Abstract
The anaerobic dehalogenation of organohalides is catalyzed by the reductive dehalogenase (RdhA) enzymes produced in phylogenetically diverse bacteria. These enzymes contain a cobamide cofactor at the active site and two iron-sulfur clusters. In this study, the tetrachloroethene (PCE) reductive dehalogenase (PceA) of the Gram-positive Desulfitobacterium hafniense strain Y51 was produced in a catalytically active form in the nondechlorinating, cobamide-producing bacterium Shimwellia blattae (ATCC 33430), a Gram-negative gammaproteobacterium. The formation of recombinant catalytically active PceA enzyme was significantly enhanced when its dedicated PceT chaperone was coproduced and when 5,6-dimethylbenzimidazole and hydroxocobalamin were added to the S. blattae cultures. The experiments were extended to D. hafniense DCB-2, a reductively dehalogenating bacterium harboring multiple rdhA genes. To elucidate the substrate spectrum of the rdhA3 gene product of this organism, the recombinant enzyme was tested for the conversion of different dichlorophenols (DCP) in crude extracts of an RdhA3-producing S. blattae strain. 3,5-DCP, 2,3-DCP, and 2,4-DCP, but not 2,6-DCP and 3,4-DCP, were reductively dechlorinated by the recombinant RdhA3. In addition, this enzyme dechlorinated PCE to trichloroethene at low rates.
INTRODUCTION
The ability for corrinoid-dependent anaerobic reductive dehalogenation of organohalides is spread among phylogenetically diverse bacterial genera that are in some cases difficult to isolate or even cultivate, e.g., Dehalococcoides mccartyi (Chloroflexi), Desulfitobacterium spp. (Firmicutes), Geobacter lovleyi (Deltaproteobacteria), and Sulfurospirillum spp. (Epsilonproteobacteria) (summarized in reference 1). These bacteria usually couple the reductive dehalogenation of organohalides catalyzed by the reductive dehalogenase enzymes (RdhAs) to energy conservation via a chemiosmotic mechanism (organohalide respiration) (2, 3). Besides physiologically versatile reductively dehalogenating bacteria, which use a variety of electron acceptors for respiratory growth, e.g., Sulfurospirillum multivorans (4) or Desulfitobacterium hafniense isolates (5), dehalogenation specialists have been isolated that are strictly dependent on organohalides as growth substrates, for example, Dehalobacter restrictus (6) or Dehalococcoides mccartyi strains (7). The latter organohalide-respiring organisms are characterized by the presence of multiple reductive dehalogenase gene homologues (rdhA) in their genome sequence (8–13). The physiological function and the substrate spectrum of the respective gene products are known for only a limited number of these enzymes. The presence of several RdhA enzymes in an organism has often hampered the assignment of a specific substrate range to a single rdhA gene product. Overlapping substrate spectra of different RdhAs might additionally impede the functional assignment. Previously, the capacity of organohalides to induce transcription of rdhA genes was investigated (14–16). However, a conclusion on the substrate spectrum of a specific RdhA enzyme could not been drawn directly from the spectra of organohalides inducing the respective rdhA expression. Furthermore, reductive dehalogenase genes found at contaminated sites and used as an indication of the presence of a dehalogenating enzyme activity cannot be investigated with respect to the substrate spectrum of the corresponding enzyme. Therefore, heterologous production of catalytically active RdhA enzymes was carried out in the study presented here to allow the analysis of specific RdhA substrate spectra.
From biochemical studies performed with purified RdhAs and in silico analyses of reductive dehalogenase gene sequences, the general requirement for two iron sulfur clusters and a cobamide (“complete” corrinoid) as cofactors for the catalytically active enzyme was deduced (2). The cytoplasmic precursors of the RdhA enzymes harbor an N-terminal Tat (twin-arginine translocation) (17) signal peptide necessary for membrane export after cofactor incorporation and folding of the enzyme. The mature form of the enzyme is located on the exoplasmic face of the cytoplasmic membrane (18, 19). An RdhA studied in detail is the cobamide-containing tetrachloroethene (PCE) reductive dehalogenase (PceA) of the anaerobe Desulfitobacterium hafniense strain Y51 (Firmicutes) (20). The enzyme enables the organism to dehalogenate PCE via trichloroethene (TCE) to cis-1,2-dichloroethene (cDCE). The PceA enzyme is encoded in the pceABCT operon (21). The pceA gene from D. hafniense Y51 was heterologously expressed in Escherichia coli; however, the PCE reductive dehalogenase was produced in an inactive and insoluble form (22). A similar result was obtained before when the heterologous expression of the pceA gene from S. multivorans was investigated (23). The formation of nonfunctional enzyme in both cases was attributed to the absence of a de novo cobamide biosynthetic pathway in E. coli and the resulting lack of a sufficient concentration of cobamide cofactor in the cells. The genome of D. hafniense Y51 harbors numerous genes encoding proteins that are predicted to be involved in de novo cobamide biosynthesis (24). The cultivation of D. hafniense Y51 with tetrachloroethene (PCE) was shown to occur independently of the presence of exogenous cobamide (19), a result pointing to functional de novo cobamide biosynthesis covering the demand for cobamide cofactor in this reductively dehalogenating organism.
In a recent survey by Leys and coworkers, the heterologous production of PceA from D. restrictus was carried out in E. coli (25). PceA was produced as a fusion protein together with the E. coli trigger factor protein. This modification led to the formation of soluble PceA apoprotein; however, no catalytically active PCE reductive dehalogenase enzyme was formed. Increased production of soluble recombinant PceA protein has also been detected when the trigger-factor-like chaperone PceT, encoded in the pce operons of reductively dehalogenating D. hafniense strains, was coproduced (26, 27). This accessory protein was shown to interact with the Tat signal peptide of the PCE reductive dehalogenase precursor protein.
In the study presented here, catalytically active cobamide-containing RdhA enzymes were formed in a nondechlorinating bacterium. The PCE reductive dehalogenase (PceA) of D. hafniense Y51 and the 3,5-dichlorophenol-induced RdhA3 of D. hafniense DCB-2 were produced in Shimwellia blattae, which is able to synthesize cobamides de novo. The amount of soluble enzyme detected in the organism was increased when the respective chaperones (PceT/RdhT) were coproduced. The presence of 5,6-dimethylbenzimidazole and hydroxocobalamin (OH-B12) in the growth medium supported the production of active enzyme. The substrate spectrum of the recombinant RdhA3 was analyzed. Besides aromatic organohalides, PCE was also converted by the enzyme.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains used in this study are listed in Table 1. Shimwellia blattae (ATCC 33430) was used as the host for heterologous expression. Strains AMN0, AMN1, AMN2, AMN3, and AMN4 are derivatives of S. blattae carrying different variants of the plasmids pASK-IBA63c-plus and pASK-IBA3C (IBA, Göttingen, Germany). All genetic constructs listed in Table 2 were verified by DNA sequencing.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant characteristic(s), plasmid(s) | Source or designation |
|---|---|---|
| Desulfitobacterium hafniense strain Y51 | Wild type | Kindly provided by Taiki Futagami (Kagoshima University, Japan) |
| Desulfitobacterium hafniense strain DCB-2 | Wild type | DSMZ 10664 |
| Shimwellia blattae | Wild type | ATCC 33430 |
| AMN0 | pASK-IBA63c-plus | This study |
| AMN1 | pASK-IBA63c-plus (pceAY51-CStrep) | This study |
| AMN2 | pASK-IBA63c-plus (NStrep-pceAY51) | This study |
| AMN3 | pASK-IBA63c-plus (NStrep-pceAY51), pASK-IBA3C (pceTY51-CStrep) | This study |
| AMN4 | pASK-IBA63c-plus (NStrep-rdhA3DCB-2), pASK-IBA3C (rdhTDCB-2-CStrep) | This study |
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristic(s), construction detailsa | Source or reference |
|---|---|---|
| pASK-IBA63c-plus | Ampr, f1 origin, tet repressor, tet promoter, multiple-cloning site includes Strep-tag II DNA sequence for production of C-terminal fusions | IBA, Göttingen, Germany |
| pASK-IBA3C | Camr, f1 origin, tet repressor, tet promoter, multiple-cloning site includes Strep-tag II DNA sequence for production of C-terminal fusions | IBA, Göttingen, Germany |
| pASK-IBA63c-plus (pceAY51-CStrep) | PCR fragment (primers, P1 and P2; template, genomic DNA of D. hafniense Y51) cut with PciI and XhoI in pASK-IBA63c-plus digested with NcoI and XhoI | This study |
| pASK-IBA63c-plus (NStrep-pceAY51) | PCR fragment (primers, P3 and P4; template, genomic DNA of D. hafniense Y51) cut with PciI and XhoI in pASK-IBA63c-plus digested with NcoI and XhoI | This study |
| pASK-IBA3C (pceTY51-CStrep) | PCR fragment (primers, P5 and P6; template, genomic DNA of D. hafniense Y51) cut with BsaI in pASK-IBA3C digested with BsaI | This study |
| pASK-IBA63c-plus (NStrep-rdhA3DCB-2) | PCR fragment (primers, P7 and P8; template, genomic DNA of D. hafniense DCB-2) cut with NcoI and XhoI in pASK-IBA63c-plus digested with NcoI and XhoI | This study |
| pASK-IBA3C (rdhTDCB-2-CStrep) | PCR fragment (primers, P9 and P10; template, genomic DNA of D. hafniense DCB-2) cut with BsaI in pASK-IBA3C digested with BsaI | This study |
Amp, ampicillin; Cam, chloramphenicol.
