension was divided into subsamples and kept at −20°C for all bioaccumulation experiments. Mengovirus (MgV) strain vMC0 (kindly provided by A. Bosch, University of Barcelona) was propagated in HeLa cells as previously described (25).
Shellfish samples. (i) Bioaccumulation experiments.
For all experiments, oysters (Crassostrea gigas), flat oysters (Ostrea edulis), mussels (Mytilus edulis), and clams (Ruditapes philippinarum) were purchased directly from the same producer and environmental data such as water temperature and salinity were recorded. Shellfish were immersed the day after collection in large tanks of seawater at the laboratory. For all bioaccumulation experiments, control tests performed on shellfish samples before bioaccumulation showed no viral (norovirus [NoV] or HEV) contamination. After 24 h of immersion at the specified temperature, only living shellfish showing filtration activity were kept for bioaccumulation experiments.
(ii) Environmental investigations.
Shellfish were collected from two sites potentially contaminated by pigs (site A) or wild boars (site B) at least every month for 2 years. At site A, naturally growing cockles, oysters, and mussels were collected, and at site B, clams and oysters were collected. Based on the bioaccumulation results, extra mussels were added to site B for a 6-month period to ensure an adequate supply for the sampling period. Additionally, shellfish (oysters, mussels, clams, or cockles) were collected from 15 other contaminated French areas (based on Escherichia coli detection; REMI surveillance network, Ifremer), and sampling occurred between 3 and 21 times at each site.
Each sample was composed of only one species of shellfish, with at least 12 individual oysters and 30 individual mussels, clams, or cockles comprising the samples. All samples were shipped to the laboratory in a refrigerated box within 24 h after collection.
Bioaccumulation experiments.
Natural seawater, collected from a clean area in which turbidity, ammonium, nitrate, phosphate, and chlorophyll A concentrations are measured (data not shown), was used for all bioaccumulation experiments. The seawater temperature was adjusted according to the season (from 10 ± 1°C in March to 16 ± 1°C in July). Aquariums were filled with seawater seeded with the HEV suspension at a concentration of around 7.37 ± 0.19 log10 RNA copies/aquarium (except in negative-control aquariums), yielding a ratio of 5 liters per kilogram of shellfish (including the shell weight). The number of shellfish used was selected based on the weight of digestive tissue (DT) recovered (30 individuals for each oyster species, 60 mussels, and 40 clams). The bioaccumulation period was 24 h under oxygenation conditions, and bioaccumulation experiments for the four shellfish species were undertaken on the same day. The experiments were repeated 5 times during different months. Half of the individuals were collected after 1 h and half after 24 h and then immediately processed.
Dissection.
All shellfish were shucked and weighed, and the DTs were recovered as previously described (26). For the first 3 bioaccumulation experiments, gills and mantle were also dissected (27).
Shellfish processing.
Mengovirus (MgV) (2 ×106 RNA copies) was added to each dissected tissue sample (1.5 g) before homogenization with 3 ml of glycine buffer. Then, the viruses were extracted by being mixed with a vortex device with equal volumes of chloroform-butanol for 30 s and Cat-Floc T (Calgon, Ellwood City, PA) (173 μl per tube) treatment for 5 min on the bench before centrifugation for 15 min at 13,500 × g. The resulting suspension was precipitated with polyethylene glycol 6000 (PEG 6000) (Sigma, St Quentin, France) for 1 h at 4°C and centrifuged for 20 min at 11,000 × g at 4°C (28). For shellfish that had bioaccumulated viruses, the DTs were extracted in triplicate, each replicate being extracted in a different batch of samples.
NA extraction and purification.
A NucliSens extraction kit (bioMérieux, Lyon, France) was used following the manufacturer's instructions, with minor modifications (26). The PEG pellet was suspended in 1 ml RNase-free water, mixed with 2 ml of lysis buffer, and incubated for 30 min at 56°C. After a brief centrifugation to eliminate particles (if needed), 50 μl of paramagnetic silica was added, and the mixture was incubated for 10 min at room temperature. The purification steps were performed with an automatic easyMAG extractor (bioMérieux). Nucleic acids (NAs) were recovered in 100 μl of elution buffer (bioMérieux) and analyzed immediately or kept frozen (−80°C).
For the stool, viral RNA was extracted from a 10% suspension using a NucliSens kit (bioMérieux, Lyon, France) as recommended by the manufacturer and was eluted in 100 μl of RNase-free water.
Primers, probes, and rRT-PCR.
Primers and probes were used as previously described for MgV, norovirus (NoV), and HEV detection (27, 29, 30), except that the HEV probe labeling was changed to minor groove binding (MGB). The rRT-PCR was carried out with an UltraSense One-Step quantitative RT-PCR system (Life Technologies, France), using adjusted concentrations of primers and probes (26). Amplifications were performed with an Mx3000P quantitative PCR (qPCR) system (Agilent Technologies, France), using 5 μl of extracted NA per well (final volume of 25 μl). Neat (undiluted) and 10-fold dilutions of the samples were analyzed in triplicate.
rRT-PCR controls and quantification.
The cycle to quantification (Cq) was defined as the cycle showing a significant increase in fluorescence. To be considered positive, a sample had to yield a Cq value of ≤40. Two negative-amplification controls (sterile, RNase-free water) were included in each amplification series, and filter tips and dedicated rooms were used to prevent false positives.
(i) Extraction efficiency.
After extraction of samples seeded with the MgV, amplifications were performed and the Cq values (undiluted and 10-fold dilutions) were compared to the Cq value of the positive control used in the extraction series and to a standard curve made by endpoint dilution. This difference (ΔCq) was used to determine the extraction efficiency and was expressed as a percentage for each sample (26). Only samples with extraction efficiencies above 10% were considered for quantification.
