Abstract
Single-chain variable fragment (scFv) antibodies are widely used as diagnostic and therapeutic agents or biosensors for a majority of human disease. However, the limitations of the present scFv antibody in terms of stability, solubility, and affinity are challenging to produce by traditional antibody screening and expression formats. We describe here a feasible strategy for creating the green fluorescent protein (GFP)-based antibody. Complementarity-determining region 3 (CDR3), which retains the antigen binding activity, was introduced into the structural loops of superfolder GFP, and the result showed that CDR3-inserted GFP displayed almost the same fluorescence intensity as wild-type GFP, and the purified proteins of CDR3 insertion showed the similar binding activity to antigen as the corresponding scFv. Among of all of the CDRs, CDR3s are responsible for antigen recognition, and only the CDR3a insertion is the best format for producing GFP-based antibody binding to specific antigen. The wide versatility of this system was further verified by introducing CDR3 from other scFvs into loop 9 of GFP. We developed a feasible method for rapidly and effectively producing a high-affinity GFP-based antibody by inserting CDR3s into GFP loops. Further, the affinity can be enhanced by specific amino acids scanning and site-directed mutagenesis. Notably, this method had better versatility for creating antibodies to various antigens using GFP as the scaffold, suggesting that a GFP-based antibody with high affinity and specificity may be useful for disease diagnosis and therapy.
INTRODUCTION
To date, antibodies are used for a very broad and still steadily expanding spectrum of applications, such as cancer therapy, disease diagnosis, and signal pathway investigation (1–3). Monoclonal antibodies (Abs) produced by hybridoma technology have been widely used to rapidly detect pathogens in food and raw seafood (4). In spite of their high affinity and specificity, monoclonal Abs have some obvious flaws, including a high molecular weight, a requirement for large-scale culture, and the instability of cell lines. Furthermore, it is difficult to create higher-affinity and higher-specificity Abs by genetic manipulation.
The single-chain variable fragment (scFv) is a class of engineered antibodies generated by the fusion of heavy (VH) and light (VL) chains of immunoglobulin gene through a short polypeptide linker (3) and still retains the binding activity to target antigen (5, 6). The smaller size of scFv fragment allows better tissue penetration, leading to improve tumor targeting and enhanced blood-brain barrier permeability for the treatment of neurodegenerative diseases (7). Because of these advantages, scFv has been used as a therapeutic agent and plays a key role in the therapy and diagnosis of a variety of human diseases (8, 9). Moreover, scFv antibody can be produced in Escherichia coli in the form of small, recombinant fragments that retain the binding property (10). In comparison to polyclonal or hybridoma antibodies, scFv antibody may be easily manipulated for improving specificity and affinity, thereby reducing the production cost (11, 12). However, these applications of scFv were limited by drawbacks, such as the formation of an inclusion body, which often leads to low binding activity and an unstable structure and is cytotoxic to host cells (3). Hence, it is very important to develop a feasible approach for reducing these limitations.
Green fluorescent protein (GFP) is a protein that exhibits bright green fluorescence when exposed to light in the blue-to-UV range (13, 14), and it has been widely used in many ways, such as in flow cytometry (15), small interfering RNA/DNA transfection (14, 16), protein delivery (17–19), peptide presentation (20), protein-protein interaction (21), and monitoring of intracellular processes (22). GFP has also been established as an in vivo marker for gene expression and protein localization (23, 24). Previous analyses demonstrated that GFP could be used as a scaffold for fluorobody development (25–30). The stable beta-barrel structure and the ability to yield multiple-color fluorescence prompted interest in developing a new class of antibody using the GFP frame as the template. Zeytun et al. developed fluorobodies by inserting diverse binding fragments of antibody into four of the exposed loops at the end of GFP, and the fluorobodies retain the binding characteristics (28). Pavoor at al. obtained a GFP-based biosensor by inserting two complementarity-determining regions 3 (CDR3s) into different loops of GFP to create a high-affinity GFP-Ab by directed evolution using a surrogate loop approach and yeast surface display (30). Other researchers also tried to insert the binding loops into various exposed loops of GFP (26, 29), but the results from those studies were not ideal, and the fluorescence of GFP was diminished substantially or eliminated when random loops were inserted. Among all the loops of GFP, loop 9 (positions 171 to 176) was considered the most tolerated site for peptide insertion (30), and it was possible to isolate a fluorescent binder against specific target using phage display.
In a previous study, we successfully screened a high-affinity antibody to a thermolabile hemolysin (TLH) named scFv-LA3 using phage display (2). In the present study, the superfolder GFP, a GFP variant that has high stability and improved folding kinetics, was used as a template for CDR insertion at loop 9. The binding activities of superfolder GFP-based antibody were determined by enzyme-linked immunosorbent assay (ELISA), and the utility of this insertion was demonstrated by inserting CDR3s of different scFv antibodies.
MATERIALS AND METHODS
Reagents.
All of the strains used in the present study were from Fujian Agriculture and Forestry University (Fujian, China). The PCR mixture and DNA restriction enzymes were purchased from Thermo Fisher Scientific (Waltham, MA). Taq DNA polymerase and T4 DNA ligase were purchased from TaKaRa (Dalian, China). Anti-6×His tag monoclonal antibody was purchased from Abgent (San Diego, CA), and horseradish peroxidase (HRP)-labeled goat anti-mouse IgG was from Boster Biological Technology Co. (Wuhan, China). All of the oligonucleotides are listed in Table 1, and all other reagents used were of analytical reagent grade.
TABLE 1.
Primers used for different scFv, CDR, and CDR3 insertions
| Target genea | Primer | DNA sequence (5′–3′) |
|---|---|---|
| NaeI variant | GFP-F | CTGCGATGCTGGTTGCCAACGATC |
| 9-NaeI-r | GAACGGATCCATCTTCGCCGGCAACGTTGTGGCGAATTTTGAAG | |
| scFv | 9-scFv/VH-F | ATAGCCGGCATGGCCCAGGTGAAACTGCAGGAG |
| 9-scFv/VL-R | CTCGGATCCCCGTTTGATTTCCAGCTTGGTCCCTC | |
| VH | 9-VH-R | TTTGGATCCTGAACCGCCTCCACCTGAGGAGACG |
| VL | 9-VL-F | TTTAGCCGGCGGATCGGACATTGAGCTCACCC |
| NotI-R | GCGGCACATCGTACGGATAACCAGAAC | |
| HCDR1 | 9-HCDR1-F | AACGGATCCGGCTACACCTTTACTAACTACACGGGATCAGTTCAACTAGCAGACCATTATCAAC |
| HCDR2 | 9-HCDR2-F | AACGGATCCATTAATCCTAGCAGTGATTATACTGGATCAGTTCAACTAGCAGACCATTATCAAC |
| LCDR1 | 9-LCDR1-F | AACGGATCCTCAAGTGTAAGTTCCAGTTATGGATCAGTTCAACTAGCAGACCATTATCAAC |
| LCDR2 | 9-LCDR2-F | AACGGATCCTATAGTACATCCAACCTGGCTGGATCAGTTCAACTAGCAGACCATTATCAAC |
| HCDR3A | 9-HCDR3-F | TTAGGATCCGCAAGATTCCCCCATCTCGAGGACTACTGGTACTTCGATGTCCTGCAG GGATCAGTTCAACTAGCAGACCATTATCAAC |
| LCDR3A | 9-LCDR3-F | TTAGGATCCCACCAGTATCATCGTTCCCCACGGACGCTGCAGGGATCAGTTCAACTAGCAGACCATTATCAAC |
| HCDR3B | 9-HCDR3B-F | AACGGATCCGTCTATTACTGTGCAAGATTCCCCCATCTCGAGGACTACTGGTACTTCGATGTCTGGGGCCAAGGGACCACGGGATCAGTTCAACTAGCAGACCATTATCAAC |
| HCDR3C | 9-HCDR3C-F | AACGGATCCGCAGTCTATTACTGTGCAAGATTCCCCCATCTCGAGGACTACTGGTACTTCGATGTCTGGGGCCAAGGGACCACGGTCGGATCAGTTCAACTAGCAGACCATTATCAAC |
| LCDR3B | 9-LCDR3B-F | AACGGATCCTACTGCCACCAGTATCATCGTTCCCCACGGACGTTCGGTGGAGGGACCGGATCAGTTCAACTAGCAGACCATTATCAAC |
| LCDR3C | 9-LCDR3C-F | AACGGATCCACTTATTACTGCCACCAGTATCATCGTTCCCCACGGACGTTCGGTGGAGGGACCAAGCTGGGATCAGTTCAACTAGCAGACCATTATCAAC |
| HCDR3A | 5-HCDR3-F | TCCAGATCTGCAAGATTCCCCCATCTCGAGGACTACTGGTACTTCGATGTCGACGGGAACTACAAGACGCGT |
| LCDR3A | 5-LCDR3-F | TCCAGATCTCACCAGTATCATCGTTCCCCACGGACGCTGCAGGACGGGAACTACAAGACGCGT |
All are scFv genes except for the NaeI variant.
