Abstract
Selection of a starter culture with excellent viability and metabolic activity is important for inoculated fermentation of traditional food. To obtain a suitable starter culture for making Chinese sesame-flavored liquor, the yeast and bacterium community structures were investigated during spontaneous and solid-state fermentations of this type of liquor. Five dominant species in spontaneous fermentation were identified: Saccharomyces cerevisiae, Pichia membranaefaciens, Issatchenkia orientalis, Bacillus licheniformis, and Bacillus amyloliquefaciens. The metabolic activity of each species in mixed and inoculated fermentations of liquor was investigated in 14 different cocultures that used different combinations of these species. The relationships between the microbial species and volatile metabolites were analyzed by partial least-squares (PLS) regression analysis. We found that S. cerevisiae was positively correlated to nonanal, and B. licheniformis was positively associated with 2,3-butanediol, isobutyric acid, guaiacol, and 4-vinyl guaiacol, while I. orientalis was positively correlated to butyric acid, isovaleric acid, hexanoic acid, and 2,3-butanediol. These three species are excellent flavor producers for Chinese liquor. Although P. membranaefaciens and B. amyloliquefaciens were not efficient flavor producers, the addition of them alleviated competition among the other three species and altered their growth rates and flavor production. As a result, the coculture of all five dominant species produced the largest amount of flavor compounds. The result indicates that flavor producers and microbial interaction regulators are important for inoculated fermentation of Chinese sesame-flavored liquor.
INTRODUCTION
Traditional fermented foods include typical products prepared by spontaneous fermentation (1, 2). However, reliance on the “nature” of spontaneous fermentation makes the quality of these products neither predictable nor controllable (3). Therefore, to optimize and control the food characteristics, most inoculated fermentations are developed by using pure cultures as starter inoculums. For example, wine fermentation is typically inoculated with selected indigenous yeasts as starters, and this approach yields homogeneous, high-quality products (2).
Knowledge of indigenous yeasts in spontaneous fermentation and selection of suitable strains are the basic requirements for the inoculated fermentation process (2). Isolation of indigenous species and study of their physiological and metabolic properties form a classical method to select strains for starter cultures (4–7). However, because most traditional fermented foods exhibit a wide range of characteristics, a mixed culture is often a better choice as a starter culture to enhance the qualities of the product (3, 8). In mixed-culture fermentations, the microbial interactions influence the growth and fermentative behavior of all of the microbes, thereby influencing the microbial community structures and functions (9–12). For example, Saccharomyces and non-Saccharomyces yeasts appear to interact in mixed fermentations of wine, and some enological traits of the non-Saccharomyces yeasts can be modulated by Saccharomyces cerevisiae (13–16). Therefore, the metabolic activity of microbial species in mixed culture can be different from that in pure culture. It is important to reveal microbial interactions and the microbial metabolic activity in the designed mixed-culture fermentation for the purpose of selecting suitable starters for inoculated and mixed-culture fermentations.
Chinese sesame-flavored liquor is one of the most popular distilled liquors in China (17). It is famous for its roasted sesame aroma and is also characterized by the solid-state and spontaneous fermentation process. Its production technique is similar to that used to produce a liquor of the strong aroma style (18) but with the additional stage of stacking fermentation. Before fermentation in the pits, newly steamed grains are mixed with Daqu (the starter), stacked in the ground, and fermented for 24 to 40 h. This stage is derived from the production technique used for soy sauce aroma-style liquor (7). The fermentation process accumulates a specific and complex community of microorganisms, including fungi, yeasts, and bacteria. Fungal species produce amylolytic enzymes to degrade the starch in the raw material to fermentable sugars (17, 18). These then become substrates for alcoholic fermentation and flavor production by yeasts and bacteria. Lactic acid bacteria mainly produce lactic acid, which becomes the substrate for esterification of yeasts (19). The fermentation process has previously been shown to be mainly dominated by Bacillus coupled with variable levels of yeast species, such as S. cerevisiae and non-Saccharomyces species. Of this native microflora, the yeasts and bacteria play an essential role in liquor making because they are responsible for the alcoholic fermentation and also for the synthesis of other minor metabolites that determine the aroma and other sensorial properties (7, 20).
Chinese sesame-flavored liquor is an example of a spontaneous fermentation food associated with various microbial species that exhibit different metabolic patterns. Due to the reliance on the “nature” of the fermentation process, the product quality is influenced by climate, location, and the temperature of the environment. This is not always the most efficient fermentation process because the microbial community tends to vary with the environment. In addition, harmful species, such as Staphylococcus or Streptomyces, may contaminate the spontaneous fermentation process and lead to unsuccessful fermentation. Therefore, it is necessary to move from spontaneous fermentation to inoculated fermentation on the basis of a comprehensive understanding of the metabolic activity of the useful microbes responsible for liquor flavor. At present, starter selection for Chinese liquor is mainly based on the metabolic characteristics of the pure cultures of these species (7, 18). The interaction of these species in mixed fermentation has not been explored in detail, and, similarly, the microbial metabolic activity of species in mixed fermentation is little known. Addressing these deficiencies should aid the development of inoculated fermentation for Chinese liquor.
The present study aimed to develop a method for selection of metabolically active yeasts and bacteria species that can direct the fermentation processes to produce high-quality liquor. We selected species that were dominant during the fermentation process, because the dominant species tend to have excellent viability and play important roles in liquor fermentation. Then we implemented mixed fermentations by different combinations of these dominant species, and the volatile compound fingerprinting of them was analyzed by mathematical analysis. The ultimate aim of this work will be to predict which strains have significant effects on sesame-flavored liquor fermentation and to aid the development of large-scale and controllable production of Chinese liquor without the loss of unique flavors and particular characteristics.
MATERIALS AND METHODS
Sampling.
Sampling of fermented grains was carried out at a factory in Jiangsu Province, China, where sesame-flavored liquor was produced. Samples were taken from the stacking and liquor fermentation stages. The pit was a cube-shaped underground cellar (about 2.6 by 2.4 by 2.9 m). Samples of fermented grains were collected from the upper layer (center of the pit, 0.2 m deep) in liquor fermentation at 5-day intervals during the 30-day fermentation process.
Enumeration, isolation, and identification of yeasts and bacteria.
Each sample (10 g) was mixed with 90 ml of sterile saline (0.85% NaCl) and soaked at 4°C for 30 min. Yeast enumeration and isolation were carried out on Wallerstein laboratory nutrient (WLN) medium as previously reported (21), which has been used in soy sauce-style liquor, another type of Chinese liquor (19). According to the macroscopic features (texture, surface, margin, elevation, and color), colonies of different types on the WLN medium were counted separately. From 10 up to 25 colonies of each representative type were picked and restreaked to obtain the pure clones. Yeast species were first identified by PCR-restriction fragment length polymorphism (RFLP) analysis, and then 1 to 7 randomly selected isolates per distinct restriction pattern were obtained for confirmation by 26S ribosomal DNA (rDNA) sequence analysis (19).
