Abstract
To evaluate the dose-response effects of endogenous indole-3-acetic acid (IAA) on Medicago plant growth and dry weight production, we increased the synthesis of IAA in both free-living and symbiosis-stage rhizobial bacteroids during Rhizobium-legume symbiosis. For this purpose, site-directed mutagenesis was applied to modify an 85-bp promoter sequence, driving the expression of iaaM and tms2 genes for IAA biosynthesis. A positive correlation was found between the higher expression of IAA biosynthetic genes in free-living bacteria and the increased production of IAA under both free-living and symbiotic conditions. Plants nodulated by RD65 and RD66 strains, synthetizing the highest IAA concentration, showed a significant (up to 73%) increase in the shoot fresh weight and upregulation of nitrogenase gene, nifH, compared to plants nodulated by the wild-type strain. When these plants were analyzed by confocal microscopy, using an anti-IAA antibody, the strongest signal was observed in bacteroids of Medicago sativa RD66 (Ms-RD66) plants, even when they were located in the senescent nodule zone. We show here a simple system to modulate endogenous IAA biosynthesis in bacteria nodulating legumes suitable to investigate which is the maximum level of IAA biosynthesis, resulting in the maximal increase of plant growth.
INTRODUCTION
Rhizobia are Gram-negative bacteria able to grow in the soil as free-living organisms and as endocellular symbionts inside root nodule cells of leguminous plants.
The association of leguminous plants and rhizobia allows the conversion of atmospheric nitrogen into ammonia in a new specific organ, the root nodule.
A complex series of events, coordinated by host and bacterial signal molecules, underlie the development of this symbiotic interaction (1, 2).
Based on nodule morphology and anatomy, two major types of nodules are distinguished: the indeterminate and determinate nodule types.
Temperate legumes generally develop indeterminate nodules, where cortical cell divisions are initiated in the inner cortex. A persistent meristem develops at the distal end of the nodule, generating an age gradient of differentiating zones in the following sequence: the meristematic zone (zone I) followed by the invasion zone (zone II), the interzone (zones II and III), the fixation zone (zone III), and the senescence zone (zone IV) at the proximal end of nodules. All developing (growing) steps, starting from infection and proceeding to senescence, coexist, and this gives indeterminate nodules their irregular oval shape (3).
The nodulation mechanism is highly specific and involves the same subset of plant phytohormones, namely, auxin, cytokinin, and ethylene, which are required for root development.
In particular, auxin is involved in multiple processes, including cell division, differentiation, and vascular bundle formation.
The most abundant form of auxin in plants is indole-3-acetic acid (IAA). It is known that many plant-associated soil bacteria are able to synthesize auxin, in particular, IAA, and that bacterially produced auxin can alter the auxin transportation and localization inside the host plant (4, 5). A number of experiments suggest that auxin transport regulation is part of the process leading to nodule initiation (1, 3, 6, 7).
Studies with Rhizobium mutants deficient in IAA synthesis have shown that nitrogen fixation can be impaired by a lack of rhizobial auxin whereas increased nodulation efficiency can be reached with IAA-overproducing strains, although this might differ between determinate and indeterminate legumes (8).
In our work, we analyzed the 85-bp bacterial promoter (9) that acts as the intron of the rolA gene of A. rhizogenes when the transfer DNA (T-DNA) is transferred to the plant nucleus (10). Because of its roles as a promoter in bacteria and as an intron in plant cells, it is described as a “promintron” (11). It contains in its DNA sequence stretches of homology to −35 and −10 consensus sequences typical of prokaryotic promoters that are properly located.
This promoter was successfully used to drive expression of a new pathway for the biosynthesis of the auxin indole-3-acetic acid (IAA), providing a powerful tool to enhance IAA levels in different rhizobial species under free-living and symbiotic conditions (12–14). Beneficial effects on N-fixation, salt tolerance, plant yield, and mineral phosphate solubilization were observed for the analyzed Rhizobium-legume systems (15–17).
The purpose of this work was to further increase the synthesis of IAA in rhizobia and to check, at this stage, if these strains can further improve medicago plant yield.
We used site-directed mutagenesis to modify the −10 and −35 regulative regions of promintron, providing 11 different mutants. For these mutations, the activity of the promoter was preliminarily tested by analyzing the expression of the uidA reporter gene. This strategy led us to select three mutations for more-detailed analysis. For the selected mutations (M6-RD65, M11-RD66, and M7-RD67), the iaaM and tms2 genes were introduced as a bicistronic unit under the control of promintron and were used to alter the synthesis of IAA in Rhizobium.
The mRNA levels of both genes were significantly induced in the RD66 mutant. This result was positively correlated to the higher IAA content measured in the bacterial supernatant and to the stronger immunofluorescence signal, detected in invasion zone II and nitrogen fixation zone III of nodules, deriving from Medicago sativa RD66 (Ms-RD66) plants than from Ms-RD64 plants. Ms-RD66 plants also showed a significant increase in shoot fresh weight and higher expression of the nifH gene than Ms-RD64 plants.
We describe here an easy-to-manipulate short promoter sequence suitable for performing studies on hormone regulation in bacteria and bacteroids. Indeed, we provide a collection of four promintron constructs that might be used to increase hormone levels during Rhizobium-legume symbiosis.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
The strain used in our study was Sinorhizobium meliloti 1021 (streptomycin resistant [Strr]) (18). The bacterial strain was grown in minimal medium as previously described (15).
Streptomycin (200 mg/liter) and spectinomycin (200 mg/liter) were included as required. The construction of 85-17Gus recombinant plasmids (Fig. 1A) was performed as previously published (9).
