Abstract
The human multidrug and toxin extrusion (MATE) transporter 1 contributes to the tissue distribution and excretion of many drugs. Inhibition of MATE1 may result in potential drug-drug interactions (DDIs) and alterations in drug exposure and accumulation in various tissues. The primary goals of this project were to identify MATE1 inhibitors with clinical importance or in vitro utility and to elucidate the physicochemical properties that differ between MATE1 and OCT2 inhibitors. Using a fluorescence assay of ASP+ uptake in cells stably expressing MATE1, over 900 prescription drugs were screened and 84 potential MATE1 inhibitors were found. We identified several MATE1 selective inhibitors including four FDA-approved medications that may be clinically relevant MATE1 inhibitors and could cause a clinical DDI. In parallel, a QSAR model identified distinct molecular properties of MATE1 versus OCT2 inhibitors and was used to screen the DrugBank in silico library for new hits in a larger chemical space.
Keywords: MATE1, MATE2-K, OCT2, SLC47A1, SLC47A2, SLC22A2, prescription drug library, HTS, iterative modeling, membrane transporters, QSAR
INTRODUCTION
Transporters on the plasma membrane of cells are major determinants of drug absorption, elimination, and accumulation in various tissues. In the past decade, many studies have been devoted to understanding and characterizing transporters involved in pharmacokinetic processes, primarily in the intestine, kidney, and liver (e.g., P-gp, OATs, OATPs).1 Information from these studies has enhanced our understanding of drug absorption and elimination phenomena.2 In addition, transporter-related drug accumulation and drug toxicity studies have formed the basis for understanding and predicting clinically important drug-drug interactions (DDIs). With respect to the kidney, several solute carrier transporters (SLCs) for both anions (e.g., OAT1 and OAT3) and cations (e.g., OCT2, MATE1, and MATE2-K) have been shown to be of clinical importance for drug elimination.1,3–5 This manuscript focuses on the human multidrug and toxin extrusion transporters MATE1 and MATE2-K as well as the organic cation transporter OCT2 which are important in the disposition of positively charged drugs.
The MATE transporter family consists of two members, namely MATE1 (SLC47A1) and MATE2 (SLC47A2), and has only relatively recently been described.6,7 Both transporters contribute to the urinary excretion of structurally diverse organic cations at the apical membrane of renal cells. MATE1 is widely expressed in body tissues, including the kidney, liver, skeletal muscle, adrenal gland, and testis. In the kidney, MATE1 is localized to the apical membrane of the proximal and distal convoluted tubule.6 Three major splice variants of MATE2 have been identified, MATE2, MATE2-B, and MATE2-K. Of these, only MATE2 and MATE2-K demonstrate functional activity in cell-based experiments.7,8 MATE2-K consists of only 566 amino acid residues (as opposed to 602 amino acid residues in MATE2) and is almost exclusively expressed in the apical membrane of human proximal tubule. Both MATE1 and MATE2-K act as electroneutral exchangers of their substrates with an oppositely directed proton gradient as a driving force.
Of the organic cation transporters OCT1 through 3 (SLC22A1, SLC22A2, and SLC22A3), OCT2 is most strongly expressed in the kidney while OCT1 is predominantly expressed in the liver and OCT3 is expressed in many different tissues.3 In epithelia, OCTs are generally localized to the basolateral membrane and mediate the uptake of organic cations into the cells by facilitated diffusion driven by an inside-negative membrane potential. In the kidney, OCTs and MATEs have been shown to act concertedly in the excretion of organic cations. Their substrate and inhibitor specificity are supposed to overlap heavily.9,10
Recent studies to identify and compare the affinity and potency of MATE and OCT substrates and inhibitors have been published (for a review see Nies et al.).3 Several articles concerning ligand selectivity for MATE1 compared to MATE2-K have been published using different methodologies, ranging from classical experimental procedures11 to pharmacophore-based ligand identification.12 These studies, which provide interesting models for predicting inhibitors of MATEs, were based on small libraries with a maximum of about 60 compounds and were not focused on prescription drugs. Previous studies to identify inhibitors of OCT1 and OCT2 are more extensive with screens that included larger numbers of inhibitors and many prescription drugs.13–15 However, little work has been performed on substrate and inhibitor overlap between OCTs and MATEs. Even though few examples of selective inhibitors have been described16 more probes for both in vitro and in vivo applications are required.
