Abstract
Marine microbes use alginate lyases to degrade and catabolize alginate, a major cell wall matrix polysaccharide of brown seaweeds. Microbes frequently contain multiple, apparently redundant alginate lyases, raising the question of whether these enzymes have complementary functions. We report here on the molecular cloning and functional characterization of three exo-type oligoalginate lyases (OalA, OalB, and OalC) from Vibrio splendidus 12B01 (12B01), a marine bacterioplankton species. OalA was most active at 16°C, had a pH optimum of 6.5, and displayed activities toward poly-β-d-mannuronate [poly(M)] and poly-α-l-guluronate [poly(G)], indicating that it is a bifunctional enzyme. OalB and OalC were most active at 30 and 35°C, had pH optima of 7.0 and 7.5, and degraded poly(M·G) and poly(M), respectively. Detailed kinetic analyses of oligoalginate lyases with poly(G), poly(M), and poly(M·G) and sodium alginate as substrates demonstrated that OalA and OalC preferred poly(M), whereas OalB preferred poly(M·G). The catalytic efficiency (kcat/Km) of OalA against poly(M) increased with decreasing size of the substrate. OalA showed kcat/Km from 2,130 mg−1 ml s−1 for the trisaccharide to 224 mg−1 ml s−1 for larger oligomers of ∼50 residues, and 50.5 mg−1 ml s−1 for high-molecular-weight alginate. Although OalA was most active on the trisaccharide, OalB and OalC preferred dimers. Taken together, our results indicate that these three Oals have complementary substrate scopes and temperature and pH adaptations.
INTRODUCTION
Algal polysaccharides constitute an important carbon and energy resource for marine organisms and have been considered as cost-competitive biomass for the production of biodiesel, bioethanol, and biohydrogen (1, 2). Macroalgal carbohydrates exhibit several features of a promising feedstock that may complement the increased global demand on energy and food production. The cultivation of macroalgae does not require arable land, freshwater, or fertilizer, circumventing adverse impacts on food supplies and resource availability (3). Moreover, algal polysaccharides are present in the form of gels rather than as crystalline fibers such as cellulose and are therefore easier to digest with enzymes than terrestrial plant biomass (4). Furthermore, most algae lack or contain only small quantities of lignin (5), a natural glue that renders cellulose from terrestrial plants recalcitrant to degradation by cellulases. The most abundant carbohydrate in brown macroalgae, such as kelps, is alginate, a major component of the cell wall matrix. Interestingly, although alginate constitutes 14 to 37% of seaweed biomass, this polysaccharide has only recently been considered a carbon source or starting material for biorefinery (6).
Alginate is a linear block copolymer of two uronic acids, β-d-mannuronate and α-l-guluronate, arranged in various sequences of poly-β-d-mannuronate [poly(M)], poly-α-l-guluronate [poly(G)], and the heteropolymer [poly(M·G)] (7). The gelling properties strongly depend on the poly(G) content of the algal polysaccharide, because the chelation of Ca2+ cations by guluronate residues creates intermolecular networks giving rise to viscous solutions and gels (8). Alginates are also found in some bacteria; Pseudomonas aeruginosa and Azotobacter vinelandii produce alginate and also express alginate lyases (9, 10). In addition to its potential for biofuels, pharmaceutical and medical uses of alginates have gained interest due to attractive physicochemical properties. For example, alginate gels can be fabricated into hydrophilic drug carriers and molded into biocompatible matrix materials for immobilized cells or growth of artificial organs (11).
To date, several microorganisms that metabolize alginate have been characterized (12). Alginate lyases belong to the polysaccharide lyases (PLs), which catalyze cleavage of the glycosidic bond via β-elimination. A total of 23 Pl families exist in the carbohydrate-active enzyme (CAZy) database (www.cazy.org) (13). Alginate lyases are present in the PL5, PL6, PL7, PL14, PL15, PL17, and PL18 families and can be further organized into alginate lyases that cleave the polymer with an endo mode of action and oligoalginate lyases, which use an exo mode of action to cleave unsaturated and saturated monomers from the nonreducing end (9, 14) (see Fig. S1 in the supplemental material). Alginate lyases belonging to the PL5 and PL7 families generally degrade alginate polymers endolytically into oligosaccharides (8). Oligoalginate lyases (Oals) in the PL15 and PL17 families degrade alginate exolytically to produce monosaccharides (15). Alginate lyases are also valuable bioanalytical tools. Recent applications of alginate lyases have included the use of an M lyase from a Photobacterium sp. to produce poly(M) and poly(M·G) blocks from acetylated and deacetylated P. aeruginosa alginate (7). These defined substrates were used to probe the specificity of other lyases (7). Similarly, a G-specific lyase from Flavobacterium multivorum, which degrades poly(G) and poly(M·G) alginate, has been applied toward the preparation of poly(M) blocks from sodium alginate for substrate specificity studies (16, 17).
Recently, an alginate degradation pathway (∼40 kbp) from the marine bacterium Vibrio splendidus, which included genes for import and metabolism of alginate, has been transferred 12BO1 (18) into an Escherichia coli host. V. splendidus is an abundant marine vibrio and appears to have relatively recently diversified into a range of ecologically defined populations (19), including some that result in complex host interactions (20). Vibrios are abundant, planktonic bacteria in coastal waters, and they frequently colonize the intestines of animals such as crustaceans or mammals (20). After the transfer of the alginate degradation pathway from V. splendidus into E. coli, this bacterium was further bioengineered, which resulted in the successful conversion of alginate into bioethanol (18). The transferred pathway contained three oligoalginate lyases belonging to the PL15 (OalA) and the PL17 (OalB and OalC) families, sharing 40 to 50% sequence identity with previously characterized oligoalginate lyases (21). The relevance of these genes for alginate conversion was confirmed by the deletion of oalA, oalB, and oalC in E. coli, which impaired growth on alginate oligomers and polymers (21). However, biochemical characterization of these three Oals has not been carried out, and their specific role for the efficient conversion of alginate remained unclear.
We report here the cloning, heterologous expression, classification, and characterization of the three oligoalginate lyases from V. splendidus 12B01, including detailed kinetic parameters and substrate scope, providing comparative new insights into the PL15 and PL17 family of alginate lyases. Our results indicate that these enzymes have different pH and temperature profiles, as well as complementary substrate scopes.
MATERIALS AND METHODS
Materials.
Restriction enzymes, factor Xa protease, and Q5 high-fidelity DNA polymerase were obtained from New England BioLabs (Ipswich, MA). Kanamycin, lysozyme, Triton X-100, guanidine hydrochloride, and bovine serum albumin were purchased from Sigma-Aldrich (St. Louis, MO). Genomic DNA purification kit was obtained from Promega (Madison, WI). Oligonucleotide primers were obtained from Integrated DNA Technologies (Coralville, IA). His-trap FF crude columns and a fast protein liquid chromatography (FPLC) system (ATKA pure M1) were purchased from GE Healthcare (Princeton, NJ). Reagents for the protein assay and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), protein markers, Sepharose Fast Flow, Bio-Gel P2, and the high-pressure liquid chromatography (HPLC) column (Hypercarb 100 by 3 mm) were obtained from Bio-Rad (Hercules, CA). All other chemicals were purchased from Thermo-Fisher Scientific (Pittsburgh, PA) unless otherwise specified. poly(M) (degree of polymerization [dp], ∼20), poly(G) (dp, ∼20), and poly(M·G) (dp, ∼20) substrates were prepared as previously described (22). Mannuronate oligosaccharides (dp, 30 to 50) were obtained from Elicityl (Crolles, France). A low-viscosity grade of sodium alginate (viscosity, 5.0 to 40.0 cP) isolated from brown algae was purchased from Sigma-Aldrich.