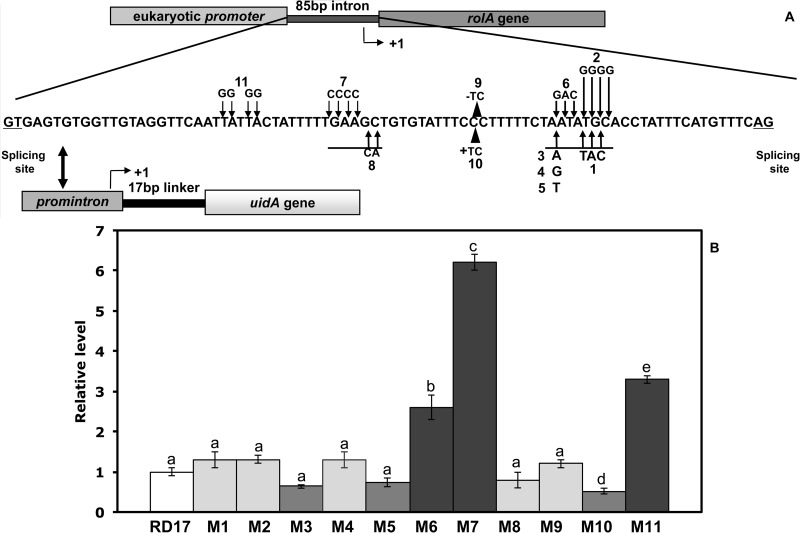
FIG 1.
(A) Schematic drawing of the chimeric reporter gene. The position of the major transcriptional start site (+1) is identified by an arrowhead, and the −10 and −35 sequences are underlined. The different mutations are also indicated. (B) Quantitative RT-PCR analysis of the uidA gene in S. meliloti cells containing the wild type (RD17) and the mutated 85-17-uidA construct. Values are the averages ± standard deviations of the results from four biological replicates conducted at different times. Relative gene expression levels determined by the comparative CT method are presented as follows: 2−ΔΔCT > 1, more highly expressed genes in cells transformed with the mutated construct (RD65, RD66, and RD67 cells); 2−ΔΔCT < 1, more highly expressed genes in cells transformed with the wild-type construct (RD64 cells). Different letters are used to indicate means that differ significantly according to Tukey's test (P < 0.05). The numbers refer to the mutations described in Table 1.
The S. meliloti IAA-overproducing strains (RD64, RD65, RD66, and RD67) were generated by introducing the promintron-iaaM-tms2 wild-type construct (RD64) and mutagenized constructs (RD65, RD66, and RD67) into S. meliloti 1021 as previously described (9; R. Defez and A. Spena, 9 November 1998, European Patent Office [EPO] application no. EP98/830674.2, extension no. PCT24190, European Patent Office, Munich, Germany).
Site-directed mutagenesis of the promintron sequence.
Site-directed mutagenesis of the promintron consensus sequences was performed in two steps by using a PCR-based QuikChange site-directed mutagenesis kit (Stratagene).
In the first step, two PCRs were run at the same time, one with the external A* primer (Table 1) and the internal mutagenic primer (Table 1) and the other with the second external B* primer (Table 1) and the mutagenic primer (Table 1).
TABLE 1.
Oligonucleotide primers used in this study
| Primer designation | Configuration | Sequence (5′→3′)a |
|---|---|---|
| A* | BamHI | actagtggatcccccggg |
| B* | SnabI | caaaaggtgatacgtacactt |
| M1 | −13/−11 ATA→TAC | ccctttttctaTACtgcaccta |
| M2 | −13/−10 ATAT→GGGG | ctttttctaGGGGgcaccta |
| M3 | −16 C→A | tccctttttAtaatatgcacct |
| M4 | −16 C→G | ttccctttttGtaatatgcacc |
| M5 | −16 C→T | tttccctttttTtaatatgcacc |
| M6 | −16/−14 CTA→GAC | tttccctttttGACatatgcacc |
| M7 | −40/−37 TTGA→CCCC | ttactatttCCCCagctgtgt |
| M8 | −36/−35 AG→CA | ttactatttttgaCActgtgt |
| M9 | Deletion of −25 and −24 bases | gaagctgtgtatt..cctttttc |
| M10 | Insertion between −25 and −24 bases | gaagctgtgtatttTCccctttttc |
| M11 | −52, −51, −49, −48 T→G | gtaggttcaaGGaGGactatt |
Sequences complementary to the target DNA are lowercased; sequences unique to the oligonucleotide primers are shown in capital and bold letters; deleted nucleotides are indicated by dots; restriction sites used for cloning are in bold letters and underlined. The sequence of A* and B* primers contains the restriction sites used in the cloning.
In the second step, to amplify the full-length mutagenized product, primers A* and B* were used as the primers and the mix of fragments obtained in the first two PCRs was used as the template.
The final product sequence was identical to the promintron region sequence between the external primers, except for the desired mutations. The wild-type and mutated 85-17Gus constructs were introduced in S. meliloti 1021, used as the model system.
Quantitative PCR (qPCR) analysis.
The isolation of RNA from bacterial cells was performed as previously described (13).
To isolate RNA from plant nodules, frozen tissue was homogenized in 500 μl QIAzol lysis reagent (Qiagen) and centrifuged at 12,000 × g for 1 min at 4°C.
Chloroform (100 μl) was then added to the clear supernatant, mixed well, and centrifuged at 12,000 × g for 15 min. The transparent upper phase was mixed with an equal volume (300 μl) of 70% ethanol, transferred to an RNeasy Mini spin column (RNeasy minikit, Qiagen), and centrifuged at 8,000 × g for 15 s. After the addition of Buffer RW1 (350 μl) to the column, the manufacturer's instruction was followed.
Residual DNA, present in the RNA preparations, was removed by using a Turbo DNA-free kit (Applied Biosystems), following the manufacturer's instruction.
After purification and quality checking, by agarose gel electrophoresis, the RNA concentration was determined, by absorbance at 260 nm, and the RNA was stored at −20°C until further use.
First-strand cDNA was synthesized from 1 μg of total RNA with a RETROscript kit (Applied Biosystems) and random decamers, according to the manufacturer's instructions.