Media and growth conditions.
Desulfitobacterium hafniense strain Y51 was cultivated anaerobically with pyruvate and PCE as reported by Reinhold et al. (19). D. hafniense strain DCB-2 was grown under the same conditions, but chlorophenols rather than PCE were added as electron acceptors. Stock solutions of the chlorophenols (100 mM) were prepared in ethanol. The maximum concentration of the chlorophenols in the growth medium was 100 μM.
Recombinant protein production in S. blattae was conducted under anoxic conditions in a mineral medium (28) amended with yeast extract (see the following paragraph). For culture maintenance, however, S. blattae strains were routinely cultivated aerobically. The medium (1 liter) contained K2HPO4 (14.0 g), KH2PO4 (6.0 g), (NH4)2SO4 (3.0 g), MgSO4·7H2O (0.2 g), CoCl2·6H2O (0.024 g), yeast extract (YE) (2 or 0.2 g [as indicated in Results]), cysteine-HCl (0.2 g), and trace element solution SL4 (1 ml) (pH 7.5). Glycerol (300 mM) served as the growth substrate. Where indicated, a sterile solution of fumarate (1 M) or 5,6-dimethylbenzimidazole (DMB) (1 mM) or hydroxocobalamin (100 μM), each dissolved in ultrapure water (UPW), was added prior to the inoculation of the cultures. The final concentration of fumarate in the growth medium was 10 mM, of DMB was 10 μM, and of hydroxocobalamin was 200 nM. For S. blattae strains harboring the expression plasmids, 100 μg/ml ampicillin and, if required, 50 μg/ml chloramphenicol were added.
For recombinant production of an RdhA enzyme, LB-grown precultures of the respective S. blattae expression strains were cultivated aerobically at 28°C for 6 to 8 h. The cultures were shaken (120 rpm). From the preculture, a 250-ml anaerobic main culture was inoculated at an optical density at 578 nm (OD578) of 0.01 to 0.02. For the main culture, the mineral medium amended with yeast extract (see above) was used. The cells were cultivated at 18°C. The cultures were shaken (100 rpm). When an OD578 of 0.25 to 0.3 was reached, recombinant protein production was induced by the addition of anhydrotetracycline (final concentration, 20 ng/ml). After several hours (for details, see Results) of cultivation, the cells were harvested by centrifugation (12,000 × g, 10 min, 4°C) under air and the cell pellets were stored at −20°C.
Construction of plasmids and transformation of S. blattae.
The pairs of oligonucleotides used for the production of the different genetic constructs in this study are given in Table 2. The DNA sequences of the primers are listed in Table 3. The plasmids were constructed as follows. A PCR fragment was generated covering the coding sequence of the pceA and rdhA3 or pceT and rdhT genes. Genomic DNA of D. hafniense Y51 or DCB-2 served as the template in the PCR. The sequences of the oligonucleotides included restriction sites for specific endonucleases (for details, see Table 3). Via ligation of the cut PCR fragment into the compatibly cut expression vector (pASK-IBA63c-plus or pASK-IBA3C), the plasmid construction was finished. All cloned DNA fragments were sequenced. Competent S. blattae cells were prepared using calcium chloride, and the transformation of the plasmids was conducted as described by Inoue et al. (29). For transformation with two plasmids, the two were mixed in equal amounts (20 ng DNA) prior to the addition of the competent cells. Selection of positive clones was conducted on agar plates containing ampicillin and chloramphenicol.
TABLE 3.
Oligonucleotides used for cloning proceduresa
| Primer | Sequence (5′–3′) |
|---|---|
| P1 | AGCTACATGTCGGGAGAAATCAACAGGAG |
| P2 | CCGCTCGAGTTGTTTTATAGACTCAGGAT |
| P3 | AGCTACATGTCGTGGAGCCACCCGCAGTTCGAAAAAGGAGAAATCAACAGGAG |
| P4 | CCGCTCGAGCTATTGTTTTATAGACTCAGGAT |
| P5 | ATAGCCGGTCTCGAATGATAATGAAGCAGTTCGA |
| P6 | CTACCAGGTCTCAGCGCTACTTGCCAAATTGATTTTTA |
| P7 | AGCTCCATGGGGTGGAGCCACCCGCAGTTCGAAAAAGAGATGAATTTAGACAG |
| P8 | CCGCTCGAGTTACTTTGTGGGCTCGGTGTCT |
| P9 | ATCTAGGGTCTCGAATGAGACAATTCGAACTTGG |
| P10 | ATCTAGGGTCTCAGCGCTCTCTCTTGCCAAGTTTTGCT |
Restriction sites are underlined.
RdhA enzyme activity measurements.
Cells of S. blattae were transferred into an anaerobic glove box (CoyLab, Grass Lake, MI) and resuspended in an anoxic buffer (50 mM Tris-HCl [pH 7.5]; 2 ml buffer per gram wet cells). The buffer was made anaerobic by alternate degassing and flushing with nitrogen. The resuspended cells were mixed with an equal volume of silica spheres (Carl Roth GmbH, Karlsruhe, Germany) (0.5 mm diameter) and were disrupted with a bead mill (Mixer Mill MM400; Retsch, Haan, Germany) (30 Hz, 10 min). The cell debris was removed by centrifugation (5,900 × g, 5 min). The reductive dehalogenase enzyme activity in the crude extract was measured in accordance with the method described by Neumann et al. (30). Activity measurements were conducted in high-performance liquid chromatography (HPLC) vials (volume 1.5 ml) flushed with nitrogen and closed with butyl rubber stoppers. The concentration of organohalides was 0.5 mM in the assay. The assay mixture was incubated for 30 min (for the C-terminal Strep-tag-fused PceA, 1 h) at room temperature. The PceA activity measurements displayed no decrease in the PCE dechlorination rate until 1 h of incubation time. The concentrations of chlorinated ethenes in the assay mixture were determined by gas chromatography (GC) and the concentrations of the chlorinated aromatic hydrocarbons by HPLC (see below). The protein concentrations of the crude extracts were determined according to the method of Bradford (31) using Roti-Nanoquant reagent (Carl Roth GmbH, Karlsruhe, Germany). The amount of protein applied in the assay ranged from 250 to 750 μg.
GC/HPLC.