Bacterial strains and culture conditions.
The Vibrio splendidus 12B01 genomic sequence (accession number NZ_AAMR00000000.1) and oligoalginate lyase sequences were accessed from the National Center for Biotechnology Information. 12B01 was obtained from C. V. Rao, University of Illinois at Urbana-Champaign. E. coli DH5α and E. coli BL21(DE3) were used as hosts for cloning and heterologous protein expression, respectively. Both E. coli strains were grown in lysogeny broth (Bacto tryptone [10 g/liter], yeast extract [5 g/liter], and NaCl [10 g/liter]) supplemented with ampicillin or kanamycin (100 and 50 μg ml−1, respectively) at 37°C.
Cloning and expression of OalA, OalB, and OalC from V. splendidus 12B01.
PCR amplification was conducted using Q5 high-fidelity DNA polymerase. The genes were amplified from the genomic DNA of V. splendidus 12B01 using the oligonucleotide primers shown in Table S1 in the supplemental material. The sequences of the oligonucleotide primers are based on the DNA sequences of the oligoalginate lyases (OalA, OalB, and OalC) from V. splendidus 12B01 (EMBL accession numbers EAP93067.1, EAP93062.1, and EAP93063.1, respectively). SacI and BamHI sites were included in the primers for cloning of oalA into pMAL-c2X. BamHI and SalI sites or NdeI and BamHI sites were incorporated into the forward and reverse primers for cloning of oalB and oalC, respectively, into the expression vector pET-28(a). The cloned genes were confirmed to be free of point mutations by DNA sequencing using the BigDye terminator sequencing method and analyzed with a 3730×L genetic analyzer (Applied Biosystems, Foster City, CA) at the Biotechnology Center at the University of Illinois at Urbana-Champaign (Urbana, IL). The oalA gene was ligated with pMAL-c2X vector to give pMAL-c2X-oalA, which is under the control of the Ptac promoter and expresses OalA as a protein fused to the C terminus of a maltose-binding protein (MBP) tag. The oalB and oalC genes were ligated with pET-28(a) vector to give pET28(a)-oalB and pET28(a)-oalC, respectively, which are under the control of the T7 promoter and express the respective proteins with an N-terminal His6 tag.
Protein expression and purification of OalA, OalB, and OalC.
E. coli BL21(DE3) containing the oalA, oalB, and oalC genes were grown overnight at 37°C on a rotary shaker at 250 rpm. Overnight culture (5 ml) was used to inoculate a fresh culture (500 ml), which was grown at 37°C with shaking at 250 rpm until the optical density at 600 nm reached 0.6 to 1.0. The expression of target proteins were then induced with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and further incubated at 16°C for 20 h. To purify the recombinant OalA, cell pellets were resuspended in 100 mM Tris-HCl buffer (pH 7) supplemented with 25 μg of DNase I ml−1. The cell suspension was incubated on ice for 30 min in the presence of 1 mg of lysozyme/ml. Cell disruption was carried out by sonication on ice for 5 min, and the lysate was centrifuged at 14,000 × g for 20 min at 4°C to remove the cell debris. The resulting crude extract was retained for purification. The isolation of OalA as a fusion protein was facilitated by MBP affinity purification as described elsewhere (23). After factor Xa cleavage, the OalA was separated from MBP, factor Xa, and trace contaminants by amylose and Sepharose ion-exchange chromatography. OalB and OalC were refolded using the flash dilution method described elsewhere (24). The refolded proteins were applied onto Ni-NTA Superflow cartridges (1 ml; Qiagen) previously equilibrated with binding buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole; pH 8.0). Unbound proteins were washed from the column with washing buffer (50 mM NaH2PO4, 300 mM NaCl, 30 mM imidazole; pH 8.0). Then, OalB and OalC were eluted from the column with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole; pH 8.0). Enzyme fractions were analyzed by SDS–12% PAGE and visualized by staining with SimplyBlue SafeStain (Invitrogen).
Protein quantification and determination of molecular mass.
Protein concentrations were determined by the Bradford method according to the manufacturer's protocol. The molecular weights of the protein were determined using a Bio-Sil SEC 250 column on a Shimadzu HPLC system (Shimadzu, Kyoto, Japan). The mobile phase consisted of 50 mM Na2HPO4, 50 mM NaH2PO4, 150 mM NaCl, and 10 mM NaN3 (pH 6.8), and the flow rate was 1.0 ml/min. The molecular weight was calculated by comparing the retention times to those of protein molecular weight standards. The quaternary structures were determined based on the molecular weight observed by HPLC, and the molecular weights of monomeric subunits were determined by SDS-PAGE analysis.
Alginate degradation activity assay.
The formation of alginate degradation products was analyzed by measuring the absorbance arising from the formation of unsaturated urinates, either at 235 nm or by the thiobarbituric acid (TBA) assay at 548 nm (25). The thiobarbituric acid test was used with the following modifications. The sample in 0.20 ml or less of solution was added to 0.25 ml of 0.025 N H2IO6 in 0.125 N H2SO4. After 20 min at room temperature, 0.50 ml of 2% sodium arsenite in 0.5 N HCl was added with shaking, and the solution was permitted to stand for 2 min. Thiobarbituric acid (pH 2, 2 ml, 0.3%) was added with stirring, and then the mixture was heated to 100°C for 10 min. Enzyme activity was determined by monitoring the increase in absorbance at 548 nm arising from the condensation of β-formyl-pyruvic acid and TBA (molar absorption coefficient ε = 29.0 × 103 M−1 cm−1). For the quantitative analysis of α-keto acid (4-deoxy-l-erythro-5-hexoseulose uronic acid), 2-deoxy-d-glucose was used as a standard, because deoxy sugars react with TBA (26).
Optimum pH, temperature, and metal ions for alginate degradation.
The pH optimum of the purified enzyme was determined using the following three buffer systems: 50 mM sodium phosphate (pH 6.0 to 7.5), 50 mM Tris-HCl (pH 7.0 to 8.0), and 50 mM Tris-glycine-NaOH (pH 8.0 to 8.5). The optimum temperature was measured in 50 mM sodium phosphate buffer (pH 6.5 or 7.0 or 7.5), at various temperatures, from 10 to 55°C. The thermostability was investigated by incubating the enzyme from 10 to 40°C in the same buffer but without substrate. Samples were removed at fixed time intervals and allowed to cool in an ice bath. Residual activity was tested under standard conditions.