Quantitative PCR was performed, as previously described (17), except that iQ SYBR green Supermix (Bio-Rad) was used.
Specific primer pairs for actin and nifH genes were those reported by Bianco and Defez (15).
Specific primer pairs for the uidA, iaaM, and tms2 genes, designed using Primer3 software, were as follows: for uidA, 5′-GGCCGCTCTAGAACTAGTGGA-3′ and 5′-GGCACAGCACATCAAAGAGA-3′; for iaaM, 5′-ATGTATGACCATTTTAATTCACCCAGT-3′ and 5′-CTGGGAGGAAAGCGCATCGCAC-3′; and for tms2, 5′-GGATTAGCGGATTCAGACCA-3′ and 5′-GTTTTCCCAGTCACGACGTT-3′.
Primers for Mtc27 (constitutively expressed gene) and the actin gene were included in all the qPCR analyses for the purpose of data normalization. qPCR amplification for each cDNA sample was performed in quadruplicate wells. During the reactions, the fluorescence signal, due to SYBR green intercalation, was monitored to quantify the double-stranded DNA product formed in each PCR cycle.
Results were recorded as relative gene expression changes, after normalizing for Mtc27 or actin gene expression, and computed using the comparative threshold cycle (CT) method (2−ΔΔCT) as previously described (19).
For free-living bacteria, the 2−ΔΔCT value was >1 for the more highly expressed genes in cells transformed with the construct containing the mutated promoter and was <1 for the more highly expressed genes in cells transformed with the construct containing the wild-type promoter.
Under symbiotic conditions, the 2−ΔΔCT value was >1 for the more highly expressed genes in plants infected with strain RD65, RD66, or RD67 and was <1 for the more highly expressed genes in plants infected with strain RD64.
Plant material and growth conditions.
M. sativa seeds were surface sterilized, germinated, and transferred into hydroponic units, as previously reported (15). Growth of rhizobia, plant infections, and greenhouse conditions were those described by Bianco and Defez (15). Bacteria were isolated from nodules, and their identities were verified by their antibiotic resistance patterns.
IAA analysis. (i) Enzyme-linked immunosorbent assay (ELISA).
The extraction of IAA prior to immunoassay was performed on both the nodules and the supernatant of bacterial cultures. For the former, the method employed has already been described (15). For the latter, the bacterial culture was centrifuged at 4,000 × g for 30 min at 4°C, and the corresponding supernatant was filtered and used for liquid-liquid extraction, as reported by Alvarez et al. (20).
The displacement immunoassay for IAA determination was carried out according to a protocol previously described (15).
(ii) HPLC–high-resolution mass spectrometry (HPLC-HRMS).
The high-pressure liquid chromatography (HPLC) column was a Synergi Polar column (Phenomenex, Torrance, CA) (4-μm pore size, 75 by 2 mm), operating at a 250 μl/min flow rate at 30°C. The mobile phases were H2O (mobile phase A) and acetonitrile (ACN) (mobile phase B), both containing 0.1% formic acid (FoAc).
For gradient elution, the percentage of mobile phase B was linearly increased from 0% to 100% in 20 min, after an isocratic step of 3 min; after 2 min at 100% mobile phase B, the initial composition was restored and the system equilibrated for 8 min.
The injection volume was 20 μl. The HPLC-HRMS instrument used was an Ultimate 3000 HPLC instrument coupled to a LTQ-Orbitrap mass spectrometer through an electrospray ionization (ESI) interface (Thermo Scientific, Bremen, Germany). The interface and MS parameters were optimized by infusion of a standard solution of IAA (Sigma-Aldrich). The settings used were the following: ESI spray voltage, 4.5 kV; capillary voltage, 10 V; tube lens, 50 V. Nitrogen was used as sheath, auxiliary, and sweep gas at 10, 5, and 2 (arbitrary units), respectively. Acquisition was done in positive-ion mode in the Orbitrap analyzer, operating at a resolution of 30,000 (at 400 m/z) and in full-scan mode in the range of 85 to 400 m/z. The ion at m/z 214.08963, due to the protonated molecule of a contaminant (molecular formula, C10H15NO2S) in HPLC solvents, was used as the internal calibrant (lock mass).
Tandem MS (MS/MS) experiments were performed to further confirm the identity of the detected signals, selecting the precursor ion of interest in an isolation window of 3 m/z in the linear trap. Product ion spectra were acquired using the Orbitrap mass spectrometer in the scan range of 50 to 300 m/z and with resolution set at 15,000 (at 400 m/z).
For HPLC-HRMS analyses, 50 μl of a solution of indole-2,4,5,6,7-d5-3-acetic-2,2-d2 acid (d7-IAA; purchased from CDN Isotopes, Pointe-Claire, Canada) (20 mg/liter in MeOH/H2O [1:1]) was added to 100 μl of samples, mixed using a vortex device, and then diluted with 850 μl MeOH containing 0.1% (FoAc).
The d7-IAA molecule was used as the internal standard for quantitative purposes. After centrifugation at 1,500 × g for 10 min, the solution was transferred into a vial for HPLC-HRMS analysis. Peak identification was accepted only if the retention time was consistent with that of the standard (± 1%) and if the m/z value was in agreement with theoretical value, with less than 4 ppm error.
The signals of the [M+H]+ ion of IAA (m/z 176.0707) and d7-IAA (m/z 183.1146) were extracted in a window of ± 0.0030 mass unit (approximately ± 18 ppm) and the peak areas measured. Their ratio (peak area ratio [PAR]) was calculated and linearly regressed over the corresponding amount of IAA added in each calibration point. The linear calibration curve was then used to calculate the amount of IAA in each sample.
(iii) IAA immunolocalization.