Therefore, the primary goal of the current study was to use high throughput screening (HTS) to identify inhibitors of MATE1 that can be used as in vitro and in vivo probes. The screen was complemented by a quantitative structure-activity relationship (QSAR) model based on the random forest (RF) methodology17 for the prediction of MATE1 inhibitors. This approach led to the identification of novel potent and selective inhibitors of MATE1. A special emphasis was placed on drugs that may potentially cause clinical drug-drug interactions. The International Transporter Consortium (ITC) has issued guidelines for selected transporters (OCT2, P-gp, BCRP, OAT1, OAT3, OATP1B1, OATP1B3) that define when a clinical DDI study should be conducted.2 According to these guidelines, if the ratio Cmax,unbound /IC50 is greater than or equal to 0.1 then a clinical DDI study should be performed. Although, to date, no guidelines for MATEs are available, the ITC is considering making similar recommendations for these transporters. Therefore, in this manuscript we used the threshold of ≥ 0.1 to identify drugs that may cause clinically significant DDIs.
A secondary goal was to compare properties of inhibitors of MATE1 with those of OCT2 which was screened in a previously published study from our laboratory.15 This study provides novel insights into the inhibitor specificity profiles of organic cation transporters, including their charge selectivity and required physicochemical properties.
RESULTS
High Throughput Screen for MATE1-Inhibitors with ASP+ as Fluorescent Probe
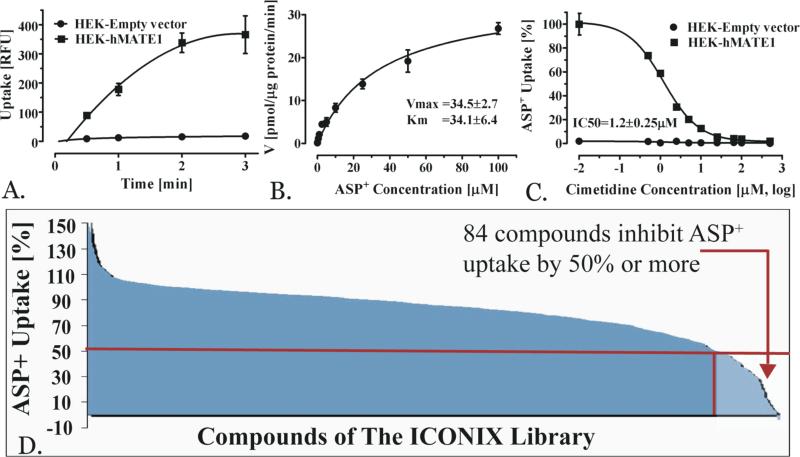
A high throughput screening (HTS) to identify inhibitors of MATE1 was performed using the fluorescent probe ASP+. The uptake assay of ASP+ in cells over-expressing MATE1 was characterized and optimal experimental conditions were derived (i.e., duration of the uptake experiment and ASP+ concentration; Methods) (Figures 1A and 1B). In particular, 1.5 minutes was selected to perform the screen because it was in the linear range of transport (Figure 1A). An ASP+ concentration of 2 μM was selected for the screening studies because it was below the Km of 34 μM (Figure 1B). The functionality of the assay for inhibition studies was confirmed by determining the IC50 value of cimetidine, a well-established MATE1 inhibitor. The IC50 was 1.2 ± 0.25 μM which is in agreement with published data6,18,19 (Figure 1C). For the screening, the probe uptake was determined in the presence of compounds of the ICONIX library which contains 910 FDA approved drugs from 124 therapeutic classes.15 We identified 84 compounds that exhibited 50% or more inhibition of ASP+ uptake at 20 μM. These compounds were considered as strong inhibitors (hits) of MATE1 (Figure 1D) and selected compounds were subjected to follow-up IC50 determinations.
Figure 1.
Methods development and results of high throughput screening. Time-course (A) and kinetics (B) of probe uptake (ASP+) in HEK-cells stably overexpressing MATE1. Uptake data are presented as mean±SEM (N=3). Inhibition of ASP+ uptake (C) by the model-inhibitor cimetidine. ICONIX library HTS results (D) in overview. The red line depicts the threshold of less than 50% ASP+ uptake at 20 μM inhibitor concentration. The red arrow marks the 84 compounds with less than 50% maximum ASP+ uptake; these compounds were considered as hits.