Before studying the effect of metal ions on OalA activity, the purified enzyme was dialyzed against 50 mM sodium phosphate (pH 6.0 to 7.5) containing 10 mM EDTA for 24 h at 4°C. Subsequently, the enzyme was dialyzed against EDTA-free sodium phosphate buffer. The enzyme activity was then assessed with 0.1% sodium alginate under standard assay conditions in the presence of various metal ions (MgCl2, MnCl2, ZnCl2, CaCl2, CuSO4, CoCl2, BaCl3, KCl, FeCl3, and NaCl) at 1 mM.
Determination of kinetic parameters.
The kinetic parameters of OalA were determined in sodium phosphate (pH 6.5). The kinetic constants were obtained from at least triplicate measurements of the initial rates at various concentrations of alginate and oligosaccharides of defined size (0.1 to 8 mg of substrate/ml). The kinetic parameters Km and Vmax were determined from nonlinear regression fitting of the Michaelis-Menten equation using Prism 5 (GraphPad Software, Inc., La Jolla, CA).
Production and purification of oligosaccharides.
The oligomers of α-l-guluronate (di-, tri-, and tetramers) used in the present study were produced from digestion of sodium alginate using Flavobacterium multivorum alginate lyase (AlgL; Sigma) and purified using a BioGel P-2 column prior to HPLC-MS analysis. Oligomers (20 mg) were loaded onto a BioGel P-2 column (1.5 cm by 50 cm) equilibrated with 1 mM phosphate buffer (pH 6.8). The column was developed at 0.5 ml min−1 using FPLC, and fractions containing oligomers, identified from their absorbance at 235 nm, were pooled and concentrated by rotary evaporation.
HPLC and ESI-MS characterization of products.
Samples were injected onto a Hypercarb column (100 mm by 3 mm) using an Agilent 1100 series HPLC system. The solvent flow rate was 0.3 ml min−1, with a temperature of 65°C. The products were eluted according to the following gradient, where solvent A is 0.2% trifluoroacetic acid in water, and solvent B is methanol: 30% B from 0 to 10 min, gradient from 30 to 90% B from 10 to 40 min. Electrospray ionization-mass spectrometry (ESI-MS) was performed on an Agilent 1100 series LC/MSD XCT Plus ion trap mass spectrometer (Agilent, Palo Alto, CA). Oligosaccharides delivered to the electrospray source using a syringe pump at a flow rate of 10 μl/min. The MS instrument was run in negative mode, with the mass scan range from 100 to 1,500 Da. The capillary temperature was kept at 250°C, and the electrospray needle voltage was 4.5 kV. Nitrogen sheath gas was provided to the source from a nearby Dewar flask of liquid nitrogen at 40 to 80 lb/in2.
Saccharification of alginate by OalA.
Batch saccharification of alginate by OalA was carried out in 1 ml of 20 mM KH2PO4 buffer at pH 6.5 in 2-ml screw-cap bottles. Alginate (0.1% [wt/vol]) was degraded in the presence of 20 μg of purified enzyme/ml at 16°C for 60 min. The progression of the batch saccharification was followed by the analysis of samples withdrawn periodically from the reaction mixture by using TBA analysis (27).
Site-directed mutagenesis and superposition of OalA on Atu3025.
Site-directed mutagenesis was carried out using the QuikChange site-directed mutagenesis kit from Stratagene. To replace Arg114, His226, Tyr280, and His446 by Ala, Ala, Phe, and Ala, respectively, the plasmid pMAL-c2X-oalA was used as a PCR template, and the oligonucleotides listed in Table S1 in the supplemental material were used as primers. Mutations were confirmed by DNA sequencing (ACGT, Inc., Wheeling, IL). Expression and purification of the mutants were conducted using the same procedures as for OalA. Multiple sequence alignment of oligoalginate lyases was performed using CLUSTAL Omega (28). An OalA homology model was built using the SWISS-MODEL online program (29). PyMOL v1.5 was used for superposition of OalA and Atu3025 (PDB 3AFL).
Gene expression analysis.
Vs12B01 was grown in Marine broth with 1% oligoalginate for 16 h with constant shaking (250 rpm). Temperature was maintained at 20 or 30°C, and the initial pH was 6.5 or 7.5. The total RNA was extracted using the RNeasy minikit (Qiagen). Reverse transcription was carried out using the ProtoScript II first-strand cDNA synthesis kit (NEB, Ipswich, MA). The qPCR experiments were carried out using the LightCycler 480 system (Roche, Indianapolis, IN) using the SYBR green-based method according to the manufacturer's instructions. Primers were designed by the online PrimerQuest tool provided by Integrated DNA Technologies. A total of 10 μl of 2× SYBR green mix, 6 μl of cDNA, 0.2 μl of each primer at a concentration of 10 pmol μl−1, and 3.6 μl of double-stranded H2O were mixed gently in each well of a LightCycler 480 96-well reaction plate. Reactions were performed using the following program: 10 min at 95°C for one cycle, followed by 10 s at 95°C, 10 s at 60°C, and 10 s at 72°C for 45 cycles, with a final cycle of 10 min at 72°C. The endogenous gene rpoA, encoding the RNA polymerase alpha subunit, was used as an internal control. The expression levels of all genes were normalized by the expression of the control.
RESULTS
Heterologous expression of oalA, oalB, and oalC genes.
OalA, OalB, and OalC were heterologously expressed in E. coli BL21(DE3). Analyses carried out with the crude lysate of E. coli BL21(DE3) harboring pMAL-c2X-oalA, pET28a-oalB, and pET28a-oalC revealed the presence of a high level of alginate lyase activity compared to the control E. coli BL21(DE3) cells harboring plasmid pMAL-c2X and pET28a. To determine whether unsaturated monomer products were produced from alginate specifically by those proteins and not by another enzyme induced in the host cell as a consequence of the overexpression of these genes, the OalA, OalB, and OalC enzymes were purified. Total protein extracts from E. coli BL21(DE3) transformed with these genes or with pET28a as a control were analyzed by SDS-PAGE. OalA, OalB, and OalC were identified in total and soluble or insoluble extracts only from cells harboring pMAL-c2X-oalA, pET28a-oalB, and pET28a-oalC and induced by IPTG (see Fig. S2 in the supplemental material). OalA, OalB, and OalC showed a lower soluble expression when cloned into pET28(a). OalA fused with MBP showed higher solubility compared to OalA with pET28(a). MBP enhanced the solubility of OalA but did not increase the solubility of OalB and OalC, so we decided to use different methods for the purification of OalB and OalC. These enzymes were purified using amylose affinity or Ni-NTA affinity chromatography. The purified OalA, OalB, and OalC protein solutions were colorless, and their UV-visible spectra showed no evidence of chromogenic cofactors. The conditions we used to resuspend the OalB and OalC were harsh; however, they were effective at solubilizing the protein and reducing non-native, inter- and intramolecular disulfide bonds. Using the standard folding/refolding protocol, we obtained approximately 7 to 10 mg of bioactive OalB and OalC per liter of cell culture. OalA, OalB, and OalC showed specific activities of 28.5, 20.0, and 20.8 U/mg, respectively, with alginate as the substrate (Table 1). These results strongly suggest that the oligoalginate lyase activities observed in crude extracts of E. coli BL21(DE3) were from OalA, OalB, and OalC.