For IAA immunolocalization, fresh nodules from 5-week-old plants were prefixed by the use of a brief vacuum treatment in freshly prepared 4% (wt/vol) 1-ethyl-3-(3-dimethyl-aminopro-pyl)-carbodiimide hydrochloride (EDAC) (Sigma)–0.1× phosphate-buffered saline (PBS) (pH 7) at room temperature for 30 min. Subsequently, nodules were postfixed in 4% paraformaldehyde–cacodylate buffer (pH 7.4) containing 2.5% sucrose (40 min at room temperature in a vacuum). After fixation, the plant materials were washed in 1× PBS and embedded in 6% agarose and 70-μm-thick sections were cut with a vibratome (Leica VT1000S). The nodule sections were transferred into well plates (Nunc) and blocked with 1× PBS, 0.1% (vol/vol) Tween 20, 1.5% (vol/vol) glycine, and 5% (wt/vol) bovine serum albumin (BSA) for 30 min at 20°C. Sections were then rinsed in PNTB solution (1× PBS, 0.88% [wt/vol] NaCl, 0.1% [vol/vol] Tween 20, and 0.8% [wt/vol] BSA]) for 5 min and briefly washed in PB solution (1× PBS and 0.8% [wt/vol] BSA).
A drop of 100 μl 1:100 (wt/vol) primary IAA antibody (raised against IAA-BSA conjugate and obtained from Agrisera) was added to each well, before the plate was covered with a lid, and then incubated overnight in a humidity chamber at 4°C. The sections were rinsed (three times for 10 min each time) first in 1× PBS–0.1% (vol/vol) Tween 20–0.1% (wt/vol) BSA–2.9% (wt/vol) NaCl and then in PNTB solution followed by two 10-min washes with PB solution. Then, 100 μl secondary antibody (1:100 [vol/vol] dilution of goat anti-rabbit IgG [H&L] DyLight 488-conjugated antibody [Agrisera]) was added to the well and the mixture was incubated overnight in a humidity chamber at 4°C. After rinsing four times in PNTB solution and once in 1× PBS, the sections were counterstained with propidium iodide (PI; Sigma-Aldrich) (2 mg/liter in 1× PBS) for approximately 1 h at room temperature. Control experiments were carried out in the absence of primary antibodies. Sections were finally rinsed (twice 10 min) in 1× PBS and mounted in deionized water for microscopy observation.
Fluorescent images were acquired with a Zeiss LSM 710 confocal microscope using a 10×/0.3 EC Plan Neofluar objective and a 488-nm laser line to excite DyLight 488 (emission range, 494 to 544 nm), a 561-nm laser line to excite propidium iodide (emission range, 562 to 680 nm), and a beamsplitter (488 and 561 nm). Whole sections were obtained with the Tile-scan macro of ZEN 2008 software that also controls the Zeiss confocal hardware.
Data analysis.
Data were subjected to statistical evaluation using one-way analysis of variance (ANOVA) and Tukey's multiple-comparison test. Data are presented as the means ± standard deviations (SD) of the results of at least four biological replicates conducted at different times.
RESULTS
Point mutations in the promintron regions: qPCR analysis of the uidA reporter gene.
To modulate IAA biosynthesis from the promintron-iaaM-tms2 construct, the 85-bp promoter sequence was mutagenized.
In the preliminary screening of mutations able to alter promoter activity, the uidA reporter gene was used and reverse transcription-PCR (RT-PCR) performed to quantify its expression in free-living cells.
The main prokaryotic promoter elements which facilitate specific transcript initiation by RNA polymerase are the −10 and −35 elements, with other elements located upstream of the −35 hexamer and in the spacer region between the −10 and −35 elements (21–23).
In our study, single-base substitutions were introduced in the −10 (mutations from M1 to M6) and −35 (mutations M7 and M8) regions, in the spacer between these elements (mutations M9 and M10), and in the region located upstream of the −35 hexamer (mutation M11).
The location of each introduced change is shown in Table 1.
Base substitution performed in the −10 region provided six mutants, including five (M1 to M5) that did not affect promintron activity and one (M6) that significantly increases promoter activity compared to the wild-type promoter (Fig. 1).
The introduction of a long stretch of cytosine in the −40 and −37 positions (M7) strongly induced the expression of the uidA reporter gene, while the second mutation performed in the −35 region (M8) did not significantly alter promoter activity (Fig. 1).
It is known that in bacteria, the length of the spacer between the −10 and −35 regions influences promoter activity. Deviation from optimal spacer length, by insertion or deletion of bases, may impair promoter function (21). The −10 and −35 promintron consensus sequences are separated by a region of 17 bp. We modified the spacer length by inserting or deleting TC nucleotides within the promoter spacer (−25 and −24 positions) and analyzed the promintron activity. For the deletion of spacer bases (−TC; 15 bp) (M9), the alterations of uidA gene expression were not statistically significant, whereas reduced promoter activity was observed when two bases were introduced (+TC; 19 bp) (M10) (Fig. 1). We also tested the effect of mutations in the sequence upstream of the −35 region (M11; see Table 1), in which elements that facilitate specific transcript initiation by RNA polymerase are often located. The substitution of the T-rich sequence with a G-rich sequence significantly enhanced the promintron activity (Fig. 1).
Promintron-iaaM-tms2 construct expression.
Three mutations, i.e., M6, M11, and M7, based on the data of uidA gene expression-induced promoter activity, have been selected to increase IAA biosynthesis. For this purpose, the corresponding modified promintron was inserted upstream of the iaaM gene from Pseudomonas syringae pv. savastanoi and the tms2 gene from Agrobacterium tumefaciens. The resulting plasmids were introduced into S. meliloti 1021 to generate RD65, RD66, and RD67 strains, respectively.
