Abstract
The ability to maintain a dual lifestyle of colonizing the ruminant gut and surviving in nonhost environments once shed is key to the success of Escherichia coli O157:H7 as a zoonotic pathogen. Both physical and biological conditions encountered by the bacteria are likely to change during the transition between host and nonhost environments. In this study, carbon starvation at suboptimal temperatures in nonhost environments was simulated by starving a New Zealand bovine E. coli O157:H7 isolate in phosphate-buffered saline at 4 and 15°C for 84 days. Recovery of starved cells on media with different nutrient availabilities was monitored under aerobic and anaerobic conditions. We found that the New Zealand bovine E. coli O157:H7 isolate was able to maintain membrane integrity and viability over 84 days and that the level of recovery depended on the nutrient level of the recovery medium as well as the starvation temperature. In addition, a significant difference in carbon utilization was observed between starved and nonstarved cells.
INTRODUCTION
Ruminants have been identified as the major animal reservoir of Escherichia coli O157:H7, with cattle being one of the main sources of human infection (1–3). Carriage of E. coli O157:H7 in calves and adult cattle is largely asymptomatic (4, 5); however, infection in humans can result in the development of hemolytic-uremic syndrome (HUS), characterized by microangiopathic hemolytic anemia, uremia, and thrombocytopenia (6, 7). The major virulence factors responsible for the most severe clinical presentations are the Shiga toxins (Stx1 and -2). Of the two Shiga toxins, Stx2 was found to be at least 400 times more toxic to mice than Stx1 (8). Although a clinically favorable prognosis may be achieved with supportive treatments applied during the acute phase of infection, the risk of long-term sequelae remains. Postinfection complications include hypertension, renal complications, and neurological defects, particularly in children in whom primary infection occurred under the age of five (9). In New Zealand, reported E. coli O157:H7 infections are sporadic, with no recorded outbreaks, and infections are predominantly linked to environmental contaminations from animal sources or familial transmission (10). In addition, stx2 is more prevalent than stx1 in New Zealand strains isolated from bovine, human, and water samples (11).
The survival and persistence of E. coli O157:H7 in a variety of environments outside an animal host have been documented, including animal hides, soil, and water, all of which pose a significant risk for transmission among cattle and to humans (12–18). Persistent subtypes of E. coli O157:H7 were found in fecal samples from feedlot cattle despite turnover of populations (19). Studies showed that E. coli O157:H7 can survive on cattle hides for up to 9 days (12), in soil for up to 105 days (17), and in water troughs for up to 245 days (20).
Low temperature and limited nutrient availability in aqueous environments can induce bacteria, including E. coli O157:H7, to enter into a viable but nonculturable (VBNC) state (21, 22). Entry into the VBNC state is characterized by low metabolic activity and decreasing recovery on standard solid culturing medium over the course of starvation (23). Detection and monitoring of food-borne pathogens, such as E. coli O157:H7, primarily depend on enumeration of colonies on agar (24). It is likely that underestimation of VBNC E. coli O157:H7 in food products may result in increased risk of transmission and human exposure.
Transitions from the host to the external environment and vice versa involve physical changes in temperature as well as changes in the level of available nutrients. To survive these changes and thrive in both environments, physiological and metabolic adjustments are required. In this study, the metabolic profiles of three New Zealand bovine E. coli O157:H7 strains isolated from young calves were compared. The growth and metabolic capabilities of a New Zealand bovine E. coli O157:H7 isolate following starvation were investigated; colony formation and changes in carbon utilization kinetics under aerobic and anaerobic growth conditions were determined during starvation.
MATERIALS AND METHODS
Bacterial strains, culture, and starvation conditions.
Three bovine Escherichia coli O157:H7 strains isolated from calf hides, N427, N635, and H11 (Meat Industry Research New Zealand culture collection), and NCTC12900 (ATCC 700728; NZRM 3614) were used in this study. Stationary-phase cultures were obtained by culturing cells in Luria-Bertani broth (LB; 5% yeast extract, 5% sodium chloride, and 10% tryptone) at 37°C without agitation for 24 h. Late-exponential-phase cultures were obtained by culturing cells in LB at 37°C with shaking at 150 rpm for 4 h to an optical density at 600 nm (OD600) of 0.7. For starvation assays, late-exponential-phase cultures were divided into two equal portions to form biological replicates. Cells were harvested, washed three times, and resuspended in phosphate-buffered saline (PBS) at pH 6.8 (Lorne Laboratory, United Kingdom). Each cell suspension was further divided into two equal portions, one maintained at 4°C and the other at 15°C for up to 84 days.
Determination of cell viability.
In this study, cells with intact membranes were deemed viable. Bacterial suspensions were stained with a LIVE/DEAD BacLight bacterial viability kit (Invitrogen Life Technologies New Zealand Ltd.) according to the manufacturer's instructions. Where necessary, bacterial suspensions were concentrated by pelleting cells using centrifugation (4,000 rpm for 10 min) and resuspending into smaller volumes. Stained cells were loaded into a Helber counting chamber with Thoma ruling (Hawksley, Sussex, United Kingdom) and photographed (Olympus BX60; Olympus America Inc.) using a Nikon DS-5Mc camera. The same field was photographed using both phase contrast and fluorescence with excitation wavelengths between 450 and 480 nm (U-MWIB filter; Olympus America Inc.). Two slides were prepared for each sample, and a total of 16 squares were photographed per slide. Image analysis and cell counting were performed using Adobe Photoshop CS3. The number of viable cells per ml was calculated as the number of green-fluorescent cells counted in the photographs. Statistical comparisons of viable cell counts were conducted using Student's t test assuming two-tailed distribution with unequal variance. P values less than 0.01 were considered significant.
Determination of cell culturability.