Bacterial enumeration and isolation were carried out on nutrient agar medium (0.5% beef extract, 1% peptone, 0.5% NaCl, 2% agar). Cultures were incubated at 37°C for 24 h. According to the macroscopic properties (texture, surface, margin, elevation, and color), colonies of different types on the medium were counted separately. Bacillus with apparent different colony properties accounted for most of the bacterial population. From 10 up to 25 colonies of each representative type were picked and restreaked to obtain the pure clones. To identify the bacterial species, the genomic DNA was extracted using a phenol-chloroform method (22) and used as a template to amplify the 16S rDNA by using the primers 27F and 1492R (23). The PCR products were subjected to BLAST analysis.
Mixed-culture fermentations.
The fermentation medium was prepared with sorghum, the material for liquor fermentation. Eight hundred grams of sorghum was added to 1.0 liter of deionized water, steamed for 2 h, and then kept at 60°C for 4 h for saccharification with the addition of glucoamylase at a final concentration of 50 U/g. The original total reducing sugar in the extract was about 90 g/kg. This medium was sealed in 2-liter beakers and autoclaved at 121°C for 15 min. A loopful of yeast and bacterial cultures was inoculated in 250-ml Erlenmeyer flasks with 50 ml of sorghum extract, respectively, which was prepared as previously reported (7). Fermentations were conducted at 150 rpm and 30°C for 48 h. Yeast cell number was determined with a hemacytometer. Bacteria cell number was determined by using a Helber counting chamber (Z30000; Helber, Hawksley, United Kingdom) viewed under a microscope (BX51; Olympus) operated in phase-contrast mode at ×100 magnification. The cell numbers of S. cerevisiae, Issatchenkia orientalis, Pichia membranaefaciens, Bacillus licheniformis, and Bacillus amyloliquefaciens in the extract fermentation were 1.8 × 108, 2.4 × 108, 1.4 × 108, 1 × 108, and 2 × 108 CFU/ml, respectively. These species were then inoculated into solid-state sorghum in beakers for mixed fermentation, with the initial cell density of each species adjusted to 1 × 106 CFU/g. The total added liquid was made the same by addition of sorghum extract. All the fermentations were conducted without agitation at 30°C for 30 days. Fermentations were conducted with single components, double combinations, and higher-order combinations. For higher-order combinations, a total of 14 trials of mixed fermentation were conducted: mixed fermentation with five species, four species (with one of the five species absent), three species (with three yeast species or one bacterium and two yeast species), and two species (with B. licheniformis and S. cerevisiae). A noninoculated sample of fermentation medium was prepared as the control. All experiments were performed in triplicate.
Enumeration of cell numbers of different species at the end of solid-state fermentation was performed as follows. Each sample (10 g) was mixed with 90 ml of sterile saline (0.85% NaCl) and soaked at 4°C for 30 min. Yeast enumeration was carried out on WLN medium. Cultures were incubated at 30°C for 4 days. Bacterial enumeration and isolation were carried out on nutrient agar medium. Cultures were incubated at 37°C for 24 h. According to the macroscopic properties, colonies of different species on the medium were counted separately.
Volatile compound analysis.
The fermented grain (5 g) was mixed with 20 ml of sterile saline (0.85% NaCl, 1% CaCl2) and soaked overnight. After ultrasonic treatment for 30 min, the mixture was centrifuged at 8,000 × g at 4°C for 10 min. The supernatant was filtered through a 0.22-μm-pore filter and stored at −20°C until analysis.
Volatile compounds in fermented sorghums were assayed by headspace solid-phase microextraction coupled with gas chromatography-mass spectrometry (HS-SPME-GC-MS). A divinylbenzene-carboxen-poly(dimethylsiloxane) (DVB/CAR/PDMS)-coated fiber (50/30 μm coating; Supelco, Bellefonte, PA) was used for volatile compound extraction. Eight milliliters of the obtained supernatant, the internal standard (4-methyl-2-pentanol; final concentration, 54.50 μg/liter), and 3 g of sodium chloride were combined in a 20-ml vial, and the vial was hermetically sealed with a polytetrafluoroethylene (PTFE)-faced silicone septum. This sample was equilibrated at 50°C for 5 min and extracted for 45 min at the same temperature with stirring in a multipurpose sampler with SPME capability (MPS 2; Gerstel, Mülheim, Germany). After extraction, the DVB/CAR/PDMS-coated fiber was inserted into the injection port of the gas chromatograph set at 250°C and left for 5 min to desorb the analytes. All analyses were carried out in triplicate.
Statistical analysis.
SPSS 19.0 software (IBM, Armonk, NY) was used for cluster analysis of flavor compounds produced by different mixed-culture fermentations. A one-way analysis of variance (ANOVA) test (SPSS 19.0) was applied to select flavor metabolites that were significantly different (P ≤ 0.05) in different mixed cultures. To establish the relationship between the significantly different flavor metabolites and microbial variables, partial least-squares (PLS) regression (SIMCA-P-11.5) was used. The predictive power of the model was assessed by cross-validation and the Q2 value (goodness of prediction); only the variance of volatile compounds with a Q2 value of >20% could be explained by microbial species. The native functions of the software were used for statistical analysis.
RESULTS
Identification of dominant species during fermentation of Chinese sesame-flavored liquor.