Construction of different binding regions of scFv into loop 9 of GFP.
In a previous study, we obtained the scFv against Vibrio parahaemolyticus TLH (2). To identify the feasibility of insertion at loop 9 of superfolder GFP, anti-TLH scFv was used as a template for amplifying the different insertion regions. For scFv, VH, and VL insertion, an NaeI restriction enzyme site was introduced into loop 9, and a superfolder GFP with NaeI at positions V-171 and E-172 of loop 9 was constructed. NaeI and BamHI were used to insert VL, VH, and scFv, and specific primers (Table 1) were used to amplify the VH, VL, and scFv genes, respectively. After PCR and gel purification, the PCR products were digested with NaeI and BamHI. The digested PCR products were ligated to the same enzyme-digested vector pGEPi-sfgfp, and the resulting plasmids were designated pGEPi-sfgfp-9-VL, pGEPi-sfgfp-9-VH, and pGEPi-sfgfp-9-scFv, respectively. For H-CDR3a (101-FPHLEDYWYFD-101) and L-CDR3a (228-HQYHRSPRT-236) insertion, the synthesized primers (Table 1) containing 11 amino acids of the HCDR3 sequence or 9 amino acids of the LCDR3 sequence from the anti-TLH scFv were used to insert at positions 175S and 176V with BamHI and NotI, and the resulting vectors were designated pGEPi-sfgfp-9-HCDR3a and pGEPi-sfgfp-9-LCDR3a, respectively. All of the expressed proteins contained a 6×His tag at the C terminus, and the proteins were named G9-VL, G9-VH, G9-scFv, G9-H3(a), and G9-L3(a), respectively.
Construction of different CDR insertions.
To determine the detailed functions of different CDRs, two heavy-chain CDRs (HCDR1, 28-GYTFTNYT-35; HCDR2, 53-INPSSDYT-60) and two light-chain CDRs (LCDR1, 165-SSVSSSY-171; LCDR2, 188-YSTSNLA-194) were amplified with the specific primers (Table 1) and inserted at positions S175 and V176 of loop 9; these constructs were designated pGEPi-sfgfp-9-HCDR1, pGEPi-sfgfp-9-HCDR2, pGEPi-sfgfp-9-LCDR1, and pGEPi-sfgfp-9-LCDR2, respectively. The expressed proteins were named as G9-H1, G9-H2, G9-L1, and G9-L2, respectively. To further demonstrate the best format of CDR3 insertions, three different formats of insertions were selected, including the loop insertion (H-CDR3a, 101-FPHLEDYWYFD-111; L-CDR3, 228-HQYHRSPRT-236), the loop and half beta sheet insertion (H-CDR3b, 96-YYCARFPHLEDYWYFDVWGQGTT-111; L-CDR3b, 226-YCHQYHRSPRTFGGGTK-242), and the loop and full beta sheet insertion (H-CDR3c, 94-AVYYCARFPHLEDYWYFDVWGQGTTV-119; L-CDR3c, 224-TYYCHQYHRSPRTFGGGTKL-243); the resulting constructs were named pGEPi-sfgfp-9-HCDR3a, pGEPi-sfgfp-9-HCDR3b, pGEPi-sfgfp-9-HCDR3c, pGEPi-sfgfp-9-LCDR3a, pGEPi-sfgfp-9-LCDR3b, and pGEPi-sfgfp-9-LCDR3c, respectively. All of the expressed proteins contained a 6×His tag at C-terminal, and the proteins were named G9-H3a, G9-H3b, G9-H3c, G9-L3a, G9-L3b, and G9-L3c, respectively.
Protein expression and purification.
For protein expression, the recombinant plasmid was transformed into E. coli BL21(DE3) by electroporation, and a single colony from the selection plate was inoculated into 5 ml of Luria-Bertani (LB) liquid medium containing 100 μg of ampicillin/ml. The culture was incubated overnight with shaking at 37°C and then transferred to a larger-scale preparation in LB medium (500 μl of culture was transferred to 50 ml of fresh LB). The expression of the target protein was induced under T7 promoter by adding 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) when the culture reached an optical density at 600 nm (OD600) of 0.8. The expressed product was first harvested by centrifugation (10,000 rpm) at 4°C for 10 min, followed by the addition of 5 ml of binding buffer (50 mM NaH2PO4, 300 mM NaCl, 1 mM imidazole, 0.05% Tween 20) after removal of the supernatant. The complex was then mixed gently, sonicated, and centrifuged at 4°C for 10 min. The supernatant was collected and loaded onto a 3-ml Ni2+-NTA column, which was equilibrated with binding buffer before the purification. After allowing the sample to flow through, the column was washed twice with wash buffer (100 ml each time). After the wash, the retained protein of interest was eluted with 0.5 ml of elution buffer four times, and the eluate was collected and analyzed by SDS-PAGE. The protein concentration was determined by using a bicinchoninic acid protein assay kit.
Fluorescence tracing and ELISA.
For fluorescence analysis, the different constructs were expressed at 30°C by IPTG (1 mmol/liter) when the culture reached an OD600 of 0.8 for overnight, and the expressed products were adjusted to the same OD600 for accurate analysis and harvested the pellet by centrifugation. The treated samples were placed on the black background and visualized the fluorescence of the protein by UV stimulation. The result was recorded by a camera under UV exposure. The activities of the purified GFP-Abs were determined by ELISA. The dialyzed TLH antigen (2.5 μg/ml [without 6×His tag]) was used to coat the 96-well plates at 4°C overnight. After blocking and washing, the purified different GFP-Abs products (1 μg/ml) derived from the different insertions described above were added to the reaction wells, followed by incubation at 37°C for 2 h. The anti-His6 tag antibody was added to the reaction wells, followed by incubation at 37°C for 2 h. The binding activity of the purified GFP-Abs was detected by using an HRP-conjugated anti-mouse IgG antibody. The enzyme reaction was then performed using tetramethylbenzidine (TMB) as the substrate, and color development was terminated with 2 M H2SO4. The absorbance at 450 nm was measured using a microplate reader. For indirect competitive ELISA (ic-ELISA), before interaction, the TLH antigen was used as the competing antigen and reacted to the purified G9-LCDR3(1 μg/ml) at 37°C for 2 h, and the other ELISA steps correspond to the steps described above. The data are presented as means and standard deviations (SD) for three separate experiments.
Specificity of G9-HCDR3 and G9-LCDR3 assay.
To identify the specificity of the purified G9-HCDR3 and G9-LCDR3, a competitive ELISA was performed. TLH, bovine serum albumin (BSA), keyhole limpet hemocyanin (KLH), and TLH-correlated antigens such as thermostable direct hemolysin (TDH), VP1694, and VP1668 were diluted and coated in 96-well plates (2.5 μg/ml). The diluted G9-HCDR3 and G9-LCDR3 antibodies were added to the reaction wells for 2 h at 37°C, the specificities of the antibodies were analyzed using anti-His tag antibody and HRP-conjugated goat anti mouse IgG antibody, and the absorbance at 450 nm was measured using a microplate reader. The data are presented as means plus the SD for three separate experiments.
ALA scanning and lysine scanning.
To find the key binding sites of CDR3, 11 amino acids of the heavy-chain CDR3 and 9 amino acids of the light-chain CDR3, regarded as the major binding activity for antigen-antibody interaction, were each individually mutated to alanine. At the same time, each of these 7 amino acids of the heavy chain (HCDR3) and 9 amino acids of the light chain (LCDR3) was individually mutated to lysine. Oligonucleotide-directed mutagenesis was performed by PCR using the former constructed plasmids as a template with the mutant primers (Tables 2 and 3), and the amplified CDR3 mutants were inserted into pGEPi-sfGFP vector with BamHI and NotI, respectively. Protein expression and purification were carried out as described above. The data are presented as means plus the SD for three separate experiments.
TABLE 2.