The chlorinated ethenes were detected using a Clarus 500 gas chromatograph (GC; PerkinElmer, Rodgau, Germany) equipped with a flame ionization detector. Headspace sampling was applied using an HS 40 Headspace Autosampler (PerkinElmer, Rodgau, Germany). For headspace sampling, the GC vial containing the probe was heated to 95°C for 6 min in the HS 40 instrument. After automatic insertion of the needle (temperature of the needle, 100°C), the vial was pressurized with nitrogen for 1 min (pressure, 0.1 kPa). The sample was then injected (duration, 0.1 min) and transferred through a tempered transfer line (temperature, 130°C) to the GC apparatus. A CP-PoraBOND Q fused silica column (Agilent Technologies, Böblingen, Germany) (25 m by 0.32 mm) was used for the stationary phase and was constantly flushed with nitrogen (pressure, 100 kPa). The temperature of the injector of the GC was 250°C, and the temperature of the detector was 300°C. The following temperature program was applied for separation: 4 min at 150°C followed by a gradient of 10°C/min increasing to 280°C. The retention times were as follows: PCE, 10.2 min; TCE, 7.2 min; cDCE, 4.9 min. Nonane served as the internal standard (retention time, 14.5 min). The detection limit for the chlorinated ethenes was 1 μM.
The chlorinated aromatic compounds and the respective degradation products were analyzed using a reversed-phase HPLC system (Merck-Hitachi, Darmstadt, Germany) (flow rate, 0.4 ml/min). An RP8 column (LiChrosphere 100; Merck, Darmstadt, Germany) (4.6 nm inner diameter [ID] by 100 nm) was used for the stationary phase. For isocratic elution of the chlorinated phenols, 50% (vol/vol) methanol–0.3% (vol/vol) orthophosphoric acid–UPW was used. For isocratic elution of 3-chloro-4-hydroxy-phenylacetate (ClOHPA) and 4-hydroxy-phenylacetate (OHPA), 25% (vol/vol) methanol–0.3% (vol/vol) orthophosphoric acid–UPW was applied. The wavelength chosen for detection was 210 nm. The retention times were as follows: 2,4,5-trichlorophenol (2,4,5-TCP), 40 min; 2,4,6-trichlorophenol (2,4,6-TCP), 38 min; 3,5-dichlorophenol (3,5-DCP), 29 min; 3,4-dichlorophenol (3,4-DCP), 21.5 min; 2,4-dichlorophenol (2,4-DCP), 18.5 min; 2,3-dichlorophenol (2,3-DCP), 15.4 min; 2,6-dichlorophenol (2,6-DCP), 12.4 min; 3-chlorophenol (3-CP), 10.8 min; 4-chlorophenol (4-CP), 10.6 min; 2-chlorophenol (2-CP), 8.25 min; ClOHPA, 20.2 min; OHPA, 10 min. The detection limit for the chlorinated aromatics was 5 μM.
Immunoblot analysis.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, 12.5%) was used to separate proteins of S. blattae crude extracts (10 μg protein/lane). The immunoblot was generated as described by John et al. (32). The Strep-tag-specific antibody solution (IBA, Göttingen, Germany) was diluted 3,000-fold, and the antibodies were detected via a secondary antibody coupled to alkaline phosphatase (Sigma-Aldrich, Munich, Germany).
Purification and analysis of cobamides.
The purification and HPLC analysis of cobamides were performed as described earlier by Keller et al. (33). For the subsequent mass spectrometric analysis, the cobamides were applied to an ultra-high-performance liquid chromatography (UHPLC) system using an Ultimate 3000 series rapid-separation liquid chromatography (RSLC) system (Dionex, Sunnyvale, CA) connected to a LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). UHPLC was performed on an Acclaim C18 column (Dionex) (150 by 2.1 mm, 2.2-μm-pore-size filter) mounted to a C18 guard column (Waters, Dublin, Ireland) (2.1 by 10 mm, 3.5-μm-pore-size filter). The sample was injected into a binary solvent system of water (solvent A) and acetonitrile (solvent B; hypergrade for LC-mass spectrometry [LC-MS]; Merck, Darmstadt, Germany), both containing 0.1% (vol/vol) formic acid (eluent additive for LC-MS; Sigma-Aldrich, Steinheim, Germany). The flow rate was set to 300 μl/min. Separation was accomplished using a gradient as follows: 5 min constant at 0% B—linear increase from 0% B to 100% B within 17 min—10 min constant at 100% B—5 min equilibration time at 0% B. Separated extracts were ionized using electrospray ionization (ESI). ESI source parameters were set to 4 kV for spray voltage and 35 V for transfer capillary voltage at a capillary temperature of 275°C. Cobamide analysis was performed in positive-ion mode using 30,000 m/Δm resolving power at a mass range of m/z 150 to 2,000 in the Orbitrap mass analyzer. XCALIBUR software (Thermo Fisher Scientific, Waltham, MA) was used for interpretation of the data.
Isolation of nucleic acids, RT, and qPCR.
Genomic DNA (gDNA) of D. hafniense strains Y51 and DCB-2 was extracted according to the protocol described by Reinhold et al. (19). Total RNA was isolated from D. hafniense DCB-2 cells using an RNeasy minikit (Qiagen, Hilden, Germany). The samples were treated with DNase I (RNase free; Roche, Mannheim, Germany) to remove residual gDNA from RNA preparations. The quality of the isolated RNA was verified by agarose gel electrophoresis. For synthesis of cDNA, 1 μg of total RNA was subjected to reverse transcription (RT) using a Qiagen OneStep RT-PCR kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The rdhA expression was tested with D. hafniense DCB-2 cells cultivated with pyruvate (40 mM) as the electron donor. The 3,5-DCP, the 2,6-DCP, and the 2,4-DCP were added (at 100 μM each) in the initial growth phase (OD578 = 0.2) to induce rdhA expression. To ensure comparability, total RNA was extracted from cultures at the same stage of bacterial growth (OD578 = 0.3). Transcript abundance levels were compared via quantitative PCR (qPCR) using Maxima SYBR green qPCR master mix (Fermentas, St. Leon Rot, Germany) and a CFX96 Real Time PCR machine (Bio-Rad, Munich, Germany). Assays were performed in triplicate, and all data were normalized to the rpoB housekeeping gene (Dhaf_0414). The rpoB endogenous reference gene showed constant expression levels under the different experimental conditions. The PCR assay reaction mixture was composed of 6 μl 2× Maxima SYBR green qPCR master mix, a 0.5 μM concentration of each primer, and 2.5 μl cDNA and filled with PCR-grade water to reach a final volume of 12 μl. Primer pairs used for reverse transcription and qPCR are listed in Table 4. Each PCR run included a no-template control with water instead of cDNA as well as an RT negative control for each rdhA gene to exclude product formation from contaminations of gDNA. To confirm that the accumulation of SYBR green-bound DNA was gene specific and not due to primer dimers or unspecific byproducts, a melting curve analysis was performed as well as agarose gel electrophoresis with qPCR products. The 2−ΔΔCT threshold cycle (CT) method was applied for the calculation of relative gene expression levels (34).
TABLE 4.
Oligonucleotides used in reverse transcription and qPCR
| Primer | Locus tag | Sequence (5′–3′) | Amplicon size (bp) |
|---|---|---|---|
| rdhA1_fw | Dhaf_0689 | CTGGCTATGGAGGGGAAATT | 114 |
| rdhA1_re | AGTATCGGAAATGGGTGCAA | ||
| rdhA3_fw | Dhaf_0696 | ATTGCCCATTATCCGTTCAA | 137 |
| rdhA3_re | ACCGACTCGAACTTCCATTG | ||
| rdhA4_fw | Dhaf_0711 | GTCCAAGTTAAAGCCCAAAGT | 107 |
| rdhA4_re | TCTTTTCAATGTTCCCGACG | ||
| rdhA5_fw | Dhaf_0713 | CGGAACAGATGTCCCAGAAT | 105 |
| rdhA5_re | GCGCCTTGTGGGAATAGTAG | ||
| rdhA6_fw | Dhaf_0737 | GGTAAAATACGCTCCAAACTTC | 131 |
| rdhA6_re | TCCGCTTCAGATGTCATTTT | ||
| rpoB_fw | Dhaf_0414 | GATTCGGGCTTTGGGTTATGC | 138 |
| rpoB_re | CGCAGACGCTTGTAGATTTCC |
RESULTS
For functional heterologous expression of reductive dehalogenase genes, Shimwellia blattae (ATCC 33430; formerly Escherichia blattae [35]) was chosen because of its ability to synthesize cobamides de novo and its accessibility to the molecular techniques and tools developed for the related Escherichia coli (28). Glycerol can be utilized as an energy substrate and carbon source by S. blattae (36), and a cobamide-dependent glycerol-dehydratase was previously described to be involved in catabolic glycerol conversion (28). To ensure cobamide production in S. blattae, all cultivations were performed with glycerol as the sole energy and carbon source. In the first part of the study, the PceA enzyme from Desulfitobacterium hafniense Y51 was analyzed for functional heterologous production in S. blattae. The enzyme has already been characterized for its properties and substrate spectrum (22). In the second part of the investigations, the reductive dehalogenase RdhA3, an enzyme whose substrate spectrum has not been reported before, from D. hafniense DCB-2 was tested for functional production.