For environmental investigation samples, as only qualitative detection was undertaken, the acceptance criteria was 5%. If the extraction efficiency percentage could not be improved after repeated extractions (up to 3), the sample was not considered further in the analysis of results.
(ii) Quantification.
The absence of inhibitors was verified for each sample by comparing the Cq values of undiluted and 10-fold-diluted extracts. No adjustment was made for rRT-PCR efficiency, as no significant inhibition was observed (less than 25%), or for extraction efficiency. The number of RNA copies was estimated by comparing the Cq value to standard curves derived from in vitro transcription plasmids containing nucleotides 5190 to 5489 of an HEV G3f strain (kindly provided by N. Pavio, Anses, Paris, France). The final concentration was then back-calculated based on the volume of NA and expressed per g of tissue. For bioaccumulation experiments, DTs were extracted 3 times and each NA extract was amplified in triplicate on the same plate. If one extracted replicate presented a difference in Cq values of more than 3 compared to the others, the extraction was repeated a fourth time.
Statistical analysis.
All concentrations obtained were log transformed, and geometric mean titers (GMT) and geometric standard deviations (SDs) were calculated. Statistical analysis was performed using the Statgraphics Centurion XV statistical package (Sigma plus, Levallois-Perret, France). Data were compared using the Student t test, and a P value of <0.05 was considered significant.
RESULTS
Conditions for the bioaccumulation experiments.
For each bioaccumulation experiment, frozen subsamples of the same stool suspension were used. Before each bioaccumulation experiment, the inoculum was quantified, and the titer was stable over the five experiments (7.37 ± 0.19 log10 RNA copies/aquarium). Allometric coefficients (body weight divided by DT weight) were 13 ± 5, 11 ± 2, 12 ± 1, and 13 ± 4 for oysters, flat oysters, mussels, and clams, respectively. The large variation observed for oysters was due to a high value in July (allometric coefficient of 22). For clams, the allometric coefficients in January and March (10 and 9) were lower than those obtained in the 3 other experiments (14, 16, 18). For the 2 other species, low variability was observed. For all experiments, no difference in pseudofeces or feces production between the different shellfish species was observed while cleaning the aquariums.
To be able to compare the different species, virus concentrations and the number of individuals were adjusted based on a maximal theoretical bioaccumulation (MTB) calculation. This was calculated using the following assumptions: (ii) that 1 log of virus would be lost (i.e., by adsorption to aquarium walls, shell, etc.), as demonstrated using radioactive virus (31); (ii) that 90% of the viruses would be concentrated in the DT, as observed during preliminary experiments and demonstrated for norovirus and hepatitis A virus (28); and (iii) that the calculation would be based on the weight of the DT recovered. For example, for oysters in January, the dose seeded in the aquarium was 2.5 × 107 RNA copies and 21 g of DT was recovered. Therefore, 2.5 × 106 RNA copies were available for uptake (assuming a 1 log loss), of which 90% were assumed to be present in the DT (2.25 × 106). Considering the weight of the DT recovered (21 g), this gave an MTB of 1.07 × 105 RNA copies/g of DT. The MTB calculated for each shellfish species for all five experiments showed average values of 5.11 ± 0.25 log10 RNA copies/g of DT for oysters, 5.17 ± 0.20 log10 RNA copies/g of DT for flat oysters, 5.18 ± 0.31 log10 RNA copies/g of DT for mussels, and 5.32 ± 0.27 log10 RNA copies/g of DT for clams. These MTB values were not statistically significantly different (P = 0.65) and therefore allowed comparisons between the different experiments and different species. The bioaccumulation efficiency was calculated by dividing the virus concentration detected in the shellfish DT by the MTB and was expressed as a percentage.
For greater confidence in quantification, after verification that no inhibition had occurred, all DTs were extracted three times in independent extraction runs, and all NA extracts were analyzed in triplicate. Among the 40 extractions performed, the average Cq variation was around 0.6 Cq units after a 1-h bioaccumulation period or 0.4 units after 24 h. The highest variability observed was for oysters in July after the 1-h bioaccumulation period (with Cq values of 38.4 ± 1.0, 38.5 ± 1.9, and 37.5 ± 1.7). The triplicate extractions were useful for flat oysters as after 1 h the concentration was low (close to the limit of detection) and some Cq values were greater than 39, the limit for quantification. For mussels, the three extractions of DT collected after 24 h gave variable Cq values (35.2 ± 0.5, 37.0 ± 1.0, and 32.2 ± 0.3), and the three extraction efficiencies were 18%, 19%, and 24%, respectively. The incorporation of triplicate extractions into the methodology was useful in demonstrating this variability.
For three of the experiments, HEV in seawater was analyzed after 24 h and represented about 1% of the inoculum. No difference in the HEV concentrations in seawater was found between shellfish species or months (data not shown). Given that the level of HEV in seawater was negligible, the residual virus in seawater was not incorporated into the MBT calculation.
Bioaccumulation experiments.