Primers used for Ala scanning in LCDR3 and HCDR3
| Target gene and primer | DNA sequence (5′–3′) |
|---|---|
| LCDR3-Ala | |
| L-H1/A-F | GATGGATCCGCCCAGTATCATCGTTCCCCACGGAC |
| L-Q/A-F | GATGGATCCCACGCTTATCATCGTTCCCCACGGAC |
| L-Y/A-F | GATGGATCCCACCAGGCTCATCGTTCCCCACGGAC |
| L-H2/A-F | GATGGATCCCACCAGTATGCTCGTTCCCCACGGAC |
| L-R1/A-F | GATGGATCCCACCAGTATCATGCTTCCCCACGGAC |
| L-S/A-F | GATGGATCCCACCAGTATCATCGTGCCCCACGGAC |
| L-P/A-F | GATGGATCCCACCAGTATCATCGTTCCGCACGGAC |
| L-R2/A-F | GATGGATCCCACCAGTATCATCGTTCCCCAGCGAC |
| L-T/A-F | GATGGATCCCACCAGTATCATCGTTCCCCACGGGCG |
| HCDR3-Ala | |
| H-F/A-F | ATTGGATCCGCCCCCCATCTCGAGGACTACTGGTACTTCGATC |
| H-P/A-F | ATTGGATCCTTCGCCCATCTCGAGGACTACTGGTACTTCGATC |
| H-H/A-F | ATTGGATCCTTCCCCGCTCTCGAGGACTACTGGTACTTCGATC |
| H-L/A-F | ATTGGATCCTTCCCCCATGCCGAGGACTACTGGTACTTCGATC |
| H-E/A-F | ATTGGATCCTTCCCCCATCTCGCGGACTACTGGTACTTCGATC |
| H-D1/A-F | ATTGGATCCTTCCCCCATCTCGAGGCCTACTGGTACTTCGATC |
| H-Y/A-F | ATTGGATCCTTCCCCCATCTCGAGGACGCCTGGTACTTCGATC |
| H-W/A-F | ATTGGATCCTTCCCCCATCTCGAGGACTACGCGTACTTCGATC |
| H-Y2/A-F | ATTGGATCCTTCCCCCATCTCGAGGACTACTGGGCCTTCGATC |
| H-F/A-F | ATTGGATCCTTCCCCCATCTCGAGGACTACTGGTACGCCGATC |
| H-D2/A-F | ATTGGATCCTTCCCCCATCTCGAGGACTACTGGTACTTCGCTC |
TABLE 3.
Primers used for Lys scanning in LCDR3 and HCDR3
| Target gene and primer | DNA sequence (5′–3′) |
|---|---|
| LCDR3-Lys | |
| L-Q/K-F | GATGGATCCCACAAGTATCATCGTTCCCCACGGAC |
| L-Y/K-F | GATGGATCCCACCAGAAACATCGTTCCCCACGGAC |
| L-H2/K-F | GATGGATCCCACCAGTATAAACGTTCCCCACGGAC |
| L-R1/K-F | GATGGATCCCACCAGTATCATAAATCCCCACGGAC |
| L-S/K-F | GATGGATCCCACCAGTATCATCGTAAACCACGGAC |
| L-P/K-F | GATGGATCCCACCAGTATCATCGTTCCAAACGGAC |
| L-R2/K-F | GATGGATCCCACCAGTATCATCGTTCCCCAAAGAC |
| HCDR3-Lys | |
| H-F/K-F | ATTGGATCCAAACCCCATCTCGAGGACTACTGGTACTTCGATC |
| H-P/K-F | ATTGGATCCTTCAAACATCTCGAGGACTACTGGTACTTCGATC |
| H-H/K-F | ATTGGATCCTTCCCCAAACTCGAGGACTACTGGTACTTCGATC |
| H-L/K-F | ATTGGATCCTTCCCCCATAAAGAGGACTACTGGTACTTCGATC |
| H-E/K-F | ATTGGATCCTTCCCCCATCTCAAAGACTACTGGTACTTCGATC |
| H-D1/K-F | ATTGGATCCTTCCCCCATCTCGAGAAATACTGGTACTTCGATC |
| H-Y/K-F | ATTGGATCCTTCCCCCATCTCGAGGACAAATGGTACTTCGATC |
| H-W/K-F | ATTGGATCCTTCCCCCATCTCGAGGACTACAAATACTTCGATC |
| H-Y2/K-F | ATTGGATCCTTCCCCCATCTCGAGGACTACTGGAAATTCGATC |
| H-F/K-F | ATTGGATCCTTCCCCCATCTCGAGGACTACTGGTACAAAGATC |
| H-D2/K-F | ATTGGATCCTTCCCCCATCTCGAGGACTACTGGTACTTCAAAC |
Mutation of key amino acids.
Once the key amino acids of LCDR3s were confirmed, the key amino acid R235 of LCDR3, which contributes most to the binding activity, was mutated to six other different hydrophilic amino acids (the primers are listed in Table 4). The expressed proteins were purified by using Ni2+ affinity chromatography, and the binding activities of mutated GFP-Abs were determined by ELISA according to the instructions described above. For the Q229 mutation (LCDR3 in loop 9), primers containing mutated sites (Table 4) were used to amplify the insertion DNA with the mutated R235K as a template, and the amplified DNA fragments were inserted into the designated site of GFP. The other steps were similar to those given above.
TABLE 4.
Primers used for site mutagenesis of key amino acids (LCDR3)
| Mutant type and primer | DNA sequence (5′–3′) |
|---|---|
| R2 mutants | |
| L-R2/D-F | GATGGATCCCACCAGTATCATCGTTCCCCAGACAC |
| L-R2/E-F | GATGGATCCCACCAGTATCATCGTTCCCCAGAGAC |
| L-R2/H-F | GATGGATCCCACCAGTATCATCGTTCCCCACACAC |
| L-R2/Q-F | GATGGATCCCACCAGTATCATCGTTCCCCACAGAC |
| L-R2/S-F | GATGGATCCCACCAGTATCATCGTTCCCCAAGCAC |
| L-R2/T-F | GATGGATCCCACCAGTATCATCGTTCCCCAACGAC |
| Q mutants | |
| L-Q/DR2/K-F | GATGGATCCCACGACTATCATCGTTCCCCAAAGAC |
| L-Q/ER2/K-F | GATGGATCCCACGAGTATCATCGTTCCCCAAAGAC |
| L-Q/HR2/K-F | GATGGATCCCACCACTATCATCGTTCCCCAAAGAC |
| L-Q/RR2/K-F | GATGGATCCCACCGGTATCATCGTTCCCCAAAGAC |
| L-Q/SR2/K-F | GATGGATCCCACAGCTATCATCGTTCCCCAAAGAC |
| L-Q/TR2/K-F | GATGGATCCCACACGTATCATCGTTCCCCAAAGAC |
| L-Q/KR2/K-F | GATGGATCCCACAAGTATCATCGTTCCCCAAAGAC |
Dual CDR3 insertions at loop 5 and loop 9.
To create the dual-insertion GFP-Abs, the HCDR3 loop (101-FPHLEDYWYFD-101) and LCDR3 (228-HQYHRSPRT-236) loop of anti-TLH scFv antibody were separately inserted in two loops (loop 5 and loop 9) by different insertion formats, including 5L9L (LCDR3 insertions in loop 5 and loop 9), 5L9H (LCDR3 in loop 5 and HCDR3 in loop 9), 5H9L (HCDR3 in loop 5 and LCDR3 in loop 9), 5H9H (HCDR3 insertions in loop5 and loop 9), and 5L9L(R235K) (LCDR3 containing R235K mutant insertions in loop 5 and loop 9). For the efficient insertion in loop 5, a GFP variant that introduced the restriction enzyme BglII site at the 101 and 102 sites of loop 5 was constructed. Primers containing H/LCDR3 or LCDR3(R235K) sequences (Table 1) were used to amplify the insertion genes with the constructed pGEPi-sfgfp-9-H/LCDR3a or pGEPi-sfgfp-9-LCDR3a(R235K) as a template, and the amplified DNA fragments were inserted into the designated sites of loop 5 with the restriction enzymes BglII and NotI to form dual CDR3 insertions. All of the constructs were sequenced and transformed into E. coli BL21 by electroporation, and the steps for protein expression and the determination of binding activity were as previously described.
Affinity determination.
After purification, the affinity of the purified GFP-Ab was determined by ELISA. The dialyzed TLH antigen (2.5 μg/ml) was used to coat the 96-well plates at 4°C for overnight. After the ELISA, the plates were blocked and washed, and the serially diluted GFP-Abs (the maximum concentration of antibody was 20 μg/ml) were added to the reaction wells, followed by incubation at 37°C for 2 h. Anti-6×His tag antibody was added to the reaction wells, followed by incubation at 37°C for 2 h. The affinities of the different antibodies were detected by using an HRP-conjugated anti-mouse IgG antibody. The enzyme reaction was then performed with TMB as a substrate, and color development was terminated with 2 M H2SO4. The absorbance at 450 nm was measured using a microplate reader. The affinity constant (Kaff) of purified GFP-Ab was determined by using the equation Kaff = n − 1/(n[Ab2] − [Ab1]) as described previously (2), where [Ab1] and [Ab2] represent the respective GFP-Ab concentrations required to achieve 50% of the maximum absorbance between two different concentrations of coated antigen ([Ag1] = n[Ag2]), and n is the dilution factor between the concentrations of antigen used. The data are presented as means plus the SD for three separate experiments.