Heterologous production of catalytically active PceA in S. blattae.
For the heterologous expression of the pceA gene from D. hafniense Y51, two different genetic constructs were generated, one encoding a fusion with a C-terminal Strep-tag II sequence and the other one with an N-terminal Strep-tag II sequence (for details, see Table 2 and Materials and Methods). The fusion of PceA to a Strep-tag sequence was generated to allow simple immunologic detection of the protein in crude extracts of S. blattae using a Strep-tag-specific antibody. In each case, the pceA sequence cloned into the respective expression plasmid included the DNA sequence encoding the Tat signal peptide, which led to the production of the Tat signal peptide-containing precursor of the PceA enzyme. The presence of both variants (i.e., PceA-CStrep and NStrep-PceA) was tested by measuring PCE dechlorination in crude extracts with reduced methylviologen as the electron donor (Fig. 1). The cells, which had been induced for protein induction, were harvested (about 1.3 g protein/250 ml) after 6 h. While no PCE-dechlorinating enzyme activity was detectable in extracts of the S. blattae strain harboring the empty expression plasmid (strain AMN0) (data not shown), the crude extracts of the S. blattae strains producing the affinity-tagged PceA variants (strains AMN1 and AMN2) exhibited conversion of PCE when pceA expression was induced by the addition of anhydrotetracycline (Fig. 1). Using gas chromatography, TCE and cDCE were identified as products of PCE dechlorination. The S. blattae AMN2 strain producing the N-terminal Strep-tagged PceA displayed significantly higher specific enzyme activity than strain AMN1 producing the C-terminal Strep-tagged variant. The level of PceA enzyme in the crude extract was analyzed via immunoblotting (Fig. 1) and shown to be increased in strain AMN2 compared to strain AMN1. This result indicated that the C-terminal Strep-tag hampered the formation of catalytically active PceA. In order to increase the level of soluble enzyme, the chaperone PceT was coproduced. Besides a minor increase in the total amount of PceA protein in cells that coexpressed pceT, a significant impact of the chaperone on the solubility of PceA was observed (see Fig. S1 in the supplemental material). However, the enzyme activity was marginally higher for the N-terminal Strep-tagged PceA when PceT was present (strain AMN3) than for the strain lacking the chaperone (strain AMN2). The PceA activity was significantly increased by the addition of exogenous 5,6-dimethylbenzimidazole (DMB; 10 μM) and hydroxocobalamin (OH-B12; 200 nM) to the growth medium, a result pointing to a limitation in the cobamide cofactor supply for the maturation of the recombinant PceA enzyme in S. blattae (Fig. 1). This implication was supported by the fact that the positive effect of DMB and OH-B12 on the PceA activity was also observed in the absence of PceT (data not shown). A control experiment was performed to exclude the cobamide-mediated abiotic dehalogenation of PCE in the crude extract of PceA-producing S. blattae cells. When denatured protein (95°C, 15 min) was applied to the assay, no conversion of PCE was detected. All attempts to purify the N- or C-terminal Strep-tagged PceA proteins via affinity chromatography in an amount allowing further biochemical characterization have failed so far, most probably due to the low affinity of the Strep-tagged proteins for the Strep-Tactin column matrix. In both cases, almost all of the Strep-tagged PceA protein remained in the flowthrough rather than bound to the column material (data not shown). Such low affinity was recently reported by Leys and coworkers for the Strep-tagged PceA protein from D. restrictus heterologously produced in E. coli and applied to affinity chromatography using Strep-Tactin (25).
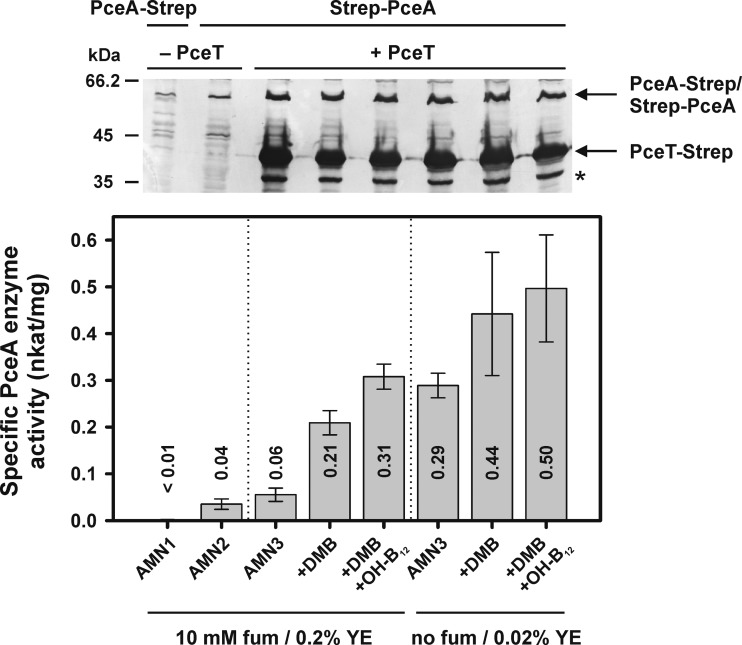
FIG 1.
Levels of the PceA and PceT protein and PceA activity in crude extracts of S. blattae strains expressing the pceA and pceT genes from D. hafniense Y51. S. blattae strains used: AMN1 (pceAY51-CStrep), AMN2 (NStrep-pceAY51), and AMN3 (NStrep-pceAY51, pceTY51-CStrep). DMB, 5,6-dimethylbenzimidazole; OH-B12, hydroxocobalamin; fum, fumarate; YE, yeast extract. The PceA enzyme activity data were obtained from at least three independent cultivation experiments. The standard deviation is given. The panel above the diagram displays an immunoblot developed with antibodies directed against the Strep-tag. Crude extracts (10 μg protein per lane) were separated using 12.5% SDS-PAGE. A degradation product of PceT-Strep is marked by an asterisk. The PceT-Strep degradation product was identified via immunoblotting using PceT-specific antibodies (data not shown).
The experiments described above were conducted with glycerol-grown S. blattae cells cultivated in the presence of fumarate (10 mM) and an elevated level of yeast extract (0.2% [wt/vol]). The addition of these supplements resulted in a more reproducible growth of the organism under the conditions applied. When only fumarate was added, no significant effect on the activity of the recombinant PceA enzyme was observed (data not shown). In contrast, cultivation with higher concentrations of yeast extract appeared to result in reduced PceA activity in a manner independent of the presence or absence of PceT. A reduction in the PceA enzyme activity was also observed when complex medium (tryptone-glucose-yeast extract-peptone [TGYEP]) was used. Hence, all cultivations described below were carried out on glycerol-containing mineral medium without fumarate and with 0.02% (wt/vol) yeast extract, since maximal PceA activity in AMN3 cells was observed under these conditions (Fig. 1). In consideration of the fact that such cultures exhibited a yield different from that seen with cells cultivated in the presence of 10 mM fumarate and 0.2% (wt/vol) yeast extract, the induced cells were harvested after 8 h (about 1.0 g protein/250 ml). The maximal PceA activity in crude extracts of S. blattae AMN3 (0.50 ± 0.11 nkat/mg) was about 50% of the activity measured under the same conditions in D. hafniense Y51 crude extracts (1.03 ± 0.15 nkat/mg), assuming that the recombinant PceA enzyme has a turnover rate similar to that of the native enzyme.