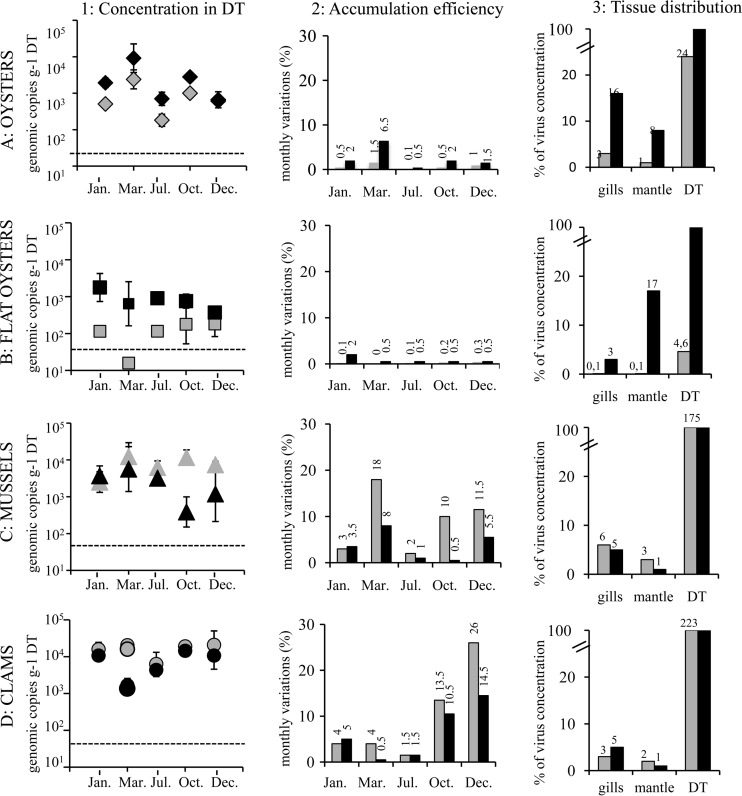
All extraction efficiencies were acceptable, the lowest one being for oysters in October and December (Table 1). No HEV was detected in shellfish in control aquariums. For oysters, the concentration of HEV detected in the DT after a bioaccumulation period of 1 h was lower than the concentration detected after 24 h, except in December (Table 1) (Fig. 1A, column 1). Concentrations observed in March were about 1 log higher (viral copies per g) than those observed in the other months. The observed variabilities were similar for the bioaccumulation periods of 1 and 24 h, showing a statistically significant difference from those determined for the other months (P = 0.003 and P = 0.001 for 1 h and 24 h, respectively). Based on the MTB and detected virus, the highest bioaccumulation efficiency was observed in March after a bioaccumulation period of 24 h (Fig. 1A, column 2), whereas the efficiency was lower than 2% for the other months. The tissue distribution studies showed that most of the HEV was located in the DT, with the mantle and gills representing 16% and 8% of the total, respectively (Fig. 1A, column 3).
TABLE 1.
HEV concentrations detected in DT after bioaccumulationa
| Month | Oysters |
Flat oysters |
Mussels |
Clams |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ext. ef. (%) | DT (mean log concn/g ± SD) |
Ext. ef. (%) | DT (mean log concn/g ± SD) |
Ext. ef. (%) | DT (mean log concn/g ± SD) |
Ext. ef. (%) | DT (mean log concn/g ± SD) |
|||||
| 1 h | 24 h | 1 h | 24 h | 1 h | 24 h | 1 h | 24 h | |||||
| January | 12 | 2.7 ± 0.1 | 3.3 ± 0.3 | 19 | 2.1* | 3.3 ± 0.4 | 51 | 3.6 ± 0.3 | 3.6 ± 0.3 | 25 | 4.2 ± 0.2 | 4.0 ± 0.2 |
| March | 23 | 3.4 ± 0.3 | 4.0 ± 0.4 | 29 | <LQ | 2.8 ± 0.6 | 33 | 4.1 ± 0.4 | 3.7 ± 0.8 | 27 | 4.3* | 3.2 ± 0.2 |
| July | 20 | 2.3 ± 0.2 | 2.9 ± 0.2 | 11 | 2.1 ± 0.1 | 3.0 ± 0.2 | 23 | 3.8 ± 0.2 | 3.5 ± 0.1 | 17 | 3.8 ± 0.3 | 3.7 ± 0.2 |
| October | 10 | 3.0 ± 0.1 | 3.5 ± 0.1 | 13 | 2.3 ± 0.5 | 2.9 ± 0.2 | 18 | 4.1 ± 0.2 | 2.6 ± 0.4 | 25 | 4.3 ± 0.1 | 4.2 ± 0.1 |
| December | 10 | 2.8 ± 0.1 | 2.8 ± 0.2 | 18 | 2.3 ± 0.3 | 2.6 ± 0.2 | 17 | 3.9 ± 0.1 | 3.1 ± 0.8 | 37 | 4.3 ± 0.4 | 4.0 ± 0.4 |
DT values are the geometric means ± standard deviations of the log concentrations of RNA/g of DT determined on the basis of three separate extractions (each analyzed in triplicate), except for two values based on one extraction (indicated by an asterisk [*]) whose results were compromised due to technical problems (extraction failure and lack of DT). The percent extraction efficiency coefficient (Ext. ef.) represents the average value from the 6 extractions. LQ, limit of quantification.
FIG 1.
Hepatitis E virus bioaccumulation and tissue distribution in four shellfish species. In column 1, the concentrations obtained in digestive tissue after 1 h (gray) or 24 h (black) of bioaccumulation are reported as numbers of genome copies per gram of DT (y axis) for the five experiments (January [Jan.], March [Mar.], July [Jul.], October [Oct.], and December [Dec.]) (x axis) for oyster (A), flat oyster (B), mussel (C), and clam (D). Each symbol and the standard variation represent mean values for 3 extractions; dotted lines represent the limit of quantification of the method. In column 2, monthly variations of the bioaccumulation efficiency in digestive tissues (viral bioaccumulation efficiency after 1 h [gray bar] or 24 h [black bar]) (x axis) are expressed as percentages of virus (y axis) recovered in DT calculated on the basis of the virus seeded into seawater and virus detected in DT, taking into account recovered weight for the five experiments (January, March, July, October, and December) and the four shellfish species. All values are given above the bars. In column 3, results representing tissue distribution following bioaccumulation are expressed as mean percent values (y axis) of recovered virus concentrations in the different tissues after 1 h (gray bar) and 24 h (back bar) (x axis) for three bioaccumulation experiments and the four shellfish species. The concentration detected after 24 h was assigned a value of 100%; actual values are noted above each bar (for mussel and clams, concentrations in DT after 1 h were higher than after 24 h, and the corresponding values are more than 100%).