TABLE 1.
Substrate specificity of OalA, OalB, and OalC for polymeric substrates
| Substrate | Mean sp act (U/mg) ± SEMa |
||
|---|---|---|---|
| OalA | OalB | OalC | |
| Sodium alginate | 29 ± 2 | 20 ± 1.3 | 21 ± 1 |
| Poly(G) | 107 ± 5 | 33 ± 2 | 3 ± 0.2 |
| Poly(M) | 121 ± 6 | 46 ± 3 | 76 ± 4 |
| Poly(M·G) | 44 ± 3 | 79 ± 7 | 28 ± 2 |
Values are means of triplicate measurements.
Determination of molecular mass and quaternary structure.
oalA, oalB, and oalC genes encode a polypeptide of 692, 735, and 717 amino acids, with a calculated molecular mass of 79,018, 82,766, and 80,500 Da, respectively. The molecular masses of OalA, OalB, and OalC were ∼80, 83, and 81 kDa, as determined by SDS-PAGE (Fig. S2A). The molecular mass of the OalA protein with MBP was 112 kDa. Using gel filtration chromatography on a Bio-Sil SEC-250 column, OalA was eluted as a symmetrical peak between γ-globulin and thyroglobulin, corresponding to a Mr of ∼160 kDa (see Fig. S2B in the supplemental material). These results indicated that the enzyme migrates as a dimer in gel filtration and thus may also be present and active as a homodimer in solution. OalB and OalC were eluted as a symmetrical peak between ovalbumin and γ-globulin and appeared monomeric.
Optimum pH, temperature, and effect of metal ions.
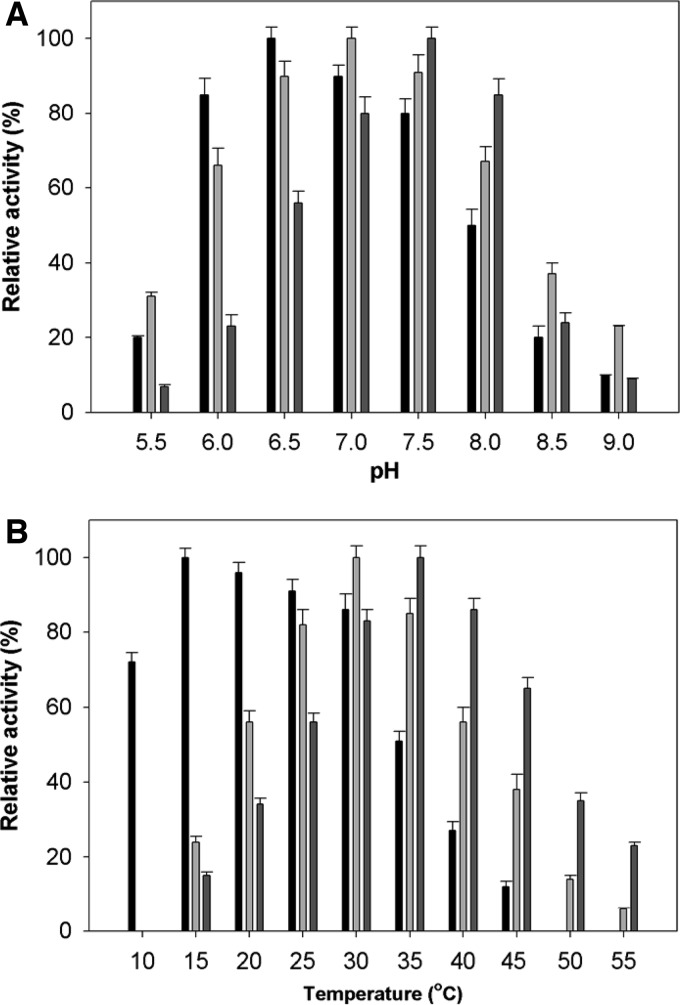
Vibrios can live in different environments (coastal water or animal guts) and are therefore exposed to a dynamic temperature range and different pHs. Hence, we reasoned that the three Oals may have different physiological adaptations. When we measured the activities at different pHs, the oligoalginate lyases OalA, OalB, and OalC showed maximal activities at pH 6.5, 7.0, and 7.5, respectively (Fig. 1A). The optimum temperatures of OalA, OalB, and OalC for the alginate lyase reaction were 16, 30, and 35°C, respectively (Fig. 1B). When tested at 25 and 30°C OalA was able to retain ∼50% of its activity for 34 and 21 min, respectively, whereas at 35°C its activity was rapidly lost leading to 50% reduced activity in 13 min. The apparent half-denaturation temperature (T1/2) of OalA was established by measuring the remaining activities at standard assay conditions after heat treatment at various temperatures. The T1/2 of OalA was 34.5°C.
FIG 1.
Effect of pH (A) and temperature (B) on the activity of OalA, OalB, and OalC. Enzyme assays were carried out under standard conditions in the presence of 0.1% alginate. Activities at the optimal temperature and pH were defined as 100%. The buffers used are 50 mM sodium phosphate (pH 6.0 to 7.5), 50 mM Tris-HCl (pH 7.0 to 8.0), and 50 mM Tris-glycine-NaOH (pH 8.0 to 8.5). The values are means of triplicate measurements. Error bars show the standard errors of the means.
For purified OalA dialyzed against phosphate buffer in the presence of 10 mM EDTA, 98% of the initial activity was left. The OalA activity was measured in the presence of various metal ions (see Table S2 in the supplemental material) and was found independent of the presence of divalent cations. The addition of 1 mM Mg2+, Mn2+, Ca2+, Fe2+, and Ba2+ showed a decrease (10 to 20%) in the initial OalA activity. The OalA activity was inhibited up to 26 and 13% in the presence of ZnCl2 and CoCl2, respectively, at 1 mM. The reduced activity might also be caused by the gelation of alginate by the cations.
Substrate specificity against polymers and oligomers.
A variety of polymers and oligomers were screened to determine the substrate scope of OalA, OalB, and OalC (Table 1). OalA showed higher activities on poly(M) and poly(G) compared to poly(M·G) and native alginate. Among the tested substrates, OalB was most active on native alginate and poly(M·G). OalC showed a preference for the degradation of poly(M).
Among the tested oligomers (di-, tri-, tetra-, and pentamers), OalA showed the highest specific activities compared to OalB and OalC (Table 2). OalA showed high specific activities on trimer (524 U/mg) and tetramer (476 U/mg), whereas OalB and OalC showed specific activities toward dimer (65 and 7.1 U/mg, respectively), but turned over this substrate faster than the longer oligomers tested.
TABLE 2.