Stimulation of cobamide biosynthesis in Shimwellia blattae.
The positive effect of exogenous DMB (10 μM) on the PceA enzyme activity may be due to the stimulation of cobamide biosynthesis resulting from providing additional cobamide lower-ligand precursor, the production of which might be limited in S. blattae (for details of the cobamide structure, see Fig. S2 in the supplemental material.). The addition of OH-B12 (200 nM) to the growth medium might sustain the cobamide requirements of the cells when PceA is produced. To test for the amount and type of cobamide produced in S. blattae, cells grown on glycerol without and with the amendment of DMB and OH-B12 were subjected to corrinoid extraction and analysis. Almost twice the amount of cobamide was extracted from cells grown in the presence of DMB and OH-B12 than was extracted from nontreated cells (data not shown). The extracted and purified cobamides were analyzed using HPLC, and the HPLC elution profiles were compared to those of a mixture of cobamide standards (Fig. 2). The chromatogram of the sample derived from cells cultivated without exogenous DMB and OH-B12 displayed a single peak with a retention time identical to that of the pseudovitamin B12 (Coα-adeninyl-Coβ-cyano-cobamide) standard. The elution profile obtained from the cobamide extract of cells grown in the presence of DMB and OH-B12 showed a single signal with a retention time identical to that of the vitamin B12 (Coα-5,6-dimethylbenzimidazolyl-Coβ-cyano-cobamide) standard. The same elution profile was observed when only DMB was present in the growth medium. When only OH-B12 was present, the cells harbored both types of cobamides (data not shown). The isolated cobamides were analyzed using UHPLC coupled with electrospray ionization high-resolution Orbitrap XL mass spectrometry (UHPLC-ESI-Orbitrap XL MS). When the cobamide extracted from S. blattae cells cultivated without DMB or OH-B12 was applied to the MS analysis, two ions at m/z 1,344.5432 [M+H]+ and 672.7755 [M+2H]2+ were detected that have been assigned to the monoisotopic masses of singly and doubly protonated pseudovitamin B12 with mass differences of 1.1 ppm (calculated for C59H84O14N17CoP, 1,344.5448) and 0.7 ppm (C59H85O14N17CoP, 672.7760), respectively. The analysis of the cobamide purified from cells grown in the presence of both additives revealed two ions at m/z 1,355.5720 [M+H]+ and 678.2897 [M+2H]2+ that coincided with the exact masses of singly protonated vitamin B12 (C63H89O14N14CoP, 1,355.5747 [Δ ppm, 1.9]) and its doubly protonated form ( C63H90O14N14CoP, 678.2910 [Δ ppm, 1.6]). Based on these unique signatures, the production of pseudovitamin B12 and vitamin B12, respectively, was confirmed. Taking the results from the PceA activity measurements and cobamide analyses together, it was concluded that both types of cobamides, the adeninyl-cobamide and the 5,6-benzimidazolyl-cobamide, are functional as cofactors of the PceA enzyme from D. hafniense Y51 produced in S. blattae.
FIG 2.
Analysis of the cobamides extracted from S. blattae cells cultivated in the absence or presence of 5,6-dimethylbenzimidazole (DMB) and hydroxocobalamin (OH-B12). (A) Schematic representation of the different cobamides used as standards in the HPLC analysis. The substituents at position C176 in the linker moiety (CH3 = methyl group or H = hydrogen atom) and the lower ligand base (adenine or 5,6-dimethylbenzimidazole [DMB]) are indicated. The presence of a cyano group as the upper ligand of the cobalt is a result of the cobamide extraction procedure. (C) HPLC elution profiles of the extracted cobamides. The standard mixture contained Norps-B12 (norpseudovitamin B12), Nor-B12 (norvitamin B12), Ps-B12 (pseudovitamin B12), and B12 (vitamin B12). Abs, absorbance; mAU, milliabsorbance units.
Substrate spectrum of recombinant RdhA3 from D. hafniense DCB-2.
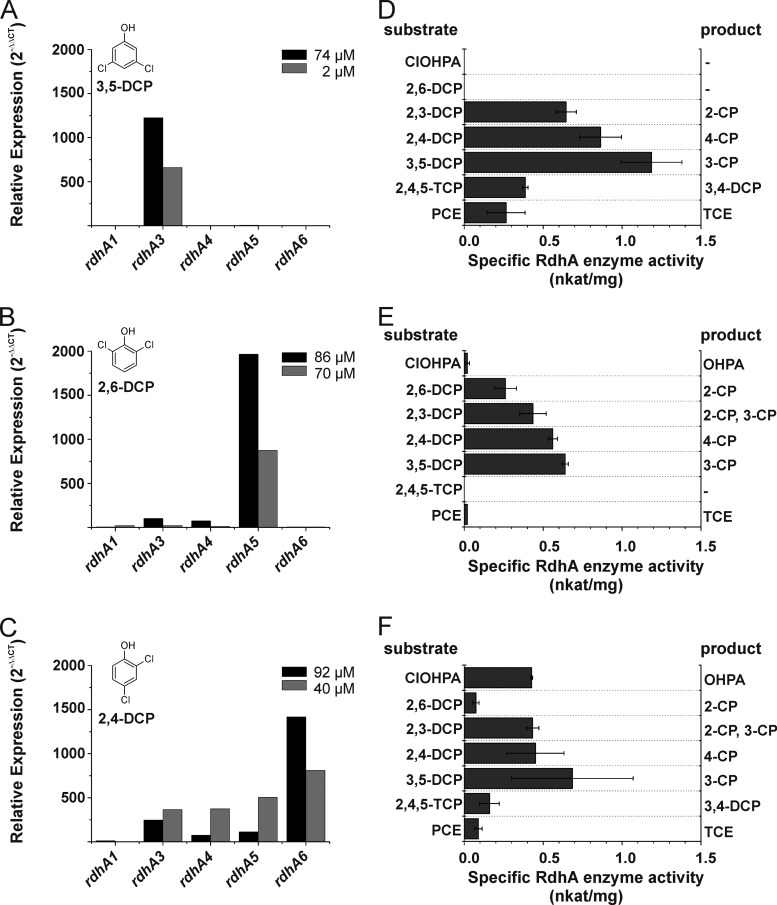
In order to demonstrate the capability of the heterologous expression system to be used for the analysis of uncharacterized rdhA genes and gene products, the rdhA3 gene from D. hafniense strain DCB-2 was expressed in S. blattae. The rdhA3 gene is one of seven rdhA genes present in the D. hafniense DCB-2 genome (37). The rdhA3 gene cluster shows organization similar to that of the pceA gene cluster in D. hafniense Y51. It also contains a gene encoding a trigger-factor-like chaperone (rdhT, gene locus tag Dhaf_0699). The expression of rdhA3 in strain DCB-2 was shown to be inducible by 3,5-dichlorophenol (3,5-DCP), a result pointing to aromatic organohalides as major substrates of the enzyme. The expression profiles of the rdhA genes in D. hafniense DCB-2 in response to the presence of different chlorophenols have been reexamined in this study. For the meta-substituted 3,5-DCP, the ortho-substituted 2,6-DCP, and the ortho/para-substituted 2,4-DCP, two independent cultures were analyzed. When the cells were harvested, the concentration of the chlorinated substrate in the culture was measured by HPLC analysis. Since the concentrations differed between the two cultures analyzed for each DCP, the results from the subsequent expression profiling experiment were not averaged (see also Fig. 3). The rdhA2 and rdhA7 genes were excluded from the analysis because a gene disruption in the case of rdhA2 and a nonsense mutation in the case of rdhA7 let the genes appear to be nonfunctional (37). From the results of the RT-qPCR experiments, it was concluded that the rdhA3 expression was specifically induced by 3,5-DCP (Fig. 3A) whereas the expression of all other rdhA genes was not. When 2,6-DCP was applied, only rdhA5 was significantly upregulated (Fig. 3B). In the presence of 2,4-DCP, the rdhA3, rdhA4, rdhA5, and rdhA6 expression was induced (Fig. 3C).