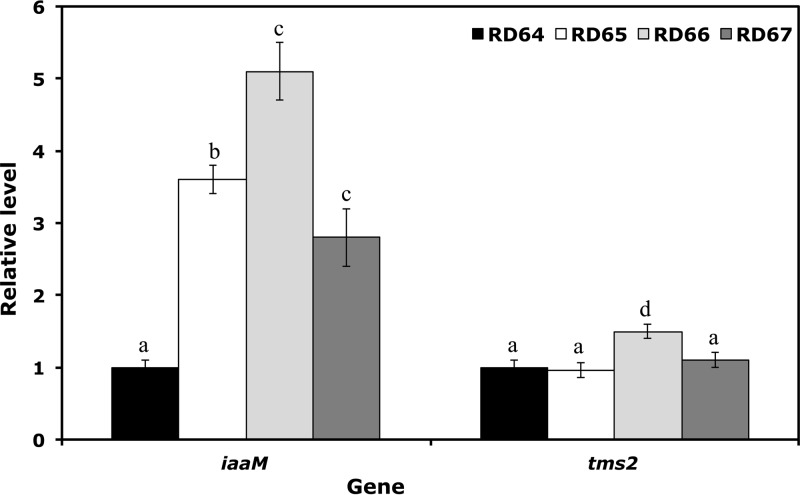
To evaluate the expression levels of the two genes which were used to increase the synthesis of IAA in Rhizobium, a quantitative RT-PCR analysis was performed on RD64, RD65, RD66, and RD67 free-living bacteria. The induced mRNA level of the iaaM gene was increased in RD65 and RD66 compared to that in RD64 (Fig. 2), with the highest increment observed for the RD66 strain. For the tms2 gene, the expression level of this gene was positively affected in RD66 cells, whereas for RD67 cells, the observed expression level was not statistically different from that seen with RD64 cells.
FIG 2.
Quantitative RT-PCR analysis of iaaM and tms2 genes involved in the synthesis of IAA. Values are the means ± SD of the results of four biological replicates conducted at different times. Relative gene expression levels determined by the comparative CT method are presented as follows: 2−ΔΔCT > 1, more highly expressed genes in cells transformed with the mutated construct (RD65, RD66, and RD67 cells); 2−ΔΔCT < 1, more highly expressed genes in cells transformed with the wild-type construct (RD64 cells). Different letters are used to indicate means that differ significantly according to Tukey's test (P < 0.05).
IAA level in free-living bacteria and nodules.
To quantify the IAA produced by free-living bacteria, liquid chromatography—coupled to high-resolution mass spectrometry in an Orbitrap—was applied. High signal-to-noise ratios and therefore enhanced sensitivity were obtained by applying this procedure. The deuterated IAA (d7-IAA) molecule was used as an internal standard for quantitative purposes. Significant increases in the production of IAA were observed in all the modified strains (RD64, RD65, RD66, and RD67) compared to the wild-type 1021 strain (Table 2). Even when the absolute values of the IAA concentrations in the modified strains were similar, it was evident that RD66 was the strain with the highest levels of IAA accumulation. The auxin biosynthetic pathway used resulted in an increase in auxin synthesis of at least 100-fold in the free-living modified bacteria. These results were consistent with the increment of mRNA levels measured in free-living rhizobia for the two genes involved in the biosynthesis of IAA (Fig. 2) and with the data of the ELISAs performed on nodules deriving from plants nodulated by the modified strains (Table 3).
TABLE 2.
LC-MS analysis of IAA in free-living S. meliloti cells
| Strain | IAA content (μM)a |
|---|---|
| 1021 | 0.60 ± 0.01 |
| RD64 | 56 ± 13 |
| RD65 | 57 ± 11 |
| RD66 | 63 ± 11 |
| RD67 | 55 ± 11 |
The values represent means ± SD of the results of three biological replicates conducted at different times.
TABLE 3.
Estimation of IAA levels in M. truncatula nodules by means of ELISA
| Plant | IAA content (μmol/g FW)a |
|---|---|
| Ms-1021 | 0.23 ± 0.03 |
| Ms-RD64 | 29 ± 2 |
| Ms-RD65 | 32 ± 3 |
| Ms-RD66 | 35 ± 3 |
| Ms-RD67 | 26 ± 1 |
The values represent means ± SD of the results of four biological replicates conducted at different times. FW, fresh weight.
Localization of IAA in root nodules.
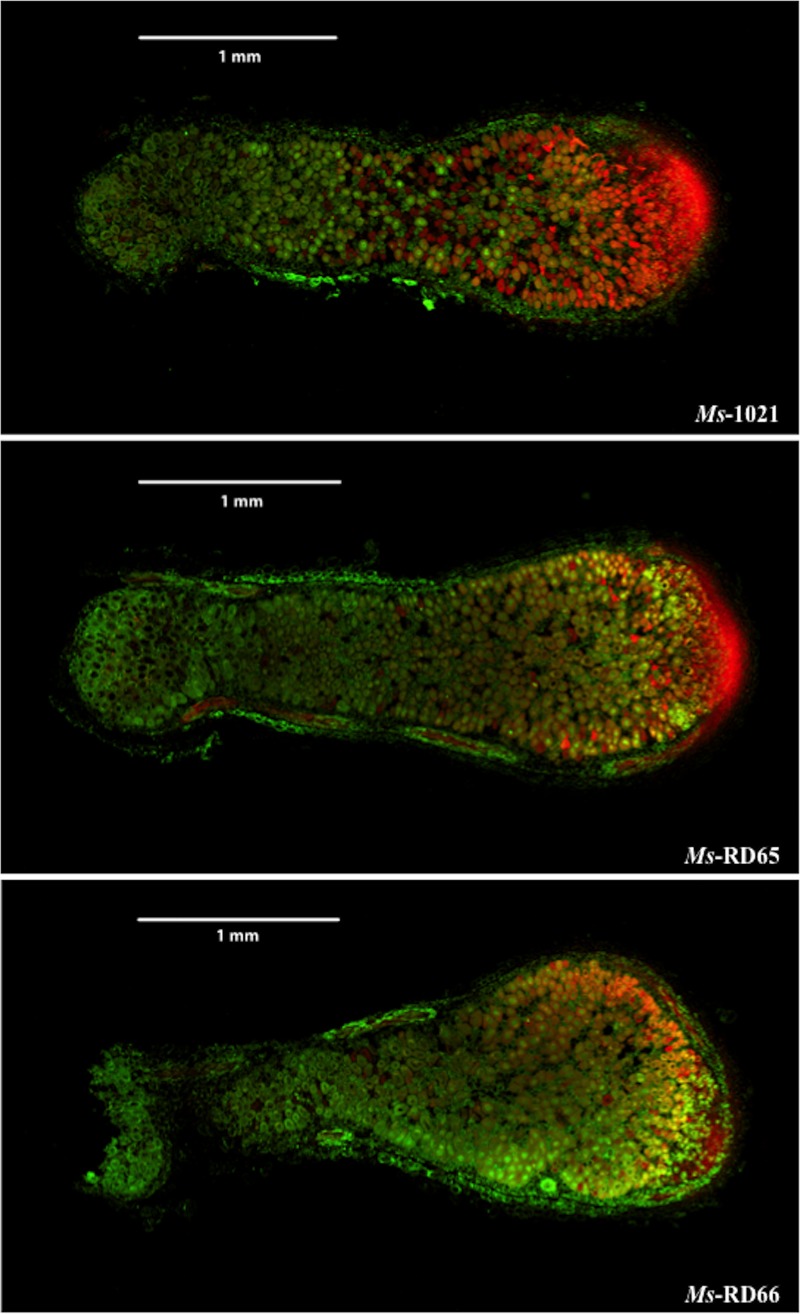
Immunohistochemical detection of IAA was carried out in nodules of Ms-1021, Ms-RD65, and Ms-RD66 plants. The EDAC reagent, which cross-links the carboxyl group of free IAA to structural proteins while preserving the antigenicity of IAA (24, 25), was used. Nodule sections were labeled with anti-IAA and counterstained with PI to show the bacteroid distribution. Representative images from six independent nodules are shown in Fig. 3.
FIG 3.