Versatility assay of GFP-Abs.
To evaluate the versatility of CDR insertion at loop 9, the HCDR3s and LCDR3s of three different scFv antibodies specific to three small-molecular-weight toxins (fumonisin [FB1], tetrodotoxin [TTX], and citreoviridin [CIT]) were amplified with specific primers (Table 5) by PCR inserted into the same sites of loop 9. All of the constructs were transformed into E. coli BL21 by electroporation, and a single colony from the selection plate was inoculated into 5 ml of LB liquid medium containing 100 μg of ampicillin/ml for the expression of mutant GFP-Ab antibody. The expressed protein contained a 6×His tag at the N terminus, and the target proteins were purified by using Ni2+ affinity chromatography. The binding activities of the purified GFP-Abs were determined by ELISA. The data are presented as means plus the SD for three separate experiments.
TABLE 5.
Primers used for CDR3 insertion of other scFv antibodies
| Target gene (toxin) | Primer | DNA sequence (5′–3′) |
|---|---|---|
| HCDR3 (FB1) | 9-F-H3-F | AACGGATCCGCTAGGATGGAATTGCAGGTTCGGGGATCAGTTCAACTAGCAGACCATTATCAAC |
| LCDR3 (FB1) | 9-F-L3-F | AACGGATCCCAGCAAAGTAGGAGGGTTCCGTACACGGGATCAGTTCAACTAGCAGACCATTATCAAC |
| HCDR3 (TTX) | 9-T-H3-F | AACGGATCCGTAAGAGTGGCCTTTGATGGTTACTACGATGACTTCGGATCAGTTCAACTAGCAGACCATTATCAAC |
| LCDR3 (TTX) | 9-T-L3-F | AACGGATCCCAAAATGGTCACAGCTTTCCTCCGACGGGATCAGTTCAACTAGCAGACCATTATCAAC |
| HCDR3 (CIT) | 9-C-H3-F | AACGGATCCAGCATGGGGGCCCTCCCTCCGTACTACTTTGACTACTGGGGATCAGTTCAACTAGCAGACCATTATCAAC |
| LCDR3 (CIT) | 9-C-L3-F | AACGGATCCCAGCAAAGGAGTAGTTACCCGTACACGGGATCAGTTCAACTAGCAGACCATTATCAAC |
RESULTS
Definition of superfolder GFP.
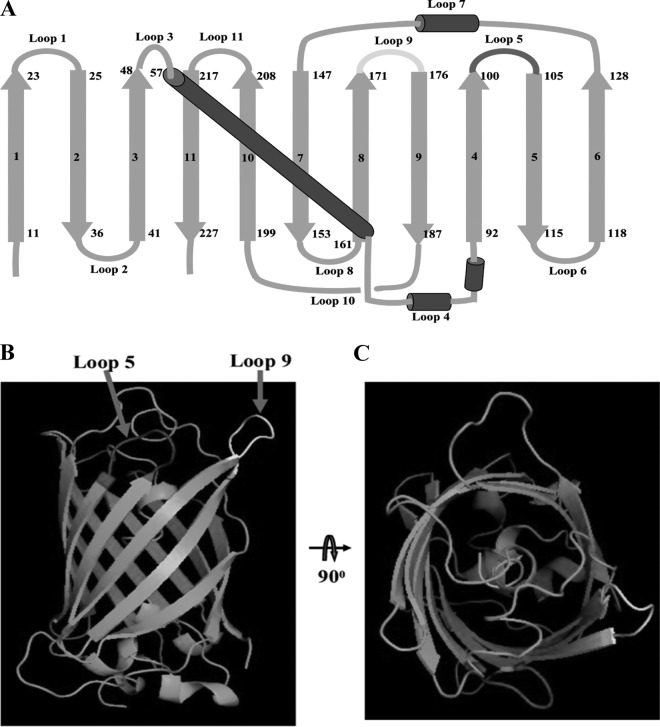
In the present study, a more stable GFP variant, superfolder GFP, was used as a scaffold to insert the CDR at the designed sites to generate a fluorobody that retains binding activity for the target antigen. As shown in Fig. 1A, there are 11 beta strands numbered in sequence and a core helix, forming 11 different loops at the end of a GFP barrel-like scaffold. Among of 11 loops, loop 9 (positions 171 to 176) is the most favorable insertion site (30), and this site is marked in yellow (Fig. 1B and C). In addition, another loop 5 (positions 101 to 105) marked in red was used as the second site to insert CDR3 for creating the dual insertions in combination with loop 9 (Fig. 1B and C).
FIG 1.
Description of superfolder GFP. (A) Topological structure of superfolder GFP. The beta-strands were numbered in sequences 1 to 11, and the loops were numbered in sequences loop 1 to loop 11 (loop 5, dark gray; loop 9, light gray). (B) Side view of superfolder GFP with loop insertion site. The different loops were colored (positions 100 to 105, dark gray for loop 5; positions 171 to 176, light gray for loop 9). (C) Top view of superfolder GFP with a loop insertion site.
Binding activity assay of different insertions.
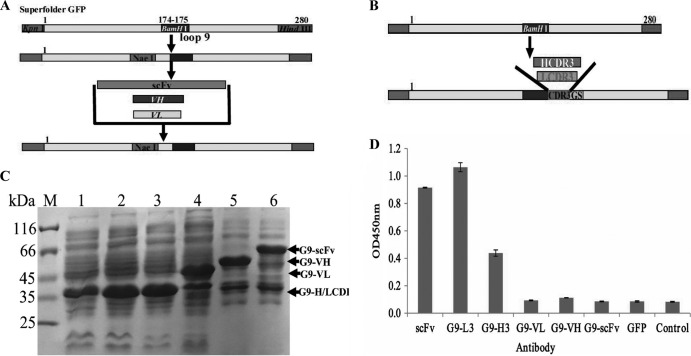
To determine which insertion has higher binding activity for the TLH antigen, full-length scFv, VH, and VL (Fig. 2A) and HCDR3 and LCDR3 (Fig. 2B) fragments were amplified and inserted into the specific site of loop 9, and the binding activity of these expressed proteins were analyzed by ELISA. At the same time, anti-TLH scFv antibody and GFP with a 6×His tag were used as controls. Protein expression and purification results showed that the proteins were highly expressed (Fig. 2C). Compared to the traditional anti-TLH scFv antibody, the G9-LCDR3 has a binding activity similar to that of TLH, with the binding activity being higher than all of the others (Fig. 2D). Although the G9-HCDR3 derived from HCDR3 insertion reacted with TLH, the binding activity is lower than that of G9-LCDR3 and scFv (Fig. 2D). In fact, the proteins expressed from VL, VH, and scFv insertions showed very low binding activity to TLH. These results demonstrated that loop 9 is feasible for the insertion of CDR3 to produce the functional GFP-Abs, whereas the other insertions are not ideal.
FIG 2.
Protein expression and ELISA detection of different antibody formats. (A) Scheme of scFv, VL, and VH insertions in loop 9. For effective insertion, the NaeI restriction enzyme site was introduced into loop 9, and NaeI and BamHI were used to insert VL, VH, and scFv. (B) Scheme of HCDR3 and LCDR3 insertion in loop 9. (C) SDS-PAGE assay of the expressed total proteins. Lane M, midrange protein molecular mass markers; lane1, GFP; lane 2, G9-L3; lane 3, G9-H3; lane 4, G9-VL; lane 5, G9-VH; lane 6, G9-scFv. (D) ELISA. The binding activities were determined with HRP-conjugated goat anti-mouse IgG antibody.
Fluorescence and specificity assay.