Culturability was determined as the ability of cells to form colonies on tryptic soy agar (TSA; 1.5% casein peptone, 0.5% soy peptone, 0.5% sodium chloride, and 1.5% agar) and plate count agar (PCA; 0.5% peptone, 0.25% yeast extract, 0.1% glucose, and 1.5% agar). Cultures were diluted in PBS and plated to achieve counts of between 30 and 300 colonies per plate. Plates were incubated aerobically unless otherwise stated. An anaerobic atmosphere of 5% H2, 10% CO2, and 85% N2 was used to test for anaerobic growth. To allow comparison of colony development, all plates were incubated at 37°C for precisely 24 h. Plates were photographed using Alliance 4.7 (Uvitech, Cambridge, United Kingdom). Image analysis was performed using Adobe Photoshop CS3 to measure the horizontal and vertical width and circularity of colonies. Threshold values were used to separate colonies from the image background. Data were analyzed using Microsoft Excel. Single colonies were distinguished from clusters of multiple colonies by excluding measurements with circularities less than 0.8. The smaller of the horizontal and vertical width measurements was used as the measure of colony diameter to prevent inclusion of doublet colonies which appeared circular. Statistical comparisons of cell culturability were conducted using Student's t test assuming a two-tailed distribution with unequal variance. P values less than 0.01 were considered significant. The distribution of colony diameter was graphed in R (version 3.0.2), a free software environment for statistical computing and graphics, using the beanplot package developed by Kampstra (25). Briefly, the colony size data were imported into R, where data were sorted by the type of medium. Vertical histograms of day 0 and day 84 distributions, as well as the mean of the distributions, were shown on each side of the bean.
Utilization of carbon sources.
Ninety-six-well carbon source phenotypic microarrays (PM1 MicroPlate, Biolog Inc.) were used to determine carbon utilization by E. coli O157:H7. Metabolic activity in each well was detected by the reduction of tetrazolium violet to formazan and was measured by monitoring the change in color. The lag time, maximum rate of utilization, and maximum level of utilization after 24 h were measured.
Cultures were adjusted to an OD600 of 0.07 in inoculating fluid (IF1-0a, Biolog Inc.) containing 1× redox dye mix A (Biolog Inc.). One hundred microliters of the resulting cell suspension was dispensed into each well of the Biolog PM1 MicroPlate (Biolog Inc.). Plates were incubated at 37°C for 24 h in a VersaMax ELISA microplate reader (Molecular Devices). Optical density measurements were taken every 15 min at both 590 nm (color development plus turbidity; A590) and 750 nm (turbidity; A750). The reaction within each well (W) was determined using equation 1 (26). The reaction in each well containing a substrate was corrected to that of the negative control, thus determining the level of metabolic activity (Aactivity) (equation 2).
| (1) |
| (2) |
Negative Aactivity values were arbitrarily set to zero. The Aactivity measured at the end of 24 hours of incubation was considered the endpoint (Aendpoint) of substrate utilization. Aendpoint from duplicate experiments were used to calculate the least significant difference with 95% confidence (LSD95). Where the average Aendpoint difference between a substrate and the negative control is greater than the LSD95, the substrate is considered to be utilized. Where comparisons between strains or conditions were made, average Aendpoint differences between strains or conditions greater than the LSD95 were considered significant. Aendpoint values for the 95 substrates utilized by the three strains were used to generate a dendrogram comparison of metabolic profiles in R using the pheatmap package developed by Kolde (27). The Euclidean distance matrix was used in clustering data for dendrogram generation.
The lag time was measured as the time at which Aactivity became higher than the Aactivity at time zero by an arbitrary threshold of 0.01. To determine the rate of substrate utilization, the maximum slope from the kinetic curves was measured using the grofit package developed for R (28). The fit of the slopes, as determined by the logistic, Gompertz, modified Gompertz, and Richards laws in grofit, was visually inspected for each curve, and manual measurements of slopes were performed where necessary.
RESULTS
Metabolic variations between bovine isolates.
The level of utilization of 95 carbon sources after 24 h (Aendpoint) was measured for each strain. The averaged Aendpoint utilization of 49 substrates significantly differed (P < 0.05) between nonstarved N427, N635, and H11 (Fig. 1; also, see Table S1 in the supplemental material). Twenty-two of the 49 substrates used differently were carbohydrates, 16 were carboxylic acids, seven were peptides, and four were nucleic acids (Fig. 1). The lag time and rate of carbon utilization were compared. Similar distribution patterns were observed on the scatter plots of the rate of utilization against lag time for the three strains (Fig. 2). Utilization of most substrates lagged for 1 to 3 h, with a rate of utilization (Aactivity) between 0.03 and 1.60 h−1 (Fig. 2; also, see Table S2 in the supplemental material). While the kinetics of substrate utilization differed between strains, a similar range of substrates were used by all strains tested. Overall, many metabolic similarities were observed between the stationary-phase cells of the three bovine strains, particularly between N635 and H11.
FIG 1.
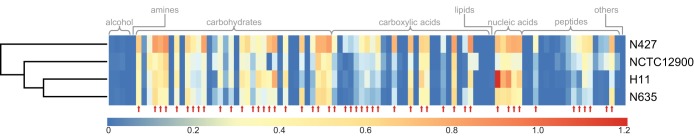
Clustering of the Aendpoint profiles of the E. coli O157:H7 strains used in this study. A hierarchical clustering analysis of the Euclidian distances of averaged Aendpoint values for 95 substrates used by stationary-phase cells was generated using the pheatmap package in R. The levels of Aendpoint are color coded from blue to red, representing lowest to highest substrate utilization after 24 h incubation at 37°C. Red arrows indicate substrates for which a significant difference (P < 0.05) in Aendpoint was observed between N247, N635, and H11.
FIG 2.
