Abstract
Acute respiratory distress syndrome (ARDS) remains a significant hazard to human health and is clinically challenging because there are no prognostic biomarkers and no effective pharmacotherapy. The lung compartment metabolome may detail the status of the local environment that could be useful in ARDS biomarker discovery and the identification of drug target opportunities. However, neither the utility of bronchoalveolar lavage fluid (BALF) as a biofluid for metabolomics nor the optimal analytical platform for metabolite identification are established. To address this, we undertook a study to compare metabolites in BALF samples from patients with ARDS and healthy controls using a newly developed liquid chromatography (LC)-mass spectroscopy (MS) platform for untargeted metabolomics. Following initial testing of three different high performance liquid chromatography (HPLC) columns, we determined that reversed phase (RP)-LC and hydrophilic interaction chromatography (HILIC), were the most informative chromatographic methods because they yielded the most and highest quality data. Following confirmation of metabolite identification, statistical analysis resulted in 37 differentiating metabolites in the BALF of ARDS compared with health across both analytical platforms. Pathway analysis revealed networks associated with amino acid metabolism, glycolysis and gluconeogenesis, fatty acid biosynthesis, phospholipids and purine metabolism in the ARDS BALF. The complementary analytical platforms of RPLC and HILIC-LC generated informative, insightful metabolomics data of the ARDS lung environment.
Keywords: biomarkers, critical illness, metabolomics, lung injury, bioinformatics, phospholipids, lactate, xanthine oxidase, hippurate, pharmacotherapy
INTRODUCTION
Sepsis annually affects more than 750,000 patients in the United States and remains a leading cause of death worldwide.1, 2 Sepsis-induced acute respiratory distress syndrome (ARDS), characterized by an exaggerated inflammatory process that leads to rapid deterioration of respiratory function that necessitates mechanical ventilation (MV)3, is a particularly clinically challenging problem because its onset, severity and duration are unpredictable and it has an associated mortality of up to ~40%.3–7 The pathogenesis and risk factors for the development and progression of ARDS and its severity remain poorly understood and effective pharmacotherapy has not been identified. An incomplete understanding of the complex pathophysiology of ARDS and extensive patient heterogeneity likely contribute to this situation. As such, continued improvement in the understanding of the mechanistic underpinnings of ARDS could lead to the identification of clinically useful biomarkers and the discovery of much needed drug target opportunities.
The application of untargeted metabolomics for biomarker discovery lends itself well to the complexity of ARDS because it detects as many as several hundred metabolites, depending on the analytical platform, from a single sample with minimal bias and no preconception of the sample composition.8–10 We have previously used untargeted 1D-1H-nuclear magnetic resonance (NMR) metabolomics to measure and differentiate endogenous metabolites in plasma samples collected from patients with ARDS and healthy subjects.9, 11 The utilization of plasma was based on the idea that this biofluid reflects a physiological average of the metabolomes of different tissues and compartments, so changes in metabolites in the blood are indicative of the metabolome of the host.10 Despite this, more detailed and timely characterization of the distress signal of the acutely injured lungs may require the use of a compartment specific biofluid like exhaled breath condensate or bronchoalveolar lavage fluid (BALF).12–14 Furthermore, it is likely that metabolomics data acquired from the lung compartment could elaborate or substantiate those acquired from the blood.
The acquisition of BALF is often routinely performed as part of the clinical care of patients with ARDS. However, its utility as a biofluid for quantitative metabolomics may be limited because of its high protein and salt content and low concentrations of metabolites.8 The latter hinders the utility of 1H-NMR, which is illustrated by a few studies that have been conducted using this approach, one of which employed a high-resolution 1-D and 2-D NMR strategy.15–18 In all cases, either no or only a few metabolites were named and to date, there are few metabolomics data from the BALF of ARDS patients. As such, the present study aimed to test the ability of a newly developed novel LC-MS untargeted metabolomics platform that was specifically designed to detect biologically relevant differentiating metabolites in existing BALF samples acquired from ARDS patients and healthy controls.
MATERIALS AND METHODS
Study samples
The samples used for this study were acquired from healthy subjects and patients enrolled in the Acute Lung Injury Specialized Center of Clinically Oriented Research randomized trial of granulocyte-macrophage colony stimulating factor in the treatment of ARDS by non-directed bronchoalveolar lavage as previously described.11 Briefly, samples from mechanicallyventilated ARDS patients were collected between 0–72 h of the diagnosis of ARDS but before randomization to study drug and from non-ventilated healthy controls following informed consent and under protocols approved by the University of Michigan Institutional Review Board. A standard technique was employed in which BALF was collected by bronchoscopy in which 150 ml of normal saline was instilled into the right middle lobe or lingula in 30 ml aliquots and was retrieved by gentle suctioning through the suction port of the bronchoscope. Non-ventilated control subjects underwent bronchoscopy with conscious sedation using the same standard technique as the study patients. At the time of collection, BALF was clarified by centrifugation and the supernatants were frozen (−70°C) until the time of assay.
Protein Electrophoresis
To characterize the heterogeneity of the ARDS BALF samples, protein concentration was measured using a bicinchoninic acid (BCA) assay (Pierce Protein Biology Products, Thermo Fisher Scientific, Rockford, IL USA). BALF proteins were separated by SDS-PAGE on a 12.5% acrylamide gel and stained with Coomassie blue as described in the Supporting Information.
LC-MS
Chemicals and Reagents
Water, methanol, acetonitrile, formic acid, ammonium bicarbonate, ammonium hydroxide solution, and ammonium acetate (99.999%) were purchased from Sigma-Aldrich (St. Louis, MO) and were LC-MS grade except as noted.
Sample preparation
Before processing, samples were assigned ID numbers which de-dentified control from ARDS subjects. At the time of assay, BALF samples were thawed on ice. For deproteinization in preparation for LC-MS analysis, 100 µL of BALF was combined with 400 µL of 9:1 methanol:chloroform containing 18 µg/mL of an 13C-labeled amino acid mix (Sigma-Aldrich part 426199) which served as an internal standard. The sample was vortexed, then centrifuged (10 min at 15,000 × g). For hydrophilic interaction chromatography (HILIC)-, the supernatant was transferred directly to an autosampler vial for analysis. For reversed phase (RPLC)-MS analysis, the supernatant was transferred to a clean vial and dried under a stream of nitrogen gas. The dried sample was reconstituted in 50 µL of water and transferred to an autosampler vial for analysis. All samples were processed in random order and were assigned to a random LC-MS run order using a computerized algorithm.
Chromatographic method evaluation
A preliminary analysis of six BALF samples from ARDS patients was performed by LC-MS using three different HPLC columns: a Waters Acquity HSS T3 1.8µ (50 × 2.1 mm ID), a Phenomenex Luna NH2 3µ column (150 mm × 2.1 mm ID) and a Cogent Diamond Hydride (150 × 2.1 mm ID). These columns were selected due to their orthogonal modes of separation and their proven utility for metabolomic analysis of other biofluids. They were initially evaluated using chromatographic conditions described previously to determine the number of features detected in BALF and to assess typical peak quality.19–21 Based upon these preliminary evaluations, chromatographic methods were further optimized for BALF analysis using the Acquity HSS T3 and Luna NH2 columns. The resulting chromatographic methods are described below.