Immunolocalization of IAA in nodules of Medicago sativa. Nodule tissues from 4-week-old plants were labeled with anti-IAA (green) and counterstained with propidium iodide (red) to visualize nucleic acids. The yellow color is due to the addition of red fluorescence and green fluorescence.
The meristematic zone in nodules deriving from all plants showed strong immunolabeling of nucleoplasm due to the PI staining of nucleoli. No signal was detected when the primary antibody was omitted, indicating that the signal is dependent on the presence of anti-IAA on the tissue section. The immunofluorescence signal that was detected in the invasion zone and in the first layer of cells of the fixation zone of nodules deriving from Ms-RD65 and Ms-RD66 plants was stronger than that detected in those deriving from Ms-1021 plants. The most intense labeling was observed for Ms-RD66 plants, in agreement with the measurement of the highest IAA concentrations in the preliminary ELISAs.
M. sativa plant growth.
The effect of inoculation of S. meliloti strains 1021, RD64, RD65, RD66, and RD67 on the growth and nitrogen-fixing ability of M. sativa was analyzed (Fig. 4 and Table 4). Four weeks after inoculation, significant increases in the fresh weight of plants infected with strains RD65 (Ms-RD65) and RD66 (Ms-RD66) were observed compared to the fresh weight of plants nodulated by strain RD64 (Ms-RD64), which contains the wild-type construct (Fig. 4). This effect was even more evident in comparisons of Ms-RD65 and Ms-RD66 plants with those infected with the wild-type 1021 strain (Ms-1021). When shoot fresh weights of plants, nodulated by strain RD67 (Ms-RD67), were compared with those of Ms-RD64 plants, a slight increase was measured, although the observed effect was not statistically significant (Fig. 4). Ms-RD65 and Ms-RD66 plants also showed root systems that were more highly branched with more secondary roots and a higher number of nodules than Ms-1021 plants (Table 4), as already found for M. truncatula RD64 (Mt-RD64) plants (13).
FIG 4.
Effect of bacterial IAA on Medicago sativa growth. The photograph shows the phenotype of 4-week-old Medicago sativa plants infected with S. meliloti strains, synthesizing different IAA levels.
TABLE 4.
Effect of rhizobial IAA on Medicago sativa growth and nitrogen fixation
| Plant | Shoot fresh wt (mg)a | Secondary root no.b | Nodule no.b | Relative levelc |
|---|---|---|---|---|
| Ms-1021 | 231 ± 51 a | 15 ± 4 a | 9 ± 3 a | |
| Ms-RD64 | 303 ± 83 b | 17 ± 2 a | 11 ± 3 a | 1.5 ± 0.1 |
| Ms-RD65 | 381 ± 87 c | 19 ± 2 b | 14 ± 3 b | 2.9 ± 0.3 |
| Ms-RD66 | 414 ± 81 c | 19 ± 2 b | 18 ± 4 c | 3.3 ± 0.5 |
| Ms-RD67 | 311 ± 70 b | 15 ± 3 a | 13 ± 4 a | 1.8 ± 0.2 |
Data represent means ± SD (n = 50), and those marked with different letters are significantly different based on the Tukey's test (P < 0.01).
Data were obtained from the analysis of equal root portions and represent means ± SD (n = 15); those marked with different letters are significantly different based on the Tukey's test (P < 0.01).
Relative gene expression levels of nifH gene from comparative CT method; 2−ΔΔCT > 1, more highly expressed genes in plants nodulated by the modified strains (RD64, RD65, RD66, and RD67); 2−ΔΔCT < 1, more highly expressed genes in plants nodulated by the wild-type strain 1021. Values represent means ± SD of the results of four biological replicates conducted at different times.
These results were connected to the higher expression of the nifH gene, encoding the Fe protein of the nitrogenase enzyme, observed for Ms-RD65 and Ms-RD66 plants than for Ms-1021 control plants (Table 4).