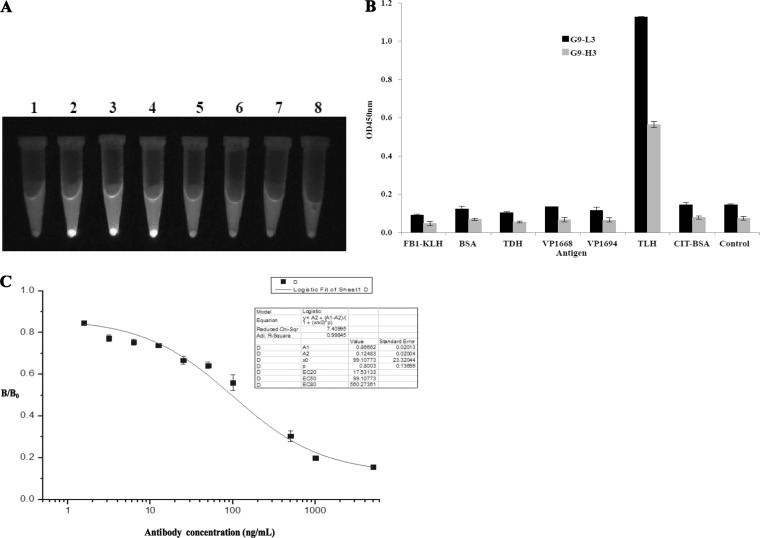
To further investigate the effect of different insertions on the folding of the GFP scaffold, the fluorescence of different GFP-Ab variants was visualized under UV exposure. In Fig. 3A, expressed G9-L3 and G9-H3 (tube 3 and 4) demonstrated much higher fluorescence and kept the same level of fluorescence as GFP (no insertion, tube 2). However, the fluorescence intensity of other proteins was significantly diminished or undetectable. The results further indicated that loop 9 is feasible for the insertion of CDR3 to produce the functional GFP-Abs. To further determine the specificity of G9-L3 to TLH antigen, ELISA and ic-ELISA were carried out. As shown in Fig. 3B, expressed G9-L3 and G9-H3 were specific to TLH, whereas there was no cross binding to any other proteins, such as BSA, KLH, TDH, Vp1668, and VP1694. Moreover, the result obtained from ic-ELISA also showed that the uncoated TLH could effectively compete with the G9-L3 antibody (Fig. 3C). These results indicated that expressed protein G9-L3 could recognize TLH specifically.
FIG 3.
Identification of GFP-based antibody. (A) Fluorescence tracking for antibodies. The different constructs were expressed at 30°C by IPTG induction, and the fluorescence levels for culture pellets after centrifugation are shown after UV exposure. Lanes: 1, scFv; 2, GFP; 3, G9-L3; 4, G9-H3; 5, G9-VL; 6, G9-VH; 7, G9-scFv; 8, BL21. (B) Specificity assay of G9-L3/H3. TLH and other correlated antigens were coated in a 96-well plate, and G9-H3 and G9-L3 antibodies were added to the wells. The specificity of the antibodies was determined using HRP-conjugated goat anti-mouse antibody. (C) ic-ELISA. The signals obtained in the presence of various inhibitor concentrations and without inhibitor (maximal signal) are referred to as “B” and “B0” (x axis), respectively. Standard curves were generated by plotting the inhibition percentage (B/B0) versus the log of free inhibitor concentrations (y axis).
G9-L3a is the major binding region to TLH.
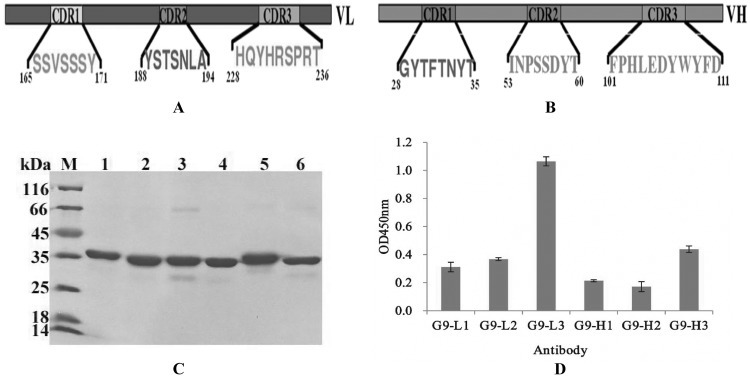
An intact antibody consists of six different CDRs: HCDR1, HCDR2, and HCDR3 for the heavy chain and LCDR1, LCDR2, and LCDR3 for the light chain. To find the major regions responsible for antigen recognition, six different CDRs were inserted into the designed sites of loop 9, respectively (Fig. 4A and B), and the binding activities of these purified antibodies were analyzed by ELISA. As shown in Fig. 4, the six different insertion proteins were expressed and purified successfully, and the purified proteins demonstrated a high purity (Fig. 4C). ELISA result showed that of these six different proteins, G9-L3a showed the greatest binding activity to TLH of the three LCDR insertions, while the G9-H3a showed the greatest binding activity of the three HCDR insertions (Fig. 4D), and the G9-L3a is the best of these. The results derived from the ELISA demonstrated that LCDR3 played a key role in this antigen recognition.
FIG 4.
Different CDR insertions in loop 9. (A and B) Construction of different CDR insertions. (A) Selected light-chain CDRs and three CDR sequences are indicated. (B) Selected heavy-chain CDRs and three CDR sequences are indicated. (C) SDS-PAGE assay of the purified target proteins. Lane M, midrange protein molecular mass markers; lane 1, G9-L1; lane 2, G9-L2; lane 3, G9-L3; lane 4, G9-H1; lane 5, G9-H2; lane 6, G9-L3. (D) ELISA. All ELISA steps were performed as described in Materials and Methods.
CDR3 loop is the best format for insertion.
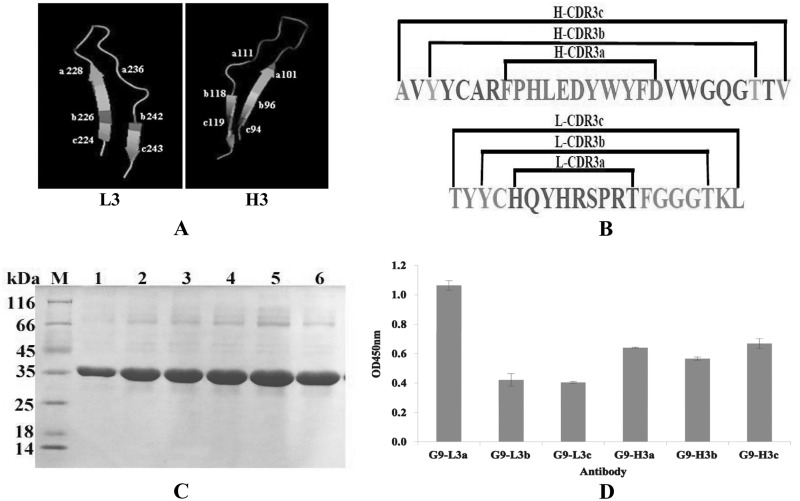
We were interested in determining whether the different lengths of CDR3 could affect the binding activities of antibody to recognize TLH antigen. To achieve this purpose, three different formats of HCDR3 or LCDR3 (L3a, L3b, L3c, H3a, H3b, and H3c) fragments (Fig. 5A and B) were inserted into the same site of loop 9 as described above, and the binding activities of purified antibodies were analyzed by ELISA. As shown in Fig. 5C and D, the expressed proteins were purified successfully, and the purified G9-L3a showed the greatest binding activity of all three LCDR3 insertions. Conversely, there were no obvious differences in the binding activities of these three HCDR3 insertions (Fig. 5D). In view of this result, L3a and H3a were regarded as the best formats for CDR3 insertion, indicating that the shorter insertion of CDR3 might be the best format.
FIG 5.
CDR3 insertion with different formats in loop 9. (A) Three-dimensional model of CDR3 insertions. (Left) LCDR3 insertion. a, b, and c indicate the different insertion formats. (Right) HCDR3 insertions. a, b, and c indicate the different insertion formats. (B) Amino acid sequence of CDR3 insertions. (C) SDS-PAGE assay of purified target proteins. Lane M, midrange protein molecular mass markers; lane 1, G9-L3a; lane 2, G9-Lb; lane 3, G9-L3c; lane 4, G9-H3a; lane 5, G9-H3b; lane 6, G9-H3c. (D) ELISA. All ELISA steps were performed as described in Materials and Methods.
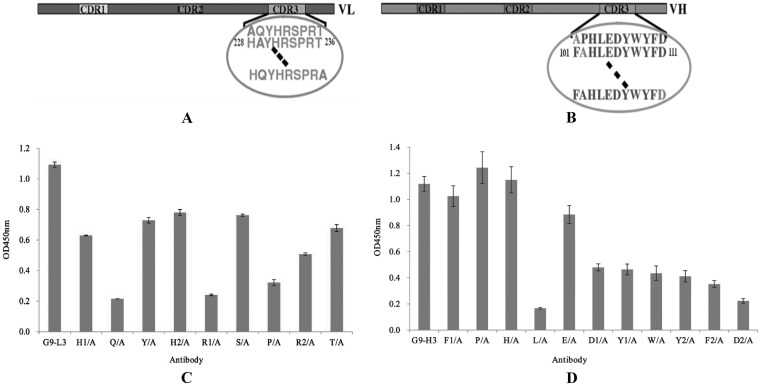
Alanine scanning of G9-L3/H3.
