Abstract
Purpose of Review
Detailed genetic and structural characterization has revealed that broadly neutralizing antibodies (bnAbs) against HIV-1 have unusually high levels of somatic hypermutation, long CDRH3 domains, and the ability to target one of four sites of vulnerability on the HIV-1 envelope (Env) glycoproteins. A current priority is to understand how bnAbs are generated during natural infection, and translate this information into immunogens that can elicit bnAb following vaccination.
Recent Findings
Strain-specific neutralizing antibodies (nAb) can acquire broad neutralizing capacity when the transmitted/founder Env or a specific Env variant is recognized by an unmutated rearranged germline that has the capacity to develop bnAb like features. This could be a relatively infrequent event, as only certain germlines appear to possess inherent features needed for bnAb activity. Furthermore, the glycosylation pattern and diversity of circulating HIV-1 Envs, as well as the state of the B cell compartment, may influence the activation and maturation of certain antibody lineages.
Summary
Collectively, studies over the last year suggest that the development of HIV-1 Env immunogens that bind and activate bnAb-like germlines is feasible. However, more information about the features of Env variants and the host factors that lead to breadth during natural infection is needed to elicit bnAbs through immunization.
Keywords: Broadly neutralizing antibodies, affinity maturation, long CDRH3, unmutated common ancestor, HIV-1 envelope evolution
Introduction
A number of excellent reviews in the last year have highlighted the abundant new knowledge gleaned from studies on HIV-infected individuals with broadly neutralizing antibodies (bnAbs) and speculated on how such information might inform vaccine design (1-7). It is generally accepted that the 4 major sites of vulnerability on the HIV-1 envelope (Env) glycoproteins are the CD4 binding site (CD4bs), glycan dependent epitopes in V1V2 and near the base of V3/C3, and linear epitopes in the membrane proximal external region (MPER) of gp41. However, the presence of unmapped epitopes for some bnAbs suggests that additional sites may exist (1). bnAbs show unusual genetic features including high levels of somatic hypermutation and selective germline gene usage for CD4bs antibodies, long CDRH3s for antibodies that penetrate the glycan shield, and polyreactivity for some MPER antibodies. bnAbs represent subdominant responses, and sera with breadth mostly comprise single specificities, although rarer cases of multiple specificities have been reported (8, 9*, 10, 11) While bnAbs are found only in a subset of people after many years of infection, almost all infected individuals develop strain-specific neutralizing antibodies (nAbs) that can neutralize autologous virus but not heterologous viruses. Previous studies have shown that these mostly target variable regions, unlike bnAbs that target more conserved sites. However, there is some overlap in these epitopes; for example, both bnAb and strain-specific nAb target the CD4bs, V1V2 and the base of the V3/C3 region (12-18). The question of how these bnAbs arise and whether they mature from earlier strain-specific nAbs is only now being revealed.
Ontogeny of broadly neutralizing antibodies
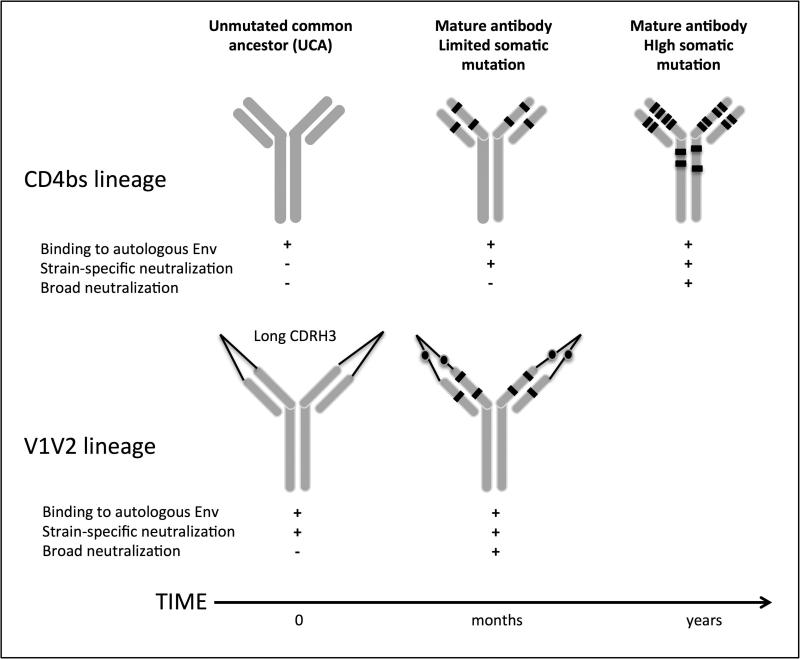
The high degree of somatic mutation in many bnAbs suggests that they undergo multiple rounds of affinity maturation to acquire breadth. It was unknown whether long CDRH3s also develop through the same gradual process or arise through immunoglobulin gene recombination. Results from 2 longitudinal studies have indicated that the pathway to neutralization breadth differs depending on the epitope being targeted (Figure 1). For CD4bs, the initial B cell recognized the infecting virus but neutralization was only achieved after a sufficient level of affinity maturation, resulting in neutralization firstly of the autologous virus and later heterologous viruses (19**). In a second study that followed the development of a V1V2 directed antibody lineage, the unmutated common ancestor (UCA) both bound and neutralized the infecting virus. Breadth required a more modest level of somatic hypermutation than seen for the CD4bs lineage, perhaps explaining how breadth developed early in this donor (20**). This study also revealed that the long CDRH3 characteristic of this class of antibody developed in this case as a result of a recombination event prior to encountering antigen.