FIG 3.
Relative transcript levels of the rdhA1, rdhA3, rdhA4, rdhA5, and rdhA6 genes in cultures of D. hafniense DCB-2 (A to C) and conversion of different organohalides in crude extracts of the cells cultivated under the same conditions (D to F). Cells were cultivated with pyruvate in the presence of 3,5-dichlorophenol (3,5-DCP) (B and E), 2,6-dichlorophenol (2,6-DCP) (A and D), or 2,4-dichlorophenol (2,4-DCP) (C and F). The black and gray columns in panels A to C represent the data obtained from cultures with high and low dechlorination rates, respectively. The concentrations of the chlorophenols present in the cultures when the cells were harvested (OD578 = 0.3) are indicated in the figure. The results shown in panels D to F were obtained from three independent cultivations. The standard deviation from the average is given. CP, chlorophenol; DCP, dichlorophenol; TCP, trichlorophenol; ClOHPA, 3-chloro-4-hydroxy-phenylacetate; OHPA, 4-hydroxy-phenylacetate; PCE, tetrachloroethene; TCE, trichloroethene.
In order to correlate the expression data of the rdhA genes with a certain RdhA substrate spectrum in D. hafniense DCB-2, the dechlorination of different chlorophenols (2,6-DCP, 2,3-DCP, 2,4-DCP, 3,5-DCP, and 2,4,5-TCP), 3-chloro-4-hydroxy-phenylacetate (ClOHPA), and PCE was tested in crude extracts of the organism cultivated with 3,5-DCP, 2,6-DCP, or 2,4-DCP. When 3,5-DCP-treated cells were analyzed, dechlorination of meta- or ortho-chlorinated dichlorophenols (3,5-DCP, 2,3-DCP, 2,4-DCP) was observed (Fig. 3D). In addition, 2,4,5-trichlorophenol (2,4,5-TCP) was dechlorinated in the ortho position, yielding 3,4-DCP, which was not further dechlorinated. PCE was dechlorinated to TCE as detected by GC measurements. ClOHPA and 2,6-DCP were not converted. The dehalogenation of the ortho-halogenated 2,6-DCP was mediated by cells cultivated in the presence of this compound or 2,4-DCP (Fig. 3E and F). A significant transformation of ClOHPA was observed only with cells grown with 2,4-DCP (Fig. 3F). Besides rdhA3, rdhA4, and rdhA5, expression of the reductive dehalogenase gene rdhA6 was induced under these conditions. The rdhA6 gene product was previously shown to convert ClOHPA (38). An unambiguous conclusion on the substrate spectrum of the different rdhA gene products can hardly be drawn from such data, since an overlap of the substrate spectra cannot be excluded (see results for the 2,4-DCP-treated cells). Hence, the substrate range of RdhA proteins from D. hafniense DCB-2 must be tested in a reductive dehalogenase-free background to draw unambiguous conclusions. From the experiments described above, first evidence was gained on the range of substrates used by RdhA3. To confirm these data, the substrate spectrum of RdhA3 was tested with recombinant enzyme produced in S. blattae. When the C- or N-terminal affinity-tagged RdhA3 fusion proteins were produced, no differences in the amounts of recombinant enzyme and levels of enzyme activity were observed (data not shown). For further analysis, the N-terminal Strep-tagged RdhA3 construct was used. The dedicated chaperone RdhT was coproduced, and the cultures were amended with DMB and OH-B12. The 3,5-DCP-dechlorinating activity of the recombinant enzyme was found to be at a maximum after 18 h of cultivation after induction of the gene expression in S. blattae AMN4. From the dichlorophenols tested in the enzyme assay, only 3,5-DCP, 2,4-DCP, and 2,3-DCP were converted (Table 5). The conversion of the meta-chlorinated 3,5-DCP led to the formation of 3-chlorophenol (3-CP) and the dehalogenation of 2,3-DCP to 2-chlorophenol (2-CP) production. The ortho-chlorinated 2,6-DCP was not dehalogenated by the enzyme, a result also obtained with 3,4-DCP. However, 2,4,6-TCP was converted via 2,4-DCP to 4-CP as the final product. When 2,4,5-TCP was applied in the assay, the formation of 3,4-DCP was observed, which was not further converted. ClOHPA was not a substrate of the enzyme. PCE was dechlorinated to TCE with low rates. The overall pattern of substrates converted by the recombinant RdhA3 was identical to that seen with the control, which was the substrate range measured for cells of D. hafniense DCB-2 cultivated in the presence of 3,5-DCP and exclusively expressing the rdhA3 gene (Fig. 3D). This result confirmed the usability of the heterologous production system for the characterization of rdhA genes and gene products.
TABLE 5.
Rates of reductive dehalogenation of different organohalides by S. blattae AMN4 crude extract containing the D. hafniense DCB-2 RdhA3 enzymea
| Substrate | Specific RdhA3 enzyme activity (nkat/mg) | Product |
|---|---|---|
| ClOHPA | 0.0 | |
| 2,6-DCP | 0.0 | |
| 2,3-DCP | 0.15 ± 0.03 | 2-CP |
| 2,4-DCP | 0.40 ± 0.09 | 4-CP |
| 3,4-DCP | 0.0 | |
| 3,5-DCP | 0.41 ± 0.08 | 3-CP |
| 2,4,5-TCP | 0.24 ± 0.07 | 3,4-DCP |
| 2,4,6-TCP | 0.17 ± 0.03 | 4-CP |
| PCE | 0.03 ± 0.01 | TCE |
The average values of the results from at least three independent cultures are given together with the standard deviations.
DISCUSSION
All previous attempts (18–20) for functional expression of rdhA genes in Escherichia coli failed, most probably due to the absence of a complete cobamide biosynthesis pathway in this organism. Instead of using the standard expression host E. coli, which is not able to synthesize cobamides de novo, the cobamide-producing Shimwellia blattae ATCC 33430 strain was chosen in the study presented here. From the production of functional RdhA enzymes in S. blattae rather than in E. coli, it can be concluded that a certain intracellular level of cobamide cofactor is crucial for the production of active enzyme. Most natural E. coli isolates lack cobamide-dependent catabolic metabolism but harbor B12-dependent anabolic enzyme functions (e.g., methionine synthase) (36). The low cobamide demand of E. coli is covered by corrinoid salvage from the environment. In S. blattae, the uptake of cobamides or precursors thereof may be surplus to the intrinsic source of cobamide cofactor, the de novo biosynthesis. The central enzyme of the glycerol catabolism in S. blattae is the cobamide-dependent glycerol dehydratase. The cellular level of enzymes involved in energy metabolism is expected to be higher than that of anabolic enzymes. Hence, the cobamide production in S. blattae might be adapted to a higher demand of cobamide cofactor, especially when cultivated with glycerol. In this study, all modifications of the cultivation conditions increasing the amount of cobamide produced by S. blattae and forcing the cobamide-dependent growth with glycerol led to an increase of the amount of active RdhA enzyme formed. Under the cultivation conditions applied, S. blattae cells produced either pseudo-B12, which contains an adeninyl moiety as the lower ligand base of the cobalt ion, or 5,6-dimethylbenzimidazolyl-cobamide. Exogenous 5,6-dimethylbenzimidazole (DMB) substantially replaced the adenine as the lower ligand base and led to an increase in cobamide production. The functionality of 5,6-dimethylbenzimidazolyl-cobamide as a cofactor in the recombinant RdhA enzymes may point to the utilization of this type of cobamide as part of the reductive dehalogenases in both Desulfitobacterium hafniense strains, a fact that has to be proven in further studies. No negative effect of exogenous DMB on the PCE-dependent growth or dechlorination rate has been observed in D. hafniense Y51 or DCB-2 cells (unpublished results) in contrast to the results seen with the PCE-dechlorinating Sulfurospirillum multivorans. In the latter organism, PCE-dependent growth was impaired by the presence of DMB (33). The presence of DMB in the growth medium of the S. blattae expression strains resulted in a higher intracellular cobamide level accompanied by higher activity of the recombinant RdhA enzyme. The amendment of DMB caused a modification in the cobamide structure, which changed from pseudo-B12 to the 5,6-dimethylbenzimidazolyl-cobamide. Based on the results obtained in this study, the possibility of a positive effect of this structural change on the RdhA activity cannot be excluded.