Substrate specificity of OalA, OalB, and OalC for oligomeric substrates
| Substrate | Mean sp act (U/mg) ± SEMa |
||
|---|---|---|---|
| OalA | OalB | OalC | |
| Dimer | 201 ± 24 | 65 ± 5 | 7.1 ± 0.8 |
| Trimer | 524 ± 51 | 11 ± 2 | 1.5 ± 0.1 |
| Tetramer | 476 ± 53 | 42 ± 3 | 2.6 ± 0.3 |
| Pentamer | 196 ± 30 | 3.7 ± 0.5 | 0.4 ± 0.1 |
Values are means of triplicate measurements.
Kinetic parameters of OalA.
Initial velocities were determined in the standard assay mixture at pH 6.0. Figures S3, S4, and S5 in the supplemental material show the kinetic parameters of OalA, OalB, and OalC activity with increasing concentrations of alginate, poly(G), poly(M), and poly(M·G) ranging from 0.1 to 8 mg ml−1 (see Fig. S3 to S5A, B, C, and D, respectively, in the supplemental material). Based on the nonlinear regression analysis, the conversion of alginate by OalA, OalB, and OalC under standard assay conditions showed Km values of 3.25 ± 0.12 mg ml−1, 27.6 ± 1.8 mg ml−1, and 34.7 ± 1.3 mg ml−1, respectively (Tables 3 to 5). The catalytic efficiencies (kcat/Km) of OalA, OalB, and OalC against alginate were 50.5 ± 0.8 mg−1 ml s−1, 49.7 ± 2.7 mg−1 ml s−1, and 89 ± 6 mg−1 ml s−1, respectively (Tables 3 to 5). The conversion of poly(M) by OalA under standard assay conditions showed a Km of 0.60 ± 0.04 mg ml−1 and a Vmax of 162 ± 9 U mg of protein−1 (Table 3). The kcat/Km values of OalB and OalC against poly(M·G) and poly(M) were 270 ± 18 mg−1 ml s−1 and 272 ± 13 mg−1 ml s−1, respectively (Tables 4 and 5).
TABLE 3.
Kinetic parameters of OalA for different substrates
| Parameter | Mean ± SEMa |
|||
|---|---|---|---|---|
| Sodium alginate | Poly(G) | Poly(M) | Poly(M·G) | |
| Vmax (U mg of protein−1) | 124 ± 7 | 185 ± 10 | 162 ± 7 | 324 ± 14 |
| Km (mg ml−1) | 3.25 ± 0.1 | 0.89 ± 0.06 | 0.60 ± 0.03 | 8.38 ± 0.3 |
| kcat (s−1) | 164 ± 14 | 246 ± 12 | 217 ± 10 | 432 ± 24 |
| kcat/Km (mg−1 ml s−1) | 50.5 ± 4.5 | 277 ± 13 | 361 ± 17 | 51.6 ± 2.6 |
Values are means of triplicate measurements.
TABLE 5.
Kinetic parameters of OalC for different substrates
| Parameter | Mean ± SEMa |
|||
|---|---|---|---|---|
| Sodium alginate | Poly(G) | Poly(M) | Poly(M·G) | |
| Vmax (U mg of protein−1) | 34.7 ± 1.3 | 5 ± 0.17 | 94.7 ± 5 | 39.3 ± 1.4 |
| Km (mg ml−1) | 0.53 ± 0.08 | 0.6 ± 0.07 | 0.48 ± 0.1 | 0.5 ± 0.07 |
| kcat (s−1) | 48 ± 1.5 | 6.8 ± 0.2 | 130 ± 15 | 54.3 ± 1.8 |
| kcat/Km (mg−1 ml s−1) | 89 ± 6 | 10.8 ± 0.3 | 272 ± 13 | 98.2 ± 3.3 |
Values are means of triplicate measurements.
TABLE 4.
Kinetic parameters of OalB for different substrates
| Parameter | Mean ± SEMa |
|||
|---|---|---|---|---|
| Sodium alginate | Poly(G) | Poly(M) | Poly(M·G) | |
| Vmax (U mg of protein−1) | 27.6 ± 1.8 | 40.8 ± 1.9 | 60.7 ± 3.9 | 94.6 ± 6.1 |
| Km (mg ml−1) | 0.76 ± 0.1 | 0.47 ± 0.08 | 0.6 ± 0.1 | 0.48 ± 0.1 |
| kcat (s−1) | 38 ± 2.3 | 56.3 ± 2.6 | 83.7 ± 5 | 130 ± 8 |
| kcat/Km (mg−1 ml s−1) | 49.7 ± 2.7 | 120 ± 6.2 | 137 ± 6 | 270 ± 18 |
Values are means of triplicate measurements.
Substrate specificity.
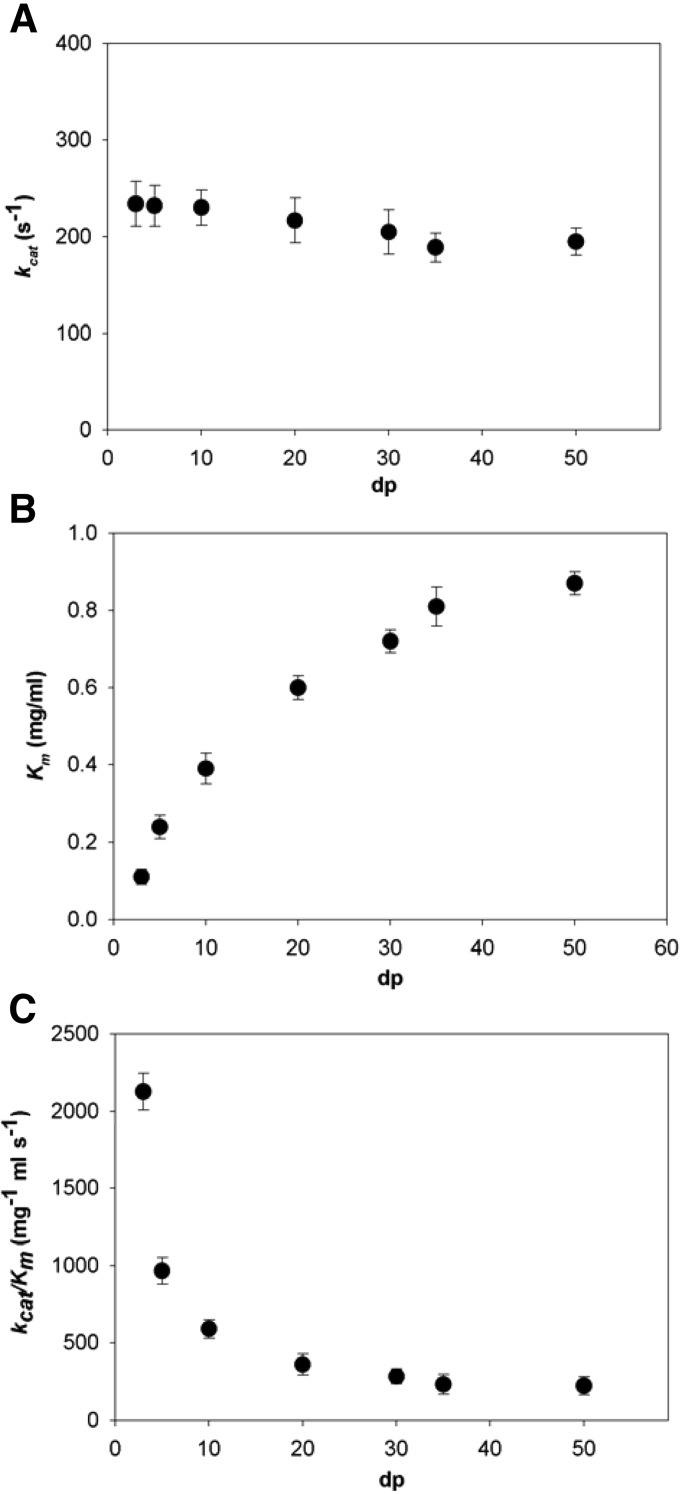
The kinetic parameters of OalA for cleavage of various sizes of poly(M) (degree of polymerization [dp], 3 to 50) were determined (Fig. 2). There was only modest variation in kcat across the size range examined (Fig. 2A); however, Km increased with increasing substrate length (Fig. 2B), with a corresponding decrease in kcat/Km. The kcat/Km of the reaction decreased nonlinearly with the number of residues in the substrate, from 2,130 mg−1 ml s−1 for the substrate containing 3 residues to 224 mg−1 ml s−1 for the substrate with 50 residues (Fig. 2C).