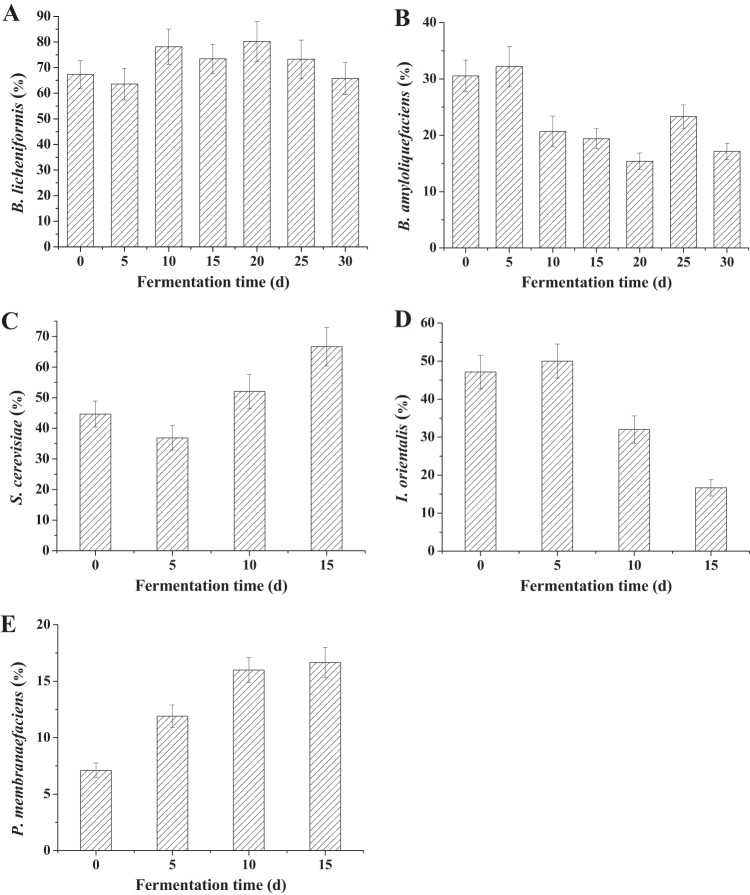
A total of 224 isolated colonies of bacteria were classified into 10 species, corresponding to B. licheniformis, B. amyloliquefaciens, Bacillus subtilis, Bacillus cereus, Bacillus pumilus, Lysinibacillus fusiformis, Staphylococcus saprophyticus, Paenibacillus lactis, Brevibacillus borstelensis, and Staphylococcus lentus (see Fig. S1 in the supplemental material). Bacillus licheniformis and B. amyloliquefaciens were the two major species according to analysis by the culture-dependent isolation method. As shown in Fig. 1A, the population of B. licheniformis ranged from 12.8 × 104 to 14.6 × 104 CFU/g before 20 days, and then it decreased to 4.6 × 104 CFU/g at the end of fermentation. As shown in Fig. 1B, the population of B. amyloliquefaciens was smaller than that of B. licheniformis. It ranged from 2.8 × 104 to 5.8 × 104 CFU/g before 25 days and then decreased to 1.2 × 104 CFU/g at the end of fermentation. The counts of these two species maintained 92.9 to 98.9% of the total number of bacterial cells before 25 days and then decreased to 82.9% of all the bacterial species at the end of liquor fermentation. Bacillus licheniformis has been reported as one of the dominant bacteria and plays an important role in the fermentation process for Chinese Maotai-flavored liquor (20). It may also be an important species in the fermentation of sesame-flavored liquor.
FIG 1.
Ratio of dominant species in liquor fermentation. (A) Ratio of B. licheniformis to the total bacterial population; (B) ratio of B. amyloliquefaciens to the total bacterial population; (C) ratio of S. cerevisiae to the total yeast population; (D) ratio of I. orientalis to the total yeast population; (E) ratio of P. membranaefaciens to the total yeast population. The total bacterial and yeast populations were obtained from the counts of all bacterial and yeast species. Yeast species were not detected after 15 days in liquor fermentation.
A total of 145 colonies of yeasts were isolated in stacking and liquor fermentation stages. They were divided into five types with the use of WLN medium (see Fig. S2 in the supplemental material) and PCR-RFLP analysis of the 5.8S internal transcribed spacer (ITS) rDNA region and were assigned to the species S. cerevisiae, I. orientalis, P. membranaefaciens, Rhodotorula mucilaginosa, and Pichia anomala.
In liquor fermentation, the total yeast population decreased, and no yeast was detected after 15 days. The low oxygen supply and the acids produced in the fermented grains in the sealed pit are probably responsible for yeast death in the later stages of liquor fermentation (19). Only three yeast species—I. orientalis, S. cerevisiae, and P. membranaefaciens—were detected at 15 days: about 66.7% of the total yeast count was S. cerevisiae, 16.7% was P. membranaefaciens, and 16.7% was I. orientalis (Fig. 1C, D, and E).
The dominant species in spontaneous fermentation have excellent viability in liquor fermentation and have the potential to play important roles in liquor fermentation, such as producing flavor compounds. Species with low population densities have an accordingly low potential for use in fermentation. Therefore, the five dominant species and their metabolic activities are highlighted in our investigation.
Interactions between the different dominant species.
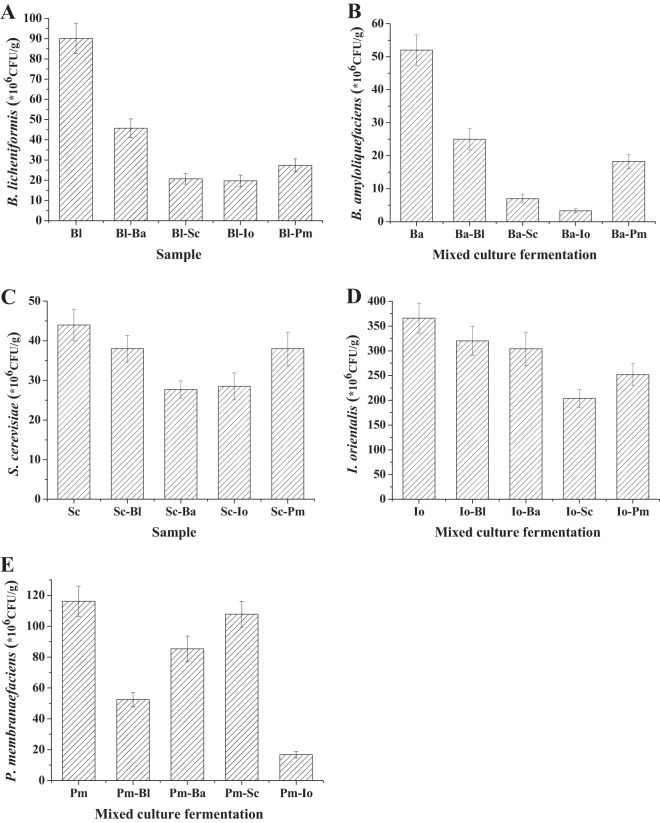
Microbial interaction between different species would influence growth and hence influence whole microbial community structures and their metabolic activities. Therefore, to reveal the interactions among the five different dominant species (B. licheniformis, B. amyloliquefaciens, S. cerevisiae, I. orientalis, and P. membranaefaciens), fermentations with single and double combinations were conducted. The growth of each species is shown in Fig. 2. Bacillus licheniformis and B. amyloliquefaciens competed with each other, and both their populations decreased by nearly 50% when they were cocultured. In addition, they were significantly inhibited by S. cerevisiae, I. orientalis, and P. membranaefaciens, and both of their populations decreased to less than 35.0% compared with those in the single fermentations, respectively (Fig. 2A and B).