In this survey, RdhA enzymes were exclusively analyzed that, according to the operon structure, involve a chaperone (PceT/RdhT; trigger factor-like chaperone) in RdhA maturation. It has to be tested if the expression system can also be applied to RdhA enzymes encoded in rdh gene clusters lacking genes for a specific folding helper protein, which is usually the case for rdh gene clusters of Dehalococcoides mccartyi (8–12). Since reductive dehalogenase activity was detectable also in the absence of the PceT/RdhT protein, a functional production of other RdhAs without the coproduction of a dedicated chaperone appears feasible. The preferential use of benzimidazolyl-cobamide cofactors for reductive dehalogenation by the cobamide-auxotroph D. mccartyi was reported recently (39–41). Hence, the production of 5,6-dimethylbenzimidazolyl-cobamide in S. blattae cells grown in the presence of DMB might be beneficial for the heterologous production of RdhAs from D. mccartyi.
Using the rdhA3 gene product from D. hafniense DCB-2, the potential use of the expression system for the characterization of RdhAs with unknown substrate spectrum was demonstrated. The rdhA3 gene was the only reductive dehalogenase gene expressed in 3,5-DCP-exposed D. hafniense DCB-2. Therefore, the substrate spectrum of the respective gene product could be analyzed in crude extracts of the organism. The same substrate range was shown for the recombinant enzyme produced in S. blattae. The native and the recombinant RdhA3 converted 3,5-DCP, 2,4-DCP, and 2,3-DCP rather than 2,6-DCP and 3,4-DCP. This approach allowed also for an assignment of the PCE conversion measured in crude extracts of 3,5-DCP-grown D. hafniense DCB-2 cells to the function of the rdhA3 gene product. The difference in the dechlorination rates measured in crude extracts of D. hafniense DCB-2 and the S. blattae strain producing RdhA3 was higher for PCE than for all the chlorophenols. This cannot be explained at the moment. It can only be speculated that a certain factor in D. hafniense DCB-2 promotes PCE dechlorination or that, e.g., the Strep-tag and/or a different cobamide cofactor may have a negative impact specifically on the PCE dechlorination activity of heterologously produced RdhA3. The RdhA3 gene product from D. hafniense DCB-2 displayed 67% sequence identity to the PCE reductive dehalogenase of D. hafniense Y51; however, a conversion of chlorophenols by the recombinant PceA enzyme could not be shown under the conditions applied in this study.
A lot of reductively dehalogenating bacteria contain several homologues of reductive dehalogenase genes (rdhA) that might be coexpressed and hard to analyze for the specific substrate range of the respective gene products. These limitations in the analyses of the substrate spectra of reductive dehalogenases might be overcome by the functional heterologous expression system for rdhA genes described in this study. In addition, it is expected to be helpful for the functional analysis of RdhA enzymes, e.g., by site-directed mutagenesis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the International Leibniz Research School (ILRS) for Microbial and Biomolecular Interactions (project 2708) and the DFG Research Unit FOR1530.
We express our gratitude to Rolf Daniel (University of Göttingen, Germany) for providing Shimwellia blattae ATCC 33430 and to Taiki Futagami (Kagoshima University, Japan) for supplying antibodies against PceT. The excellent technical assistance by Stefanie Kröckel is acknowledged.
Footnotes
Published ahead of print 9 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00881-14.
REFERENCES
- 1.Hug LA, Maphosa F, Leys D, Löffler FE, Smidt H, Edwards EA, Adrian L. 2013. Overview of organohalide-respiring bacteria and a proposal for a classification system for reductive dehalogenases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20120322. 10.1098/rstb.2012.0322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holliger C, Wohlfarth G, Diekert G. 1998. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 22:383–398. 10.1111/j.1574-6976.1998.tb00377.x [DOI] [Google Scholar]
- 3.Leys D, Adrian L, Smidt H. 2013. Organohalide respiration: microbes breathing chlorinated molecules. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20120316. 10.1098/rstb.2012.0316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scholz-Muramatsu H, Neumann A, Meßmer M, Moore E, Diekert G. 1995. Isolation and characterization of Dehalospirillum multivorans gen. nov., sp. nov., a tetrachloroethene-utilizing, strictly anaerobic bacterium. Arch. Microbiol. 163:48–56. 10.1007/BF00262203 [DOI] [Google Scholar]
- 5.Villemur R, Lanthier M, Beaudet R, Lépine F. 2006. The Desulfitobacterium genus. FEMS Microbiol. Rev. 30:706–733. 10.1111/j.1574-6976.2006.00029.x [DOI] [PubMed] [Google Scholar]
- 6.Holliger C, Hahn D, Harmsen H, Ludwig W, Schumacher W, Tindall B, Vazquez F, Weiss N, Zehnder AJ. 1998. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch. Microbiol. 169:313–321. 10.1007/s002030050577 [DOI] [PubMed] [Google Scholar]
- 7.Löffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT, Müller JA, Fullerton H, Zinder SH, Spormann AM. 2013. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 63(Pt 2):625–635. 10.1099/ijs.0.034926-0 [DOI] [PubMed] [Google Scholar]
- 8.Seshadri R, Adrian L, Fouts DE, Eisen JA, Phillippy AM, Methe BA, Ward NL, Nelson WC, Deboy RT, Khouri HM, Kolonay JF, Dodson RJ, Daugherty SC, Brinkac LM, Sullivan SA, Madupu R, Nelson KE, Kang KH, Impraim M, Tran K, Robinson JM, Forberger HA, Fraser CM, Zinder SH, Heidelberg JF. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105–108. 10.1126/science.1102226 [DOI] [PubMed] [Google Scholar]
- 9.Kube M, Beck A, Zinder SH, Kuhl H, Reinhardt R, Adrian L. 2005. Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23:1269–1273. 10.1038/nbt1131 [DOI] [PubMed] [Google Scholar]
- 10.Sung Y, Ritalahti KM, Apkarian RP, Löffler FE. 2006. Quantitative PCR confirms purity of strain GT, a novel trichloroethene-to-ethene-respiring Dehalococcoides isolate. Appl. Environ. Microbiol. 72:1980–1987. 10.1128/AEM.72.3.1980-1987.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMurdie PJ, Behrens SF, Müller JA, Göke J, Ritalahti KM, Wagner R, Goltsman E, Lapidus A, Holmes S, Löffler FE, Spormann AM. 2009. Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet. 5:e1000714. 10.1371/journal.pgen.1000714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pöritz M, Goris T, Wubet T, Tarkka MT, Buscot F, Nijenhuis I, Lechner U, Adrian L. 2013. Genome sequences of two dehalogenation specialists - Dehalococcoides mccartyi strains BTF08 and DCMB5 enriched from the highly polluted Bitterfeld region. FEMS Microbiol. Lett. 343:101–104. 10.1111/1574-6968.12160 [DOI] [PubMed] [Google Scholar]