FIG 2.
Kinetic parameters for OalA-catalyzed degradation of poly(M) as a function of substrate length (dp). (A) kcat; (B) Km; (C) kcat/Km. The values are means of triplicate measurements. Error bars show the standard errors of the means.
Saccharification of alginate.
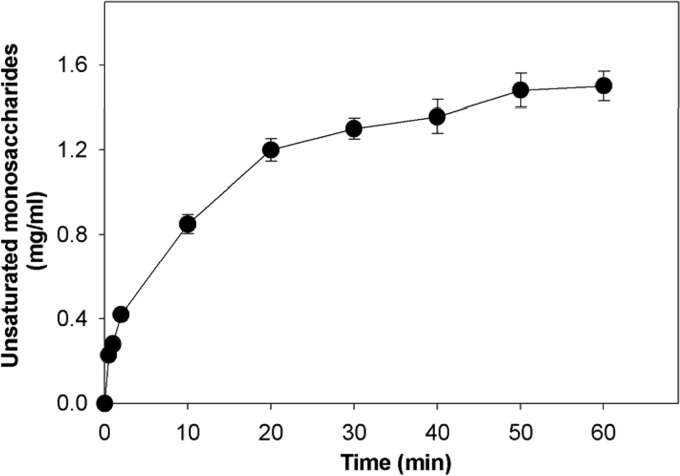
The degradation rates at the beginning of the reaction increased with increasing enzyme concentrations. The time courses of the unsaturated monosaccharides production against enzyme concentrations were obtained as saturation curves. For quantitative analysis of production of unsaturated monosaccharides, we represented the concentration of unsaturated monosaccharides as 2-deoxy-d-glucose equivalents. About 1.6 ± 0.1 mg of monosaccharides ml−1 was obtained from 0.1% (wt/vol) alginate by OalA (Fig. 3) after 60 min of incubation with the purified recombinant oligoalginate lyase. The alginate substrate was almost completely depolymerized into monosaccharides by OalA, clearly indicating that the recombinant oligoalginate lyase could degrade alginate into monosaccharides. The molecular mass ([M-H]−) of the resulting unsaturated uronic acid was determined to be 175 Da by ESI-MS analysis (see Fig. S6 in the supplemental material).
FIG 3.
Effect of recombinant lysate concentration on initial alginate degradation rate using the recombinant OalA. Alginate at 0.1% (wt/vol) in 20 mM phosphate buffer (pH 6.5) with recombinant OalA as the biocatalyst was incubated at 16°C for 60 min. The values are mean of triplicate measurements. Error bars show the standard errors of the means.
Site-directed mutagenesis.
To investigate the importance of highly conserved regions around the substrate binding pocket, site-directed mutagenesis was performed with OalA. All of the mutant enzymes were successfully overexpressed (see Fig. S7 in the supplemental material). To identify the catalytic residues and structural determinants for the exolytic mode of action in OalA, a homology model of OalA was prepared. Superposition with the crystal structure of Atu3025 from A. tumefaciens strain C58 (PDB 3AFL) in complex with an oligoalginate trimer (ΔGGG) allows positioning of the substrate with respect to the catalytic residues in OalA (see Fig. S8 in the supplemental material). OalA/ΔGGG revealed the binding mode of the substrate ΔGGG molecule in the active cleft. Subsites are labeled so that −n represents the nonreducing terminus and n the reducing terminus (see Fig. S8 in the supplemental material), and cleavage occurs between the −1 and +1 sites (30). The catalytic His and Tyr residues, which have been identified in PL5, PL7, and PL15, families that correspond to His226 and Tyr280 in OalA (30). His226 comprises a component of the active pocket, and the mutant H226A shows a significantly lower activity than the wild-type enzyme. The significant decrease in the specific activity of H226A and Y280F (Table 6) suggested that His226 and Tyr280 function as a catalytic base and acid, respectively. The reduced activity of the H446A and R114A mutant indicates their role in stabilizing or neutralizing the negative charge of the carboxylate anion of G+1 and G−1 (Table 6) (30).
TABLE 6.
Specific activity of wild type and OalA mutants
| Enzyme | Sp act (U/mg) | Relative activity (%) |
|---|---|---|
| Wild type | 28.5 | 100 |
| H226A | 0.01 | 0.03 |
| Y280F | 0.14 | 0.5 |
| H446A | 0.19 | 0.7 |
| R114A | 1.2 | 4.21 |
Oligoalginate lyase gene expression analysis.
To test how temperature and pH would affect expression of the Oals in 12B01, we incubated this bacterium under different physiological conditions and monitored expression levels via qPCR. The expression was varied under different temperatures and pHs. OalA and OalC gene expression levels were higher at 20°C and pH 7.5 (see Fig. S9 in the supplemental material). The relative expression level of OalC was 4-fold higher at 20°C and pH 7.5 than at 30°C and pH 6.5. The gene expression levels of OalA and OalC were decreased at 30°C and pH 6.5. OalB and OalC gene expression levels were similar since they are both part of an operon structure in the alginate degradation pathway of 12B01.
DISCUSSION
V. splendidus 12B01 degrades alginate into oligomers with extracellular, endo-acting, and exo-acting alginate lyases of family PL7 and PL6 such as A1yA, AlyB, AlyC, and AlyD (30). The oligomers are imported through conserved porins situated in the genome in close proximity to the Aly and Oal lyases within the alginate degradation pathway (30). The imported oligosaccharides are further cleaved within the cell into unsaturated monosaccharides by OalA, OalB, and OalC. The oalA, oalB, and oalC genes were overexpressed in E. coli, and exo-type oligoalginate activity was confirmed. OalA, OalB, and OalC are noteworthy in having broad substrate scope for oligomers, including di-, tri-, tetra-, and pentasaccharides. OalA has a unique broad oligomer substrate specificity compared to reported oligoalginate lyases from V. cyclitrophicus, A. tumefaciens strain C58, R. lupini HPC, and Sphingomonas sp. strain A1 (26, 27). Moreover, OalA appeared to have the highest specific activity (28.5 U/mg) for alginate among all previously characterized oligoalginate lyases from family PL15 and PL17 (see Table S3 in the supplemental material).