Optimized LC-MS methods
For RPLC-MS, samples were analyzed on an Agilent 1200 LC / 6530 qTOF MS system (Agilent Technologies, Inc., Santa Clara, CA USA) using the Waters Acquity HSS T3 1.8 µ column (Waters Corporation, Milford, MA). Each sample was analyzed twice, once in positive and once in negative ion mode. For positive ion mode runs, mobile phase A was 100% water with 0.1% formic acid and mobile phase B was 100% methanol with 0.1% formic acid. For negative ion mode runs, the formic acid was replaced with 0.1% (m/v) ammonium bicarbonate. The gradient for both positive and negative ion modes was as follows: 0–0.5 min 1%B, 0.5–2 min 1–99%B, 2–6 min 99%B, 6–6.1 min 99-1%B, hold 1%B until 9 min. The flow rate was 0.35 mL/min and the column temperature was 40°C. The injection volume for positive and negative mode was 5 µL and 8 µL, respectively. Mass spectrometry was performed by electrospray ionization with an Agilent Jetstream ion source, with full-scan mass spectra acquired over the m/z range 50–1500 Da. Source parameters were: drying gas temperature 350°C, drying gas flow rate 10 L/min, nebulizer pressure 30 psig, sheath gas temp 350°C and flow 11 l/min, and capillary voltage 3500V, with internal reference mass correction enabled.
For HILIC-MS, samples were analyzed on an Agilent 1200 LC / 6520 qTOF MS with the Phenomenex Luna NH2 column. Mobile phase A was acetonitrile. Mobile phase B was 5 mM ammonium acetate in water, adjusted to pH 9.9 with ammonium hydroxide. The gradient was 0–15 min 20–100%B, 15–18 min 100%B, 18-18.1min 100-20%B, 18.1–30 min 20%B. The flow rate was 0.25 mL/min and the column temperature was 25°C. The injection volume was 10 µL. Mass spectrometry was performed in negative ion mode using source parameters identical to the RPLC method, except that a standard electrospray source was used in place of the Jetstream source and sheath gas was not used.
Data analysis workflow
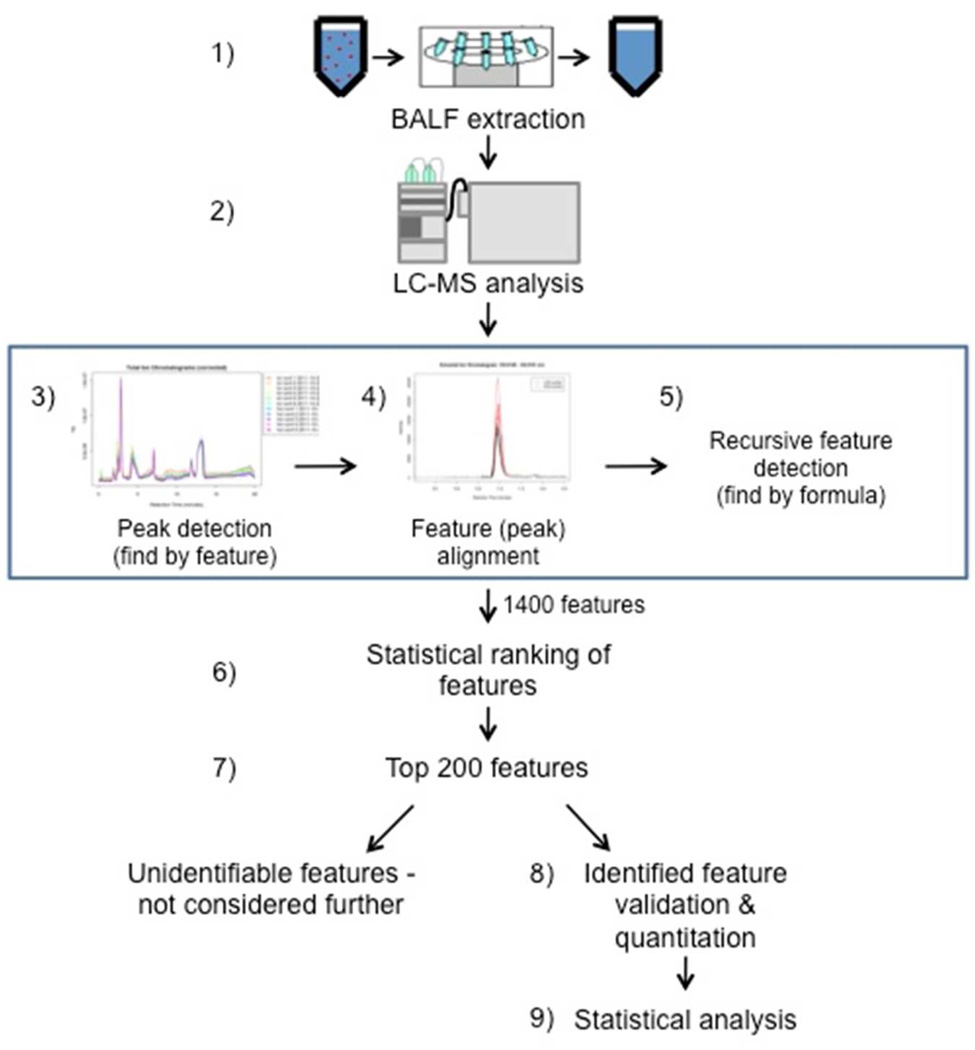
Scheme 1 shows the details of the LC-MS data analysis workflow. Chromatographic peaks that represent metabolites – which are henceforth termed “features” – were detected in the data using the automated “Find by Feature” algorithm in Agilent Masshunter Qualitative Analysis Software. Feature alignment between samples was performed using an in-house written software package, which matched features with the same mass and retention time between samples. The software was configured to allow up to a 0.5 min RT shift and/or a 20 ppm mass shift between LC-MS runs. To reduce gaps in the data, recursive feature identification was then performed by searching the data a second time with the list of aligned features using the “Find by Formula” algorithm. Then, features that were present in over 70% of ARDS and control samples were analyzed by partial least squares-discriminant analysis (PLS-DA) as described below in Statistical Analysis section and ranked by loading scores. The lists of features obtained from positive and negative modes were ranked separately. The top 200 features from each mode were selected for further analysis. This threshold was chosen to allow a manageable number of features to be validated in a timely manner, while still retaining the vast majority of the features responsible for differentiating BALF from control and ARDS patients. To improve data quality, these top 200 features were re-quantitated and all peaks were visually inspected using Agilent MassHunter Quantitative Analysis software, with the analyst blinded to the identity of the subjects.
Scheme 1.
Schematic of the data processing workflow. Following sample extraction (step 1) and either RP- or HILIC-MS (step 2), features were identified using an in-house metabolite library. An attempt to identify remaining unknown features was carried out by manual validation using the “Find by Feature” algorithm in Agilent Masshunter Qualitative Analysis Software (Santa Clara, CA), peak alignment between samples by mass and retention time and the “Find by Formula” feature in Masshunter (steps 3–5). To minimize gaps in the data, recursive feature identification was performed by searching the data a second time with the list of aligned features using the “Find by Formula” algorithm. The resulting features were statistically ranked (step 6) and the top 200 ranked features were utilized for additional analysis (step 7). At this point, unidentifiable features were not considered further and features with putative identifications were analytically validated using either reference standards or MS/MS (step 8). The resulting data sets were used to statistically compare ARDS BALF to healthy control BALF metabolites (step 9). BALF = bronchoalveolar lavage fluid; ARDS = acute respiratory distress syndrome; RP = reverse phase; HILIC = hydrophilic interaction chromatography.
Once the list of features was generated, in order to identify metabolites present in the BALF samples, the features were searched against an in-house library of 800 known metabolite standards which had been previously analyzed under identical LC-MS conditions. Features not identified by standards were searched for possible matches using the online Metlin database (http://metlin.scripps.edu) and Human Metabolome Database (HMDB; http://www.hmdb.ca). In many cases, the database searches resulted in multiple possible matches for each feature within a 10 ppm mass error window. Metabolite matches were ranked in order of ascending mass error, and among matches with equivalent mass error, in order of ascending Metlin or HMDB identification (ID) number. At this point, remaining features which did not match any database entries were not considered for further evaluation. Putative feature IDs from the database matches were validated or rejected by co-injection of BALF samples with authentic standards, or by acquisition of targeted MS/MS data on the q-TOF MS followed by comparison with MS/MS spectra in the online databases. Data generated by HILIC-MS were processed the same way.