Figure 1. The pathway to neutralization breadth.
Maturation of a CD4bs lineage required somatic hypermutation for both strain-specific and broad neutralizing capacity (19). This is consistent with high levels of somatic hypermutation seen in other CD4bs bnAbs e.g. VRC01. In contrast, V1V2 directed bnAbs selected a germline with preexisting long CDRH3s and had the inherent ability to bind and neutralize the founder virus. Additional levels of somatic hypermutation were needed to mediate broad neutralizing activity (20).
The use of deep sequencing has expanded our ability to interrogate the antibody response to HIV-1 even without the benefit of longitudinal sampling. Analysis of the patient from whom the CD4bs bnAb VRC01 was isolated identified clonal relatives that were less somatically mutated (21). Testing of these archived members of the lineage demonstrated the need for extensive affinity maturation for neutralization. Similarly for the V3 glycan PGT121-like antibodies, the degree of somatic mutation was associated with neutralization although antibodies with half the level of somatic hypermutation still showed significant neutralization (22). bnAbs also show high levels of mutation outside the antigen-binding sites with these mutations in the framework regions required for broad neutralizing activity (23).
While the focus has been on subjects with significant breadth, analysis of large cohorts reveals that most infected individuals develop some level of cross-neutralizing activity suggesting breadth occurs as a continuum rather than an extreme phenotype (17, 24-26). The immune system is capable of making potent neutralizing antibodies, but why all strain-specific nAbs do not eventually mature to acquire breadth remains an important and unanswered question. This may be related to unsuitable germline gene usage, auto-reactivity, suboptimal angle of approach due to glycan interference or insufficient long-term antigenic stimulation.
Role of viral evolution in shaping broadly neutralizing antibody responses
Several studies have suggested that viral factors contribute to the development of breadth, beyond simply providing sufficient antigenic stimulation in the form of high viral loads. During the last year, there has been progress in defining the precise viral mechanisms driving improved breadth, through detailed longitudinal studies. Moore et al. described the evolution of a N332 glycan-dependent bnAb epitope in the V3/C3 region, absent on the infecting virus, through immune escape from earlier strain-specific nAb responses that targeted the same region of the envelope (27). This study showed neutralization escape from strain-specific nAbs resulted in viral convergence towards conserved glycan motifs, creating the epitope for later broad nAbs (28). In a subsequent study breadth was similarly associated with a reduced probability of the N332 residue being glycosylated on the infecting virus (29).
A second mechanism was identified in an infected subject who developed sequential broad neutralizing antibodies to both V2 and the CD4bs. Here, viral escape drove the deletion of a highly conserved glycan at N160 in V2 (9*) resulting in exposure of the CD4bs, which immediately became the target for the next wave of bnAbs. Thus viral escape may force the exposure of otherwise occluded conserved epitopes, facilitating the development of breadth. Intriguingly, in three of four studies where multiple bnAbs emerge, the antibodies target V2 and the CD4bs, suggesting a common developmental pathway for consecutive specificities (8, 9*, 10, 30).