For flat oysters, contamination occurred slowly, and after a bioaccumulation period of 1 h, concentrations were around 100 copies before reaching approximately 1,000 copies per g of DT after 24 h (Fig. 1B, column 1). The highest accumulation efficiency (2%) was observed in January (Fig. 1B, column 2). Comparisons of the different tissues (gills and mantle) showed that the concentrations were even lower in these organs following a 1-h bioaccumulation period (Fig. 1B, column 3).
Mussels showed markedly different results, with higher concentrations detected after 1 h than after 24 h in the October and December experiments (Fig. 1C, column 1). Comparison of the concentrations obtained in mussels after 1 h to oyster concentrations after 1 h (excluding March, for which a different behavior was observed) showed a significant difference (P < 0.01). For the five experiments, the concentrations detected in the DT were around 104 RNA copies/g and the variability between experiments was lower than 0.5 log (Table 1). After 24 h, concentrations were similar for the first 3 experiments, whereas lower concentrations were observed in October and December (Fig. 1C, column 1). It may be important that greater variability in concentrations was observed during these 2 months (October and December). As mentioned above, in December, large variations were observed between the three extractions. A higher bioaccumulation efficiency was observed after a period of 1 h compared to 24 h for four experiments, and similar efficiencies were noted for one experiment (Fig. 1C, column 2). The virus concentration in seawater following bioaccumulation was in the same range for all experiments, which suggests that faster release of HEV by mussels may not account for the lower concentrations observed following a 24-h bioaccumulation period. The production of pseudofeces or feces appeared identical to that seen with other species (visual observation), but no viral analysis was undertaken. A comparison of levels in the mantle, gills, and DT showed that, as for oysters, most of the viruses were detected in the DT. However, the concentrations after a 1-h bioaccumulation period appeared to be similar to the concentrations observed after 24 h for all tissues (Fig. 1C, column 3).
Clams displayed behavior similar to that seen with mussels, with very fast contamination and even greater concentrations than those observed for mussels. Detected concentrations were quite similar after 1 h and 24 h for all experiments except for the one conducted in March (Fig. 1D, column 1). However, this result needs to be considered with caution as the extraction procedure failed for two replicates (Table 1). For all the other extractions, the concentrations obtained were very similar within replicates. This shellfish species displayed the highest bioaccumulation efficiency, with a maximum observed in December (Fig. 1D, column 2), and viruses were predominantly located in the DT, with less than 5% detected in the gills and mantle (Fig. 1D, column 3).
Environmental investigations.
A total of 286 samples were collected, and 274 were successfully analyzed (Table 2). Indeed, most of the samples gave acceptable extraction efficiencies; however, 46 failed the acceptance criteria. After repeated extractions, all mussel samples were acceptable. Two clam samples (about 1% of samples) were still unacceptable after 3 extractions, whereas the other 14 samples that initially failed, as well as most of the cockle samples, were acceptable. The highest failure rate (9.6%) was observed for oyster samples (Table 2). Considering these results, 12 samples were excluded from the analysis, and the average overall extraction efficiency was above 13%. Similar numbers of clam, mussel, and oyster samples were collected, with cockles being less frequently collected. This species presented the lowest average extraction efficiency. Mussels represented 34% of collected shellfish and showed good extraction efficiencies (the lowest being 16%).
TABLE 2.
Extraction efficiency for environmental samples
| Shellfish | No. of samples collected | No. of samples with indicated extraction efficiency quality (%) |
No. of samples considered | Avg extraction efficiency ± SD | ||
|---|---|---|---|---|---|---|
| Acceptable (>5) | Poor (≤5) | Poor with repeated extractions (≤5)a | ||||
| Clams | 67 | 51 | 16 | 2 | 65 | 24 ± 12 |
| Cockles | 52 | 42 | 10 | 3 | 49 | 13 ± 6 |
| Mussels | 94 | 90 | 4 | 0 | 94 | 30 ± 14 |
| Oysters | 73 | 57 | 16 | 7 | 66 | 26 ± 19 |
| Total | 286 | 240 | 46 | 12 | 274 | |
Data represent samples for which repeated extractions (2 or 3) presented low extraction efficiencies and thus were not considered.
All of these samples were collected from class B production areas according to European Union regulations (more than 4,600 E. coli bacteria/100 g of shellfish flesh and intervalvular liquid). However, despite several assays for each sample having been undertaken, all samples were found to be below the limit of detection for HEV contamination. Even shellfish collected in areas that are known to be impacted by pig (site A) or wild-boar (site B) fecal contamination were below the limit of detection for HEV (Table 3). For site A, NoVs were detected in 3.6% of the samples collected (mainly in March 2013), showing a rather low human contamination impact. For site B, NoVs were detected in 8% of collected samples. At this site, only clams and oysters were growing. Considering the results from the bioaccumulation experiments, mussels were added to this site but were found to be below the limit of detection for HEV. After 1 year of monitoring these two sites, 128 additional samples were collected from 15 different locations along the French coastline, including Corsica, in order to increase the likelihood of detecting HEV-contaminated shellfish. None of these additional samples were positive for HEV, while NoVs were detected in about 13% of them (Table 3).
TABLE 3.
Results obtained considering shellfish species and site collection
| Site | Clam samples |
Cockle samples |
Mussel samples |
Oyster samples |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Avg ext. eff. ± SD (%) | No. NoV positive | No. | Avg ext. eff. ± SD (%) | No. NoV positive | No. | Avg ext. eff. ± SD (%) | No. NoV positive | No. | Avg ext. eff. ± SD (%) | No. NoV positive | |
| A | None | 28 | 14 ± 6 | 1 | 28 | 25 ± 15 | 1 | 28 | 23 ± 19 | 1 | ||
| B | 24 | 27 ± 17 | 2 | None | 13 | 37 ± 11 | 1 | 25 | 35 ± 21 | 2 | ||
| Ca | 41 | 23 ± 10 | 3 | 21 | 12 ± 5 | 5 | 53 | 32 ± 14 | 5 | 13 | 17 ± 10 | 0 |
C samples were collected from different locations all around the coasts of France.