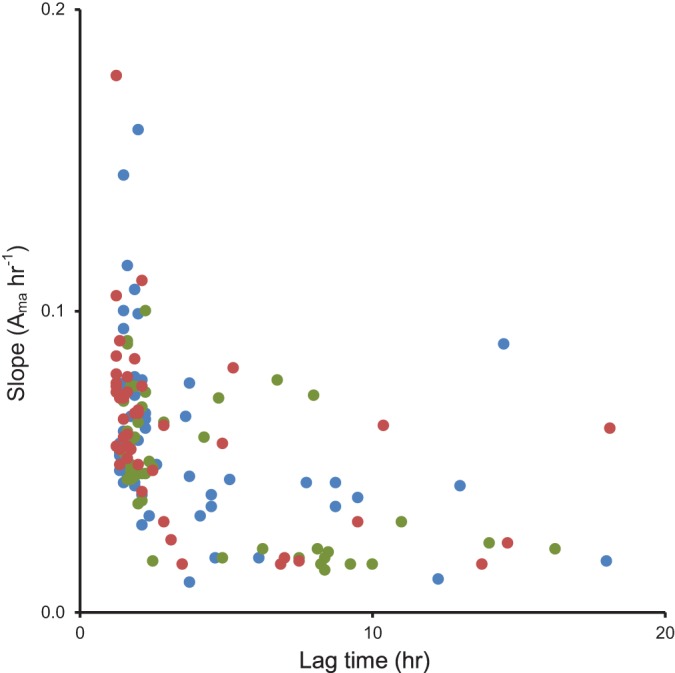
Comparison of the patterns in lag and rate of substrate utilization between N427, N635, and H11. A scatter plot of the rate of substrate utilization against the lag time of substrates utilized by stationary-phase N427 (blue), N635 (green), and H11 (red) is shown.
Survival and recovery of starved cells.
For H11, the model strain used in this study, no significant change in the number of viable cells, determined by quantification of cells with intact membranes, was detected over 84 days for cells starved at 4 or 15°C (Fig. 3). When starved at 4°C, recovery of the cells after 84 days of starvation on TSA was 6.9 log units, significantly higher (P < 0.01) than the 5.2 log units recovered on PCA (Fig. 3A). However, no significant difference in recovery on PCA and TSA was observed from cells maintained at 15°C, with 7.1 and 7.2 log CFU ml−1 recovered on PCA and TSA, respectively, after 84 days of starvation (Fig. 3B).
FIG 3.
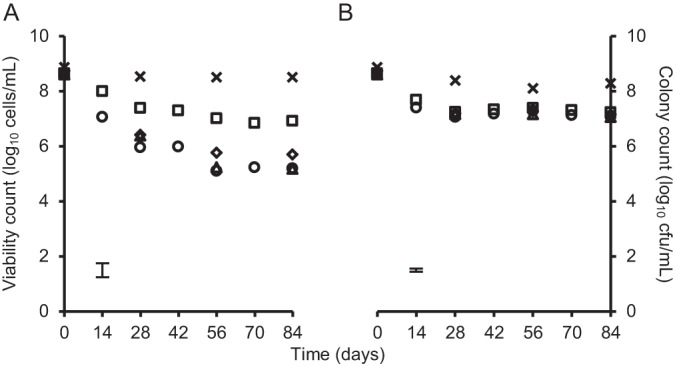
Viability and colony counts of E. coli O157:H7 H11 during nutritional starvation at 4 and 15°C. Viability count (crosses) and colony count on PCA (circles), PCA with 0.02% sodium pyruvate (diamonds), PCA with 0.02% sodium acetate (triangles), and TSA (squares) cells starved at 4°C (A) and 15°C (B) are shown. Each data point of the viability count represents the average for two independently prepared counting chamber slides; 16 squares were photographed and counted per slide. Data points of the colony counts are the averages for four technical repeats. The maximum standard deviations of all data points are indicated by the error bars.
The role of two specific nutrients, sodium pyruvate and sodium acetate, known to influence recovery of starved cells, was investigated. Supplementing PCA with 0.2% (wt/vol) sodium acetate had no significant effect on the number of colonies recovered from cells starved at 4 or 15°C (Fig. 3). Although recovery of cells starved at 15°C on PCA was not affected by supplementation with 0.2% (wt/vol) sodium pyruvate (Fig. 3B), recovery of those starved at 4°C was significantly higher (P < 0.01) on pyruvate-supplemented PCA than on PCA alone (Fig. 3A).
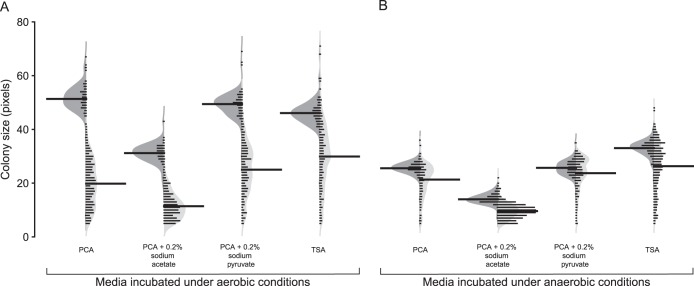
In addition to the observed decrease in the number of colonies recovered after 84 days of starvation at 4°C on all recovery media, changes in colony size were also observed. By standardizing the duration of incubation as well as the method of photography, variations in colony diameter on different recovery media were detected. No significant change in colony diameter was observed for cells starved at 15°C (data not shown), but for cells starved at 4°C, a gradual decrease in colony diameter over the starvation period was observed.
Unimodal distributions in diameter were observed from colonies recovered on day 0 (Fig. 4). The mean diameters of colonies recovered on PCA, TSA, and sodium pyruvate-supplemented PCA were similar on day 0 (51, 49, and 46 pixels, respectively), while the mean diameter of colonies on PCA supplemented with sodium acetate was 31 pixels, significantly lower (P < 0.05) than on other media (Fig. 4A). After 84 days of starvation at 4°C, colony diameter on all four media was significantly (P < 0.05) smaller than that on day 0 (Fig. 4A). The greatest decrease was observed for colonies recovered on PCA with sodium acetate, where the mean colony diameter decreased by 63% over 84 days. For PCA, PCA with sodium pyruvate, and TSA, the decreases in mean colony diameter were 61, 49, and 35%, respectively, over 84 days. In addition to the overall decrease in mean colony diameter over the duration of 4°C starvation, greater variance in the distribution of colony diameter was observed for colonies recovered on PCA, TSA, and PCA with pyruvate (Fig. 4A). No significant change in variance was observed for colonies recovered on PCA with sodium acetate between day 0 and day 84 cultures (Fig. 4A).