Neutralization breadth is known to be associated with increased viral diversity (25, 31, 32). Recent studies have extended these analyses to examine the contribution of dual or superinfection to breadth. Powell et al. showed that dual infection resulted in enhanced breadth, though in this study confounding factors such as duration of infection could not be excluded (33). A more recent study reported both increased breadth and potency in superinfected individuals, with breadth emerging within a year of superinfection, independently of viral load and CD4+ T cell counts (34*). Interestingly, in this study, both intersubtype superinfection and persistence of both infecting viruses was associated with breadth, suggesting that a more divergent circulating viral population was a major contributing factor. Finally, in two studies describing the maturation of bnAbs from binding/strain-specific antibodies (see ontogeny section above) (19**, 20**), extensive viral diversification preceded the development of breadth, further supporting the notion that multiple circulating versions of epitopes may drive breadth.
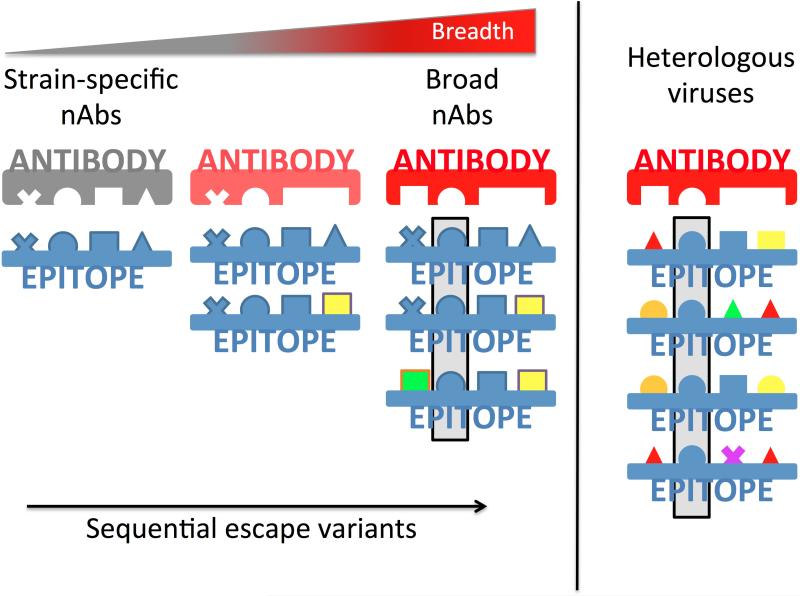
In the first study to propose a mechanism for these observations, Murphy et al. showed that two antibody light chain variants paired with a single heavy chain displayed differential neutralization of autologous viruses containing escape mutations in the base of V3/C3 region. This suggested that viral evolution drives strain-specific nAbs to recognize amino acid variants within a given epitope, implying that escape variants increase breadth (15*). However in that study, bnAbs did not develop against this early epitope. Wibmer et al. subsequently detailed the development of both broadly neutralizing V2 and CD4bs nAbs in response to the sequential accumulation of escape mutations, with later plasma responses able to tolerate new epitope variants (9*). This ability to recognize multiple immunotypes, emerging as a result of neutralization escape was also associated with increased breadth in other studies (20**, 35, 36), providing a model for the maturation of bnAbs that is consistent with the viral diversification observed prior to breadth (Figure 2) (19**, 20**). The increasing appreciation of how such antibodies mature and acquire breadth provides a template for the design of novel sequential immunization strategies (1, 9*).
Figure 2. Model for the maturation of strain-specific plasma responses to acquire breadth.
Strain-specific antibodies (gray) recognize defined epitopes (shown in blue, in a lock-and-key schematic). These antibodies drive viral escape mutations (represented as multi-colored shapes) within epitopes. The emergence of sequential escape mutations creates multiple immunotypes (or epitope variants) within a single infected subject. Maturation of nAbs to tolerate multiple residues at a given position (e.g. either a triangle or a square) or to recognize a smaller core epitope (e.g. the invariant sphere at the second position within the epitope, highlighted in the gray box) thereby better encompasses the global epitope variants in heterologous viruses (shown on the right), and enables the development of broad neutralization antibodies (red).