Physicochemical Properties of MATE1 HTS Hits and Comparison to OCT2 HTS Hits
The MATE1 inhibitors identified in the present study were compared to the OCT2 inhibitors identified previously using the same fluorescent probe and the same library.15 Interestingly, OCT2 exhibited a broader inhibition pattern (Figure 2A) and considerably fewer compounds were found to inhibit MATE1 as compared to OCT2 (Figure 2B). Figure 2A shows the amount of compounds per percent inhibition and is thus reflective of the distribution of inhibitors and non-inhibitors over the dataset. For example, it becomes apparent that not only there are more OCT2-inhibitors but also that they have more variable percent-inhibition compared to MATE1 inhibitors. 45 selective MATE1 inhibitors that did not affect OCT2 transport were identified based on the screening results. In contrast, 205 selective OCT2 inhibitors and 39 dual MATE1/OCT2 inhibitors were found (Figure 2B).
Figure 2.
Comparison of MATE1 and OCT2 inhibitors. A) Density-plot of number of inhibitors per % inhibition. B) Number of MATE1 selective (red) and OCT2 selective (blue) inhibitors as well as inhibitors of both transporters (overlap; green) as identified by screening the ICONIX library (898 compounds in total for which properties could be computed). C) Principal component analysis of the complete ICONIX library (black circles), MATE1 selective inhibitors (red circles), OCT2 selective inhibitors (blue squares), and dual inhibitors (green triangles). The first principal component is governed by polarity, size and hydrogen bonding while the second principal component is described by lipophilicity.
The existence of distinct groups of transporter inhibitors suggests that elucidating differences in structural and physicochemical properties may help in understanding the properties that lead to transporter-selectivity. We therefore used several computational methods to characterize and differentiate MATE1 or OCT2 selective inhibitors as well as inhibitors of both transporters (dual inhibitors) and non-inhibitors.
First, to examine differences between these groups, we performed a principal component analysis (PCA) and compared OCT2 and MATE1 selective inhibitors to dual inhibitors and to the whole ICONIX-library (Figure 2C). More specifically, we analyzed various physicochemical properties, such as molecular weight, molecular volume, number of heavy atoms, number of rotatable bonds, number of hydrogen bond donors and acceptors, SLogP, topological polar surface area and charge at pH 7.4 (Supplementary Table 2). The first two principal components (PC1 and PC2 in Figure 2C) explain ~70% of variability in physicochemical properties between transporter selective inhibitors and dual inhibitors or non-inhibitors. The first principal component is governed by polarity, size and hydrogen bonding, the second principal component is mainly represented by lipophilicity. The bottom right quadrant in Figure 2C, which contained large, hydrophobic, positively charged compounds, contained a large number of dual and MATE1 selective inhibitors. Smaller, positively charged compounds cluster in the bottom left quadrant. Compounds in the upper left quadrant are negatively charged. The PCA analysis proved helpful in identifying interesting compounds for further testing and, in particular, compounds that showed to be selective MATE1 or OCT2 inhibitors. Many promising inhibitors with strong inhibition values in the screen, such as indinavir, dihydroergotamine, and vecuronium bromide, clustered in the lower right quadrant. Leucomycin was selected for further testing based on proximity to these compounds. Follow-up testing revealed that this drug was not only a MATE1 inhibitor but was also selective over MATE2-K and OCT2 (Table 1). The cluster in the lower left quadrant contained many compounds with medium to high IC50 values, such as ethinyl estradiol, but also some highly active inhibitors like rimantadine. Finally, the upper left quadrant contained many true negative and false positive drugs with respect to MATE1 inhibition, including niacin and ciclopirox. The latter was selected for a confirmatory experiment based on its association with this cluster.
Table 1.
Overview of IC50 Results for Selected HTS Hits.