FIG 4.
Distribution of colony diameters of cultures maintained at 4°C grown on PCA, PCA with 0.2% sodium acetate, PCA with 0.2% sodium pyruvate, and TSA. Beanplot visualizations of the distribution of colony diameters measured from day 0 (dark gray) and day 84 (light gray) cultures recovered under aerobic (A) and anaerobic (B) conditions are shown. The bean lines for each half of a bean are vertical histograms of the data distribution; the thicker, longer bean lines indicate the mean of each distribution. The shape of each half of the asymmetric bean represents the Gaussian density estimation of the distribution. Data presented are from combined colony measurements of biological duplicate cultures.
Effect of anaerobic conditions on the recovery of starved cells.
E. coli is a facultative anaerobe. To test the hypothesis that the growth phenotype exhibited by starved cells during anaerobic metabolism differs from that observed under aerobic conditions, recovery of starved and nonstarved cells was compared under anaerobic conditions. On day 0, prior to starvation, no significant difference in the number of colonies recovered from aerobic and anaerobic incubations was observed (Table 1). Under anaerobic growth conditions, colony count on PCA decreased by 3 log units over 84 days of starvation at 4°C. This was similar to the decrease observed for aerobic recovery. Likewise, no significant difference was observed between recovery of day 84 cells under aerobic and anaerobic growth conditions on acetate supplemented PCA or TSA. However, recovery of day 84 cells on pyruvate-supplemented PCA was significantly higher (P < 0.01) after anaerobic growth than aerobic growth.
TABLE 1.
Colony count of E. coli O157:H7 H11 starved at 4°C recovered under aerobic and anaerobic conditions
| Day | Colony count (log CFU ml−1)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| PCA |
PCA + sodium acetate |
PCA + sodium pyruvate |
TSA |
|||||
| Ae | An | Ae | An | Ae | An | Ae | An | |
| 0 | 8.6 | 8.6 | 8.6 | 8.6 | 8.7 | 8.7 | 8.6 | 8.7 |
| 84 | 5.2 | 5.6 | 5.2 | 5.4 | 5.7 | 6.8b | 7.0 | 7.2 |
Ae, aerobic incubation; An, anaerobic incubation.
Colony count of anaerobically incubated cells significantly (P < 0.01) higher than those under aerobic incubation.
Both the average and distribution of colony diameter differed significantly (P < 0.01) between aerobically and anaerobically grown colonies regardless of the medium used (Fig. 4). On day 0, the average diameter was consistently lower for colonies grown under anaerobic conditions than for aerobically grown colonies (Fig. 4). Under anaerobic growth conditions, the average colony diameters of day 0 cells recovered on PCA and PCA with pyruvate were the same. However, the average colony diameter on PCA with sodium acetate was significantly lower than that on PCA, while the average colony diameter on TSA was significantly higher (Fig. 4B). The average diameter of aerobically grown colonies decreased by 35 to 63% over 84 days of starvation at 4°C; the reduction was markedly less for anaerobically recovered colonies, where a decrease from 8 to 32% was observed (Fig. 4).
Carbon utilization by starved cells during recovery.
While nutrient levels had no effect on the recovery of cells starved at 15°C, high nutrient levels and supplementation of PCA with pyruvate significantly improved the recovery of cells starved at 4°C. To examine the metabolic versatility of starved H11, cells starved at 4°C for 38 and 42 days were used to inoculate PM1 MicroPlate. Where the average lag time or rate of substrate utilization was similar for both stationary-phase and starved cultures, data points on the scatter plots lie close to the r = 1 line within LSD95 boundaries (Fig. 5). Data points on the left of the LSD95 boundaries indicate longer lag times or higher utilization rates for the starved cells than for the stationary-phase cells, and vice versa for data points on the right of the LSD95 boundaries.
FIG 5.
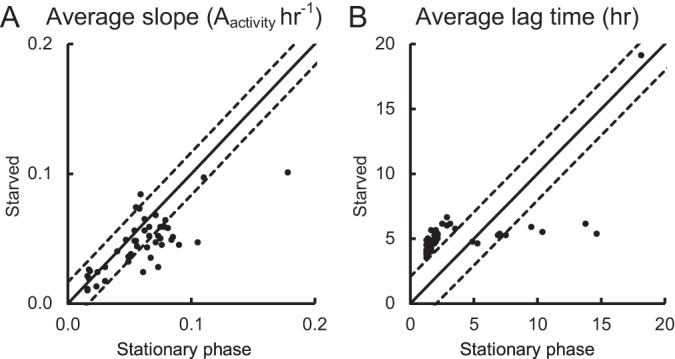
Substrate utilization kinetics for H11 stationary-phase and 4°C-starved cultures. Scatter graph comparisons of average rate of substrate utilization (A) and lag time (B) of stationary-phase and starved cultures are shown. Data points are the averages for two independent replicates. Solid lines mark the expected location of data points, assuming equal utilization kinetics. Dotted lines mark the LSD95 boundaries.
Under aerobic growth conditions, endpoint utilization for most substrates was similar between the stationary-phase and starved cells (Table 2). Of the 95 substrates tested, 15 were used differently by the two cultures (Table 2). No significant difference in the rate of substrate utilization was observed between the stationary-phase and starved cells for the majority of substrates tested (Fig. 5A). The lag time for most substrates was significantly longer for the starved cells than the stationary-phase cells (Fig. 5B), except for uridine, l-malic acid, d-malic acid, d,l-malic acid, fumaric acid, l-alanine, d-alanine, and sucrose (see Table S2 in the supplemental material).