Statistical and bioinformatics analysis
Data were auto-scaled (mean-centered and divided by the standard deviation of each feature) and log transformed; PLS-DA of the features present in 70% of the control and ARDS samples was performed using SIMCA-P (version 13, UMETRICS AB, Box 7960, SE90719 Umeå, Sweden). Two groups were used as dependent (Y) variables. The PLS-DA models were validated by random permutation of the Y variable and comparison of the goodness of fit (R2Y and Q2).22, 23 For random permutation tests, 100 models were calculated and the goodness of fit was compared with the original model in a validation plot. Variable Importance in the Projection (VIP) scores from the PLS-DA model were used to rank metabolite features. To generate a manageable data set, the top 200 ranked features of each analytical mode were compared between ARDS and control by an unpaired Student’s t-test with unequal variance and p-values were corrected for false discovery.24, 25 KEGG IDs were assigned when possible and metabolites were rank ordered by ascending p value to generate a final dataset for each analytical method. Pathway analysis was performed in Metscape.26 Metabolites with fold change Q-values were loaded into Metscape and networks graphs were created.
RESULTS
To accomplish the study’s goals, BALF samples were obtained from twenty patients with ARDS from a completed clinical trial11 and eight healthy control individuals. From ARDS patients, two samples were collected at time 0, five at 24h, two at 48h and nine at 72h. As indicated in Table 1, there was a range of etiologies of ARDS which is typical of this heterogeneous patient population which often undergoes broncheoalveolar lavage as a part of their clinical care. Of the 18 ARDS patients, one had mild, six had moderate and 11 had severe ARDS according to the Berlin definition.7 Additional demographic data about these patients has been previously reported.11 The control group had fewer patients than the ARDS group due to limitations in the ability to recruit subjects for this relatively invasive procedure, but the groups were not different in terms of age (p = 0.29 by unpaired Student’s t-test) and gender (Table 1).
Table 1.
Demographic characteristics of ARDS patients and healthy control subjects
| ARDS | Healthy | |
|---|---|---|
| Sample size (n) | 18 | 8 |
| Sex (%) | ||
| Female | 44 | 50 |
| Age (mean ±S.D.) | 46.1 ± 14.9 | 39.8 ± 11.0 |
| ARDS etiology (%) | N/A | |
| sepsis | 39 | |
| aspiration | 22 | |
| pneumonia | 33 | |
| other/unknown | 5 | |
| PaO2/FiO2 ratioa (mean ± S.D.) | 103.3 ± 49.1 | N/A |
| APSb (mean ± S.D.) | 62.3 ± 13.1 | N/A |
PaO2 is partial pressure of oxygen in arterial blood and FiO2 is the fraction of inspired oxygen;
APS is the acute physiology score and is an indicator of illness severity.9 See the text for additional information about ARDS severity.
Protein Assay and SDS-PAGE analysis
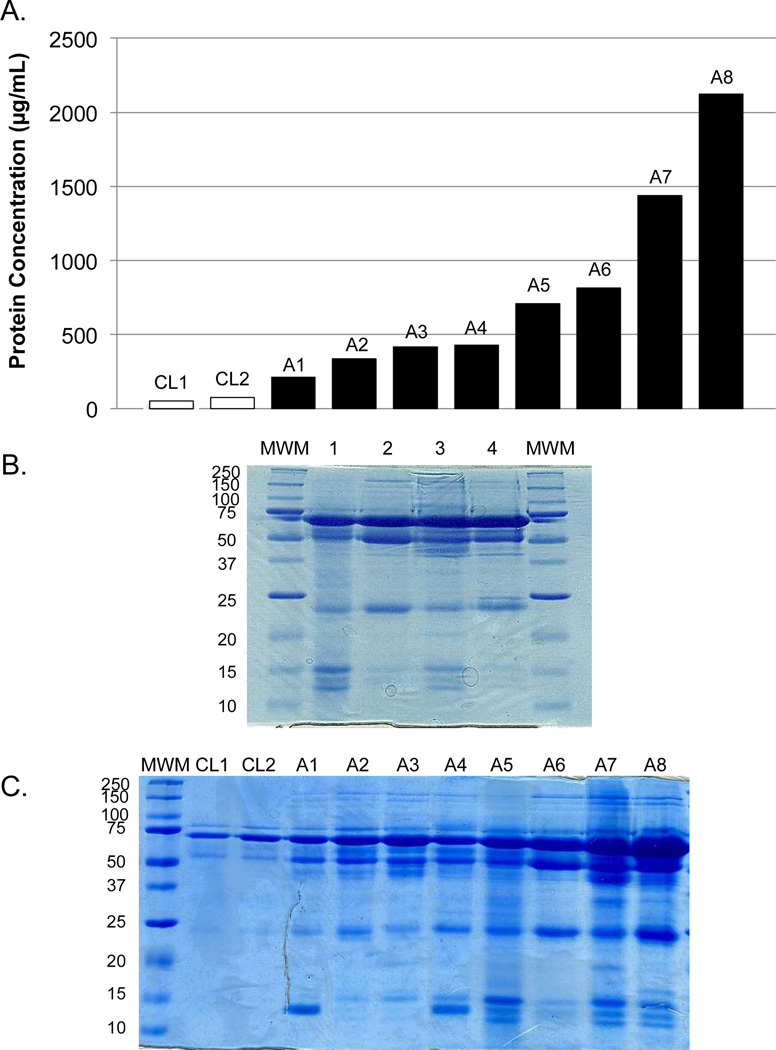
BALF has been described as a relatively protein-rich, metabolite-poor fluid.8 The total protein content of BALF varied as much as 10-fold between ARDS patients (Figure 1A), but in all cases the ARDS samples had measurably higher protein concentrations than control samples. Using the measured total protein values, a SDS-PAGE gel was loaded with a constant amount of protein per lane for four different ARDS samples (Figure 1B). Although some proteins were present in all samples, (e.g., serum albumin at approximately 70 kDa), the profiles were clearly heterogeneous in the distribution of molecular weight and abundance of proteins among patients. When samples were loaded by constant volume (Figure 1C), heterogeneity was also evident and the total protein abundance was closely mirrored by the total protein concentration of each sample (Figure 1A).
Figure 1.
Gel electrophoresis (12.5%) of BALF proteins illustrates heterogeneity across samples. (A) The associated protein concentration of each sample. (B) Coomassie-stained gel of representative ARDS BALF samples (1–4). An equal amount of protein (30µg) was loaded for each sample. (C) Coomassie-stained gel of representative control and ARDS BALF samples loaded by volume. MWM = molecular weight marker; A = ARDS.
Chromatographic method evaluation
From the chromatographic data it was determined that a reversed phase column (Waters HSS T3 C18) offered the best coverage, with a mixed-mode HILIC/anion-exchange column (Phenomenex Luna NH2) next-best, while a silica-based HILIC column (Cogent Diamond Hydride) offered the least coverage of BALF. We therefore elected to use the C18 RPLC column as the primary method for metabolomics in this study and chose to include data from a pilot analysis using the mixed mode HILIC/AEX column as a complementary approach.