In the present study, we reported the detailed kinetic parameters of OalA for cleavage of various sizes of poly(M). The variation in the kinetic parameters for the OalA reaction as a function of substrate length suggests that the enzyme is well adapted to process the oligomers that are imported into the cell. Although kcat varies only slightly with polymer length, kcat/Km increases with decreasing polymer length, and Km decreases. The relationship between kcat/Km and polymer length is noteworthy; kcat/Km increases nonlinearly with decreasing substrate length. The observed kinetic data can be elucidated by considering the meaning of kcat/Km. Thus, kcat/Km is seen to be a measure of the frequency with which the substrate is productively captured by the enzyme, where productive capture refers to binding events that lead to product formation. The ratio of rate constants that moderates k1 has a maximal value of 1 in the case that binding of one substrate molecule gives rise to more equivalents of product (31). The increasing kcat/Km with decreasing substrate length may be explained by an exo-processive mode of action splitting unsaturated monosaccharide from the substrate terminus.
The catalytically important residues for OalA were identified through site-directed mutagenesis as Arg114, His226, and Tyr280 based on the crystal structure of Atu3025 with inactivating mutations for each of these residues resulting in nearly complete loss of OalA activity toward alginate (Table 6). Thus, our results are most consistent with a mechanism previously suggested (32) in which: (i) a positive residue stabilizes the negative charge of the carboxyl group, (ii) a residue acting as a general base abstracts the proton from C-5 of the sugar ring, and (iii) a general acid residue protonates the glycosidic bond oxygen. In the PL15 family these residues have been previously identified (33) and here confirmed for OalA. The residues for mutation were assumed to play an important role in recognizing the acidic polysaccharide and/or catalyzing the enzyme reaction based on the structure of Atu3025/ΔGGG. The positively charged residues Arg114 is supposed to be crucial for binding acidic polysaccharides and/or neutralizing the negative charge of the carboxyl groups in ΔG−1 and ΔG+1 (33). Moreover, His226 and Tyr280 were proposed to function as a catalytic base and acid, respectively.
Alginate degradation products such as di- or trisaccharides were not observed, likely because the recombinant OalA is an exo-processive enzyme. In terms of conversion, around 1.6 mg of saccharification products ml−1 was obtained from 0.1% (wt/vol) initial alginate through oligoalginate lyase-catalyzed saccharification. The saccharification of alginate is a prerequisite, and oligoalginate lyases can play important roles in the production of unsaturated monosaccharides, which could be used as a renewable source for the production of biofuels. The fundamental process designs for a brown macroalga biorefinery have already been established (3). In this process, because of their high viscosity, moisture, and salt-retaining properties, alginate and oligoalginate must be degraded into DEHU (4-deoxy-l-erythro-5-hexoseulose uronic acid) before concentration and subsequent fermentation. Especially at low temperatures OalA has a good potential for production of these unsaturated monosaccharides that are converted into DEHU (3).
In addition to the complementary substrate scope for OalA, OalB, and OalC, we observed a broad range in temperature and pH adaptations. Vibrios are marine species adapted to temperate coastal waters, which represent a highly dynamic environment. Thus, the temperature and pH range that we observed for the three Oals may promote efficient alginate degradation in and represent an adaptation to different environmental conditions. Our results also suggest that alginate degradation pathways, which function at elevated temperatures, must be identified bioengineered for more efficient and rapid conversion of alginate into biofuels. Notably, the conversion of alginate into bioethanol in the bioengineered E. coli strain was only efficient up to 30°C and likely constrained by the low thermostability of the Oals (especially OalA) and or other proteins in the pathway. For example our experiments showed OlaA, which was the most active of the three enzymes, rapidly lost its activity at temperatures above 30°C.
In summary, the successful overexpression of OalA, OalB, and OalC allowed us to characterize novel oligoalginate lyases that showed broad substrate scope for oligomers, bifunctionality, higher turnover for oligomers than polymers, and activity at a comparatively low temperature. The saccharification of alginate yields raw materials that could be used as a renewable source for the production of biofuels and commodities. Structural and mechanistic studies to clarify the molecular determinants for this broad substrate scope are under way.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the U.S. Department of Energy (DE-SC0008743). This work was partially supported by the Energy Efficiency & Resources Core Technology Program of KETEP, granted financial resources from the Ministry of Trade, Industry & Energy, Republic of Korea (201320200000420). This work was also supported by WTU joint research grants from Konkuk University.
Footnotes
Published ahead of print 2 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01285-14.
REFERENCES
- 1.Beer LL, Boyd ES, Peters JW, Posewitz MC. 2009. Engineering algae for biohydrogen and biofuel production. Curr. Opin. Biotechnol. 20:264–271. 10.1016/j.copbio.2009.06.002 [DOI] [PubMed] [Google Scholar]
- 2.Chisti Y. 2008. Biodiesel from microalgae beats bioethanol. Trends Biotechnol. 26:126–131. 10.1016/j.tibtech.2007.12.002 [DOI] [PubMed] [Google Scholar]
- 3.Enquist-Newman M, Faust AM, Bravo DD, Santos CN, Raisner RM, Hanel A, Sarvabhowman P, Le C, Regitsky DD, Cooper SR, Peereboom L, Clark A, Martinez Y, Goldsmith J, Cho MY, Donohoue PD, Luo L, Lamberson B, Tamrakar P, Kim EJ, Villari JL, Gill A, Tripathi SA, Karamchedu P, Paredes CJ, Rajgarhia V, Kotlar HK, Bailey RB, Miller DJ, Ohler NL, Swimmer C, Yoshikuni Y. 2013. Efficient ethanol production from brown macroalga sugars by a synthetic yeast platform. Nature 505:239–243. 10.1038/nature12771 [DOI] [PubMed] [Google Scholar]
- 4.Horn SJ, Aasen IM, Østgaard K. 2000. Ethanol production from seaweed extract. J. Ind. Microbiol. Biotechnol. 25:249–254. 10.1038/sj.jim.7000065 [DOI] [Google Scholar]
- 5.Martone PT, Estevez JM, Lu F, Ruel K, Denny MW, Somerville C, Ralph J. 2009. Discovery of lignin in seaweed reveals convergent evolution of cell wall architecture. Curr. Biol. 19:169–175. 10.1016/j.cub.2008.12.031 [DOI] [PubMed] [Google Scholar]