FIG 2.
Populations of single and double combinations of five dominant species at the end of different mixed-culture fermentations. The initial cell density of each species was adjusted to 1 × 106 CFU/g. (A) B. licheniformis; (B) B. amyloliquefaciens; (C) S. cerevisiae; (D) I. orientalis; (E) P. membranaefaciens. Bl, B. licheniformis; Ba, B. amyloliquefaciens; Sc, S. cerevisiae; Io, I. orientalis; Pm, P. membranaefaciens.
Saccharomyces cerevisiae and I. orientalis were not significantly inhibited by bacteria. However, P. membranaefaciens was significantly inhibited, and its population decreased from 116.2 × 106 to 52.5 × 106 CFU/g and to 85.4 × 106 CFU/g when it was cocultured with B. licheniformis and B. amyloliquefaciens, respectively (Fig. 2C, D, and E).
Competition between three yeast species also existed. Saccharomyces cerevisiae decreased from 44.0 × 106 to 28.5 × 106 CFU/g, and I. orientalis decreased from 366.2 × 106 to 204.0 × 106 CFU/g when they were mixed. However, they were both less sensitive to P. membranaefaciens. In addition, P. membranaefaciens was not influenced by S. cerevisiae but was significantly inhibited by I. orientalis (Fig. 2C, D, and E).
Differential profiling of higher-order mixed-culture fermentations.
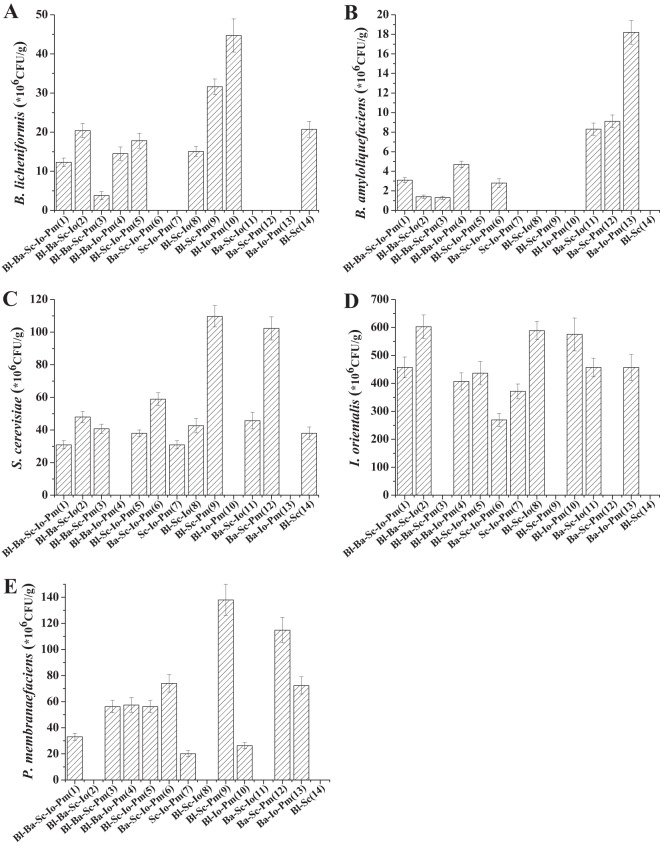
Based on the interactions among the five species, we investigated the metabolic activities of the five species in liquor fermentation in depth, using fermentations with higher-order combinations. A total of 14 mixed-culture fermentations were performed, and the population of each species was determined at the end of fermentation. The initial inoculation concentration of each species was 1.0 × 106 CFU/g. After fermentation for 30 days, nearly all of the inoculated species had propagated in different mixed cultures.
As shown in Fig. 3A, the population of B. licheniformis decreased from 31.6 × 106 (trial 9) to 3.8 × 106 (trial 3) CFU/g and from 44.7 × 106 (trial 10) to 14.5 × 106 (trial 4) CFU/g when it was inoculated with B. amyloliquefaciens. As shown in Fig. 3B, growth of B. amyloliquefaciens was the weakest among all the inoculated species in each mixed-fermentation trial. Its count decreased from 9.1 × 106 (trial 12) to 1.3 × 106 CFU/g (trial 3), from 18.2 × 106 (trial 13) to 4.7 × 106 CFU/g (trial 4), and from 8.3 × 106 (trial 11) to 1.4 × 106 CFU/g (trial 2) when it was cultured with B. licheniformis. This indicates that B. licheniformis and B. amyloliquefaciens competed with each other when cocultured and reflects the similar result when they were used in a double-combination fermentation.
FIG 3.
Populations of five dominant species at the end of different mixed-culture fermentations. The initial cell density of each species was adjusted to 1 × 106 CFU/g. (A) B. licheniformis; (B) B. amyloliquefaciens; (C) S. cerevisiae; (D) I. orientalis; (E) P. membranaefaciens. Bl, B. licheniformis; Ba, B. amyloliquefaciens; Sc, S. cerevisiae; Io, I. orientalis; Pm, P. membranaefaciens. Numbers in parentheses and below the x axis refer to trial numbers.
Inhibition of S. cerevisiae against the bacteria also occurred in the higher-order combinations: when S. cerevisiae was inoculated, the population of B. licheniformis decreased from 44.7 × 106 (trial 10) to 17.8 × 106 (trial 5) CFU/g (Fig. 3A), and B. amyloliquefaciens decreased from 18.2 × 106 (trial 13) to 2.8 × 106 (trial 6) CFU/g (Fig. 3B). However, although S. cerevisiae was slightly inhibited in the double combination with each bacterium, it was strongly inhibited when it was cultured with both species of bacteria. The population of S. cerevisiae in the B. licheniformis-B. amyloliquefaciens-S. cerevisiae-P. membranaefaciens (trial 3) culture was much lower than that in the B. amyloliquefaciens-S. cerevisiae-P. membranaefaciens (trial 12) or B. licheniformis-S. cerevisiae-P. membranaefaciens (trial 9) culture (Fig. 3C).
Although bacteria showed no significant effect on I. orientalis, I. orientalis inhibited bacterial growth. The growth of both B. licheniformis (compare trials 9 and 5 and compare trials 14 and 8) (Fig. 3A) and B. amyloliquefaciens (compare trials 12 and 6) (Fig. 3B) decreased when cocultured with I. orientalis.
Pichia membranaefaciens significantly inhibited the growth of B. amyloliquefaciens in the higher-order combination (compare trials 6 and 11) (Fig. 3B), which was consistent with the result in their double combination. However, it did not inhibit B. licheniformis in the higher-order combination (compare trials 5 and 8) (Fig. 3A).