DISCUSSION
Shellfish are known to accumulate human pathogens such as human enteric viruses as demonstrated for rotavirus in mussels (32) and enterovirus (33), hepatitis A virus (34), and norovirus (35) in oysters. We previously studied norovirus bioaccumulation and tissue distribution in oysters, comparing different levels of concentrations (27, 35).
The first aim of this study was evaluation of the capacity of four different shellfish species to accumulate HEV and analysis of tissue distribution. As hepatitis E infection in humans, unlike winter gastroenteritis due to norovirus, is not a seasonal disease, bioaccumulation experiments were performed throughout the year. Two oyster species, Crassostrea gigas and Ostrea edulis, the main shellfish that are consumed in France during winter months, as well as mussels, Mytilus edulis, which are consumed mainly during the spring and summer months, were selected. A recent environmental study demonstrated that contamination of clams was more frequent than contamination of oysters; therefore, we also considered a clam species, Ruditapes phillipinarum, in the bioaccumulation comparison experiments (36). However, our virus bioaccumulation protocol did not consider that clams normally live in contact with the marine sediment, which represents a limitation to our observations. Comparing different shellfish species of different body sizes, which display different physiological parameters, such as filtration activity, was challenging and required careful consideration during the design and analysis of the experiments. Preliminary studies were undertaken (data not shown) in which experimental conditions were adjusted to get similar maximal theoretical bioaccumulation (MTB) values in the DT, allowing comparison between the different species. Additionally, we considered the physiological status of the shellfish by measuring the allometric coefficient, which is obtained from the ratio of DT weight to body weight. This approach is commonly used to account for the physiological status of shellfish (i.e., growing or reproduction phase). In July, oysters displayed an allometric coefficient that was different from that seen in other experiments, and this may explain the low accumulation of virus. Similarly, for clams, lower allometric coefficients were observed in January and March, which were associated with the lowest concentrations of accumulated HEV. While these results suggest an association between the allometric coefficient and viral accumulation, we could not clearly demonstrate the influence of the coefficient on the viral bioaccumulation efficiency. Nonetheless, we suggest that it may be useful for future experiments to consider this parameter further, along with other physiological indicators of shellfish health such as filtration rate and fecal production. Consideration of these physiological parameters in relation to viral accumulation may facilitate and improve comparisons between shellfish species and results obtained by different laboratories in the future. The volume of seawater compared to the number of animals per aquarium reduced influences related to the filtration activity of the four shellfish species. Based on the filtration activity, the highest virus concentrations should have been observed in July. Instead, the lowest bioaccumulation efficiencies and the lowest viral concentrations were actually observed that month, confirming that the design of our experiments correctly avoided the potential bias introduced by such physiological variations between the five assays and between species. The last methodological point considered important for this work was the quantitative approach by rRT-PCR. All precautions were taken to monitor extraction efficiencies and to test for inhibitors, as routinely undertaken in our laboratory for several years (26, 35–37) and in the CEN/ISO reference method (38). To take into account errors due to virus extraction and variability in rRT-PCR, all samples were extracted three times and all rRT-PCRs were conducted in triplicate. We also introduced an acceptance criterion for standard curves, rejecting runs with values displaying a Cq difference of greater than 1 for more than one point of the standard curve. This allows a better comparison over time and avoids possible variations in quantification. Such controls are important to be fully confident in the quantitative approach by rRT-PCR and to allow us to compare results from the different shellfish species.
Our data confirm that most of the HEV accumulated in the DT of all four species considered. This organ is frequently used in the recently developed methods, suggesting that most of these methods should be able to detect HEV (38). Some differences in tissue distributions were observed among the four species. For example, HEV was still detected in oyster gill tissue after 24 h but was not detected (less than 5%) in that of the three other species. The virus concentration in the mantle was negligible compared to observed concentrations in the DT for all species except flat oysters. This species showed the lowest bioaccumulation efficiency, with a maximum of 2% of the seeded virus bioaccumulated by flat oysters in January after a 24-h period. After 1 h, only low concentrations of HEV were detected in the DT, suggesting that this shellfish species may be less sensitive to sporadic contamination events, consistent with previously reported differences between the susceptibilities of oyster species to virus contamination (39). Our results additionally suggest that clams and mussels may be more sensitive to sporadic contamination events, as after 1 h, the detected viral concentration was already high. This is similar to results we previously obtained for Moroccan shellfish which suggested that mussels and clams readily accumulate viruses (36); however, it differs from results reported for Italian shellfish (40).
The second aim of this study was to analyze shellfish potentially impacted by pig and wild-boar fecal contamination. HEVs have been demonstrated to circulate in some French pig farms and in wild boars, and some clinical cases have been described (8, 20, 22, 41, 42). HEV was also reported in sewage and river waters, confirming the virus circulation in environmental waters in some countries (7, 43–45). Considering the zoonotic potential of distribution of this virus from various animal species, environmental transmission needs to be considered (5, 8, 46, 47). Despite our effort to select contaminated areas, we failed to detect any positive samples for HEV. The bioaccumulation experiments showed that this failure was not caused by a methodological issue. The detection of norovirus contamination in some of the samples confirmed the efficiency of the methods, and the sensitivity limit of the methods for HEV, as estimated by previous bioaccumulation studies, was quite similar to that of norovirus detection (26). The primers and probe used were found to be efficient in different studies as well as for the analysis of clinical cases in France (30, 48). Several hypotheses may explain the absence of detectable HEV in environmental samples. First, pig farms in France are not located directly on the coast; these areas are primarily reserved for tourist activities and the local population. The site selected was about 1.3 km from the closest pig farm. However, the catchment basin of about 123 km2 comprised about 59 farms breeding about 100,500 animals, representing the highest density of pig farms for France. In Scotland, HEV was detected in mussel samples directly under the influence of a pig slaughterhouse (12); however, a similar situation could not be found on the French coastline. The other site was selected based on observations of wild boars refreshing themselves in the sea. The area is a wild-life protection zone, and consumption of naturally grown shellfish is forbidden. No data were available on the presence of HEV in these wild-boar populations, and the few stools that were collected when shellfish were sampled were found to be negative (data not shown). Finally, to improve the likelihood of detecting HEV-contaminated shellfish, we extended the sampling to 15 additional sites, albeit without success. While we were unable to detect HEV in shellfish from France, HEV has been detected in some European shellfish. For example, HEV was detected in Italian mussels that were being utilized as bioindicators of marine pollution (11) and in marketplace mussels in Spain (49). In Thailand, none of 213 samples analyzed were found to be positive for HEV (50), and in Korea, about 9% of 161 oyster samples were positive (14).