TABLE 2.
Comparison of Aendpoint observed after 24 h of aerobic or anaerobic incubation between H11 cells harvested during stationary phase (SP) and after starvation at 4°C
| Chemical class | Substrate |
Aendpointa |
|||
|---|---|---|---|---|---|
| Aerobic |
Anaerobic |
||||
| SP | Starved | SP | Starved | ||
| Ethylene glycols | Tween 20 | — | — | 0.510 | 0.176b |
| Tween 40 | — | — | 0.113 | 0.093 | |
| Tween 80 | — | — | 0.352 | 0.130b | |
| Glycosides | α-Methyl-d-glucosidec | 0.620 | 0.563 | 0.187 | 0.157 |
| β-Methyl-d-glucoside | — | — | 0.141 | —b | |
| Monosaccharides | d-Fructose | 0.484 | 0.460 | 0.278 | 0.188b |
| d-Galactose | 0.721 | 0.554b | 0.134 | —b | |
| d-Galacturonic acidc | 0.674 | 0.716 | 0.312 | 0.274 | |
| d-Gluconic acid | 0.754 | 0.619 | 0.402 | 0.212b | |
| d-Mannose | 0.673 | 0.504b | 0.264 | 0.122b | |
| d-Ribose | 0.555 | 0.597 | 0.191 | —b | |
| d-Xylose | 0.357 | 0.484 | 0.230 | —b | |
| l-Arabinose | 0.335 | 0.375 | 0.204 | 0.096b | |
| l-Fucosec | 0.486 | 0.508 | 0.205 | 0.134 | |
| N-Acetyl-d-glucosamine | 0.598 | 0.452 | 0.411 | 0.172b | |
| N-Acetyl-β-d-mannosamine | — | 0.082 | 0.091 | —b | |
| α-d-Glucose | 0.532 | 0.368 | 0.339 | 0.091b | |
| Oligosaccharides | d-Melibiose | 0.699 | 0.552b | 0.260 | 0.144b |
| d-Trehalose | 0.645 | 0.496b | 0.234 | 0.215 | |
| Lactulose | 0.312 | 0.142b | — | — | |
| Maltose | 0.758 | 0.525 | 0.306 | 0.142b | |
| Maltotriose | 0.651 | 0.422 | 0.318 | 0.220b | |
| Sucrose | 0.619 | 0.594 | 0.270 | 0.109b | |
| α-d-Lactose | 0.888 | 0.533 | 0.363 | 0.263b | |
| Sugar alcohols | d-Mannitol | 0.718 | 0.440b | 0.271 | —b |
| Dulcitol | — | — | 0.138 | —b | |
| Glycerol | 0.233 | 0.555d | — | — | |
| Sugar phosphates | d,l-α-Glycerol-phosphate | — | 0.340d | 0.112 | —b |
| Fructose-6-phosphatec | 0.665 | 0.520 | 0.335 | 0.293 | |
| Glucose-1-phosphatec | 0.750 | 0.634 | 0.303 | 0.233 | |
| Glucose-6-phosphate | 0.626 | 0.603 | 0.472 | 0.256b | |
| Acetic acids | Acetic acid | 0.344 | 0.188b | 0.178 | —b |
| Dicarboxylic acids | Bromo succinic acid | — | 0.124 | 0.104 | —b |
| d,l-Malic acid | 0.279 | 0.374 | — | — | |
| d-Malic acid | 0.451 | 0.299b | 0.201 | —b | |
| Fumaric acid | 0.253 | 0.286 | 0.186 | —b | |
| l-Malic acid | 0.338 | 0.324 | 0.184 | —b | |
| Monomethyl succinate | — | — | 0.165 | —b | |
| Succinic acid | 0.229 | 0.333 | — | — | |
| Hydroxy acids | l-Lactic acid | 0.665 | 0.515 | 0.132 | —b |
| Keto acids | Methyl pyruvate | 0.799 | 0.456 | 0.228 | 0.093b |
| Pyruvic acidc | 0.533 | 0.506 | 0.313 | 0.253 | |
| Propionates | Propionic acid | — | 0.111 | — | — |
| Sugar acids | d-Glucuronic acidc | 0.727 | 0.609 | 0.249 | 0.175 |
| Glucuronamide | — | 0.324d | 0.135 | 0.083 | |
| l-Galactonic acid-γ-lactonec | 0.733 | 0.612 | 0.242 | 0.185 | |
| Mucic acid | — | — | 0.117 | —b | |
| Fatty acids | Formic acid | — | — | 0.195 | —b |
| Nucleosides | 2-Deoxyadenosine | 1.160 | 0.623b | 0.256 | 0.164b |
| Adenosine | 0.879 | 0.561b | 0.227 | 0.216 | |
| Inosine | 0.958 | 0.629b | 0.238 | 0.279 | |
| Thymidinec | 0.669 | 0.659 | 0.340 | 0.321 | |
| Uridine | 0.794 | 0.557 | 0.327 | 0.127b | |
| Amino acids | d-Alanine | 0.253 | 0.225 | 0.168 | —b |
| Glycine | — | — | 0.107 | —b | |
| Glycyl-l-glutamic acid | — | — | 0.110 | —b | |
| Glycyl-l-proline | — | — | 0.141 | —b | |
| l-Alanine | 0.197 | 0.210 | 0.126 | —b | |
| l-Alanyl-glycine | — | 0.289d | — | — | |
| l-Asparagine | 0.536 | 0.428 | — | — | |
| l-Aspartic acid | 0.407 | 0.382 | — | — | |
| l-Leucine | — | — | 0.203 | —b | |
| l-Phenylalanine | — | — | 0.111 | —b | |
| l-Proline | — | — | 0.107 | —b | |
| l-Pyroglutamic acid | — | — | 0.117 | —b | |
| l-Serine | 0.375 | 0.610 | — | — | |
Results are the averages from duplicates. —, substrate was tested but not utilized.