Yeasts grew better than bacteria in nearly all the trials. When the three yeast species were cocultured (trials 1 and 7), the growth of S. cerevisiae and P. membranaefaciens was the weakest, indicating they might also compete with each other (Fig. 3C and E). Issatchenkia orientalis was the most vigorous among the three yeast species, because its growth was less influenced by the other two yeast species (Fig. 3D).
Another interesting phenomenon was observed, as shown in Fig. 3A and B, where the inhibition of yeast against bacteria weakened in the mixed fermentation of all five species (trial 1); the growth of B. licheniformis and B. amyloliquefaciens remained the same when S. cerevisiae was added to the B. licheniformis-B. amyloliquefaciens-I. orientalis-P. membranaefaciens culture (trial 4). In addition, the competition between the two species of bacteria also weakened in trial 1, because the growth rates of B. licheniformis in trials 5 and 1 were about the same, as was the growth of B. amyloliquefaciens between trials 6 and 1. This indicates that coculture of the five species would coordinate the microbial interactions and weaken the competition among the five species, which would be beneficial for food fermentation.
Differential profiling of different mixed-culture fermentations.
The complex interactions among the five species not only would influence microbial growth, but also would influence the metabolic activity of each species in the mixed-culture fermentation. To discriminate the 14 different mixed-culture groups, and to determine which flavor compounds most significantly contributed to the distinction of the groups, the volatile compounds present in each culture after fermentation were detected by GC–MS (see Table S1 in the supplemental material). Hierarchical clustering analysis was performed based on the volatile compounds in different cultures. The tree represented 42 mixed-fermentation samples (14 different mixed fermentations, each with three parallel experiments). Distance was indicative of a uniquely close association of different samples, and the samples appearing close in the tree were those that had a close proximity in the cluster. As shown in Fig. 4A, different cultures were differentially discriminated, and the three parallel samples of each culture were also assigned to the same cluster, except the combination B. licheniformis-S. cerevisiae-I. orientalis-P. membranaefaciens (trial 5), but they were still in the close clusters. The S. cerevisiae-I. orientalis-P. membranaefaciens culture (trial 7) was in a cluster far from all the other clusters, indicating it was significantly different from the other mixed cultures. This might be because of the weakest growth of the yeast species. The B. licheniformis-S. cerevisiae-P. membranaefaciens (trial 9) and B. licheniformis-B. amyloliquefaciens-S. cerevisiae-P. membranaefaciens (trial 3) cultures were assigned to the same cluster, indicating that the two cultures were in close proximity to each other, which suggested that B. amyloliquefaciens might have no significant contribution to the flavor compounds in the mixed culture. The assignment of B. licheniformis-S. cerevisiae-I. orientalis (trial 8) and B. licheniformis-B. amyloliquefaciens-S. cerevisiae-I. orientalis (trial 2) cultures to the same cluster also demonstrated this point. Similar phenomena were found in the two groups of cultures consisting of the combinations B. amyloliquefaciens-S. cerevisiae-I. orientalis (trial 11) and B. amyloliquefaciens-S. cerevisiae-I. orientalis-P. membranaefaciens (trial 6) and B. licheniformis-S. cerevisiae-I. orientalis (trial 8) and B. licheniformis-S. cerevisiae-I. orientalis-P. membranaefaciens (trial 5), which indicated that P. membranaefaciens had no significant effect on production of flavor compounds.
FIG 4.
Differential profiling of different mixed-culture fermentations. (A) Hierarchical clustering by using ANOVA test samples of volatile compounds. The tree represents 42 mixed fermentations of the five selected strains after 30 days of fermentation at 30°C, based on the ANOVA-filtered volatile compounds detected by GC-MS. (B) Pair plot showing the differences in levels of volatile compounds detected in different mixed fermentations. The compounds displayed are the flavor compounds most important in discriminating sample types by PLS. Names of compounds in the squares on the diagonal indicate the compound displayed on the axes in the rows or columns of graphs intersecting with that square. The level increases from left to right and from bottom to top in the squares. The pair plot visualizes which compounds allow discrimination of different culture groups. The pair plot showed, for instance, that the purple solid circle (trial 1) was in the top of the squares in the row and in the right of the squares in the phenethyl butyrate column (as shown in the red dotted box). This indicates that a high concentration of phenethyl butyrate was highly discriminative for the combination B. licheniformis-B. amyloliquefaciens-S. cerevisiae-I. orientalis-P. membranaefaciens (trial 1), whereas the black solid diamond (trial 7) was at the bottom of the squares in the row and in the left of the squares in the guaiacol column (as shown in the green dotted box). This indicates that a low concentration of guaiacol was highly discriminative for the mixed culture of S. cerevisiae, I. orientalis, and P. membranaefaciens (trial 7). Bl, B. licheniformis; Ba, B. amyloliquefaciens; Sc, S. cerevisiae; Io, I. orientalis; Pm, P. membranaefaciens.
A pair plot of 14 relevant compounds was generated, which was most significant for discriminating the different groups. From this plot, the low or high concentrations of compounds responsible for distinction of the flavor profiles in the 14 different cocultures were identified. As shown in Fig. 4B, most of these metabolites were important flavor compounds in the Chinese liquors (24, 25), indicating the importance of these species in liquor fermentation. The lower levels of acetoin, guaiacol, phenol, 4-vinyl guaiacol were only discriminated in the mixed-culture fermentation of the combination S. cerevisiae-I. orientalis-P. membranaefaciens (trial 7) without bacterial species. This indicated that these metabolites might be associated with bacteria. The levels of isobutyric acid and isovaleric acid were relatively lower in B. amyloliquefaciens-S. cerevisiae-P. membranaefaciens cultures (trial 12) without I. orientalis and B. licheniformis, which revealed a positive correlation between these acids and I. orientalis or B. licheniformis.
The higher levels of dimethylglyoxal, ethyl hexanoate, 2,3-butanediol, phenethyl butyrate, and 4-vinyl guaiacol were only discriminated in the presence of all five species (B. licheniformis-B. amyloliquefaciens-S. cerevisiae-I. orientalis-P. membranaefaciens; trial 1). It appeared that the mixed fermentation would be beneficial for production of various flavor compounds. An exception was noted for the B. amyloliquefaciens-S. cerevisiae-I. orientalis culture (trial 11), which was related to a higher level of nonanal. This result indicated that nonanal might be associated with one of the component species of this coculture (B. amyloliquefaciens, S. cerevisiae, or I. orientalis), and that P. membranaefaciens or B. licheniformis, which were not added in this coculture, might metabolize this compound.
Metabolic activity of different species in mixed fermentation.