After verifying that the approach applied for performing the bioaccumulation experiments and the rRT-PCR assay generates reliable results, we demonstrated here that different shellfish species can bioaccumulate HEV, with significant differences in terms of uptake kinetics and concentrations. This study also demonstrated that methods targeting the DT, as developed for norovirus and hepatitis A virus, are appropriate for HEV, which is also predominantly detected in these organs. Finally, despite evidence that HEV is circulating in the French population and in pig farms, we could not detect any HEV-contaminated shellfish, suggesting that the number of HEV particles released into the environment may be low or may be under the detection limit of our method. Further work on HEV persistence in pig manure and human waste may be warranted to further evaluate this point.
ACKNOWLEDGMENTS
This work was supported by grant 2010 CESA 010 03 (HEVECoDyn) from the Agence Nationale pour la Recherche (ANR).
We are grateful to Nicole Pavio (ANSES, Paris) and Albert Bosch (University of Barcelona, Spain) for providing strains and plasmid and to Joel Haure (Ifremer, SG2M) for helpful advice on shellfish physiology. We thank colleagues from Ifremer LER N (Port en Bessin), BO (Concarneau), MPL (La Trinité sur Mer), LR (Sète), and PAC (Bastia) for their help with sampling.
Footnotes
Published ahead of print 2 May 2014
REFERENCES
- 1.Meng XJ. 2010. Hepatitis E virus: animal reservoirs and zoonotic risk. Vet. Microbiol. 140:256–265. 10.1016/j.vetmic.2009.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith DB, Purdy MA, Simmonds P. 2013. Genetic variability and the classification of hepatitis E virus. J. Virol. 87:4161–4169. 10.1128/JVI.02762-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng XJ, Anderson DA, Arankalle VA, Emerson SU, Harrison TJ, Jameel S, Okamoto H. 2012. Hepeviridae, p 1021–1028 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy: ninth report of the international committee on taxonomy of viruses. Academic Press, London, United Kingdom [Google Scholar]
- 4.Yugo DM, Meng X-J. 2013. Hepatitis E virus: foodborne, waterborne and zoonotic transmission. Int. J. Environ. Res. Public Health 10:4507–4533. 10.3390/ijerph10104507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van der Poel W. 2014. Food and environmental routes of hepatitis E virus transmission. Curr. Opin. Virol. 4:91–96. 10.1016/j.coviro.2014.01.006 [DOI] [PubMed] [Google Scholar]
- 6.Wilhelm BJ, Rajić A, Greig J, Waddell L, Trottier G, Houde A, Harris J, Borden LN, Price C. 2011. A systematic review/meta-analysis of primary research investigating swine, pork or pork products as a source ofzoonotic hepatitis E virus. Epidemiol. Infect. 139:1127–1144. 10.1017/S0950268811000677 [DOI] [PubMed] [Google Scholar]
- 7.Masclaux FG, Hotz P, Friedli D, Savova-Bianchi D, Oppliger A. 2013. High occurrence of hepatitis E virus in samples from wastewater treatment plants in Switzerland and comparison with other enteric viruses. Water Res. 47:5101–5109. 10.1016/j.watres.2013.05.050 [DOI] [PubMed] [Google Scholar]
- 8.Bouquet J, Tessé S, Lunazzi A, Eloit M, Rose N, Nicand E, Pavio N. 2011. Close similarity between sequences of hepatitis E virus recovered from humans and swine, France, 2008–2009. Emerg. Infect. Dis. 17:2018–2025. 10.3201/eid1711.110616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Guyader FS, Miura T, Atmar RL. 2013. Prevalence and control of norovirus and hepatitis A virus in shellfish, p 137–168 In Foodborne viruses and prions and their significance for public health, vol 6 Food safety assurance and veterinary public health Wageningen Academic Publishers, Wageningen, The Netherlands [Google Scholar]
- 10.EFSA. 2011. Scientific opinion on an update on the present knowledge on the occurrence and control of foodborne viruses. EFSA J. 9:2190. 10.2903/j.efsa.2011.2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donia D, Dell'Amico MC, Petrinca AR, Martinucci I, Mazzei M, Tolari F, Divizia M. 2012. Presence of hepatitis E RNA in mussels used as bio-monitors of viral marine pollution. J. Virol. Methods 186:198–202. 10.1016/j.jviromet.2012.06.007 [DOI] [PubMed] [Google Scholar]
- 12.Crossan C, Baker PJ, Craft J, Takeuchi Y, Dalton HR, Scobie L. 2012. Hepatitis E virus genotype 3 in shellfish, United Kingdom. Emerg. Infect. Dis. 18:2085–2087. 10.3201/eid1812.120924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li TC, Miyamura T, Takeda N. 2007. Detection of hepatitis E virus RNA from the bivalve Yamato-Shijimi (Corbicula japonica) in Japan. Am. J. Trop. Med. Hyg. 76:170–172 [PubMed] [Google Scholar]
- 14.Song Y-J, Jeong H-J, Kim Y-J, Lee SW, Park S-Y, Song C-S, Park HM, Choi I-S. 2010. Analysis of complete genome sequences of swine hepatitis E virus and possible risk factors for transmission of HEV to humans in Korea. J. Med. Virol. 82:583–591. 10.1002/jmv.21730 [DOI] [PubMed] [Google Scholar]