Host factors
In addition to viral evolution, host factors have been implicated in the development of breadth. A genome-wide association study (GWAS) revealed a decreased prevalence of the protective HLA allele B*57 in individuals with neutralization breadth, while the unfavorable HLA allele B*07 was enriched in this population (37). CSF-1R, the receptor for colony stimulating factor-1 and a regulator of macrophages, also showed a potential association with neutralization breadth. While no B cell-specific genetic markers were identified in this GWAS study, the functional state of the B cell compartment is likely to influence the development of breadth. One study reported that more peripheral naïve B cells, but less tissue-like and activated memory B cells (a phenotype more like healthy individuals) favored neutralization breadth (38). In contrast, Boliar et al. found that neutralization breadth was present despite marked dysregulation of peripheral B cell subsets, and that hypergammaglobulinemia, a measure of B cell dysfunction, correlated directly with neutralization breadth (39). Additionally, in vitro binding of gp120 to peripheral B cells via the α4β7 integrin may contribute to suppression of B cell activation and proliferation in vivo through TGF-β1 production and other mechanisms that could interfere with the development of robust neutralizing activity (40). Current approaches to enhance B cell/antibody responses during immunization include incorporation of adjuvants and stimulatory cytokines such as GM-CSF, APRIL, and IL-21 directly into gp120 (41-43). Adjuvants will undoubtedly also be incorporated into human vaccine trials to enhance antibody responses (44, 45) and see the review on “Modulation of HIV-1 Immunity by Adjuvants” in this same issue. There is also evidence that a higher frequency of a functional memory subset of T follicular helper cells in the periphery in early infection, and maintenance of this population over time, may contribute to the development of bnAb activity (46*). Taken together, these studies illuminate avenues to enhance T and B cell responses that could promote the development of bnAbs.
Activation of germline B cells
A significant obstacle to generating bnAbs during natural infection or by immunization could be that Env is poorly recognized by the germline-like (GL) versions of the B cell receptors (BCRs) for these antibodies. To investigate this, various methods to infer the reverted GL version of mature bnAbs (or UCA) have been employed in the absence of longitudinal data. Despite the ability of bnAbs to neutralize diverse HIV-1 Envs, the corresponding GL antibodies, soluble or expressed on the surface of a B cell, often do not recognize those same Envs produced as recombinant proteins (47, 48). However as pointed out above, in cases where bnAb lineages have been investigated from early infection, reverted GL versions of the mature antibodies do recognize an early autologous Env variant (19**, 20**).
Targeted modification of Env proteins can increase recognition by GL versions of bnAbs. Removal of conserved glycans known to modulate sensitivity to CD4bs bnAbs facilitated activation of B cells expressing the GL BCRs of CD4bs bnAbs (49*). Removal of glycan N276, which is conserved in 95% of HIV-1 strains, eliminated the steric constraints of the GL antibody versions. Computational design has been used to develop an optimized Env immunogen that is recognized by CD4bs bnAbs and their GL precursors (50**). The final construct had high affinity for the mature and GL derived CD4bs bnAbs. In a parallel study, a chimeric CD4bs antibody containing the GL VH and mature VL showed a similar angle of approach and outer domain contacts as the mature antibody, but lacked critical inner domain and bridging sheet contacts (51). Together these studies suggest that it is difficult to identify naturally occurring Envs that will elicit bnAbs, but that immunogens can indeed be rationally designed to potentially elicit CD4bs bnAbs.
Given the potential to develop and test Env immunogens that are recognized by GL-bnAbs, it will be important to understand the pathways to bnAb in nonhuman primates. To this end, Sundling et al. showed that VH gene family structure is similar between humans and rhesus macaques (RM), with high homology (average of 92%) between VH genes (52*). These investigators also recovered CD4bs mAbs from an immunized RM that had high affinity for gp120 and tier 1 neutralizing capacity, modest somatic hypermutation levels (~5% from germline), and relatively longer CDRH3 domains. However, these CD4bs mAbs were derived from RM VH3 and VH4 families, as opposed to the human VH1 germlines that give rise to most CD4bs bnAbs. Env immunogens designed to interact with GL-versions of human bnAbs may therefore not activate the expected germline in RMs (50**). Thus, using nonhuman primates to model the development of bnAbs will require a thorough understanding of the biological similarities and limitations.
Vaccine induced responses in human subjects
The ultimate goal of eliciting bnAbs following immunization is still elusive. The RV144 human vaccine trial elicited cross-reactive but weakly neutralizing antibodies directed against linear and conformational epitopes in V2 that were inversely correlated with the rate of HIV-1 infection (53). Liao et al. found that mAbs from a single vaccinated RV144 subject targeted residue 169 in the V2 loop, had tier 1 neutralizing activity, and mediated killing of HIV-1 infected CD4 T cells in vitro (54). Further studies suggest that the vaccine elicited V2 mAbs recognize a conformation of V1V2 that is distinct from that recognized by V1V2 bnAbs (54, 55). This has led to the development of V1V2 glycopeptide immunogens that bind with high affinity to GL and mature V1V2 bnAbs, but with much lower affinity to the vaccine elicited, strain-specific V2 mAbs (56*). It remains unclear whether these vaccine-induced antibodies are the precursors of bnAbs, or whether they can be coaxed along the pathway to breadth with novel immunization strategies.