To identify the in situ metabolic activity of five dominant species in mixed-culture fermentation and their contribution to flavor compounds, covariation between metabolic profiling and microbial species was analyzed by using PLS regression. The predictive power of the model was assessed by cross-validation and the Q2 value. Only the variance of volatile compounds with a Q2 value of >20% could be explained by microbial species.
Nine flavor compounds could be predicted to have significant difference among the 14 different cultures (Table 1). Bacillus amyloliquefaciens was positively associated with only 4-vinyl guaiacol, while P. membranaefaciens was only negatively associated with isoamyl alcohol, indicating it was unfavorable for isoamyl alcohol production. This confirmed the result in Fig. 4A that B. amyloliquefaciens and P. membranaefaciens made little contribution to flavor production. Saccharomyces cerevisiae was positively correlated with nonanal and negatively associated with 2,3-butanediol, indicating it was beneficial for nonanal production, which is consistent with the behavior of wine yeasts (26). In addition, S. cerevisiae might metabolize 2,3-butanediol and decrease its concentration.
TABLE 1.
Covariation between flavor compounds and microbial species analyzed by PLS regression
| Flavor compound | Q2 valuea | Microbial species (association)b |
|---|---|---|
| Isoamyl alcohol | 0.4656 | P. membranaefaciens (−) |
| Nonanal | 0.3388 | S. cerevisiae (+) |
| B. licheniformis (−) | ||
| 2,3-Butanediol | 0.2833 | I. orientalis (+) |
| B. licheniformis (+) | ||
| S. cerevisiae (−) | ||
| Isobutyric acid | 0.4319 | B. licheniformis (+) |
| Butyric acid | 0.3676 | I. orientalis (+) |
| Isovaleric acid | 0.4012 | I. orientalis (+) |
| Hexanoic acid | 0.4423 | I. orientalis (+) |
| Guaiacol | 0.2651 | B. licheniformis (+) |
| 4-Vinyl guaiacol | 0.2484 | B. licheniformis (+) |
| B. amyloliquefaciens (+) |
Only the variance of volatile compounds with Q2 values of >20% could be explained by microbial species.
+ and −, positively and negatively associated, respectively, with the formation of flavor compounds.
Positive correlations between I. orientalis and four flavor compounds (butyric acid, isovaleric acid, hexanoic acid, and 2,3-butanediol) indicate that I. orientalis produces these volatile acids in mixed fermentation. Bacillus licheniformis was positively associated with 2,3-butanediol, isobutyric acid, guaiacol, and 4-vinyl guaiacol, and this result is consistent with the behavior of this species in Maotai-flavored liquor fermentation (20). Such consistency in experimental results supports the validity and accuracy of PLS analysis and suggests that mixed fermentation might have little influence on the metabolic activity of B. licheniformis. However, B. licheniformis was negatively associated with nonanal, and this explained why the Bacillus amyloliquefaciens-S. cerevisiae-I. orientalis culture (trial 11) without B. licheniformis was unique for a higher level of nonanal (Fig. 4B). The negative correlation might be due to metabolism of nonanal by B. licheniformis.
Our results suggested that not all the dominant strains in the fermentation process played important roles in developing the flavor of the product. It appears that only B. licheniformis, I. orientalis, and S. cerevisiae are important flavor producers in the fermentation of Chinese sesame-flavored liquor.
DISCUSSION
Traditional fermented foods always exhibit a wide range of features and characteristics, and a mixed culture is often a better choice as a starter culture. In mixed-culture fermentations, the microbial interactions influence the overall microbial growth and fermentative behavior and hence influence the whole microbial community structure and its functions. Therefore, the metabolic activity of microbial species in mixed culture is different from that in pure culture. This underlines the importance of revealing the microbial interactions and the microbial metabolic activity in the designed mixed-culture fermentation for the purpose of selecting suitable starters for inoculated and mixed-culture fermentation.
This work investigated the in situ metabolic activities of different species in mixed fermentation. Only three dominant species showed significant in situ metabolic activity: B. licheniformis, S. cerevisiae, and I. orientalis. In contrast, P. membranaefaciens and B. amyloliquefaciens had little effect on production of flavor compounds, implying that not all the dominant species were flavor-producing active species. The low metabolic activity of the pure culture of P. membranaefaciens has been reported previously in studies of Chinese Maotai-flavored liquor (7). It indicated that the weak in situ metabolic activity of this species was mainly due to its intrinsic metabolic activity. The metabolic activity of the pure culture of B. amyloliquefaciens was also investigated in our laboratory. It produced a few types of flavor compounds, such as isobutyric acid, hexanoic acid, and guaiacol, and their concentrations were less than those produced by B. licheniformis. In addition, the growth of B. amyloliquefaciens was also the weakest in mixed-culture fermentation. Therefore, the weak in situ metabolic activity of this species might be due to both the weak intrinsic flavor-producing activity and cell growth, which were different from those of P. membranaefaciens.
Issatchenkia orientalis was predicted to produce several acids in mixed fermentation, including butyric acid, isovaleric acid, and hexanoic acid, which was consistent with the report that I. orientalis produced various types of acids regardless of the mixed culture (7). However, it was discovered that I. orientalis could not produce 2,3-butanediol in pure culture fermentation (7). This suggests that the coculture with other species was beneficial for producing 2,3-butanediol by I. orientalis. Bacillus licheniformis was an important flavor producer in liquor fermentation. It was reported that B. licheniformis produced various flavor compounds, such as 2,3-butanedione, acetoin, and guaiacol, in pure culture fermentation, and the contribution of this species to liquor flavor was noted when it was inoculated in the Maotai-flavored liquor-making process (20). This confirmed the vigorous growth and metabolic activity of this strain in mixed-culture fermentation. In addition, it also revealed that B. licheniformis was unfavorable for nonanal formation, which might be due to the metabolism of nonanal by B. licheniformis. This should be further investigated.
Microbial interaction also plays an important role in the activity of the whole microbial community. One of the main interaction types between the five species is the inhibition of S. cerevisiae against Bacillus. In our previous work, we found that S. cerevisiae inhibited the growth of B. licheniformis, with their inoculation ratio ranging from 1:1 to 1:10,000. In addition, B. licheniformis was inhibited by the acids and some peptides produced by S. cerevisiae (27).