- 15.Arankalle VA, Chada MS, Tsarev SA, Emerson SU, Risbud AR, Banerjee K, Purcell RH. 1994. Seroepidemiology of water-borne hepatitis in India and evidence for a third enterically-transmitted hepatitis agent. Proc. Natl. Acad. Sci. U. S. A. 91:3428–3432. 10.1073/pnas.91.8.3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torné J, Miralles R, Tomás S, Saballs P. 1988. Typhoid fever and acute non A non B hepatitis after shellfish consumption. Eur. J. Clin. Microbiol. Infect. Dis. 7:581–582. 10.1007/BF01962622 [DOI] [PubMed] [Google Scholar]
- 17.Koizumi Y, Isoda N, Sato Y, Iwaki T, Ono K, Ido K, Sugano K, Takahashi M, Nishoizawa T, Okamoto H. 2004. Infection of a Japanese patient by genotype 4 hepatitis E virus while traveling in Vietnam. J. Clin. Microbiol. 42:3883–3885. 10.1128/JCM.42.8.3883-3885.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Said B, Ijaz S, Kafatos G, Booth L, Thomas L, Walsh A, Ramssay M, Morgan D. 2009. Hepatitis E outbreak on cruise ship. Emerg. Infect. Dis. 15:1738–1743. 10.3201/eid1511.091094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chau TN, Lai ST, Tse C, Ng TK, Leung VKS, Lim W, Ng MH. 2006. Epidemiology and clinical features of sporadic hepatitis E as compared with hepatitis A. Am. J. Gastroenterol. 101:292–296. 10.1111/j.1572-0241.2006.00416.x [DOI] [PubMed] [Google Scholar]
- 20.Colson P, Romanet P, Moal V, Borentain P, Purgus R, Benezech A, Motte A, Gérolami R. 2012. Autochthonous infections with hepatitis E virus genotype 4, France. Emerg. Infect. Dis. 18:1361–1364. 10.3201/eid1808.111827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renou C, Pariente A, Cadranel J-F, Nicand E, Pavio N. 2011. Clinically silent forms may partly explain the rarity of acute cases of autochthonous genotype 3c hepatitis E infection in France. J. Clin. Virol. 51:139–141. 10.1016/j.jcv.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 22.Abravanel F, Sandres-Saune K, Lhomme S, Dubois M, Mansuy JM, Izopet J. 2012. Genotype 3 diversity and quantification of hepatitis E virus RNA. J. Clin. Microbiol. 50:897–902. 10.1128/JCM.05942-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berto A, Grierson S, Hakze-van der Honing R, Martelli F, Johne R, Reetz J, Ulrich RG, Pavio N, van der Poel WHM, Banks M. 2013. Hepatitis E virus in pork liver sausage, France. Emerg. Infect. Dis. 19:264–266. 10.3201/eid1902.121255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walachowski S, Dorenlor V, Lefevre J, Lunazzi A, Eono F, Merbah T, Eveno E, Pavio N, Rose N. 28 November 2013. Risk factors associated with the presence of hepatitis E virus in livers and seroprevalence in slaughter-age pigs: a retrospective study of 90 swine farms in France. Epidemiol. Infect. 10.1017/S0950268813003063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin LR, Duke GM, Osorio JE, Hall DJ, Palmenberg AC. 1996. Mutational analysis of the mengovirus poly(C) tract and surrounding heteropolymeric sequences. J. Virol. 70:2027–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Guyader FS, Parnaudeau S, Schaeffer J, Bosch A, Loisy F, Pommepuy M, Atmar RL. 2009. Detection and quantification of noroviruses in shellfish. Appl. Environ. Microbiol. 75:618–624. 10.1128/AEM.01507-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maalouf H, Schaeffer J, Parnaudeau S, Le Pendu J, Atmar RL, Crawford SE, Le Guyader FS. 2011. Strain-dependent norovirus bioaccumulation in oysters. Appl. Environ. Microbiol. 77:3189–3196. 10.1128/AEM.03010-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atmar RL, Neill FH, Romalde JL, Le Guyader F, Woodley CM, Metcalf TG, Estes MK. 1995. Detection of Norwalk virus and hepatitis A virus in shellfish tissues with the PCR. Appl. Environ. Microbiol. 61:3014–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miura T, Parnaudeau S, Grodzki M, Okabe S, Atmar RL, Le Guyader FS. 2013. Environmental detection of genogroup I, II and IV noroviruses by using a generic real-time reverse transcription-PCR assay. Appl. Environ. Microbiol. 79:6585–6592. 10.1128/AEM.02112-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jothikumar N, Cromeans TL, Robertson BH, Meng XJ, Hill VR. 2006. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods 131:65–71. 10.1016/j.jviromet.2005.07.004 [DOI] [PubMed] [Google Scholar]
- 31.Bedford AJ, Williams G, Bellamy AR. 1978. Virus accumulation by the rock oyster Crassotrea glomerata. Appl. Environ. Microbiol. 35:1012–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bosch A, Pinto RM, Abad FX. 1995. Differential accumulation and depuration of human enteric viruses by mussels. Water Sci. Tech. 31:447–451. 10.1016/0273-1223(95)00310-J [DOI] [Google Scholar]