Conclusion
The last year has seen significant progress in defining the pathway to breadth and identifying important viral and host factors. However, additional longitudinal studies on infected individuals that develop bnAbs are needed to determine whether there is a limited number of pathways to breadth, and whether these are epitope-specific. In particular, given the frequency of V3/C3 as a target, and the ability of these bnAbs to use multiple germline genes, this class of antibodies should be a focus (11, 24, 57). It may also be important to examine individuals with limited or no breadth, to determine if subdominant bnAb responses (i.e. aborted lineages) can be detected by deep sequencing. Existing studies have highlighted the difficulties in eliciting bnAbs. These roadblocks would need to be overcome in order to stimulate effective and broad neutralizing antibody responses by vaccination. For example, how do we stimulate particular germline alleles? How do we stimulate low frequency B cells with long CDRH3? Will it be possible to achieve the required level of somatic hypermutation by vaccination? Given the rapid pace of new discoveries in this field, we anticipate that the coming year will make inroads into answering many of these questions.
Key Points.
The pathway to neutralization breadth differs depending on the epitope targeted, but all require significant somatic hypermutation.
Long CDRH3 regions, formed through germline immunoglobulin gene recombination, may be particularly well suited for developing bnAbs that penetrate the glycan shield.
Viral diversity plays a key role in the development of neutralization breadth, regardless of the epitope targeted, with specific glycan changes near the base of the V3/C3 region and in V1V2 playing key roles.
Functional memory T follicular helper cells may favor the development of breadth, but more studies are needed to define the role of additional host factors.
Env immunogens can be designed to better engage germline-like B cell receptors for bnAbs derived from humans, but testing them in nonhuman primates may be complicated by germline differences.
Acknowledgements
We thank Jinal Bhiman and Kurt Wibmer for help with preparing the figures. CAD gratefully acknowledges the National Institutes for Health (NIH) for grants R01 AI58706 and U19 AI96187, the Rwanda Zambia HIV Research Group (RZHRG), and the International AIDS Vaccine Initiative (IAVI) for their support. LM and PLM gratefully acknowledge funding from CAPRISA, the National Institute of Allergy and Infectious Diseases (NIAID) Center for HIV/AIDS Vaccine Immunology (CHAVI) grant AI067854, the Center for AIDS Vaccine Discovery (CAVD) of the Bill and Melinda Gates Foundation, the South African HIV/AIDS Research and Innovation Platform of the South African Department of Science and Technology and by a HIVRAD NIH grant AI088610. CAPRISA was supported by NIAID, NIH, U.S. Department of Health and Human Services (Grant U19 AI51794). PLM is a Wellcome Trust Intermediate Fellow in Public Health and Tropical Medicine (Grant 089933/Z/09/Z).
References
- 1.Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341(6151):1199–204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwong PD, Mascola JR, Nabel GJ. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat Rev Immunol. 2013;13(9):693–701. doi: 10.1038/nri3516. [DOI] [PubMed] [Google Scholar]
- 3.Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature's pathways. Immunol Rev. 2013;254(1):225–44. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu Rev Immunol. 2013;31:705–42. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- 5.Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37(3):412–25. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Gils MJ, Sanders RW. Broadly neutralizing antibodies against HIV-1: templates for a vaccine. Virology. 2013;435(1):46–56. doi: 10.1016/j.virol.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, et al. A Blueprint for HIV Vaccine Discovery. Cell Host Microbe. 2012;12(4):396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mikell I, Stamatatos L. Evolution of cross-neutralizing antibody specificities to the CD4-BS and the carbohydrate cloak of the HIV Env in an HIV-1-infected subject. PloS one. 2012;7(11):e49610. doi: 10.1371/journal.pone.0049610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9*.Wibmer CK, Bhiman JN, Gray ES, Tumba N, Abdool Karim SS, Williamson C, et al. Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS pathogens. 2013;9(10):e1003738. doi: 10.1371/journal.ppat.1003738. [This longitudinal study of an infected subject who developed three sequential bnAb specificities enabled the description of multiple mechanisms in which viral evolution shaped breadth. The study highlighted the maturation of type-specific antibodies to acquire breadth by recognition of multiple immunotypes, and the forced exposure of otherwise occluded conserved epitopes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonsignori M, Montefiori DC, Wu X, Chen X, Hwang KK, Tsao CY, et al. Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: implications for vaccine design. Journal of virology. 2012;86(8):4688–92. doi: 10.1128/JVI.07163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomaras GD, Binley JM, Gray ES, Crooks ET, Osawa K, Moore PL, et al. Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. Journal of virology. 2011;85(21):11502–19. doi: 10.1128/JVI.05363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore PL, Ranchobe N, Lambson BE, Gray ES, Cave E, Abrahams MR, et al. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS pathogens. 2009;5(9):e1000598. doi: 10.1371/journal.ppat.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rong R, Li B, Lynch RM, Haaland RE, Murphy MK, Mulenga J, et al. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS pathogens. 2009;5(9):e1000594. doi: 10.1371/journal.ppat.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynch RM, Rong R, Boliar S, Sethi A, Li B, Mulenga J, et al. The B cell response is redundant and highly focused on V1V2 during early subtype C infection in a Zambian seroconverter. Journal of virology. 2011;85(2):905–15. doi: 10.1128/JVI.02006-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15*.Murphy MK, Yue L, Pan R, Boliar S, Sethi A, Tian J, et al. Viral Escape from Neutralizing Antibodies in Early Subtype A HIV-1 Infection Drives an Increase in Autologous Neutralization Breadth. PLoS pathogens. 2013;9(2):e1003173. doi: 10.1371/journal.ppat.1003173. [This was the first study showing that the recognition of multiple amino acid variants, arising as a consequence of neutralization escape, impacted on neutralization.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray ES, Moody MA, Wibmer CK, Chen X, Marshall D, Amos J, et al. Isolation of a monoclonal antibody that targets the alpha-2 helix of gp120 and represents the initial autologous neutralizing-antibody response in an HIV-1 subtype C-infected individual. Journal of virology. 2011;85(15):7719–29. doi: 10.1128/JVI.00563-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mikell I, Sather DN, Kalams SA, Altfeld M, Alter G, Stamatatos L. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS pathogens. 2011;7(1):e1001251. doi: 10.1371/journal.ppat.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch RM, Tran L, Louder MK, Schmidt SD, Cohen M, Dersimonian R, et al. The development of CD4 binding site antibodies during HIV-1 infection. Journal of virology. 2012;86(14):7588–95. doi: 10.1128/JVI.00734-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19**.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013 doi: 10.1038/nature12053. [This was the first study to trace the development of a CD4bs bNab lineage. Importantly, they showed that the UCA bound, but did not neutralizing the infecting virus. Moderate breadth was achieved with less somatic hypermutation than had been thought necessary for this class of antibody. Extensive viral diversification preceded breadth.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, Staupe RP, et al. Developmental Pathway for Potent V1V2-directed HIV-1 Neutralizing Antibodies. Nature. 2013 doi: 10.1038/nature13036. In press. [This was the first study to describe the development of a V1V2 directed bnAb lineage. In contrast to CD4bs bnAbs described in reference 18, this lineage was initiated by a UCA with a 35 amino acid CDRH3 fully formed by germline immunoglobulin gene rearrangement. The UCA both bound and neutralized the superinfecting virus, and acquired breadth rapidly through limited somatic hypermutation. Viral diversification within the V2 region preceded breadth, and contributed to maturation of the lineage.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333(6049):1593–602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sok D, Laserson U, Laserson J, Liu Y, Vigneault F, Julien JP, et al. The Effects of Somatic Hypermutation on Neutralization and Binding in the PGT121 Family of Broadly Neutralizing HIV Antibodies. PLoS pathogens. 2013;9(11):e1003754. doi: 10.1371/journal.ppat.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153(1):126–38. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, et al. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. Journal of virology. 2011;85(10):4828–40. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Euler Z, van den Kerkhof TL, van Gils MJ, Burger JA, Edo-Matas D, Phung P, et al. Longitudinal analysis of early HIV-1-specific neutralizing activity in an elite neutralizer and in five patients who developed cross-reactive neutralizing activity. Journal of virology. 2012;86(4):2045–55. doi: 10.1128/JVI.06091-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. Aids. 2014;28(2):163–9. doi: 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore PL, Gray ES, Wibmer CK, Bhiman JN, Nonyane M, Sheward DJ, et al. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med. 2012;18(11):1688–92. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langedijk JP, Schuitemaker H. A sweet surprise for HIV broadly neutralizing antibodies. Nat Med. 2012;18(11):1616–7. doi: 10.1038/nm.2993. [DOI] [PubMed] [Google Scholar]