RPLC-MS metabolomics of BALF samples
A total of over 1742 and 1314 features were detected in positive ion and negative ion modes, respectively. Supporting Information Figure S1 shows cross-validated PLS-DA score plots for positive and negative mode data (R2Y = 0.99 and Q2 = 0.87 for positive mode; R2Y = 0.98 and Q2 = 0.78 for negative mode). The top 200 features from each mode were selected and processed as described in the Methods. Identified features from both positive and negative ion modes of ARDS and healthy controls were statistically compared. This resulted in 26 significant endogenous metabolites (Table 2) of which 23 were positively identified, and the remaining three were assigned putative identities. Extracted ion chromatograms for these metabolites are shown in Supporting Information Figure S2.
Table 2.
Confirmed and putative BALF metabolites of ALI patients compared with healthy control subjects detected by RPLC-MS
| Name | KEGG ID |
LC-MS POS ion mode | LC-MS NEG ion mode | ||||
|---|---|---|---|---|---|---|---|
| Confirmed ID | p value | q value | Fold change |
p value | q value | Fold change |
|
| Hypoxanthine | C00262 | 6.93E-10 | 7.07E-08 | 40.96 | |||
| L-Leucine | C00123 | 5.34E-07 | 3.40E-06 | 20.42 | 6.62E-08 | 1.01E-06 | 15.58 |
| Creatinine | C00791 | 7.16E-07 | 3.84E-06 | 9.06 | |||
| Creatine | C00300 | 1.52E-06 | 6.59E-06 | 10.26 | |||
| L-Proline | C00148 | 1.55E-06 | 6.59E-06 | 6.78 | |||
| L-Glutamate | C00025 | 2.49E-06 | 9.07E-06 | 7.94 | |||
| O-Acetylcarnitine | C02571 | 1.82E-04 | 4.20E-04 | 7.63 | |||
| Guanosine | C00387 | 1.85E-04 | 4.20E-04 | 3.70 | |||
| L-phenylalanine | C00079 | 2.41E-03 | 4.64E-03 | 2.06 | 8.60E-03 | 3.50E-02 | 1.54 |
| Methionine | C01733 | 6.31E-03 | 1.13E-02 | 2.30 | |||
| Bis(2-ethylhexyl)phthalate | C03690 | 3.21E-02 | 5.12E-02 | 1.22 | |||
| L-Threonate | C01620 | 1.33E-10 | 7.28E-09 | 19.11 | |||
| Uridine | C00299 | 1.79E-10 | 7.28E-09 | 7.63 | |||
| Xanthine | C00385 | 1.05E-09 | 3.19E-08 | 18.59 | |||
| Hippurate | C01586 | 1.86E-08 | 3.78E-07 | 35.35 | |||
| L-Tyrosine | C00082 | 9.56E-08 | 1.30E-06 | 9.02 | |||
| L-Tryptophan | C00078 | 1.63E-06 | 1.81E-05 | 3.98 | |||
| 2-Oxoglutarate | C00026 | 4.30E-06 | 4.26E-05 | 8.63 | |||
| cis-Aconitate | C00417 | 4.54E-06 | 4.26E-05 | 3.12 | |||
| L-Lactate | C00186 | 5.54E-06 | 4.83E-05 | 4.35 | |||
| Citrate | C00158 | 2.40E-05 | 1.63E-04 | 2.61 | |||
| Inosine | C00294 | 3.02E-02 | 1.08E-01 | 1.75 | |||
| Palmitoleic acid | C08362 | 5.58E-02 | 1.89E-01 | 1.45 | |||
| Putative ID Only | |||||||
| 1b,3a,7a,12a- Tetrahydroxy-5b-cholanoic acid | - | 2.83E-06 | 9.96E-06 | 114.30 | |||
| 4,7,10,13-hexadecatetraenoic acid | - | 3.77E-05 | 9.61E-05 | 3.56 | |||
| 1,25-dihydroxy-3-deoxy-19-norcholecalciferol | - | 3.00E-02 | 4.86E-02 | 1.42 | |||
The detected compounds represent a range of different endogenous metabolite classes, including a number of amino acids, other polar metabolites, and lipids. Also present were a number of exogenous drug metabolites, arising from the administration of compounds associated with the lavage procedure (i.e., lidocaine) and the treatment of sepsis-induced ARDS (i.e., penicilloic acid V and propofol glucuronide); these compounds were excluded from the analysis. The most significantly different endogenous metabolites between healthy control and ARDS groups were elevated in BALF samples, with fold-change values ranging from 1.22 to 41.
HILIC-MS metabolomics of BALF samples
A subset of the BALF samples (12 of the total 26; 8 of which were from ARDS patients) were also analyzed by HILIC-MS in negative ion mode as described in the Methods. The same procedure for feature selection, statistical analysis and metabolite identification was performed as for RPLC analyses. A total of 134 features were detected in the resulting data. Of these, 18 identifiable metabolites (Table 3, Supporting Information Figure S2) were determined to be statistically different between ARDS and control groups (p < 0.05 after false discovery correction). These metabolites were dominated by two classes of molecules: free fatty acids and various phosphatidylcholine species. In most cases, phosphatidylcholine species were decreased and free fatty acids were elevated in BALF from ARDS patients.
Table 3.
Confirmed and putative BALF metabolites of ARDS patients compared with healthy control subjects detected by HILIC-MS
| KEGG ID |
HILIC-MS | |||
|---|---|---|---|---|
| Probable/Confirmed ID | p value | q value | Fold change |
|
| Phosphatidylcholine (16:0/16:0) | 4.40E-05 | 1.59E-03 | 0.13 | |
| Phosphatidylcholine (16:1/16:0) | 7.90E-04 | 1.72E-02 | 0.22 | |
| Phosphatidylcholine (16:0/14:0) | 1.87E-03 | 2.91E-02 | 0.26 | |
| Stearic acid | C01530 | 2.29E-03 | 3.12E-02 | 0.44 |
| Palmitoleic acid | C08362 | 4.92E-03 | 4.87E-02 | 4.65 |
| Arachidonic acid | C00219 | 7.67E-03 | 6.43E-02 | 6.55 |
| Palmitic acid | C00249 | 1.92E-02 | 1.23E-01 | 0.58 |
| Phosphatidylcholine (16:1/18:2) | 2.19E-02 | 1.27E-01 | 0.47 | |
| Phosphatidylcholine (16:0/18:1) | 2.21E-02 | 1.27E-01 | 0.43 | |
| Phosphatidylcholine (16:0/18:0) | 2.50E-02 | 1.30E-01 | 0.38 | |
| Phosphatidylcholine (16:1/20:4) | 3.34E-02 | 1.52E-01 | 0.54 | |
| L-Lactate | C00186 | 4.37E-02 | 1.70E-01 | 3.49 |
| D-Glucose | C00267 | 4.75E-02 | 1.70E-01 | 4.51 |
| Linoleic acid | C01595 | 4.84E-02 | 1.70E-01 | 6.68 |
| Putative ID only | ||||
| Norethindrone acetate | 3.85 E-05 | 1.59E-03 | 0.27 | |
| 2-Hydroxyethinylestradiol | 5.82E-05 | 1.59E-03 | 0.29 | |
| 2-Hydroxymestranol | 4.74E-03 | 4.87E-02 | 0.37 | |
| Monoacylglycerol (18:0) | 5.91E-03 | 5.37E-02 | 0.28 | |
Pathway analysis
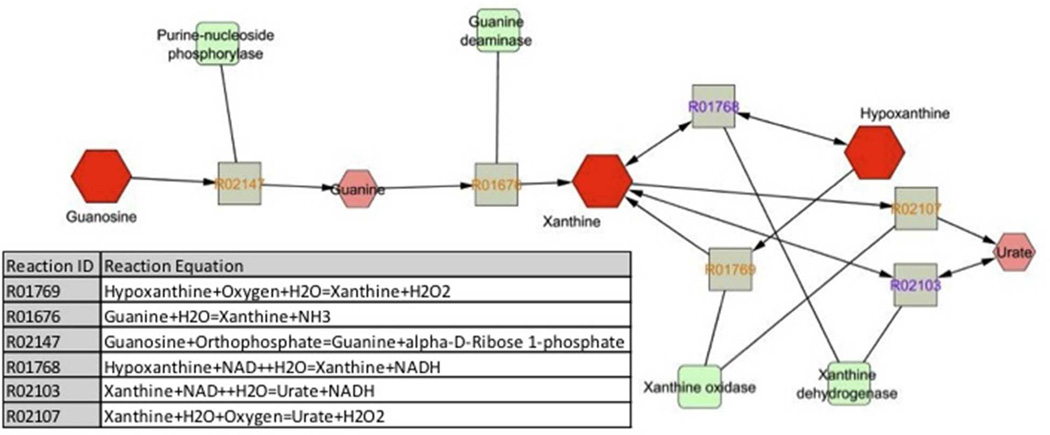
Table 4 shows the list of metabolic pathways associated with metabolites with KEGG IDs that were identified by both analytical methods; Metscape generated networks are shown in Supporting Information Figure S3.26 Notably, the amino acids and their intermediates, as well as compounds involved in carbohydrate and nucleotide metabolism were increased in ARDS samples compared to control. This included the guanosine network (Figure 2) of which the abundant ARDS BALF metabolites, hypoxanthine and xanthine, belong (Table 2). A number of lipid metabolites including arachidonic, palmitoleic and linoleic acids (Table 3), were also increased. In contrast, one of the main components of pulmonary surfactant, phosphatidylcholine, was decreased in ARDS.27
Table 4.