- 33.McLeod C, Hay B, Grant C, Greening G, Day D. 2009. Localization of norovirus and poliovirus in Pacific oysters. J. Appl. Microbiol. 106:1220–1230. 10.1111/j.1365-2672.2008.04091.x [DOI] [PubMed] [Google Scholar]
- 34.Kingsley DH, Richards GP. 2003. Persistence of hepatitis A virus in oysters. J. Food Prot. 66:331–334 [DOI] [PubMed] [Google Scholar]
- 35.Maalouf H, Zakhour M, Le Pendu J, Le Saux J-C, Atmar RL, Le Guyader FS. 2010. Norovirus genogroup I and II ligands in oysters: tissue distribution and seasonal variations. Appl. Environ. Microbiol. 76:5621–5630. 10.1128/AEM.00148-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benabbes L, Ollivier J, Schaeffer J, Parnaudeau S, Rhaissi H, Nourlil J, Le Guyader FS. 2013. Norovirus and other human enteric viruses in Moroccan shellfish. Food Environ. Virol. 5:35–40. 10.1007/s12560-012-9095-8 [DOI] [PubMed] [Google Scholar]
- 37.Sima LC, Schaeffer J, Le Saux J-C, Parnaudeau S, Elimelech M, Le Guyader FS. 2011. Calicivirus removal in a membrane bioreactor wastewater treatment plant. Appl. Environ. Microbiol. 77:5170–5177. 10.1128/AEM.00583-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.International Organization for Standardization. 2013. Microbiology of food and animal feed—horizontal method for determination of hepatitis A virus and norovirus in food using real-time RT-PCR, part 1: method for quantification. ISO/TS 15216-1. International Organization for Standardization, Geneva, Switzerland [Google Scholar]
- 39.Nappier SP, Graczyk TK, Schwab KJ. 2008. Bioaccumulation, retention, and depuration of enteric viruses by Crassostrea virginica and Crassostrea ariakensis oysters. Appl. Environ. Microbiol. 74:6825–6831. 10.1128/AEM.01000-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suffredini E, Magnabosco C, Civettini M, Rosseti E, Arcangeli G, Croci L. 2012. Norovirus contamination in different shellfish species harvested in the same production areas. J. Appl. Microbiol. 113:686–692. 10.1111/j.1365-2672.2012.05356.x [DOI] [PubMed] [Google Scholar]
- 41.Rose N, Lunazzi A, Dorenlor V, Merbah T, Eono F, Eloit M, Madec F, Pavio N. 2011. High prevalence of hepatitis E virus in French domestic pigs. Comp. Immunol. Microbiol. Infect. Dis. 34:419–427. 10.1016/j.cimid.2011.07.003 [DOI] [PubMed] [Google Scholar]
- 42.Kaba M, Davoust B, Marié J-L, Colson P. 2010. Detection of hepatitis E virus in wild boar (Sus scrofa) livers. Vet. J. 186:259–261. 10.1016/j.tvjl.2009.08.008 [DOI] [PubMed] [Google Scholar]
- 43.Kitajima M, Matsubara K, Sour S, Haramoto E, Katayama H, Ohgaki S. 2009. First detection of genotype 3 hepatitis E virus RNA in river water in Cambodia. Trans. R. Soc. Trop. Med. Hyg. 103:955–957. 10.1016/j.trstmh.2009.04.004 [DOI] [PubMed] [Google Scholar]
- 44.La Rosa G, Pourshaban M, Iaconelli M, Vennarucci VS, Muscillo M. 2010. Molecular detection of hepatitis E virus in sewage samples. Appl. Environ. Microbiol. 76:5870–5873. 10.1128/AEM.00336-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hazam RK, Singla R, Kishore J, Singh S, Gupta RK, Kar P. 2010. Surveillance of hepatitis E virus in sewage and drinking water in a resettlement colony of Delhi: what has been the experience? Arch. Virol. 155:1227–1233. 10.1007/s00705-010-0707-z [DOI] [PubMed] [Google Scholar]
- 46.Dell'Amico MC, Cavallo A, Gonzales JL, Bonelli SI, Valda Y, Pieri A, Segundo H, Ibanez R, Mantella A, Bartalesi F, Tolari F, Bartoloni A. 2011. Hepatitis E virus genotype 3 in humans and swine, Bolivia. Emerg. Infect. Dis. 17:1488–1490. 10.3201/eid1708.100769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Izopet J, Dubois M, Bertagnoli S, L'homme S, Marchandeau S, Boucher S, Kamar N, Abravanel F, Guerin JL. 2012. Hepatitis E virus strains in rabbits and evidence of a closely related strain in humans, France. Emerg. Infect. Dis. 18:1274–1281. 10.3201/eid1808.120057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abravanel F, Chapuy-Regaud S, Lhomme S, Dubois M, Peron JM, Alric L, Rostaing L, Kamar N, Izopet J. 2013. Performance of two commercial assays for detecting hepatitis E virus RNA in acute or chronic infections. J. Clin. Microbiol. 51:1913–1916. 10.1128/JCM.00661-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diez-Valcarce M, Kokkinos P, Soderberg K, Bouwknegt M, Willems K, de Roda-Husman A-M, von Bonsdorff C-H, Bellou M, Hernandez M, Maunula L, Vantarakis A, Rodriguez-Lazaro D. 2012. Occurrence of human enteric viruses in commercial mussels at retail level in three European countries. Food Environ. Virol. 4:73–80. 10.1007/s12560-012-9078-9 [DOI] [PubMed] [Google Scholar]
- 50.Namsai A, Louisirirotchanakul S, Wongchinda N, Siripanyaphinyo U, Virulhakul P, Puthavanatha P, Myint KS, Gannarong M, Ittapong R. 2011. Surveillance of hepatitis A and E viruses contamination in shellfish in Thailand. Lett. Appl. Microbiol. 53:608–613. 10.1111/j.1472-765X.2011.03152.x [DOI] [PubMed] [Google Scholar]