- 29.van den Kerkhof TL, Feenstra KA, Euler Z, van Gils MJ, Rijsdijk LW, Boeser-Nunnink BD, et al. HIV-1 envelope glycoprotein signatures that correlate with the development of cross-reactive neutralizing activity. Retrovirology. 2013;10(1):102. doi: 10.1186/1742-4690-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein F, Gaebler C, Mouquet H, Sather DN, Lehmann C, Scheid JF, et al. Broad neutralization by a combination of antibodies recognizing the CD4 binding site and a new conformational epitope on the HIV-1 envelope protein. The Journal of experimental medicine. 2012;209(8):1469–79. doi: 10.1084/jem.20120423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piantadosi A, Panteleeff D, Blish CA, Baeten JM, Jaoko W, McClelland RS, et al. Breadth of neutralizing antibody response to human immunodeficiency virus type 1 is affected by factors early in infection but does not influence disease progression. Journal of virology. 2009;83(19):10269–74. doi: 10.1128/JVI.01149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rademeyer C, Moore PL, Taylor N, Martin DP, Choge IA, Gray ES, et al. Genetic characteristics of HIV-1 subtype C envelopes inducing cross-neutralizing antibodies. Virology. 2007;368(1):172–81. doi: 10.1016/j.virol.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 33.Powell RL, Kinge T, Nyambi PN. Infection by discordant strains of HIV-1 markedly enhances the neutralizing antibody response against heterologous virus. Journal of virology. 2010;84(18):9415–26. doi: 10.1128/JVI.02732-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34*.Cortez V, Odem-Davis K, McClelland RS, Jaoko W, Overbaugh J. HIV-1 superinfection in women broadens and strengthens the neutralizing antibody response. PLoS pathogens. 2012;8(3):e1002611. doi: 10.1371/journal.ppat.1002611. [This study demonstrated enhanced breadth after superinfection that was independent of viral load and CD4 counts. Breadth was particularly associated with intersubtype superinfection and persistence of both infecting viruses, suggesting that a more divergent circulating viral population resulted in increased breadth.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore PL, Gray ES, Sheward D, Madiga M, Ranchobe N, Lai Z, et al. Potent and broad neutralization of HIV-1 subtype C by plasma antibodies targeting a quaternary epitope including residues in the V2 loop. Journal of virology. 2011;85(7):3128–41. doi: 10.1128/JVI.02658-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore PL, Sheward D, Nonyane M, Ranchobe N, Hermanus T, Gray ES, et al. Multiple pathways of escape from HIV broadly cross-neutralizing V2-dependent antibodies. Journal of virology. 2013 doi: 10.1128/JVI.03424-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Euler Z, van Gils MJ, Boeser-Nunnink BD, Schuitemaker H, van Manen D. Genome-wide association study on the development of cross-reactive neutralizing antibodies in HIV-1 infected individuals. PloS one. 2013;8(1):e54684. doi: 10.1371/journal.pone.0054684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferreira CB, Merino-Mansilla A, Llano A, Perez I, Crespo I, Llinas L, et al. Evolution of Broadly Cross-Reactive HIV-1-Neutralizing Activity: Therapy-Associated Decline, Positive Association with Detectable Viremia, and Partial Restoration of B-Cell Subpopulations. Journal of virology. 2013;87(22):12227–36. doi: 10.1128/JVI.02155-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boliar S, Murphy MK, Tran TC, Carnathan DG, Armstrong WS, Silvestri G, et al. BLymphocyte Dysfunction in Chronic HIV-1 Infection Does Not Prevent Cross-Clade Neutralization Breadth. Journal of virology. 2012;86(15):8031–40. doi: 10.1128/JVI.00771-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jelicic K, Cimbro R, Nawaz F, Huang da W, Zheng X, Yang J, et al. The HIV-1 envelope protein gp120 impairs B cell proliferation by inducing TGF-beta1 production and FcRL4 expression. Nat Immunol. 2013;14(12):1256–65. doi: 10.1038/ni.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Isik G, van Montfort T, Boot M, Cobos Jimenez V, Kootstra NA, Sanders RW. Chimeric HIV-1 envelope glycoproteins with potent intrinsic granulocyte-macrophage colony-stimulating factor (GM-CSF) activity. PloS one. 2013;8(4):e60126. doi: 10.1371/journal.pone.0060126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Isik G, Chung NP, van Montfort T, Menis S, Matthews K, Schief WR, et al. An HIV-1 envelope glycoprotein trimer with an embedded IL-21 domain activates human B cells. PloS one. 2013;8(6):e67309. doi: 10.1371/journal.pone.0067309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melchers M, Bontjer I, Tong T, Chung NP, Klasse PJ, Eggink D, et al. Targeting HIV-1 envelope glycoprotein trimers to B cells by using APRIL improves antibody responses. Journal of virology. 