The value for stationary-phase cells is significantly greater than that for starved cells (P < 0.05).
Utilization did not differ significantly between stationary-phase and starved cells regardless of the atmospheric growth condition.
The value for stationary-phase cells is significantly less than that for starved cells (P < 0.05).
As expected, carbon utilization profiles of stationary-phase E. coli O157:H7 H11 during anaerobic growth differed from those observed during aerobic growth. During anaerobic incubation, the 26 substrates utilized by stationary-phase cells were not utilized by starved cells, and 17 were utilized to a significantly higher endpoint level by stationary-phase cells than starved cells (Table 2). Of the 95 substrates tested, 13 were utilized differently by cells grown under aerobic and anaerobic conditions (Table 2). The average Aendpoint for Tween 20, Tween 40, β-methyl-d-glucoside, 2-amino-2-deoxy-d-gluconate, and d-glucarate was significantly higher (P < 0.05) after 24 h of anaerobic incubation than after aerobic incubation (see Table S1 in the supplemental material). Endpoint utilization of nine substrates did not differ significantly between stationary-phase and starved cells regardless of the atmospheric growth condition (Table 2). Since it was possible to determine endpoints only from anaerobically incubated phenotypic arrays, the delay in utilization or the rate of utilization could not be determined.
DISCUSSION
E. coli O157:H7 is a virulent zoonotic pathogen capable of causing severe disease in humans with the possibility of long-term sequelae subsequent to the acute infection phase. Fecal shedding of E. coli O157:H7 from ruminants has been shown to be the main source of contamination in farm environments, food, food processing facilities, and water (3). The maintenance of a dual lifestyle by E. coli O157:H7 involves surviving the decrease in temperature and scarcity of nutrients when the bacterium is outside animal hosts, as well as the initiation of growth once conditions become optimal for growth.
In aqueous environments, E. coli O157:H7 from microcosm sediments is culturable for at least 245 days (20), while Zhao and Matthews (29) reported that at least 3 log units of cells with intact membranes remained after 300 days of starvation in PBS at 4, 10, and 22°C from a starting culture of 6 log units. In this study, the quantification method used to determine the number of cells with an intact membrane per ml allowed rapid and accurate enumeration of viable cells within the starved culture. To mirror the average water temperature of New Zealand freshwater in warmer months, cells were held at 15°C, and they were held at 4°C to simulate the freshwater temperature in winter (30) as well as the maximum refrigeration temperature used for storage of meat products. In line with the literature, we found that the number of E. coli O157:H7 cells with intact membranes in aqueous environments with limiting nutrient and suboptimal temperatures was unaffected by starvation in PBS at 4 and 15°C for at least 84 days (Fig. 3).
Once ingested by a potential host, E. coli O157:H7 must initiate cellular processes required to recover from starvation and maximize growth opportunities under aerobic and/or anaerobic conditions it will encounter during the passage through the gastrointestinal tract. No difference in the number of colonies under aerobic and anaerobic incubation at 37°C was detected after starvation at 4°C for 84 days, except on PCA supplemented with 0.2% sodium pyruvate (Table 1), where anaerobic recovery on day 84 in the presence of sodium pyruvate was 1.1 log units higher than that obtained after aerobic incubation. Sodium pyruvate has been shown to “resuscitate” cells from the VBNC state (31–33) and is present in human saliva (34). Our results suggested that the combination of anaerobic conditions such as those found in the intestinal tract and the presence of pyruvate would increase the likelihood of poststarvation recovery in a host.
Assuming that each colony recovered arose from a single bacterium, the size of colonies developed within the 24-hour incubation period may be indicative of direct effects on the cell cycle, with the combined effect of the lag in growth initiation and the rate of cell division in the early phases of recovery. Changes in colony size in response to starvation at 4°C over 84 days were more evident under aerobic conditions, where the reduction in average colony diameter ranged between 35 and 63%, compared to an 8 to 32% reduction under anaerobic conditions (Fig. 4). These results suggest that this initial recovery phase under anaerobic conditions was less affected by starvation. It is worth noting that during starvation, cells were held under aerobic conditions.
Utilization of individual carbon sources was used as a measure of the metabolic capability of stationary-phase and starved cells under both aerobic and anaerobic incubation. Since E. coli O157:H7 is likely to be in the culture equivalent of stationary phase in the ruminant gut prior to shedding, metabolic profiles of stationary-phase cells were used to determine the metabolic capability of the test strain.
Analysis of the data presented here clearly showed that starvation altered the utilization of specific carbon sources by E. coli O157:H7. As expected, the majority of the 95 substrates tested were metabolized by both stationary-phase and starved cells at a similar rate (Fig. 5A); however, the lag time before detectable metabolic activity was observed was significantly longer for starved cells (Fig. 5B). Cells entering the VBNC state have been found to reduce cellular activities, including the transport of nutrients and synthesis of macromolecules (22). We hypothesized that the transporters and/or enzymes required for carbon transport and/or catabolism in the starved cells must be synthesized and that this results in the longer lag times observed for starved-cell carbon utilization. It is likely that this observed increase in the lag time after starvation at 4°C may also contribute to the observed decrease in colony size, with starved cells taking longer to increase in mass and be able to undergo cell division.
Comparisons of aerobic and anaerobic endpoint carbon utilization by nonstarved stationary-phase cells revealed that more substrates were utilized during anaerobic growth than during aerobic growth, although the level of substrate utilization during anaerobic recovery was generally lower than that under aerobic conditions (Table 2). However, in comparison to aerobic growth, where the Aendpoint values for most substrates were similar for stationary-phase and starved cells, substrate utilization endpoints in general for anaerobic recovery of starved cells were lower (Table 2). The limitation of incubating Biolog plates in an anaerobic chamber to ensure consistency of anaerobic condition was the inability to physically place the spectrophotometer inside chamber to collect kinetic carbon utilization data. While it is possible that the energy generated from the carbon sources during anaerobic growth was insufficient to initiate the synthesis of transporters and/or enzymes required for substrate catabolism by starved cells, it is difficult to extrapolate the underlying explanation of the difference observed in anaerobic carbon utilization between stationary-phase and starved cells.