Metabolic ARDS pathways and associated compounds
| Pathway category | Pathway | Associated compound(s) | ||
|---|---|---|---|---|
| Amino acid metabolism | Aromatic amino acid metabolism | Hippurate ↑ | L-Phenylalanine ↑ | L-Tyrosine ↑ |
| Amino acid metabolism | Glycine, serine, alanine and threonine metabolism | Creatine ↑ | ||
| Amino acid metabolism | Methionine and cysteine metabolism | Methionine ↑ | ||
| Amino acid metabolism | Urea cycle and metabolism of arginine, proline, glutamate, aspartate and asparagines | L-Glutamate ↑ | O-Acetylcarnitine ↑ | Creatinine ↑ |
| L-Proline ↑ | ||||
| Amino acid metabolism | Valine, leucine and isoleucine degradation | L-Leucine ↑ | ||
| Carbohydrate metabolism | Ascorbate and aldarate metabolism | L-Threonate ↑ | ||
| Carbohydrate metabolism | Glycolysis and Gluconeogenesis | L-Lactate ↑ | D-Glucose ↑ | |
| Carbohydrate metabolism | TCA cycle | cis-Aconitate ↑ | Citrate ↑ | 2-Oxoglutarate ↑ |
| Lipid metabolism | Arachidonic acid metabolism | Arachidonic acid ↑ | ||
| Lipid metabolism | Fatty acid biosynthesis | Palmitic acid ↓ | Stearic acid ↓ | |
| Palmitoleic acid ↑ | ||||
| Lipid metabolism | Linoleate metabolism | Linoleic acid ↑ | ||
| Lipid metabolism | Phospholipid, cell membrane | PI (36:1) ↑ | PC (16:0/16:0) ↓ | PC (16:1/16:0) ↓ |
| PC (16:0/14:0) ↓ | PC (16:1/18:2) ↓ PC | (16:0/18:1) ↓ | ||
| PC (16:0/18:0) ↓ | PC (16:1/20:4) ↓ | |||
| Nucleotide metabolism | Purine metabolism | Hypoxanthine ↑ | Xanthine ↑ | Guanosine ↑ |
| Nucleotide metabolism | Pyrimidine metabolism | Uridine ↑ | ||
Figure 2.
Guanosine network generated by Metscape 2 in which the abundant metabolites of ARDS BALF, xanthine and hypoxanthine, are components and urate (uric acid) is a product. Metabolites are depicted by octagons, with differentiating metabolites of ARDS BALF in dark red. Squares (grey) represent reactions with KEGG reaction identification (ID) numbers and round cornered squares (green) are the associated enzymes.
DISCUSSION
Our findings from this feasibility study indicate that BALF contains a wide range of metabolites amenable to detection by LC-MS. One characteristic of BALF which differentiates it from other samples typically studied using metabolomics is that BALF is a non-endogenous fluid, which is introduced into and removed from the lungs as a part of a medical procedure. As such, its contents are not physiologically regulated, and the recovery of metabolites from the environment inside the lungs is widely variable. In addition, factors such as the magnitude of lung injury, the period of time the BALF is in contact with lung tissue, and the volume of fluid recovered from the lungs, may influence metabolite concentration in BALF. Furthermore, alveolar epithelial and inflammatory cells28, 29, invading pathogens as well as the microbiome30, 31 likely contribute to the pool of metabolites in ARDS BALF. Overall, metabolite concentrations in BALF appear to be characterized more by variation than by consistency. Nevertheless, our data show that when compared with BALF from healthy volunteers, this strategy yielded nearly 50 total features that were significantly altered in ARDS patients and greater than the number of metabolites previously identified by NMR.15–18
The two analytical methods that we used (HILIC and RPLC) yielded complementary results. Of the 44 total identified compounds that were significantly different between control and ARDS groups, only one compound, L-lactate, was identified by both of the two methods. RPLC resulted in the detection of a much larger total number of features (1312 vs. 134 in negative ion mode), as well as a greater number of significantly different features between ARDS and control patients (26 versus 18 identified features with HILIC). The high salt content of BALF relative to the concentration of most metabolites in the fluid is likely partially responsible for this disparity. In the RPLC separation, the salts elute as a single peak with the void volume of the column, reducing their interference with metabolites which elute over the majority of the chromatogram. In HILIC, chloride anions and other salts are retained by the column and elute over a broad retention time range (from 6–17 minutes of the 20 minute run), thereby causing substantial ion suppression of later-eluting peaks. This may explain the somewhat atypical observation that the majority of the analytes detected by HILIC were relatively non-polar compounds such as phospholipids and free fatty acids, which elute early during the separation. On the other hand, there was a relative paucity of non-polar species such as phospholipids and free fatty acids in the RPLC separation. This can be attributed to the process of drying and reconstituting the extracted BALF samples in 100% water, which was performed only for the RPLC and not for the HILIC analysis. This step was necessary to ensure good retention of moderately polar species on the RPLC column, but likely prevented non-polar species from re-dissolving and being detected. So while our analytical approach resulted in the detection of biologically relevant metabolites of ARDS in the BALF, we recognize that it is not a comprehensive or exhaustive read-out of the ARDS metabolome.
Despite the certain technical limitations of our analytical approach, we identified potential differentiating metabolites of ARDS as evidenced by the over four-fold increase in L-lactate and changes in other metabolites associated with energy metabolism (e.g., citrate, creatine, creatinine).32 Citrate, creatine and creatinine are three metabolites that were also found to be increased in ARDS plasma samples by 1D-1H-NMR.9, 26 The remainder of the identified metabolites were specific to BALF. We hypothesized that the amplification of the distressed lung may be better detected in the BALF because it is more representative of the local lung environment.9, 26 This was illustrated, as an example, by the metabolite, L-lactate. The measurement of blood lactate is often and widely used as a biomarker of the severity of critical illnesses, including sepsis.33 While there are multiple sources of lactate represented in the blood during critical illness and because lactate is normally detected in the blood, blood lactate is not a specific marker of lung injury and the precise range of lactate concentrations associated with overall illness severity has not been defined.34, 35 Like the blood, lactate levels in the lung during normal physiology are low. However, lung damage results in increased lactate production which has been shown to be inversely related to the partial pressure of oxygen in arterial blood (PaO2):fraction of inspired oxygen (FiO2) ratio which is indicative of the lung injury severity.14, 36
Despite the potential utility of BALF lactate as a marker of lung injury severity, it is unlikely that any one metabolite can serve as a prognostic biomarker of ARDS due to the complexity and heterogeneity of this disease. This complexity is illustrated by significant alterations in a number of different metabolic pathways including a reduction in phosphatidylcholines (PC), the primary phospholipid of pulmonary surfactant.27 The PC content of surfactant has been shown to be inversely associated with inflammatory cell-mediated lung injury in ARDS.37 In addition, PC is important for the maintenance of alveolar structure and function that are needed for effective gas exchange which is compromised in ARDS.27 As such, the reduction in PC likely reflects a lower alveolar pool of surfactant38 which contributes to these ARDS-induced detriments in the lungs.