- 13.Kruse T, Maillard J, Goodwin L, Woyke T, Teshima H, Bruce D, Detter C, Tapia R, Han C, Huntemann M, Wei CL, Han J, Chen A, Kyrpides N, Szeto E, Markowitz V, Ivanova N, Pagani I, Pati A, Pitluck S, Nolan M, Holliger C, Smidt H. 2013. Complete genome sequence of Dehalobacter restrictus PER-K23T. Stand. Genomic Sci. 30:375–388. 10.4056/sigs.3787426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner A, Adrian L, Kleinsteuber S, Andreesen JR, Lechner U. 2009. Transcription analysis of genes encoding homologues of reductive dehalogenases in “Dehalococcoides” sp. strain CBDB1 by using terminal restriction fragment length polymorphism and quantitative PCR. Appl. Environ. Microbiol. 75:1876–1884. 10.1128/AEM.01042-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bisaillon A, Beaudet R, Lépine F, Villemur R. 2011. Quantitative analysis of the relative transcript levels of four chlorophenol reductive dehalogenase genes in Desulfitobacterium hafniense PCP-1 exposed to chlorophenols. Appl. Environ. Microbiol. 77:6261–6264. 10.1128/AEM.00390-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner A, Segler L, Kleinsteuber S, Sawers G, Smidt H, Lechner U. 2013. Regulation of reductive dehalogenase gene transcription in Dehalococcoides mccartyi. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20120317. 10.1098/rstb.2012.0317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer T, Berks BC. 2012. The twin-arginine translocation (Tat) protein export pathway. Nat. Rev. Microbiol. 10:483–496. 10.1038/nrmicro2814 [DOI] [PubMed] [Google Scholar]
- 18.John M, Schmitz RP, Westermann M, Richter W, Diekert G. 2006. Growth substrate dependent localization of tetrachloroethene reductive dehalogenase in Sulfurospirillum multivorans. Arch. Microbiol. 186:99–106. 10.1007/s00203-006-0125-5 [DOI] [PubMed] [Google Scholar]
- 19.Reinhold A, Westermann M, Seifert J, von Bergen M, Schubert T, Diekert G. 2012. Impact of vitamin B12 on formation of the tetrachloroethene reductive dehalogenase in Desulfitobacterium hafniense strain Y51. Appl. Environ. Microbiol. 78:8025–8032. 10.1128/AEM.02173-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suyama A, Iwakiri R, Keiichirou K, Tokunaga T, Sera N, Furukawa K. 2001. Isolation and characterization of Desulfitobacterium sp. strain Y51 capable of efficient dehalogenation of tetrachloroethene and polychloroethanes. Biosci. Biotechnol. Biochem. 65:1474–1481. 10.1271/bbb.65.1474 [DOI] [PubMed] [Google Scholar]
- 21.Futagami T, Tsuboi Y, Suyama A, Goto M, Furukawa K. 2006. Emergence of two types of nondechlorinating variants in the tetrachloroethene-halorespiring Desulfitobacterium sp. strain Y51. Appl. Microbiol. Biotechnol. 70:720–728. 10.1007/s00253-005-0112-9 [DOI] [PubMed] [Google Scholar]
- 22.Suyama A, Yamashita M, Yoshino S, Furukawa K. 2002. Molecular characterization of the PceA reductive dehalogenase of Desulfitobacterium sp. strain Y51. J. Bacteriol. 184:3419–3425. 10.1128/JB.184.13.3419-3425.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumann A, Wohlfarth G, Diekert G. 1998. Tetrachloroethene dehalogenase from Dehalospirillum multivorans: cloning, sequencing of the encoding genes, and expression of the pceA gene in Escherichia coli. J. Bacteriol. 180:4140–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nonaka H, Keresztes G, Shinoda Y, Ikenaga Y, Abe M, Naito K, Inatomi K, Furukawa K, Inui M, Yukawai H. 2006. Complete genome sequence of the dehalorespiring bacterium Desulfitobacterium hafniense Y51 and comparison with Dehalococcoides ethenogenes 195. J. Bacteriol. 188:2262–2274. 10.1128/JB.188.6.2262-2274.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sjuts H, Fisher K, Dunstan MS, Rigby SE, Leys D. 2012. Heterologous expression, purification and cofactor reconstitution of the reductive dehalogenase PceA from Dehalobacter restrictus. Protein Expr. Purif. 85:224–229. 10.1016/j.pep.2012.08.007 [DOI] [PubMed] [Google Scholar]
- 26.Morita Y, Futagami T, Goto M, Furukawa K. 2009. Functional characterization of the trigger factor protein PceT of the tetrachloroethene-dechlorinating Desulfitobacterium hafniense Y51. Appl. Microbiol. Biotechnol. 83:775–781. 10.1007/s00253-009-1958-z [DOI] [PubMed] [Google Scholar]
- 27.Maillard J, Genevaux P, Holliger C. 2011. Redundancy and specificity of multiple trigger factor chaperones in Desulfitobacteria. Microbiology 157:2410–2421. 10.1099/mic.0.050880-0 [DOI] [PubMed] [Google Scholar]
- 28.Andres S, Wiezer A, Bendfeldt H, Waschkowitz T, Toeche-Mittler C, Daniel R. 2004. Insights into the genome of the enteric bacterium Escherichia blattae: cobalamin (B12) biosynthesis, B12-dependent reactions, and inactivation of the gene region encoding B12-dependent glycerol dehydratase by a new mu-like prophage. J. Mol. Microbiol. Biotechnol. 8:150–168. 10.1159/000085788 [DOI] [PubMed] [Google Scholar]
- 29.Inoue H, Nojima H, Okayama H. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23–28. 10.1016/0378-1119(90)90336-P [DOI] [PubMed] [Google Scholar]
- 30.Neumann A, Wohlfarth G, Diekert G. 1996. Purification and characterization of tetrachloroethene reductive dehalogenase from Dehalospirillum multivorans. J. Biol. Chem. 271:16515–16519. 10.1074/jbc.271.28.16515 [DOI] [PubMed] [Google Scholar]
- 31.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 32.John M, Rubick R, Schmitz RPH, Rakoczy J, Schubert T, Diekert G. 2009. Retentive memory of bacteria: long-term regulation of dehalorespiration in Sulfurospirillum multivorans. J. Bacteriol. 191:1650–1655. 10.1128/JB.00597-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller S, Ruetz M, Kunze C, Kräutler B, Diekert G, Schubert T. 6 October 2013. Exogenous 5,6-dimethylbenzimidazole caused production of a non-functional tetrachloroethene reductive dehalogenase in Sulfurospirillum multivorans. Environ. Microbiol. 10.1111/1462-2920.12268 [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 35.Burgess NR, McDermott SN, Whiting J. 1973. Aerobic bacteria occurring in the hind-gut of the cockroach, Blatta orientalis. J. Hyg. (Lond) 71:1–7. 10.1017/S0022172400046155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrence JG, Roth JR. 1996. Evolution of coenzyme B12 synthesis among enteric bacteria: evidence for loss and reacquisition of a multigene complex. Genetics 142:11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim SH, Harzman C, Davis JK, Hutcheson R, Broderick JB, Marsh TL, Tiedje JM. 2012. Genome sequence of Desulfitobacterium hafniense DCB-2, a Gram-positive anaerobe capable of dehalogenation and metal reduction. BMC Microbiol. 12:21. 10.1186/1471-2180-12-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christiansen N, Ahring BK, Wohlfarth G, Diekert G. 1998. Purification and characterization of the 3-chloro-4-hydroxy-phenylacetate reductive dehalogenase of Desulfitobacterium hafniense. FEBS Lett. 436:159–162. 10.1016/S0014-5793(98)01114-4 [DOI] [PubMed] [Google Scholar]
- 39.Yan J, Ritalahti KM, Wagner DD, Löffler FE. 2012. Unexpected specificity of interspecies cobamide transfer from Geobacter spp. to organohalide-respiring Dehalococcoides mccartyi strains. Appl. Environ. Microbiol. 78:6630–6636. 10.1128/AEM.01535-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi S, Seth EC, Men YJ, Stabler SP, Allen RH, Alvarez-Cohen L, Taga ME. 2012. Versatility in corrinoid salvaging and remodeling pathways supports corrinoid-dependent metabolism in Dehalococcoides mccartyi. Appl. Environ. Microbiol. 78:7745–7752. 10.1128/AEM.02150-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan J, Im J, Yang Y, Löffler FE. 2013. Guided cobalamin biosynthesis supports Dehalococcoides mccartyi reductive dechlorination activity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20120320. 10.1098/rstb.2012.0320 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.