2012;86(5):2488–500. doi: 10.1128/JVI.06259-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Excler JL, Tomaras GD, Russell ND. Novel directions in HIV-1 vaccines revealed from clinical trials. Current opinion in HIV and AIDS. 2013;8(5):421–31. doi: 10.1097/COH.0b013e3283632c26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grimm SK, Ackerman ME. Vaccine design: emerging concepts and renewed optimism. Current opinion in biotechnology. 2013;24(6):1078–88. doi: 10.1016/j.copbio.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46*.Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, et al. Human circulating PD-(+)1CXCR3(−)CXCR5(+) memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39(4):758–69. doi: 10.1016/j.immuni.2013.08.031. [This study found a correlation between the early frequency of population of functional memory T follicular helper cells in the periphery and the development of later neutralization breadth in a large cohort of HIV-1 infected individuals. Maintenance of this stable memory population may also be important for breadth.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoot S, McGuire AT, Cohen KW, Strong RK, Hangartner L, Klein F, et al. Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS pathogens. 2013;9(1):e1003106. doi: 10.1371/journal.ppat.1003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGuire AT, Glenn JA, Lippy A, Stamatatos L. Diverse recombinant HIV-1 Envs fail to activate B cells expressing the germline B cell receptors of the broadly neutralizing anti-HIV-1 antibodies PG9 and 447-52D. Journal of virology. 2013 doi: 10.1128/JVI.03228-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49*.McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. The Journal of experimental medicine. 2013;210(4):655–63. doi: 10.1084/jem.20122824. [This study demonstrated that specific glycans on gp120 interfere with recognition of a subtype C HIV-1 Env by germline-like versions of two potent CD4bs bnAbs. This finding suggests that the failure of an infected individual to make CD4bs bnAbs could result from a lack of B cell activation of the VH1 germline because glycans restrict access to the CD4bs epitope.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50**.Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340(6133):711–6. doi: 10.1126/science.1234150. [This study combined computational design, large scale mutagenesis, and structural characterization to create an outer domain nanoparticle Env imunogen that binds to mature and germline-like CD4bs bnAbs and activates B cells expressing the germline-like IgM B cell receptor versions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scharf L, West AP, Jr., Gao H, Lee T, Scheid JF, Nussenzweig MC, et al. Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(15):6049–54. doi: 10.1073/pnas.1303682110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52*.Sundling C, Li Y, Huynh N, Poulsen C, Wilson R, O'Dell S, et al. High-resolution definition of vaccine-elicited B cell responses against the HIV primary receptor binding site. Sci Transl Med. 2012;4(142):142ra96. doi: 10.1126/scitranslmed.3003752. [This study demonstrated that the VH gene family structure is similar between humans and rhesus macaques, with high homology (average of 92%) between rhesus and human VH genes. They isolated CD4bs mAbs from Env immunized macaques that had high affinity for gp120 and tier 1 neutralizing capacity, modest somatic hypermutation levels, and longer CDRH3 domains, but the macaque mAbs were not derived from VH1-like germline as are many of the human CD4bs bnAbs.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zolla-Pazner S, deCamp AC, Cardozo T, Karasavvas N, Gottardo R, Williams C, et al. Analysis of V2 antibody responses induced in vaccinees in the ALVAC/AIDSVAX HIV-1 vaccine efficacy trial. PloS one. 2013;8(1):e53629. doi: 10.1371/journal.pone.0053629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao HX, Bonsignori M, Alam SM, McLellan JS, Tomaras GD, Moody MA, et al. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38(1):176–86. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480(7377):336–43. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56*.Alam SM, Dennison SM, Aussedat B, Vohra Y, Park PK, Fernandez-Tejada A, et al. Recognition of synthetic glycopeptides by HIV-1 broadly neutralizing antibodies and their unmutated ancestors. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(45):18214–9. doi: 10.1073/pnas.1317855110. [This study capitalized on the fact that that V1V2 directed bnAbs and type-specific anti-V2 antibodies may recognize different V1V2 conformations, and that the V1V2 bnAbs also recognize glycans as part of their epitope. They designed V1V2 glycopeptide immunogens that bind with high affinity to the germline-like and mature V1V2 bnAbs, but with much lower affinity to a type-specific RV144 elicited mAb.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477(7365):466–70. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]