Interestingly, despite the differences in the overall stationary-phase utilization and starved-cell utilization observed between aerobic and anaerobic growth, endpoint utilization for nine substrates, including fructose-6-phosphate, glucose-1-phosphate, pyruvic acid, and thymidine, was not affected by starvation regardless of whether the cells were recovered aerobically or anaerobically (Table 2). This showed that the ability to catabolize these substrates was preserved by starved cells, allowing prompt utilization of these substrates when they became available. Together with the physical signal of optimal growth temperature, the presence of these substrates may act as a nutritional signal detected by both nonstarved and starved cells that the environment is optimal for growth.
Based on current findings, we hypothesize that E. coli O157:H7 can maintain a dual lifestyle, for surviving outside a host and establishing growth inside the host gut, by lowering metabolic activity to preserve energy in external environments, and by rapidly reinitiating growth by detecting chemical and physical environmental changes when internalized by a suitable host. The longer the bacterium remains in environments with low nutrient levels and low temperatures, the lower the energy reservoir becomes, which contributes to the lowered recovery and decreased metabolic activity observed in this study. Furthermore, the difference between the metabolic activity of stationary-phase and starved cells observed under anaerobic incubation was greater than that observed under aerobic conditions. This suggested that initial growth occurred during the early stages of ingestion before arrival in the anaerobic environment of the intestine. It is worth noting that when the bacterium is associated with the intestinal epithelium, the availability of oxygen is likely to be greater than that in the milieu of the gut contents (35), which may assist in maximizing energy production, thus promoting colonization. If the nine substrates utilized consistently by both aerobic and anaerobically grown stationary-phase and starved cells were present in the gastrointestinal tract of the host, the presence of these substrates may further stimulate cell recovery even under anaerobic conditions. Since there was a high similarity in metabolic profiles observed for the three bovine strains tested in this study, the patterns observed in the survival of, recovery from, and metabolic response to starvation at suboptimal temperatures exhibited by H11 are likely to be similar for N427, N635, and other bovine E. coli O157:H7 strains.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Ministry of Science and Innovation program grant. Ron N. Xavier was funded by the University of Waikato Doctoral Scholarship.
Footnotes
Published ahead of print 9 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00045-14.
REFERENCES
- 1.Blanco M, Blanco JE, Blanco J, Gonzalez EA, Mora A, Prado C, Fernandez L, Rio M, Ramos J, Alonso MP. 1996. Prevalence and characteristics of Escherichia coli serotype O157:H7 and other verotoxin-producing E. coli in healthy cattle. Epidemiol. Infect. 117:251–257. 10.1017/S0950268800001424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hussein HS. 2007. Prevalence and pathogenicity of Shiga toxin-producing Escherichia coli in beef cattle and their products. J. Anim. Sci. 85:E63–E72. 10.2527/jas.2006-421 [DOI] [PubMed] [Google Scholar]
- 3.Pennington H. 2010. Escherichia coli O157. Lancet 376:1428–1435. 10.1016/S0140-6736(10)60963-4 [DOI] [PubMed] [Google Scholar]
- 4.Brown CA, Harmon BG, Zhao T, Doyle MP. 1997. Experimental Escherichia coli O157:H7 carriage in calves. Appl. Environ. Microbiol. 63:27–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cray WC, Jr, Moon HW. 1995. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:1586–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mead PS, Griffin PM. 1998. Escherichia coli O157:H7. Lancet 352:1207–1212. 10.1016/S0140-6736(98)01267-7 [DOI] [PubMed] [Google Scholar]
- 7.Tarr PI, Gordon CA, Chandler WL. 2005. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365:1073–1086. 10.1016/S0140-6736(05)71144-2 [DOI] [PubMed] [Google Scholar]
- 8.Tesh VL, Burris JA, Owens JW, Gordon VM, Wadolkowski EA, O'Brien AD, Samuel JE. 1993. Comparison of the relative toxicities of Shiga-like toxins type I and type II for mice. Infect. Immun. 61:3392–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheiring J, Andreoli SP, Zimmerhackl LB. 2008. Treatment and outcome of Shiga-toxin-associated hemolytic uremic syndrome (HUS). Pediatr. Nephrol. 23:1749–1760. 10.1007/s00467-008-0935-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Institute of Environmental Science and Research Ltd. 2004. Editorial: E. coli O157 and other VTEC in New Zealand. N. Z. Public Health Surveill. Rep. 2:2 https://surv.esr.cri.nz/PDF_surveillance/NZPHSR/2004/NZPHSR2004June.pdf [Google Scholar]
- 11.Irshad H, Cookson AL, Hotter G, Besser TE, On SLW, French NP 2012. Epidemiology of Shiga toxin-producing Escherichia coli O157 in very young calves in the North Island of New Zealand. N. Z. Vet. J. 60:21–26. 10.1080/00480169.2011.627063 [DOI] [PubMed] [Google Scholar]