Other significant metabolites included guanosine, xanthine and hypoxanthine. Levels of all three compounds were significantly higher in ARDS BALF (Table 2). There was a nearly four-fold increase in guanosine and 41 and 19 fold increases in hypoxantine and xanthine, respectively. Interestingly, uric acid, which is the product of guanosine metabolism (Figure 2) has been shown to be a major “danger signal” in the lung, contributing to cell-death induced acute inflammation.39, 40 While we did not detect uric acid, it has previously been detected in ARDS BALF41 and the presence of the precursor metabolites suggest that the pathway is activated. This could be of significant consequence to the lungs given that, in addition to uric acid production, xanthine oxidase (XO) is a known superoxide anion producing enzyme via its conversion of xanthine to uric acid.42 Surprisingly, little is known about XO activity in the ARDS BALF of humans but it has been detected in the BALF of chronic obstructive pulmonary disease patients.43 In an experimental model of lipopolysaccharide-induced indirect lung injury, XO activity was increased and was augmented by hypoxia.44 In another experimental model, MV contributed to increased enzyme activity.45 Collectively, these data suggest a role of the guanosine network in ARDS, which could be a potential avenue for future investigation.
Another compound significantly increased in ARDS BALF, hippurate, is a known metabolite of the gut microbiome, an abundant urine metabolite, and a uremic toxin.46, 47 Hippurate levels are influenced by diet and the microbiome of the gut because its precursor, benzoic acid, is produced by the conversion of low molecular weight aromatic compounds and dietary polyphenols by colon microbes.48 Hippurate is subsequently generated by the conjugation of benzoic acid and glycine in the liver. However, recent findings from a study of patients with colectomies showed that in humans, hippurate is derived from precursors absorbed in the small intestine and its production may not be specific to the gut.49 The physiological consequence of elevated levels of hippurate in the lung are not known but in the kidney, in the setting of chronic kidney disease, hippurate overload accelerates renal damage50 and it has been shown to stimulate reactive oxygen species production in endothelial cells.51 Interestingly, hippurate levels in the urine have been shown to correlate with lung function in COPD patients.52 While it is not possible to pinpoint the exact origin of this metabolite in BALF, its potential relationship to lung function and role in augmenting lung injury is intriguing and merits further investigation.
We acknowledge the limitations of our study. In particular, we recognize the use of a small sample size. These were existing samples of convenience that were used for the development of an LC-MS for BALF metabolomics assays. Acquisition of BALF is an invasive procedure which is not easily accomplished, particularly in critically ill patients and healthy volunteers. While it is often performed in patients with ARDS, it may not routinely be performed at all centers which may limit the feasibility of BALF as a biofluid for metabolomics studies. We also recognize, and have described, the heterogeneity of ARDS BALF. This is not unexpected since the sample volume is not predictable, there was a broad range of lung injury severity (as evidenced by the standard deviation of the oxygenation index in Table 1) and there were a number of different underlying etiologies of ARDS in our patient population (Table 1). The control samples displayed less variation than ARDS. Variation in the ARDS BALF may be furthered by the influence of pharmacotherapy. The onset of ARDS necessitates MV which makes it inseparable from the ARDS metabolome. However, we acknowledge that the duration of MV likely has an impact. To minimize this variable, we utilized BALF samples that were collected early in the course of ARDS. These critically ill patients also received a number of drugs and we cannot, with complete confidence, rule out the possibility that pharmacotherapy contributed to metabolite levels in the ARDS BALF. Parodoxically, higher levels of lidocaine were found in control samples. This is explained by the fact that healthy control subjects received more local lidocaine anesthetic as part of the lavage procedure than the already anesthetized, mechanically ventilated ARDS patients. Conversely, penicilloic acid V and propofol glucuronide, were only detected in ARDS patients and represent exogenous metabolites of penicillin antibiotics and propofol, respectively.53, 54
To the best of our knowledge, these are the most elaborate human ARDS BALF metabolomics data to date and demonstrate the complex, multifaceted processes involved in ARDS. We have also shown the feasibility of a complementary RPLC- and HILIC-MS analytical platform that detects a broad range of lung metabolites. These results suggest potential promising avenues for future research in larger scale studies but also raise the question as to whether the lung compartment is more informative than or complementary to the periphery for the detection of the distress signal of the lungs in ARDS. Ultimately, it is our goal to construct a longitudinal metabolomics strategy to more fully characterize the ARDS metabolome that will drive biomarker discovery and the identification of drug target opportunities to improve the outcome of patients with ARDS.
Supplementary Material
ACKNOWLEDGMENT
We acknowledge and thank Jennifer Racz for her assistance with the protein assay and SDS-PAGE analysis. This work was presented in part at the American Thoracic Society 2012 International Conference, May 18–23, 2012 in San Francisco, CA, USA.
Funding Sources
This research was supported by the Michigan Nutrition Obesity Research Center and the Michigan Regional Comprehensive Metabolomics Research Core which are funded by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK; P30 DK089503 and DK097153, respectively, to C.F.B., and K25 DK092558 to C.R.E.) and by the National Heart, Lung and Blood Institute (NHLBI; P50 HL074924, to T.J.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIDDK, the NHLBI or the National Institutes of Health.
ABBREVIATIONS
- ARDS
acute respiratory distress syndrome
- BALF
bronchoalveolar lavage fluid
- MV
mechanical ventilation
- NMR
nuclear magnetic resonance
- LC-MS
liquid chromatography-mass spectroscopy
- HPLC
high performance liquid chromatography
- HILIC
hydrophilic interaction chromatography
- RP
reverse phase
- BCA
bicinchoninic acid.