To understand the interaction between G9-L3/H3 and TLH antigen, all of the CDR3 amino acids were mutated to alanine by site-directed mutagenesis to scan the key amino acids responsible for binding (Fig. 6A and B). As shown in Fig. 6C, compared to the wild-type G9-L3, the binding activity of the four mutated proteins (Q/A, H1/A, P/A, and R2/A) decreased greatly, showing very low binding activity to TLH in all of the L3 mutants. In Fig. 6D, wild-type G9-H3 showed the highest binding activity, whereas the binding activity of the two mutated proteins was significantly reduced (L/A and D2/A), only showing a little binding activity to the TLH antigen (Fig. 6D). This result demonstrated that amino acids Q, H1, P, and R2 of LCDR3 were critical for the interaction between G9-L3 and TLH antigen and that amino acids L and D2 of HCDR3 were critical for the interaction between G9-H3 and TLH antigen.
FIG 6.
Alanine scanning test. (A) Schematic diagram of LCDR3. There are 9 amino acids in LCDR3, and all of the amino acids were mutated to alanine. The circled sequences indicate amino acids of LCDR3, and the dashed lines indicate the clipped section. (B) Schematic diagram of HCDR3. There are 11 amino acids in LCDR3, and all of the amino acids were mutated to alanine. (C) ELISA of alanine scanning in LCDR3. (D) ELISA of alanine scanning in HCDR3. All ELISA steps were performed as described in Materials and Methods.
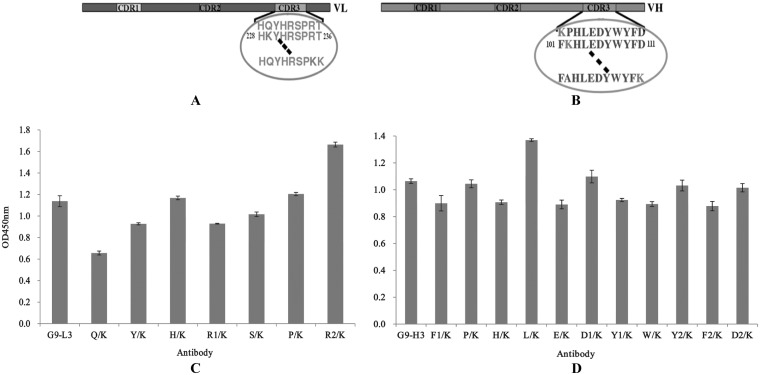
Lysine scanning of G9-L3/H3.
To further enhance the binding activity of G9-L3/H3 to TLH antigen, all of the amino acids of the CDR3 insertion regions were mutated to lysine by site-directed mutagenesis (Fig. 7A and B), and the binding activities of all G9-L3/H3 mutated variants were determined by ELISA. As shown in Fig. 7C, compared to the wild-type G9-L3 insertion, the binding activity of the mutated G9-L3-R2/K increased greatly, showing the greatest binding activity to TLH of all the L3 mutants. For the H3 mutants, mutated G9-H3-L/K showed the greatest binding activity, while the other mutated proteins did not change significantly (Fig. 7D). These results demonstrated that the G9-L3-R2/K and G9-H3-L/K mutants were critical for improving the binding activities of G9-L3 and G9-H3, respectively.
FIG 7.
Lysine scanning test. (A) Schematic diagram of LCDR3. Seven amino acids were mutated to lysine. The circled sequence indicates the amino acids of LCDR3, and the dashed lines indicate the clipped section. (B) Schematic diagram of HCDR3. Eleven amino acids were mutated to lysine. (C) ELISA of lysine scanning in LCDR3. (D) ELISA of lysine scanning in HCDR3. All ELISA steps were performed as described in Materials and Methods.
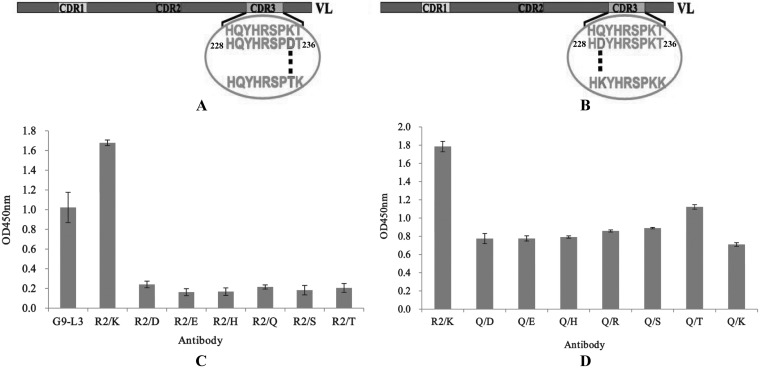
Mutation of key LCDR3 amino acids.
After scanning alanine and lysine, we found that the binding activity of G9-L3-R2/A decreased greatly, whereas the binding activity of G9-L3-R2/K increased significantly, indicating that amino acid R2 might be an important site for antigen-antibody interaction. To further determine the effect of other amino acid mutants (at the R2 site) on the binding activity of G9-L3, the key amino acid R2 (LCDR3) was mutated to six hydrophilic amino acids (Fig. 8A). As shown in Fig. 8C, of seven G9-L3-R2 variants, mutated G9-L3-R2/K has the greatest binding activity, and all of the other mutants only showed very low binding activity to TLH. The result indicated that the R2/K mutant is the best for R2 site-directed mutagenesis. On the basis of the R2/K construct, the key amino acid Q (LCDR3) was also mutated to seven other different hydrophilic amino acids (Fig. 8B), and the binding activities of these mutants were evaluated by ELISA. Compared to the G9-L3-R2/K, the binding activity of all mutants decreased greatly (Fig. 8D), and this phenomenon implied that amino acid Q is very important and immutable. Therefore, the amino acids R2 and Q were considered critical for LCDR3 binding to the TLH antigen.
FIG 8.
Mutation of key amino acids. (A) Schematic diagram of R2 mutants in LCDR3. The key amino acid R235 was mutated to the other six hydrophilic amino acids (D, E, H, Q, S, and T), respectively. (B) Schematic diagram of Q229 mutants in LCDR3. The key amino acid Q229 was mutated to the other six hydrophilic amino acids (D, E, H, R, K, S, and T), respectively. (C) ELISA of R235 mutants. The x axis indicates the concentration of different mutated GFP-Abs (1 μg/ml). (D) ELISA of Q229 mutants. All ELISA steps were performed as described in Materials and Methods.
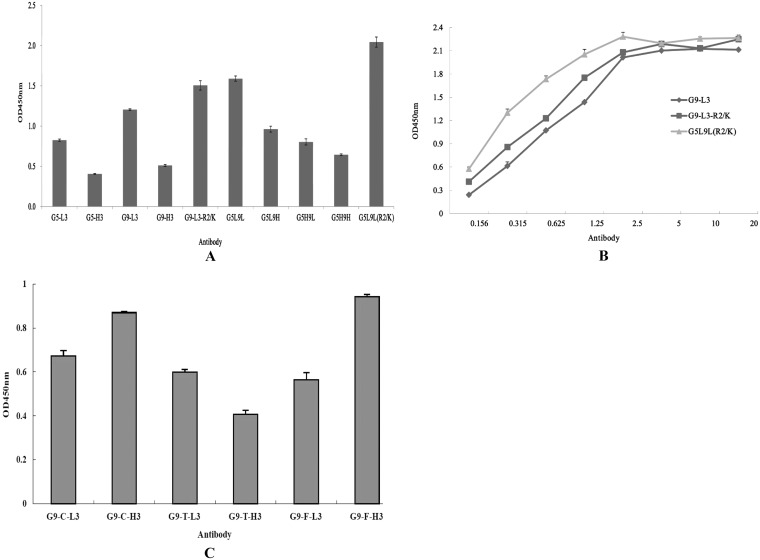
Dual CDR3 insertion at loops 5 and 9 and affinity determination.
To achieve dual-loop insertion, the loops of LCDR3 and HCDR3 from anti-TLH scFv and the loop of CDR3 containing R2/K mutant were inserted into the positions of loop 5 and loop 9 in different formats, respectively, and the binding activities of expressed proteins were analyzed by ELISA. Although the dual-insertion mutants G5L9L and G5L9L(R2/K) retain higher binding activities than the single-loop insertion, the dual-insertion mutant G5L9L(R2/K) showed the greatest binding activity to TLH. However, the other dual insertions showed low binding activity to TLH (Fig. 9A), and the fluorescence intensity of these mutants were diminished substantially (data not shown). To further probe the interaction of CDR3 insertions and TLH antigen, the affinities of G9-L3, G9-L3-R2/K, and G5L9L(R2/K) were studied by ELISA (Fig. 9B), and the measured data were used for the quantitative determination of the affinity constant. The calculated affinity constant of G9-L3, G9-L3-R2/K, and G5L9L(R2/K) were 1.1 × 108 liters/mol, 3.7 × 108 liters/mol, and 1.06 × 109 liters/mol, respectively.