DISCUSSION
In the present work, we used the promintron-iaaM-tms2 construct to increase IAA levels during Rhizobium-legume symbiosis.
The same promintron-iaaM-tms2 construct was already used by Camerini et al. (14) and by Imperlini et al. (13) to modify R. leguminosarum and S. meliloti bacteria, generating the RD20 and RD64 strains, respectively. In both cases, the authors observed increased N-fixation, nodule development, and plant dry weight production. For R. leguminosarum RD20, a 14-fold increase in the IAA concentration was measured in free-living bacteria (14). Root nodules of Vicia hirsuta plants infected with this strain showed an even higher (up to 60-fold) increase in IAA levels, especially when conjugated IAA was evaluated (up to 80-fold).
For tropical legumes developing the determinate type of nodules, such as bean, soybean, and peanut, we also have preliminary data indicating the positive effects triggered by the specific rhizobial strains modified with the promintron-iaaM-tms2 construct (12).
We are thus investigating a broad-spectrum phenomenon that shows similar characteristics in temperate and tropical legumes.
The goal of the present study was to investigate the biological effects of IAA concentrations higher than those measured in the previous work (13–15).
To increase IAA biosynthesis, the main prokaryotic promoter elements located in the promintron sequence were mutagenized.
We show here that, in the biological system described, the IAA levels produced are such that dose-response effects were observed when the concentration of free IAA measured by mass spectrometry increased from 93% in RD64 to 95% and 105% in RD65 and RD66, respectively, compared to 1021 wild-type strain results.
We found that continuous ectopic biosynthesis of endogenous IAA positively affects plant growth, root nodule development, and nitrogenase gene (nifH) expression.
We also found that IAA staining colocalizes with the bacteroids and that biosynthesis occurs all along the root nodule even in the oldest tissue of zone IV, consistent with the transcription of the promintron-iaaM-tms2 construct in mature nodules as reported by Pii et al. (8).
Considering auxins as a component of endogenous developmental programs for the shape of the final root architecture and root nodule, they remain the focus of many active research programs.
The role of auxins in nodule initiation and development was mainly hypothesized by considering the effects of exogenous IAA application or through the modulation of IAA transport (26, 27), while a smaller body of data concerns the endogenous IAA and the fact that part of the IAA found in nodules is of prokaryotic origin.
Indeed, the endogenous pool of plant IAA may be altered by the acquisition of IAA that has been secreted by soil bacteria. In plant roots, endogenous IAA may be suboptimal or optimal for growth, and additional IAA that is taken up from bacteria could alter the IAA to either an optimal or a supraoptimal level, resulting in plant growth promotion or inhibition, respectively.
The observation of more-developed nodules in Ms-RD65 and Ms-RD66 plants indicates that rhizobium-derived auxin positively affects not only nodule formation but also nodule development.
It is known that the increase in lateral root growth is a characteristic trait observed in mutants that overproduce auxin (28). The more-branched root system observed for Ms-RD65 and Ms-RD66 plants is most probably a consequence of the increased synthesis of IAA in the nodules induced by RD65 and RD66 strains as well as of a redistribution of the phytohormone in the root tissue, as already shown for plants nodulated by the RD64 strain (15).
It is known that root meristem size is influenced by the balance of two competing processes, namely, cell division and cell differentiation, and that plant growth hormones, including auxin, play an important role in regulating the balance of these two processes in root.
It would thus be interesting to understand whether the higher accumulation of IAA observed in the nodules of Ms-RD66 plants could lead to meristems that are even more enlarged and that are active for a longer time than those observed by Camerini et al. (14) and Imperlini et al. (13) in M. trucatula and V. hirsuta plants, respectively.
It would also be intriguing to investigate whether the RD66 strain might show a dose-response effect even in the presence of abiotic stress or nutrient deficiency.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rubino Stefano for technical assistance.
This work was partially supported by a dedicated grant from the Italian Ministry of Economy and Finance to the National Research Council for the project CISIA “Innovazione e Sviluppo del Mezzogiorno—Conoscenze Integrate per Sostenibilità ed Innovazione del Made in Italy Agroalimentare—Legge n. 191/2009.” This work was also partially supported by the European Commission for funding the ABSTRESS project (FP7 KBBE-2011-289562).
Footnotes
Published ahead of print 9 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00597-14.