Another type of interaction is the inhibition of S. cerevisiae against non-Saccharomyces yeast species. Many researchers have investigated this phenomenon and the mechanism. For example, S. cerevisiae produced certain toxic compounds that inhibited the growth of Hanseniaspora guilliermondii and Hanseniaspora uvarum during mixed fermentations (28). Kluyveromyces thermotolerans and Torulaspora delbrueckii showed arrested growth due to a cell-cell contact-mediated mechanism in the presence of viable S. cerevisiae cells (29). In our work, we also found that S. cerevisiae inhibited I. orientalis, while it had no effect on the growth of P. membranaefaciens. The inhibition mechanism should be investigated further. Moreover, the interaction between Saccharomyces and non-Saccharomyces yeasts appears to influence the fermentation behavior of the mixed culture; for example, it appears to enhance the glycerol content (15), increase the production of polysaccharides, and modulate the final concentrations of acetic acid and volatile compounds, such as ethyl acetate, phenyl-ethyl acetate, 2-phenyl ethanol, and 2-methyl-1-butanol (14, 16).
Another interaction between the five species was alleviation of the above inhibitions by P. membranaefaciens and B. amyloliquefaciens, which was also important to shape the community structure and function. It was discovered that the amount of flavor compounds in the mixture of the three flavor-producing species (B. licheniformis, S. cerevisiae, and I. orientalis) was not the highest among the 14 mixed-culture fermentations. The coculture of the five dominant species actually produced the highest level of flavor compounds. Given that P. membranaefaciens and B. amyloliquefaciens are essentially inactive in producing flavor compounds, the increased variety and amounts of flavor compounds produced by the five-component mixture was caused by improved activity in flavor production by B. licheniformis, S. cerevisiae, and I. orientalis. This is probably a result of interaction of these species and regulation of microbial function. For example, as shown in Fig. 3A, B. licheniformis showed the weakest growth when cultured with B. licheniformis, S. cerevisiae, and I. orientalis (trial 8). However, when B. amyloliquefaciens was added to the medium, inhibition of B. licheniformis by the yeast was slightly alleviated, and the population of B. licheniformis increased from 15.1 × 106 (trial 8) to 20.4 × 106 (trial 2) CFU/g. This would likely be beneficial for flavor production by B. licheniformis in the coculture fermentation. This result shows that although some dominant species had little effect on producing flavor, they had some significant function in regulating the microbial community. For example, P. membranaefaciens was reported to produce pectin methylesterase and extracellular proteases (e.g., β-1,3-glucanase and chitinase) (30, 31), which might be useful for influencing the microbial community structure and function. This interaction phenomenon has also been detected among several lactic acid bacteria. For example, Lactobacillus delbrueckii and Lactobacillus plantarum are strongly inhibited when they are cocultured with Streptococcus thermophilus CNRZ1066, respectively. However, when the three species were cocultured together, the inhibitions of Streptococcus thermophilus CNRZ1066 against L. delbrueckii and L. plantarum were alleviated or even disappeared (9).
It is apparent that dominant fermentation species that are not active as flavor producers are also important for inoculated fermentation of Chinese liquor. While the selection of flavor producers is important, other species must also be chosen as community structure regulators for inoculated fermentation. By these means, the microbial structure and the flavor of the product can be improved. This work proposes an original method for selecting the starter culture for inoculated fermentation and is expected to play an important role in the development of Chinese sesame-flavored liquor production.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National High Technology Research and Development Program of China (2012AA021301 and 2013AA102108), the National Natural Science Foundation of China (31371822 and 31271921), and the Program of Introducing Talents of Discipline to Universities (111 Project) (111-2-06).
Footnotes
Published ahead of print 9 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00905-14.
REFERENCES
- 1.Fleet GH. 1999. Microorganisms in food ecosystems. Int. J. Food Microbiol. 50:101–117. 10.1016/S0168-1605(99)00080-X [DOI] [PubMed] [Google Scholar]
- 2.Rodríguez-Lerma GK, Gutiérrez-Moreno K, Cárdenas-Manríquez M, Botello-Álvarez E, Jiménez-Islas H, Rico-Martínez R, Navarrete-Bolaños JL. 2011. Microbial ecology studies of spontaneous fermentation: starter culture selection for prickly pear wine production. J. Food Sci. 76:M346–M352. 10.1111/j.1750-3841.2011.02208.x [DOI] [PubMed] [Google Scholar]
- 3.Navarrete-Bolaños JL. 2012. Improving traditional fermented beverages: how to evolve from spontaneous to directed fermentation. Eng. Life Sci. 12:410–418. 10.1002/elsc.201100128 [DOI] [Google Scholar]
- 4.Cordero-Bueso G, Esteve-Zarzoso B, Cabellos JM, Gil-Dıáz M, Arroyo T. 2013. Biotechnological potential of non-Saccharomyces yeasts isolated during spontaneous fermentations of Malvar (Vitis vinifera cv.L.). Eur. Food Res. Technol. 236:193–207. 10.1007/s00217-012-1874-9 [DOI] [Google Scholar]
- 5.Lefeber T, Gobert W, Vrancken G, Camu N, De Vuyst L. 2011. Dynamics and species diversity of communities of lactic acid bacteria and acetic acid bacteria during spontaneous cocoa bean fermentation in vessels. Food Microbiol. 28:457–464. 10.1016/j.fm.2010.10.010 [DOI] [PubMed] [Google Scholar]
- 6.Suárez-Lepe JA, Morata A. 2012. New trends in yeast selection for winemaking. Trends Food Sci. Technol. 23:39–50. 10.1016/j.tifs.2011.08.005 [DOI] [Google Scholar]
- 7.Wu Q, Xu Y, Chen L. 2012. Diversity of yeast species during fermentative process contributing to Chinese Maotai-flavour liquor making. Lett. Appl. Microbiol. 55:301–307. 10.1111/j.1472-765X.2012.03294.x [DOI] [PubMed] [Google Scholar]