Footnotes
ASSOCIATED CONTENT
Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
REFERENCES
- 1.Wenzel RP. Treating sepsis. The New England journal of medicine. 2002;347(13):966–967. doi: 10.1056/NEJMp020096. [DOI] [PubMed] [Google Scholar]
- 2.Angus DC. The lingering consequences of sepsis: a hidden public health disaster? JAMA. 2010;304(16):1833–1834. doi: 10.1001/jama.2010.1546. [DOI] [PubMed] [Google Scholar]
- 3.Matthay MA, Ware LB, Zimmerman GA. The acute respiratory distress syndrome. J Clin Invest. 2012;122(8):2731–2740. doi: 10.1172/JCI60331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sevransky JE, Martin GS, Shanholtz C, Mendez-Tellez PA, Pronovost P, Brower R, Needham DM. Mortality in sepsis versus non-sepsis induced acute lung injury. Crit Care. 2009;13(5):R150. doi: 10.1186/cc8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hudson LD, Milberg JA, Anardi D, Maunder RJ. Clinical risks for development of the acute respiratory distress syndrome. Am J Respir Crit Care Med. 1995;151(2 Pt 1):293–301. doi: 10.1164/ajrccm.151.2.7842182. [DOI] [PubMed] [Google Scholar]
- 6.Rubenfeld GD, Caldwell E, Peabody E, Weaver J, Martin DP, Neff M, Stern EJ, Hudson LD. Incidence and outcomes of acute lung injury. The New England journal of medicine. 2005;353(16):1685–1693. doi: 10.1056/NEJMoa050333. [DOI] [PubMed] [Google Scholar]
- 7.Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, Fan E, Camporota L, Slutsky AS. Acute respiratory distress syndrome: the Berlin Definition. JAMA : the journal of the American Medical Association. 2012;307(23):2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 8.Serkova NJ, Standiford TJ, Stringer KA. The emerging field of quantitative blood metabolomics for biomarker discovery in critical illnesses. Am J Respir Crit Care Med. 2011;184(6):647–655. doi: 10.1164/rccm.201103-0474CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stringer KA, Serkova NJ, Karnovsky A, Guire K, Paine R, 3rd, Standiford TJ. Metabolic consequences of sepsis-induced acute lung injury revealed by plasma (1)H-nuclear magnetic resonance quantitative metabolomics and computational analysis. Am J Physiol Lung Cell Mol Physiol. 2011;300(1):L4–L11. doi: 10.1152/ajplung.00231.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ala-Korpela M. Critical evaluation of 1H NMR metabonomics of serum as a methodology for disease risk assessment and diagnostics. Clin Chem Lab Med. 2008;46(1):27–42. doi: 10.1515/CCLM.2008.006. [DOI] [PubMed] [Google Scholar]
- 11.Paine R, 3rd, Standiford TJ, Dechert RE, Moss M, Martin GS, Rosenberg AL, Thannickal VJ, Burnham EL, Brown MB, Hyzy RC. A randomized trial of recombinant human granulocyte-macrophage colony-stimulating factor for patients with acute lung injury. Crit Care Med. 2011 doi: 10.1097/CCM.0b013e31822d7bf0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bos LD, Sterk PJ, Schultz MJ. Measuring metabolomics in acute lung injury: choosing the correct compartment? Am J Respir Crit Care Med. 2012;185(7):789. doi: 10.1164/ajrccm.185.7.789. [DOI] [PubMed] [Google Scholar]
- 13.Slutsky AS, Tremblay LN. Multiple system organ failure. Is mechanical ventilation a contributing factor? Am J Respir Crit Care Med. 1998;157(6 Pt 1):1721–1725. doi: 10.1164/ajrccm.157.6.9709092. [DOI] [PubMed] [Google Scholar]
- 14.De Backer D, Creteur J, Zhang H, Norrenberg M, Vincent JL. Lactate production by the lungs in acute lung injury. Am J Respir Crit Care Med. 1997;156(4 Pt 1):1099–1104. doi: 10.1164/ajrccm.156.4.9701048. [DOI] [PubMed] [Google Scholar]
- 15.Wolak JE, Esther CR, Jr, O'Connell TM. Metabolomic analysis of bronchoalveolar lavage fluid from cystic fibrosis patients. Biomarkers. 2009;14(1):55–60. doi: 10.1080/13547500802688194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carraro S, Rezzi S, Reniero F, Heberger K, Giordano G, Zanconato S, Guillou C, Baraldi E. Metabolomics applied to exhaled breath condensate in childhood asthma. Am J Respir Crit Care Med. 2007;175(10):986–990. doi: 10.1164/rccm.200606-769OC. [DOI] [PubMed] [Google Scholar]
- 17.Izquierdo-Garcia JL, Nin N, Ruiz-Cabello J, Rojas Y, de Paula M, Lopez-Cuenca S, Morales L, Martinez-Caro L, Fernandez-Segoviano P, Esteban A, Lorente JA. A metabolomic approach for diagnosis of experimental sepsis. Intensive Care Med. 2011 doi: 10.1007/s00134-011-2359-1. [DOI] [PubMed] [Google Scholar]
- 18.Rai RK, Azim A, Sinha N, Sahoo JN, Singh C, Ahmed A, Saigal S, Baronia AK, Gupta D, Gurjar M, Poddar B, Singh RK. Metabolic profiling in human lung injuires by high-resolution nuclear magnetic resonance spectroscopy for bronchoalveolar lavage fluid (BALF) Metabolomics. 2013;9:667–676. [Google Scholar]
- 19.Sha W, da Costa KA, Fischer LM, Milburn MV, Lawton KA, Berger A, Jia W, Zeisel SH. Metabolomic profiling can predict which humans will develop liver dysfunction when deprived of dietary choline. The FASEB Journal. 2010;24(8):2962–2975. doi: 10.1096/fj.09-154054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorenz MA, Burant CF, Kennedy RT. Reducing Time and Increasing Sensitivity in Sample Preparation for Adherent Mammalian Cell Metabolomics. Analytical Chemistry. 2011;83(9):3406–3414. doi: 10.1021/ac103313x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pesek JJ, Matyska MT, Fischer SM, Sana TR. Analysis of hydrophilic metabolites by high-performance liquid chromatography-mass spectrometry using a silica hydride-based stationary phase. J Chromatogr A. 2008;1204(1):48–55. doi: 10.1016/j.chroma.2008.07.077. [DOI] [PubMed] [Google Scholar]
- 22.Westerhuis JA, van Velzen EJJ, Hoefsloot HCJ, Smilde AK. Discriminant Q(2) (DQ(2)) for improved discrimination in PLSDA models. Metabolomics. 2008;4(4):293–296. [Google Scholar]
- 23.Lindgren F, Hansen B, Karcher W, Sjostrom M, Eriksson L. Model validation by permutation tests: Applications to variable selection. J Chemometrics. 1996;10(5–6):521–532. [Google Scholar]
- 24.Storey JD. A direct approach to false discovery rates. J R Stat Soc Series B. 2002;64:479–498. [Google Scholar]
- 25.Storey JD, Taylor JE, Siegmund D. Strong control, conservative point estimation, and simultaneous conservative consistency of false discovery rates: A unified approach. J R Stat Soc Series B. 2004;66:187–205. [Google Scholar]
- 26.Karnovsky A, Weymouth T, Hull T, Tarcea VG, Scardoni G, Laudanna C, Sartor MA, Stringer KA, Jagadish HV, Burant C, Athey B, Omenn GS. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012;28(3):373–380. doi: 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agassandian M, Mallampalli RK. Surfactant phospholipid metabolism. Biochim Biophys Acta. 2013;1831(3):612–625. doi: 10.1016/j.bbalip.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vulimiri SV, Misra M, Hamm JT, Mitchell M, Berger A. Effects of mainstream cigarette smoke on the global metabolome of human lung epithelial cells. Chem Res Toxicol. 2009;22(3):492–503. doi: 10.1021/tx8003246. [DOI] [PubMed] [Google Scholar]