- 6.Lee SI, Choi SH, Lee EY, Kim HS. 2012. Molecular cloning, purification, and characterization of a novel polyMG-specific alginate lyase responsible for alginate MG block degradation in Stenotrophomonas maltophilia KJ-2. Appl. Microbiol. Biotechnol. 95:1643–1653. 10.1007/s00253-012-4266-y [DOI] [PubMed] [Google Scholar]
- 7.Wong TY, Preston LA, Schiller NL. 2000. Alginate lyase: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 54:289–340. 10.1146/annurev.micro.54.1.289 [DOI] [PubMed] [Google Scholar]
- 8.Thomas F, Lundqvist LC, Jam M, Jeudy A, Barbeyron T, Sandström C, Michel G, Czjzek M. 2013. Comparative characterization of two marine alginate lyases from Zobellia galactanivorans reveals distinct modes of action and exquisite adaptation to their natural substrate. J. Biol. Chem. 288:23021–23037. 10.1074/jbc.M113.467217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gimmestad M, Ertesvåg H, Heggeset TM, Aarstad O, Svanem BI, Valla S. 2009. Characterization of three new Azotobacter vinelandii alginate lyases, one of which is involved in cyst germination. J. Bacteriol. 191:4845–4853. 10.1128/JB.00455-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiller NL, Boyd CM, Keen NT, Ohman DE. 1993. Characterization of the Pseudomonas aeruginosa alginate lyase gene (algL): cloning, sequencing, and expression in Escherichia coli. J. Bacteriol. 155:4780–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skaugrud O, Borgerson HAB, Dornish M. 1999. Biomedical and pharmaceutical applications of alginate and chitosan. Biotechnol. Genet. Eng. Rev. 16:23–40. 10.1080/02648725.1999.10647970 [DOI] [PubMed] [Google Scholar]
- 12.Cao L, Xie L, Xue X, Tan H, Liu Y, Zhou S. 2007. Purification and characterization of alginate lyase from Streptomyces species strain A5 isolated from banana rhizosphere. J. Agric. Food Chem. 55:5113–5117. 10.1021/jf0704514 [DOI] [PubMed] [Google Scholar]
- 13.Murata K, Kawai S, Mikami B, Hashimoto W. 2008. Superchannel of bacteria: biological significance and new horizons. Biosci. Biotechnol. Biochem. 72:265–277. 10.1271/bbb.70635 [DOI] [PubMed] [Google Scholar]
- 14.Suzuki H, Suzuki K, Inoue A, Ojima T. 2006. A novel oligoalginate lyase from abalone, Haliotis discus hannai, that releases disaccharide from alginate polymer in an exolytic manner. Carbohydr. Res. 341:1809–1819. 10.1016/j.carres.2006.04.032 [DOI] [PubMed] [Google Scholar]
- 15.Yamasaki M, Ogura K, Hashimoto W, Mikami B, Murata K. 2005. A structural basis for depolymerization of alginate by polysaccharide lyase family-7. J. Mol. Biol. 352:11–21. 10.1016/j.jmb.2005.06.075 [DOI] [PubMed] [Google Scholar]
- 16.Ochi Y, Takeuchi T, Murata K, Kawabata Y, Kusakabe I. 1995. A simple method for preparation of poly-mannuronate using poly-guluronate lyase. Biosci. Biotechnol. Biochem. 59:1560–1561. 10.1271/bbb.59.1560 [DOI] [Google Scholar]
- 17.Park D, Jagtap SS, Nair SK. 2014. Structure of a PL17 family alginate lyase demonstrates functional similarities among exotype depolymerases. J. Biol. Chem. 289:8645–8655. 10.1074/jbc.M113.531111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Roux F, Zouine M, Chakroun N, Binesse J, Saulnier D, Bouchier C, Zidane N, Ma L, Rusniok C, Lajus A, Buchrieser C, Médigue C, Polz MF, Mazel D. 2009. Genome sequence of Vibrio splendidus: an abundant planktonic marine species with a large genotypic diversity. Environ. Microbiol. 11:1959–1970. 10.1111/j.1462-2920.2009.01918.x [DOI] [PubMed] [Google Scholar]
- 19.Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. 2008. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320:1081–1085. 10.1126/science.1157890 [DOI] [PubMed] [Google Scholar]
- 20.Bishop AL, Patimalla B, Camilli A. 2014. Vibrio cholerae-induced inflammation in the neonatal mouse cholera model. Infect. Immun. 82:2434–2447. 10.1128/IAI.00054-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wargacki AJ, Leonard E, Win MN, Regitsky DD, Santos CN, Kim PB, Cooper SR, Raisner RM, Herman A, Sivitz AB, Lakshmanaswamy A, Kashiyama Y, Baker D, Yoshikuni Y. 2012. An engineered microbial platform for direct biofuel production from brown macroalgae. Science 335:308–313. 10.1126/science.1214547 [DOI] [PubMed] [Google Scholar]
- 22.Arne H, Larsen B, Smidsrød O, Eriksson G, Blinc R, Paušak S, Ehrenberg L, Dumanović J. 1967. Studies on the sequence of uronic acid residues in alginic acid. Acta Chem. Scand. 21:691–704. 10.3891/acta.chem.scand.21-0691 [DOI] [Google Scholar]
- 23.Riggs P. 2001. Expression and purification of maltose-binding protein fusions. Curr. Protoc. Mol. Biol. Chapter 16:Unit 16.6. 10.1002/0471142727.mb1606s28 [DOI] [PubMed] [Google Scholar]
- 24.Burgess RR. 2009. Refolding solubilized inclusion body proteins. Methods Enzymol. 463:259–282. 10.1016/S0076-6879(09)63017-2 [DOI] [PubMed] [Google Scholar]
- 25.Tøndervik A, Klinkenberg G, Aarstad OA, Drabløs F, Ertesvåg H, Ellingsen TE, Skjåk-Bræk G, Valla S, Sletta H. 2010. Isolation of mutant alginate lyases with cleavage specificity for di-guluronic acid linkages. J. Biol. Chem. 285:35284–35292. 10.1074/jbc.M110.162800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ochiai A, Hashimoto W, Murata K. 2006. A biosystem for alginate metabolism in Agrobacterium tumefaciens strain C58: molecular identification of Atu3025 as an exotype family PL-15 alginate lyase. Res. Microbiol. 157:642–649. 10.1016/j.resmic.2006.02.006 [DOI] [PubMed] [Google Scholar]
- 27.Ryu M, Lee EY. 2011. Saccharification of alginate by using exolytic oligoalginate lyase from marine bacterium Sphingomonas sp. MJ-3. J. Ind. Eng. Chem. 17:853–858. 10.1016/j.jiec.2011.08.001 [DOI] [Google Scholar]
- 28.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22:195–201. 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- 30.Henrissat BD, Wilson KS. 1997. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 321:557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farrell EK, Tipton PA. 2012. Functional characterization of AlgL, an alginate lyase from Pseudomonas aeruginosa. Biochemistry 51:10259–10266. 10.1021/bi301425r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linker A, Meyer K, Hoffman P. 1960. The production of hyaluronate oligosaccharides by leech hyaluronidase and alkali. J. Biol. Chem. 235:924–927 [PubMed] [Google Scholar]
- 33.Ochiai A, Yamasaki M, Mikami B, Hashimoto W, Murata K. 2010. Crystal structure of exotype alginate lyase Atu3025 from Agrobacterium tumefaciens. J. Biol. Chem. 285:24519–24528. 10.1074/jbc.M110.125450 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.