FIG 9.
Dual insertion, affinity determination, and versatility assay. (A) ELISA of dual insertions. (B) The affinity constants of the dual insertions were determined by ELISA using the protocol described in the text. (C) Versatility assay. BSA-FB1, BSA-TTX, and KLH-CIT antigens were used to coat a 96-well plate, respectively, and the corresponding antibodies were added to the wells. The versatility of the GFP-based antibody was analyzed by ELISA.
Versatility assay of CDR3 insertions.
We were interested in determining whether this system is also suitable for CDR3 insertions of other scFv and IgG antibody molecules and whether the expressed GFP-Abs also retain the binding activity to their target antigen. To test this hypothesis, L3/H3 regions from three different scFvs that are specific to three toxins (FB1, TTX, and CIT), respectively, were inserted into loop 9 by the same method to create antibodies G9-F-L3, G9-F-H3, G9-T-L3, G9-T-H3, G9-C-L3, and G9-C-H3, respectively. These proteins were expressed and purified successfully (data not shown), and the purified GFP-Abs also retain the binding activity to their antigen (Fig. 9C). These results demonstrate that this system is feasible for the generation of a functional antibody-based GFP scaffold.
DISCUSSION
Over the past several decades, a variety of methods for producing a stable, soluble, and higher-affinity scFv antibody have been developed, including coexpression with molecular chaperons (3, 31), expression in different host systems (32), genetic fusion to specific proteins (33), and GFP-based binders (29, 30). Perhaps the most effectively used method is the use of a new type of antibody based on a GFP scaffold. To effectively identify the feasibility of a GFP-based binder, five different regions of antibody molecules were first inserted into the loop 9 of GFP, and the binding activities of the resultant proteins were evaluated by ELISA and ic-ELISA. Since the GFP contained a BamHI digestion site at positions 175 and 176 that encoded a “GS” peptide as a linker for CDR3 insertions, a GGATCA DNA sequence was introduced 3′ of the CDR3 for encoding another “GS” to ensure the correct folding of inserted CDR3. Compared to the VL, VH, and scFv insertions, the H/LCDR3 insertion showed higher GFP fluorescence, and the LCDR3 demonstrated greater binding activity to TLH, whereas the GFP fluorescence of the VL, VH, and scFv insertions diminished and the proteins showed low affinity. Although loop 9 is the most tolerated site for peptide insertion, this does not mean that it is compatible for all of the different sizes of protein insertions. Protein insertions with larger molecular structures (such as VH, VL, and scFv [>30 Å]) seriously affect the folding of GFP, leading to the disappearance of GFP fluorescence. Thus, this directly affects the correct folding of the inserted fragment of antibody, making the resultant antibody lose binding activity to the target antigen. This result indicates that the insertion of CDR3s does not affect the folding of GPF scaffold, and it was possible to directly create an antibody with high affinity based on GFP scaffold using only one CDR3 insertion.
CDR1 and CDR2 (but not CDR3) were inserted into the same sites of loop 9. As expected, ELISA showed that H/LCDR3 was the major region for antigen-antibody recognition. Although CDR1 and CDR2 regions exist in all antibody molecules, their functions in the interactions of antibody and antigen may be less important. In fact, antibody CDR3s have been widely used to construct binders specific to the target antigen or as active vaccines using different protein scaffolds (25–30, 34). Moreover, some published studies have shown that “nanoantibody” formed by only one variable domain proved to be a useful tool for research, diagnostics, and therapy (35, 36), and this thesis added further support that the scheme of design in the present study was feasible. We are interested in determining whether the GFP-Abs containing CDR3 insertions have specificity as good as the scFv antibody, and so the specificities of GFP-Abs were evaluated by ELISA and ic-ELISA. It is very exciting that purified GFP-Abs show specificities similar to that of the scFv antibody. Hence, these results show that only CDR3 insertions do not affect the specificity of GFP-Abs.
To develop an effective CDR3 insertion, the effects of the topological structure of CDR3 loops and the amino acid adjutant to CDR3 on the binding efficiency of the formed antibody were further investigated. It was very interesting that the LCDR3A insertion showed the greatest binding efficiency, whereas the HCDR3A insertion retained an affinity similar to that of HCDR3C and higher than that of HCDR3B. This phenomenon suggested that only the CDR3 loop insertion is sufficient for the generation of high-affinity antibody. For further verification, similar experiments were performed by insertion of the CDR3 loops of other scFvs against different toxins. Entire target proteins were expressed and purified successfully, and the resulting GFP-based Abs displayed high affinity to their respective antigens. Hence, the method developed here for the creation of GFP-based Abs shows general applicability.
After identification, the key amino acids of L/HCDR3 were determined by alanine scanning, and four LCDR3 mutants (Q/A, R1/A, P/A, and R2/A) and two HCDR3 mutants (L/A and D2/A) were found to significantly reduce the binding activity of antibody to TLH. To further enhance this binding activity, selected amino acids were mutated to lysine, and ELISA was used to evaluate the activities of these mutants. The result showed that only mutant R2/K of LCDR3 and mutant L/K had greater binding activities than their parents, but the binding activity of Q/K mutant is still declining. After the mutation described above, amino acids Q and R2 of LCDR3 were mutated to seven other hydrophilic amino acids to select the best mutants. However, the results obtained from ELISA were not very exciting, and the binding activities of all the mutants had decreased greatly, indicating that Q and R2/K is the optimal combination for LCDR3.
In the present study, the main object was to create a higher-affinity antibody by using CDR3 dual-insertion loops, so five CDR3 dual insertions [G5L9L, G5L9H, G5H9L, G5H9H and G5L9L(R2/K)] were constructed, and the affinity of the dual-insertion G5L9L(R2/K) was increased significantly, with an affinity of G5L9L(R2/K) of 1.06 × 109 liters/mol. These results indicate that the production of CDR3 single insertion or dual-insertion GFP-Abs was feasible, and the expressed proteins have better fluorescence intensity and higher binding activity. Pavoor et al. developed a GFP-based biosensor possessing the binding properties of antibody (30), but the resulting biosensor showed no fluorescence and low affinity and needed to improve its affinity by several rounds of directed evolution using a surrogate loop approach and yeast surface display.
In conclusion, our results demonstrate that it is feasible to produce a high-affinity antibody by inserting CDR3s into GFP loops. Further, this affinity can be enhanced by specific amino acid scanning and site-directed mutagenesis. Notably, this method had better versatility for creating antibodies to various antigens using GFP as the scaffold, suggesting that a GFP-based antibody may be useful for disease diagnosis and therapy.
ACKNOWLEDGMENTS
This study was supported by the National 973 Program of the Ministry of Science and Technology of China (grant 2013CB127802), the Program of Natural Science from Fujian province (grant 2011J05049), the Key Scientific and Technology Project of Fujian Province of China (grant 2012Y002), and the Fund from Fujian Development and Reform Commission.
We are grateful to Mengfei Ho (University of Illinois at Urbana-Champaign) for the donation of the pGEPi-GFP vector and technical discussions.
R.W, S.X., and S.W. designed the experiments. R.W., S.X., and Y.Z. performed the experiments. Q.C. and Y.Z. contributed when this project was initiated. R.W., S.X., and S.W. analyzed the data and prepared the manuscript. All authors read and approved the final manuscript.