- 29.Dalli J, Serhan CN. Specific lipid mediator signatures of human phagocytes: microparticles stimulate macrophage efferocytosis and pro-resolving mediators. Blood. 2012;120(15):e60–e72. doi: 10.1182/blood-2012-04-423525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickson RP, Erb-Downward JR, Huffnagle GB. The role of the bacterial microbiome in lung disease. Expert Rev Respir Med. 2013;7(3):245–257. doi: 10.1586/ers.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang YJ, Charlson ES, Collman RG, Colombini-Hatch S, Martinez FD, Senior RM. The role of the lung microbiome in health and disease. A national heart, lung, and blood institute workshop report. Am J Respir Crit Care Med. 2013;187(12):1382–1387. doi: 10.1164/rccm.201303-0488WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho WE, Xu YJ, Xu F, Cheng C, Peh HY, Tannenbaum SR, Wong WS, Ong CN. Metabolomics reveals altered metabolic pathways in experimental asthma. Am J Respir Cell Mol Biol. 2013;48(2):204–211. doi: 10.1165/rcmb.2012-0246OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones AE, Shapiro NI, Trzeciak S, Arnold RC, Claremont HA, Kline JA. Lactate clearance vs central venous oxygen saturation as goals of early sepsis therapy: a randomized clinical trial. Jama. 2010;303(8):739–746. doi: 10.1001/jama.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borthwick HA, Brunt LK, Mitchem KL, Chaloner C. Does lactate measurement performed on admission predict clinical outcome on the intensive care unit? A concise systematic review. Ann Clin Biochem. 2012;49(Pt 4):391–394. doi: 10.1258/acb.2011.011227. [DOI] [PubMed] [Google Scholar]
- 35.Iscra F, Gullo A, Biolo G. Bench-to-bedside review: lactate and the lung. Crit Care. 2002;6(4):327–329. doi: 10.1186/cc1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Routsi C, Bardouniotou H, Delivoria-Ioannidou V, Kazi D, Roussos C, Zakynthinos S. Pulmonary lactate release in patients with acute lung injury is not attributable to lung tissue hypoxia. Crit Care Med. 1999;27(11):2469–2473. doi: 10.1097/00003246-199911000-00024. [DOI] [PubMed] [Google Scholar]
- 37.Tonks A, Parton J, Tonks AJ, Morris RH, Finall A, Jones KP, Jackson SK. Surfactant phospholipid DPPC downregulates monocyte respiratory burst via modulation of PKC. Am J Physiol Lung Cell Mol Physiol. 2005;288(6):L1070–L1080. doi: 10.1152/ajplung.00386.2004. [DOI] [PubMed] [Google Scholar]
- 38.Wacharasint P, Nakada TA, Boyd JH, Russell JA, Walley KR. Normal-range blood lactate concentration in septic shock is prognostic and predictive. Shock. 2012;38(1):4–10. doi: 10.1097/SHK.0b013e318254d41a. [DOI] [PubMed] [Google Scholar]
- 39.Gasse P, Riteau N, Charron S, Girre S, Fick L, Petrilli V, Tschopp J, Lagente V, Quesniaux VF, Ryffel B, Couillin I. Uric acid is a danger signal activating NALP3 inflammasome in lung injury inflammation and fibrosis. Am J Respir Crit Care Med. 2009;179(10):903–913. doi: 10.1164/rccm.200808-1274OC. [DOI] [PubMed] [Google Scholar]
- 40.Kono H, Chen CJ, Ontiveros F, Rock KL. Uric acid promotes an acute inflammatory response to sterile cell death in mice. J Clin Invest. 2010;120(6):1939–1949. doi: 10.1172/JCI40124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuipers MT, Aslami H, Vlaar AP, Juffermans NP, Tuip-de Boer AM, Hegeman MA, Jongsma G, Roelofs JJ, van der Poll T, Schultz MJ, Wieland CW. Pre-treatment with allopurinol or uricase attenuates barrier dysfunction but not inflammation during murine ventilator-induced lung injury. PLoS One. 2012;7(11):e50559. doi: 10.1371/journal.pone.0050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olson JS, Ballow DP, Palmer G, Massey V. The reaction of xanthine oxidase with molecular oxygen. J Biol Chem. 1974;249(14):4350–4362. [PubMed] [Google Scholar]
- 43.Pinamonti S, Muzzoli M, Chicca MC, Papi A, Ravenna F, Fabbri LM, Ciaccia A. Xanthine oxidase activity in bronchoalveolar lavage fluid from patients with chronic obstructive pulmonary disease. Free Radic Biol Med. 1996;21(2):147–155. doi: 10.1016/0891-5849(96)00030-5. [DOI] [PubMed] [Google Scholar]
- 44.Hassoun PM, Yu FS, Cote CG, Zulueta JJ, Sawhney R, Skinner KA, Skinner HB, Parks DA, Lanzillo JJ. Upregulation of xanthine oxidase by lipopolysaccharide, interleukin-1, and hypoxia. Role in acute lung injury. Am J Respir Crit Care Med. 1998;158(1):299–305. doi: 10.1164/ajrccm.158.1.9709116. [DOI] [PubMed] [Google Scholar]
- 45.Abdulnour RE, Peng X, Finigan JH, Han EJ, Hasan EJ, Birukov KG, Reddy SP, Watkins JE, 3rd, Kayyali US, Garcia JG, Tuder RM, Hassoun PM. Mechanical stress activates xanthine oxidoreductase through MAP kinase-dependent pathways. Am J Physiol Lung Cell Mol Physiol. 2006;291(3):L345–L353. doi: 10.1152/ajplung.00453.2005. [DOI] [PubMed] [Google Scholar]
- 46.Kikuchi K, Itoh Y, Tateoka R, Ezawa A, Murakami K, Niwa T. Metabolomic analysis of uremic toxins by liquid chromatography/electrospray ionization-tandem mass spectrometry. J Chromatogr B. 2010;878(20):1662–1668. doi: 10.1016/j.jchromb.2009.11.040. [DOI] [PubMed] [Google Scholar]
- 47.Vanholder R, De Smet R, Glorieux G, Argiles A, Baurmeister U, Brunet P, Clark W, Cohen G, De Deyn PP, Deppisch R, Descamps-Latscha B, Henle T, Jorres A, Lemke HD, Massy ZA, Passlick-Deetjen J, Rodriguez M, Stegmayr B, Stenvinkel P, Tetta C, Wanner C, Zidek W. Review on uremic toxins: classification, concentration, and interindividual variability. Kidney Int. 2003;63(5):1934–1943. doi: 10.1046/j.1523-1755.2003.00924.x. [DOI] [PubMed] [Google Scholar]
- 48.Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3(5):431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 49.Aronov PA, Luo FJ, Plummer NS, Quan Z, Holmes S, Hostetter TH, Meyer TW. Colonic contribution to uremic solutes. J Am Soc Nephrol. 2011;22(9):1769–1776. doi: 10.1681/ASN.2010121220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watanabe H, Miyamoto Y, Otagiri M, Maruyama T. Update on the pharmacokinetics and redox properties of protein-bound uremic toxins. J Pharm Sci. 2011;100(9):3682–3695. doi: 10.1002/jps.22592. [DOI] [PubMed] [Google Scholar]
- 51.Itoh Y, Ezawa A, Kikuchi K, Tsuruta Y, Niwa T. Protein-bound uremic toxins in hemodialysis patients measured by liquid chromatography/tandem mass spectrometry and their effects on endothelial ROS production. Anal Bioanal Chem. 2012;403(7):1841–1850. doi: 10.1007/s00216-012-5929-3. [DOI] [PubMed] [Google Scholar]
- 52.McClay JL, Adkins DE, Isern NG, O'Connell TM, Wooten JB, Zedler BK, Dasika MS, Webb BT, Webb-Robertson BJ, Pounds JG, Murrelle EL, Leppert MF, van den Oord EJ. (1)H nuclear magnetic resonance metabolomics analysis identifies novel urinary biomarkers for lung function. J Proteome Res. 2010;9(6):3083–3090. doi: 10.1021/pr1000048. [DOI] [PubMed] [Google Scholar]
- 53.Cole M, Kenig MD, Hewitt VA. Metabolism of penicillins to penicilloic acids and 6-aminopenicillanic acid in man and its significance in assessing penicillin absorption. Antimicrob Agents Chemother. 1973;3(4):463–468. doi: 10.1128/aac.3.4.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Favetta P, Degoute CS, Perdrix JP, Dufresne C, Boulieu R, Guitton J. Propofol metabolites in man following propofol induction and maintenance. Br J Anaesth. 2002;88(5):653–658. doi: 10.1093/bja/88.5.653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.