- 8.King ES, Swiegers JH, Travis B, Francis IL, Bastian SEP, Pretorius IS. 2008. Coinoculated fermentations using Saccharomyces yeasts affect the volatile composition and sensory properties of Vitis vinifera L. cv. Sauvignon Blanc wines. J. Agric. Food Chem. 56:10829–10837. 10.1021/jf801695h [DOI] [PubMed] [Google Scholar]
- 9.de Bok FA, Janssen PW, Bayjanov JR, Sieuwerts S, Lommen A, van Hylckama Vlieg JET, Molenaar D. 2011. Volatile compound fingerprinting of mixed-culture fermentations. Appl. Environ. Microbiol. 77:6233–6239. 10.1128/AEM.00352-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadoudi M, Tourdot-Maréchal R, Rousseaux S, Steyer D, Gallardo-Chacón JJ, Ballester J, Vichi S, Guérin-Schneider R, Caixach J, Alexandre H. 2012. Yeast-yeast interactions revealed by aromatic profile analysis of Sauvignon Blanc wine fermented by single or co-culture of non-Saccharomyces and Saccharomyces yeasts. Food Microbiol. 32:243–253. 10.1016/j.fm.2012.06.006 [DOI] [PubMed] [Google Scholar]
- 11.Sieuwerts S, de Bok FAM, Hugenholtz J, van Hylckama Vlieg JET. 2008. Unraveling microbial interactions in food fermentations: from classical to genomics approaches. Appl. Environ. Microbiol. 74:4997–5007. 10.1128/AEM.00113-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smid EJ, Lacroix C. 2013. Microbe-microbe interactions in mixed culture food fermentations. Curr. Opin. Biotechnol. 24:148–154. 10.1016/j.copbio.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 13.Comitini F, Gobbi M, Domizio P, Romani C, Lencioni L, Mannazzu I, Ciani M. 2011. Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 28:873–882. 10.1016/j.fm.2010.12.001 [DOI] [PubMed] [Google Scholar]
- 14.Domizio P, Romani C, Lencioni L, Comitini F, Gobbi M, Mannazzu I, Ciani M. 2011. Outlining a future for non-Saccharomyces yeasts: selection of putative spoilage wine strains to be used in association with Saccharomyces cerevisiae for grape juice fermentation. Int. J. Food Microbiol. 147:170–180. 10.1016/j.ijfoodmicro.2011.03.020 [DOI] [PubMed] [Google Scholar]
- 15.Gobbi M, Comitini F, Domizio P, Romani C, Lencioni L, Mannazzu I, Ciani M. 2013. Lachancea thermotolerans and Saccharomyces cerevisiae in simultaneous and sequential co-fermentation: a strategy to enhance acidity and improve the overall quality of wine. Food Microbiol. 33:271–281. 10.1016/j.fm.2012.10.004 [DOI] [PubMed] [Google Scholar]
- 16.Howell KS, Cozzolino D, Bartowsky EJ, Fleet GH, Henschke PA. 2006. Metabolic profiling as a tool for revealing Saccharomyces interactions during wine fermentation. FEMS Yeast Res. 6:91–101. 10.1111/j.1567-1364.2005.00010.x [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Zhang X, Zhao L, Xu Y. 2008. Analysis and comparison of the bacterial community in fermented grains during the fermentation for two different styles of Chinese liquor. J. Ind. Microbiol. Biotechnol. 35:603–609. 10.1007/s10295-008-0323-z [DOI] [PubMed] [Google Scholar]
- 18.Wu Z, Zhang W, Zhang Q, Hu C, Wang R, Liu Z. 2009. Developing new sacchariferous starters for liquor production based on functional strains isolated from the pits of several famous Luzhou-flavor liquor brewers. J. Inst. Brew. 115:111–115. 10.1002/j.2050-0416.2009.tb00354.x [DOI] [Google Scholar]
- 19.Wu Q, Chen L, Xu Y. 2013. Yeast community associated with the solid state fermentation of traditional Chinese Maotai-flavor liquor. Int. J. Food Microbiol. 166:323–330. 10.1016/j.ijfoodmicro.2013.07.003 [DOI] [PubMed] [Google Scholar]
- 20.Zhang R, Wu Q, Xu Y. 2013. Aroma characteristics of Moutai-flavour liquor produced with Bacillus licheniformis by solid-state fermentation. Lett. Appl. Microbiol. 57:11–18. 10.1111/lam.12087 [DOI] [PubMed] [Google Scholar]
- 21.Pallmann CL, Brown JA, Olineka TL, Cocolin L, Mills DA, Bisson LF. 2001. Use of WL medium to profile native flora fermentations. Am. J. Enol. Vitic. 52:198–203 http://ajevonline.org/content/52/3/198.short [Google Scholar]
- 22.Flynn S, van Sinderen D, Thornton GM, Holo H, Nes IF, Collins JK. 2002. Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology 148:973–984 http://mic.sgmjournals.org/content/148/4/973.short [DOI] [PubMed] [Google Scholar]
- 23.Han S, Liu Y, Zhou Z, He S, Cao Y, Shi P, Yao B, Ring E. 2010. Analysis of bacterial diversity in the intestine of grass carp (Ctenopharyngodon idellus) based on 16S rDNA gene sequences. Aquac. Res. 42:47–56. 10.1111/j.1365-2109.2010.02543.x [DOI] [Google Scholar]
- 24.Fan W, Qian MC. 2006. Characterization of aroma compounds of Chinese “Wuliangye” and “Jiannanchun” liquors by aroma extract dilution analysis. J. Agric. Food Chem. 54:2695–2704. 10.1021/jf052635t [DOI] [PubMed] [Google Scholar]
- 25.Fan W, Qian MC. 2006. Identification of aroma compounds in Chinese ‘Yanghe Daqu' liquor by normal phase chromatography fractionation followed by gas chromatography/olfactometry. Flavour Fragr. J. 21:333–342. 10.1002/ffj.1621 [DOI] [Google Scholar]
- 26.Mauriello G, Capece A, D'Auria M, Garde-Cerdán T, Romano P. 2009. SPME-GC method as a tool to differentiate VOC profiles in Saccharomyces cerevisiae wine yeasts. Food Microbiol. 26:246–252. 10.1016/j.fm.2009.01.003 [DOI] [PubMed] [Google Scholar]
- 27.Ling J, Wu Q, Xu Y, Fan W. 2013. Interactions between Bacillus licheniformis and Saccharomyces cerevisiae in the fermentation of soy-sauce flavor liquor. Microbiol. China 40:2014–2021 (In Chinese.) [Google Scholar]
- 28.Pérez-Nevado F, Albergaria H, Hogg T, Girio F. 2006. Cellular death of two non-Saccharomyces wine-related yeasts during mixed fermentations with Saccharomyces cerevisiae. Int. J. Food Microbiol. 108:336–345 [DOI] [PubMed] [Google Scholar]
- 29.Nissen P, Nielsen D, Arneborg N. 2003. Viable Saccharomyces cerevisiae cells at high concentrations cause early growth arrest of non-Saccharomyces yeasts in mixed cultures by a cell-cell contact-mediated mechanism. Yeast 20:331–341. 10.1002/yea.965 [DOI] [PubMed] [Google Scholar]
- 30.Mamede M, Pastore G. 2006. Study of methods for the extraction of volatile compounds from fermented grape must. Food Chem. 96:586–590. 10.1016/j.foodchem.2005.03.013 [DOI] [Google Scholar]
- 31.Masih E, Paul B. 2002. Secretion of β-1,3-glucanases by the yeast Pichia membranifaciens and its possible role in the biocontrol of Botrytis cinerea causing grey mold disease of the grapevine. Curr. Microbiol. 44:391–395. 10.1007/s00284-001-0011-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.