REFERENCES
- 1.Oldroyd GED, Downie JA. 2008. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59:519–546. 10.1146/annurev.arplant.59.032607.092839 [DOI] [PubMed] [Google Scholar]
- 2.Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J. 2010. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibson KE, Kobayashi H, Walzer GC. 2008. Molecular determinants of a symbiotic chronic infection. Annu. Rev. Genet. 42:413–441. 10.1146/annurev.genet.42.110807.091427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spaepen S, Vanderleyden J. 17 November 2010. Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 10.1101/cshperspect.a001438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glick BR. 2012. Plant growth-promoting bacteria: mechanisms and applications. Scientifica (Cairo) 2012:963401. 10.6064/2012/963401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desbrosses GJ, Stougaard J. 2011. Root nodulation: a paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe 10:348–358. 10.1016/j.chom.2011.09.005 [DOI] [PubMed] [Google Scholar]
- 7.Turner M, Nizampatnam NR, Baron M, Coppin S, Damodaran S, Adhikari S, Arunachalam SP, Yu O, Subramanian S. 2013. Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol. 162:2042–2055. 10.1104/pp.113.220699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pii Y, Crimi M, Cremonese G, Spena A, Pandolfini T. 2007. Auxin and nitric oxide control indeterminate nodule formation. BMC Plant Biol. 7:21. 10.1186/1471-2229-7-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandolfini T, Storlazzi A, Calabria E, Defez R, Spena A. 2000. The spliceosomal intron of the rolA gene of Agrobacterium rhizogenes is a prokaryotic promoter. Mol. Microbiol. 35:1326–1334. 10.1046/j.1365-2958.2000.01810.x [DOI] [PubMed] [Google Scholar]
- 10.Magrelli A, Langenkemper K, Dehio C, Schell J, Spena A. 1994. Splicing of the rolA transcript of Agrobacterium rhizogenes in Arabidopsis. Science 266:1986–1988. 10.1126/science.7528444 [DOI] [PubMed] [Google Scholar]
- 11.Priefer UB, Aurag J, Boesten B, Bouhmouch I, Defez R, Filali-Maltouf A, Miklis M, Moawad H, Mouhsine B, Prell J, Schluter A, Senatore B. 2001. Characterisation of Phaseolus symbionts isolated from Mediterranean soils and analysis of genetic factors related to pH tolerance. J. Biotechnol. 91:223–236. 10.1016/S0168-1656(01)00329-7 [DOI] [PubMed] [Google Scholar]
- 12.Bianco C, Rotino GL, Campion B, Anas I, Defez R. 2010. How to improve legume production under severe environmental stresses. J. Biotechnol. 150:119. 10.1016/j.jbiotec.2010.08.309 [DOI] [Google Scholar]
- 13.Imperlini E, Bianco C, Lonardo E, Camerini S, Cermola M, Moschetti G, Defez R. 2009. Effect of indole-3-acetic acid on Sinorhizobium meliloti survival and on symbiotic nitrogen fixation and stem dry weight production. Appl. Microbiol. Biotechnol. 83:727–738. 10.1007/s00253-009-1974-z [DOI] [PubMed] [Google Scholar]
- 14.Camerini S, Senatore B, Lonardo E, Imperlini E, Bianco C, Moschetti G, Rotino GL, Campion B, Defez R. 2008. Introduction of novel pathway for IAA biosynthesis to rhizobia alters vetch root nodule development. Arch. Microbiol. 190:67–77. 10.1007/s00203-008-0365-7 [DOI] [PubMed] [Google Scholar]
- 15.Bianco C, Defez R. 2009. Medicago truncatula improves salt tolerance when nodulated by an indole-3-acetic acid-overproducing Sinorhizobium meliloti strain. J. Exp. Bot. 60:3097–3107. 10.1093/jxb/erp140 [DOI] [PubMed] [Google Scholar]
- 16.Bianco C, Imperlini E, Defez R. 2009. Legume likes more IAA. Plant Signal. Behav. 4:763–765. 10.4161/psb.4.8.9166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bianco C, Defez R. 2010. A Sinorhizobium meliloti IAA-overproducing strain improves phosphate solubilization and Medicago plant yield. Appl. Environ. Microbiol. 76:4626–4632. 10.1128/AEM.02756-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galibert F, Finan TM, Long SR, Puhler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P, Bothe G, Boutry M, Bowser L, Buhrmester J, Cadieu E, Capela D, Chain P, Cowie A, Davis RW, Dreano S, Federspiel NA, Fisher RF, Gloux S, Godrie T, Goffeau A, Golding B, Gouzy J, Gurjal M, Hernandez-Lucas I, Hong A, Huizar L, Hyman RW, Jones T, Kahn D, Kahn ML, Kalman S, Keating DH, Kiss E, Komp C, Lelaure V, Masuy D, Palm C, Peck MC, Pohl TM, Portetelle D, Purnelle B, Ramsperger U, Surzycki R, Thebault P, Vandenbol M, et al. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668–672. 10.1126/science.1060966 [DOI] [PubMed] [Google Scholar]
- 19.Bianco C, Imperlini E, Calogero R, Senatore B, Pucci P, Defez R. 2006. Indole-3-acetic acid regulates the central metabolic pathways in Escherichia coli. Microbiology 152(Pt 8):2421–2431. 10.1099/mic.0.28765-0 [DOI] [PubMed] [Google Scholar]
- 20.Alvarez R, Nissen SJ, Sutter EG. 1989. Relationship between indole-3-acetic acid levels in apple (Malus pumila Mill) rootstocks cultured in vitro and adventitious root formation in the presence of indole-3-butyric acid. Plant Physiol. 89:439–443. 10.1104/pp.89.2.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Typas A, Hengge R. 2006. Role of the spacer between the -35 and -10 regions in σS promoter selectivity in Escherichia coli. Mol. Microbiol. 59:1037–1051. 10.1111/j.1365-2958.2005.04998.x [DOI] [PubMed] [Google Scholar]
- 22.Browning FD, Busby SJW. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2:57–65. 10.1038/nrmicro787 [DOI] [PubMed] [Google Scholar]
- 23.Lee DJ, Minchin SD, Busby SJW. 2012. Acting transcription in Bacteria. Annu. Rev. Microbiol. 66:125–152. 10.1146/annurev-micro-092611-150012 [DOI] [PubMed] [Google Scholar]
- 24.Aloni R, Schwalm K, Langhans M, Ullrich CI. 2003. Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta 216:841–853 [DOI] [PubMed] [Google Scholar]
- 25.Thomas C, Bronner R, Molinier J, Prinsen E, van Onckelen H, Hahne G. 2002. Immuno-cytochemical localization of indole-3-acetic acid during induction of somatic embryogenesis in cultured sunflower embryos. Planta 215:577–583. 10.1007/s00425-002-0791-8 [DOI] [PubMed] [Google Scholar]
- 26.Reed RC, Brady SR, Muday GK. 1998. Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol. 118:1369–1378. 10.1104/pp.118.4.1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Taketa S, Ichii M, Xu L, Kai Xia Zhou X. 2003. Lateral root formation in rice (Oryza Sativa L.): differential effects of indole-3-acetic acid and indole-3-butyric acid. Plant Growth Regul. 41:41–47. 10.1023/A:1027384528993 [DOI] [Google Scholar]
- 28.Overvoorde P, Fukaki H, Beeckman T. 2010. Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2:a001537. 10.1101/cshperspect.a001537 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.