Footnotes
Published ahead of print 2 May 2014
REFERENCES
- 1.Koerber JT, Thomsen ND, Hannigan BT, Degrado WF, Wells JA. 2013. Nature-inspired design of motif-specific antibody scaffolds. Nat. Biotechnol. 31:916–922. 10.1038/nbt.2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang RZ, Fang S, Wu DL, Lian JW, Fan J, Zhang YF, Wang SH, Lin WX. 2012. Screening of a single chain variable fragment antibody that can neutralize effectively the cytotoxicity of Vibrio parahaemolyticus TLH. Appl. Environ. Microbiol. 78:4967–4975. 10.1128/AEM.00435-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang RZ, Xiang SS, Feng YJ, Srinivas S, Zhang YH, Lin MS, Wang SH. 2013. Engineering production of functional scFv antibody in Escherichia coli by coexpressing the molecule chaperone Skp. Front. Cell Infect. Microbiol. 10.3389/fcimb.2013.00072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pei SC, Zhang YY, Eremin SA, Lee WJ. 2009. Detection of aflatoxin M1 in milk products from China by ELISA using monoclonal antibodies. Food Control 20:1080–1085. 10.1016/j.foodcont.2009.02.004 [DOI] [Google Scholar]
- 5.Wang SH, Zhang JB, Zhang ZP, Zhou YF, Yang RF, Chen J. 2006. Construction of single chain variable fragment (scFv) and biscFv-alkaline phosphatase fusion protein for detection of Bacillus anthracis. Ana. Chem. 78:997–1004. 10.1021/ac0512352 [DOI] [PubMed] [Google Scholar]
- 6.Hu XJ, Hortigüela MJ, Robin S, Lin H, Li Y, Moran AP, Wang W, Wall JG. 2013. Covalent and oriented immobilization of scFv antibody fragments via an engineered glycan moiety. Biomacromolecules 14:153–159. 10.1021/bm301518p [DOI] [PubMed] [Google Scholar]
- 7.Hansel TT, Kropshofer H, Singer T, Mitchell JA, George AJ. 2010. The safety and side effects of monoclonal antibodies. Nat. Rev. Drug Discov. 9:325–338. 10.1038/nrd3003 [DOI] [PubMed] [Google Scholar]
- 8.Sakai K, Yuasa N, Tsukamoto K. 2010. Isolation and characterization of antibodies against three consecutive Tn-antigen clusters from a phage library displaying human single chain variable fragments. J. Biochem. 147:809–817. 10.1093/jb/mvq014 [DOI] [PubMed] [Google Scholar]
- 9.Sommaruga S, Lombardi A, Salvade A, Mazzucchelli S, Corsi F, Galeffi P. 2011. Highly efficient production of anti-HER2 scFv antibody variant 881 for targeting breast cancer cells. Appl. Microbiol. Biotechnol. 91:613–621. 10.1007/s00253-011-3306-3 [DOI] [PubMed] [Google Scholar]
- 10.Sørensen HP, Mortensen KK. 2011. Advanced genetic strategies for recombinant protein expression in Escherichia coli. J. Biotechnol. 115:113–128. 10.1016/j.jbiotec.2004.08.004 [DOI] [PubMed] [Google Scholar]
- 11.Cesaro-Tadic S. 2003. Turnover-based in vitro selection and evolution of biocatalysts from a fully synthetic antibody library. Nat. Biotechnol. 21:679–685. 10.1038/nbt828 [DOI] [PubMed] [Google Scholar]
- 12.Coia G, Hudson PJ, Irving RA. 2001. Protein affinity maturation in vivo using Escherichia coli mutator cells. J. Immunol. Methods 251:187–193. 10.1016/S0022-1759(01)00300-3 [DOI] [PubMed] [Google Scholar]
- 13.Tsien RY. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509–544. 10.1146/annurev.biochem.67.1.509 [DOI] [PubMed] [Google Scholar]
- 14.Paige JS, Wu KY, Jaffrey SR. 2011. RNA mimics of green fluorescent protein. Science 333:642–646. 10.1126/science.1207339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Campbell RE, Ting AY, Tsien RY. 2002. Creating new fluorescent probes for cell biology. Nat. Rev. Mol. Cell. Biol. 3:906–918. 10.1038/nrm976 [DOI] [PubMed] [Google Scholar]
- 16.McNaughton BR, Cronican J, David B, Thompson DB, Liu DR. 2009. Mammalian cell penetration, siRNA transfection, and DNA transfection by supercharged proteins. Proc. Natl. Acad. Sci. U. S. A. 106:6111–6116. 10.1073/pnas.0807883106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawrence MS, Phillips KJ, Liu DR. 2007. Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc. 129:10110–10112. 10.1021/ja071641y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim CK, Chung JD, Park SH, Burrell AM, Kamo KK, Byrne DH. 2004. Agrobacterium tumefaciens-mediated transformation of Rosa hybrida using the green fluorescent protein (GFP) gene. Plant Cell 78:107–111. 10.1023/B:TICU.0000022529.16697.90 [DOI] [Google Scholar]
- 19.Cronican JJ, Thompson DB, Beier KT, McNaughton BR, Cepko CL, Liu DR. 2010. Potent delivery of functional proteins into mammalian cells in vitro and in vivo using a supercharged protein. ACS Chem. Biol. 5:747–752. 10.1021/cb1001153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paschke M, Tiede C, Hohne W. 2007. Engineering a circularly permuted GFP scaffold for peptide presentation. J. Mol. Recognit. 20:367–378. 10.1002/jmr.844 [DOI] [PubMed] [Google Scholar]
- 21.Hu CD, Kerppola TK. 2003. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21:539–545. 10.1038/nbt816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsienet RY. 1997. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388:882–887. 10.1038/42264 [DOI] [PubMed] [Google Scholar]
- 23.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. 1999. Rapid protein folding assay using green fluorescent protein. Nat. Biotechnol. 17:691–695. 10.1038/10904 [DOI] [PubMed] [Google Scholar]
- 24.Wiehler J, Jung G, Seebacher C, Zumbusch A, Steipe B. 2003. Mutagenic stabilization of the photocycle intermediate of green fluorescent protein (GFP). ChemBioChem 4:1164–1171. 10.1002/cbic.200300595 [DOI] [PubMed] [Google Scholar]
- 25.Abedi MR, Caponigro G, Kamb A. 1998. Green fluorescent protein as a scaffold for intracellular presentation of peptides. Nucleic Acids Res. 26:623–630. 10.1093/nar/26.2.623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doi N, Yanagawa H. 1999. Design of generic biosensors based on green fluorescent proteins with allosteric sites by directed evolution. FEBS Lett. 453:305-307. 10.1016/S0014-5793(99)00732-2 [DOI] [PubMed] [Google Scholar]
- 27.Baird GS, Zacharias DA, Tsien RY. 1999. Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl. Acad. Sci. U. S. A. 96:11241–11246. 10.1073/pnas.96.20.11241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeytun A, Jeromin A, Scalettar BA, Waldo GS, Bradbury ARM. 2003. Fluorobodies combine GFP fluorescence with the binding characteristics of antibodies. Nat. Biotechnol. 21:1473–1481. 10.1038/nbt911 [DOI] [PubMed] [Google Scholar]
- 29.Dai M, Temirov J, Pesavento E, Kiss C, Velappan N, Pavlik P, Werner JH, Bradbury ARM. 2008. Using T7 phage display to select GFP-based binders. Protein Eng. Des. Sel. 21:413–424. 10.1093/protein/gzn016 [DOI] [PubMed] [Google Scholar]
- 30.Pavoor TV, Cho YK, Shusta EV. 2009. Development of GFP-based biosensors possessing the binding properties of antibodies. Proc. Natl. Acad. Sci. U. S. A. 106:11895–11900. 10.1073/pnas.0902828106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonoda H, Kumada Y, Katsuda T, Yamaji H. 2011. Effects of cytoplasmic and periplasmic chaperones on secretory production of single chain Fv antibody in Escherichia coli. J. Biosci. Bioeng. 111:465–470. 10.1016/j.jbiosc.2010.12.015 [DOI] [PubMed] [Google Scholar]
- 32.Guild K, Zhang Y, Stacy R, Mundt E, Benbow S, Green A. 2011. Wheat germ cell-free expression system as a pathway to improve protein yield and solubility for the SSGCID pipeline. Acta Crystallogr. F 67:1027–1031. 10.1107/S1744309111032143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jurado P, de Lorenzo V, Fernández LA. 2006. Thioredoxin fusions increase folding of single chain Fv antibodies in the cytoplasm of Escherichia coli: evidence that chaperone activity is the prime effect of thioredoxin. J. Mol. Biol. 357:49–61. 10.1016/j.jmb.2005.12.058 [DOI] [PubMed] [Google Scholar]
- 34.Chen SS, Barankiewicz T, Yang YM, Zanetti M, Hill P. 2008. Protection of IgE-mediated allergic sensitization by active immunization with IgE loops constrained in GFP protein scaffold. J. Immunol. Methods 333:10–23. 10.1016/j.jim.2007.10.007 [DOI] [PubMed] [Google Scholar]
- 35.Tillib SV. 2011. “Camel nanoantibody” is an efficient tool for research, diagnostics, and therapy. Mol. Biol. 45:66–73. 10.1134/S0026893311010134 [DOI] [PubMed] [Google Scholar]
- 36.Tillib SV, Ivanova TI, Lyssuk EY, Larin SS, Kibardin AV, Korobko EV, Vikhreva PN, Gnuchev NV, Georgiev GP, Korobko IV. 2012. Nanoantibodies for detection and blocking of bioactivity of human vascular endothelial growth factor A. Biochemistry 77:659–665. 10.1134/S0006297912060132 [DOI] [PubMed] [Google Scholar]