- 12.Arthur TM, Nou X, Kalchayanand N, Bosilevac JM, Wheeler T, Koohmaraie M. 2011. Survival of Escherichia coli O157:H7 on cattle hides. Appl. Environ. Microbiol. 77:3002–3008. 10.1128/AEM.02238-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avery LM, Williams AP, Killham K, Jones DL. 2008. Survival of Escherichia coli O157:H7 in waters from lakes, rivers, puddles and animal-drinking troughs. Sci. Total Environ. 389:378–385. 10.1016/j.scitotenv.2007.08.049 [DOI] [PubMed] [Google Scholar]
- 14.Elder RO, Keen JE, Siragusa GR, Barkocy-Gallagher GA, Koohmaraie M, Laegreid WW. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. U. S. A. 97:2999–3003. 10.1073/pnas.97.7.2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibekwe AM, Watt PM, Shouse PJ, Grieve CM. 2004. Fate of Escherichia coli O157:H7 in irrigation water on soils and plants as validated by culture method and real-time PCR. Can. J. Microbiol. 50:1007–1014. 10.1139/w04-097 [DOI] [PubMed] [Google Scholar]
- 16.Jenkins MB, Fisher DS, Endale DM, Adams P. 2011. Comparative die-off of Escherichia coli O157:H7 and fecal indicator bacteria in pond water. Environ. Sci. Technol. 45:1853–1858. 10.1021/es1032019 [DOI] [PubMed] [Google Scholar]
- 17.Ogden ID, Hepburn NF, MacRae M, Strachan NJC, Fenlon DR, Rusbridge SM, Pennington TH. 2002. Long-term survival of Escherichia coli O157 on pasture following an outbreak associated with sheep at a scout camp. Lett. Appl. Microbiol. 34:100–104. 10.1046/j.1472-765x.2002.01052.x [DOI] [PubMed] [Google Scholar]
- 18.Russell JB, Jarvis GN. 2001. Practical mechanisms for interrupting the oral-fecal lifecycle of Escherichia coli. J. Mol. Microbiol. Biotechnol. 3:265–272 http://www.horizonpress.com/backlist/jmmb/v/v3/v3n2/18.pdf [PubMed] [Google Scholar]
- 19.LeJeune JT, Besser TE, Rice DH, Berg JL, Stilborn RP, Hancock DD. 2004. Longitudinal study of fecal shedding of Escherichia coli O157:H7 in feedlot cattle: predominance and persistence of specific clonal types despite massive cattle population turnover. Appl. Environ. Microbiol. 70:377–384. 10.1128/AEM.70.1.377-384.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LeJeune JT, Besser TE, Hancock DD. 2001. Cattle water troughs as reservoirs of Escherichia coli O157. Appl. Environ. Microbiol. 67:3053–3057. 10.1128/AEM.67.7.3053-3057.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Gilchrist A, Zhang J, Li XF. 2008. Detection of viable but nonculturable Escherichia coli O157:H7 bacteria in drinking water and river water. Appl. Environ. Microbiol. 74:1502–1507. 10.1128/AEM.02125-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliver JD. 2005. The viable but nonculturable state in bacteria. J. Microbiol. 43:93–100 [PubMed] [Google Scholar]
- 23.Oliver JD. 2010. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 34:415–425. 10.1111/j.1574-6976.2009.00200.x [DOI] [PubMed] [Google Scholar]
- 24.Dinu LD, Delaquis P, Bach S. 2009. Nonculturable response of animal enteropathogens in the agricultural environment and implications for food safety. J. Food Prot. 72:1342–1354 [DOI] [PubMed] [Google Scholar]
- 25.Kampstra P. 2008. Beanplot: a boxplot alternative for visual comparison of distributions. J. Stat. Softw. (Code Snippet 1) 28:1–9 http://www.jstatsoft.org/v28/c01 [Google Scholar]
- 26.Classen AT, Boyle SI, Haskins KE, Overby ST, Hart SC. 2003. Community-level physiological profiles of bacteria and fungi: plate type and incubation temperature influences on contrasting soils. FEMS Microbiol. Ecol. 44:319–328. 10.1016/S0168-6496(03)00068-0 [DOI] [PubMed] [Google Scholar]
- 27.Kolde R. 2011. Package ‘pheatmap'. http://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf
- 28.Kahm M, Hasenbrink G, Lichtenberg-Fraté H, Ludwig J, Kschischo M. 2010. grofit: fitting biological growth curves with R. J. Stat. Softw. 33:1–21 http://www.jstatsoft.org/v33/i0720808728 [Google Scholar]
- 29.Zhao L, Matthews KR. 2000. Influence of starvation, temperature, and pH on culturability of Escherichia coli O157:H7. J. Food Saf. 20:193–208. 10.1111/j.1745-4565.2000.tb00297.x [DOI] [Google Scholar]
- 30.National Institute of Water and Atmospheric Research. 2011. Water quality maps and information, July to September 2011. National Institute of Water and Atmospheric Research, Auckland, New Zealand [Google Scholar]
- 31.Bjergbæk LA, Roslev P. 2005. Formation of nonculturable Escherichia coli in drinking water. J. Appl. Microbiol. 99:1090–1098. 10.1111/j.1365-2672.2005.02706.x [DOI] [PubMed] [Google Scholar]
- 32.Mizunoe Y, Wai SN, Takade A, Yoshida SI. 1999. Restoration of culturability of starvation-stressed and low-temperature-stressed Escherichia coli O157 cells by using H2O2-degrading compounds. Arch. Microbiol. 172:63–67. 10.1007/s002030050741 [DOI] [PubMed] [Google Scholar]
- 33.Na SH, Miyanaga K, Unno H, Tanji Y. 2006. The survival response of Escherichia coli K12 in a natural environment. Appl. Microbiol. Biotechnol. 72:386–392. 10.1007/s00253-005-0268-3 [DOI] [PubMed] [Google Scholar]
- 34.Silwood CJL, Lynch E, Claxson AWD, Grootveld MC. 2002. 1H and 13C NMR spectroscopic analysis of human saliva. J. Dent. Res. 81:422–427. 10.1177/154405910208100613 [DOI] [PubMed] [Google Scholar]
- 35.Marteyn B, West NP, Browning DF, Cole JA, Shaw JG, Palm F, Mounier J, Prévost MC, Sansonetti P, Tang CM. 2010. Modulation of Shigella virulence in response to available oxygen in vivo. Nature 465:355–358. 10.1038/nature08970 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.