| Compound | ICONIX Screen [% inhibition at 20 μM±SD] | prIC50 [μM] | MATE1 [IC50 μM, (95% CI)] | MATE2K [IC50 μM, (95% CI)] | OCT2 [IC50 μM, (95% CI)] |
|---|---|---|---|---|---|
| Epinastine | 97±3 | 0.6 | 1.1 (0.8-1.6) | 29.8 (22.4-39.6) | 4.3 (2.2-8.3) |
| Gabaxate | 96±1 | 0.8 | 0.5 (0.4-0.7) | 10.8 (7.9-14.7) | 0.9 (0.6-1.5) |
| Tubocurarine | 95±7 | 1.1 | 9.4 (6.2-14.4) | 55.5 (36.8-83.8) | 78.8 (35.4-175.5) |
| Bithionol | 87±7 | 3.0 | 5.2 (3.1-7.2) | 6.6 (3.5-12.4) | 1.9 (1.3-2.8) |
| Chlorhexidine | 86±1 | 3.1 | 0.7 (0.6-1.0) | 0.5 (0.4-0.6) | 0.4 (0.2-0.6) |
| Vecuronium bromide | 85±2 | 3.4 | 1.9 (1.5-2.5) | 25.2 (20.8-30.4) | 3.5 (2.7-4.6) |
| Camostat | 77±7 | 5.9 | 2.9 (2.3-3.8) | 12.7 (9.1-17.7) | 16.5 (13.0-21.0) |
| Rimantadine | 74±12 | 7.0 | 7.3 (5.3-10.1) | 288 (149-559) | 4.4 (3.2-6.1) |
| Prazosin | 74±1 | 7.0 | 1.6 (1.3-2.0) | 38.4 (23.6-62.5) | 13.6 (8.5-21.6) |
| Dihydroergotamine | 72±2 | 8.0 | 2.8 (2.2-3.7) | 12.6 (10.5-15.0) | 49.9 (33.0-75.4) |
| Buspirone | 70±9 | 8.4 | 1.7 (1.2-2.4) | 46 (29.8-72.1) | 12.0 (9.3-15.5) |
| Granisetron | 67±7 | 9.9 | 5.0 (3.7-6.9) | 311 (208-467) | 4.3 (3.3-5.8) |
| Domperidone | 66±2 | 10.2 | 2.3 (1.8-2.9) | 14.8 (11.9-18.4) | 7.9 (5.2-11.8) |
| Ethinyl estradiol | 60±19 | 13.5 | 21.1 (13.7-32.4) | 20.2 (13.6-30.1) | 2.2 (0.99-4.7) |
| Leucomycin | 56±1 | 16.0 | 11.1 (8.3-14.8) | 125 (52.0-298) | 182 (113-294) |
| Zafirlukast | 52±12 | 19 | 4.3 (3.1-5.8) | 7.6 (4.8-12.2) | 9.7 (7.6-12.4) |
| Topotecan | 50±6 | 19.7 | 1.3 (1.0-1.7) | 8.6 (7.4-10.1) | 61 (28.3-132) |
Overview of screening hits that were tested based on predicted IC50 values (prIC50) and their expected Cmax, unbound /IC50 as well as on their pharmacological interest and/or their supposed propensity to inhibit other cation transporters (based on published data if available). Note that these compounds are not expected to inhibit MATE1 at their clinical concentrations.
As a second analysis, we examined the physicochemical and structural properties of the inhibitors of MATE1, OCT2 or dual inhibitors, compared with the non-inhibitors (Figure 3A-E). No statistically significant differences were observed for the numbers of H-bond donors, H-bond acceptors or rotatable bonds (Supplementary Figure 1). With respect to molecular weight, the number of heavy atoms and the molecular volume, MATE1 selective inhibitors (but not OCT2 selective inhibitors or inhibitors of both transporters) showed statistically significant (p < 0.05) higher values compared to non-inhibitors. Highly statistically significant differences (p < 0.001) were observed for SLogP, topological polar surface area (TPSA), and charge. In particular, all three groups exhibited significantly higher values of SLogP, a measure of lipophilicity, in comparison to non-inhibitors. Interestingly, TPSA values were lower for OCT2 selective inhibitors compared to both MATE1 selective inhibitors and non-inhibitors. Finally, at pH 7.4 positively charged compounds appeared more frequently in the groups of OCT2-selective and dual inhibitors compared to the non-inhibitor group.
Figure 3.
Comparison of physicochemical parameters for different groups of inhibitors. A-E: Comparison of physicochemical parameters calculated for MATE1 selective inhibitors (red), OCT2 selective inhibitors (blue), dual inhibitors (OCT2/MATE1, green), and non-inhibitors (gray). # marks a statistically significant difference in comparison to the non-inhibitor group as described in the text. F-J: Distribution plots of basic (BpKa > 5.4), acidic (ApKa < 9.4), neutral (ApKa > 9.4 and BpKa < 5.4), zwitterionic (ApKa < 9.4 and BpKa > 5.4), and unknown drugs within the groups described above.
For the third analysis, we binned the compounds of each group (i.e., MATE1 selective inhibitors, OCT2 selective inhibitors, dual inhibitors, and non-inhibitors) into bases, acids, zwitterions, neutral, and unknown (Figure 3F-J). As expected for cation transporters, such as MATE1 and OCT2, bases were overrepresented in the inhibitor groups compared to the whole ICONIX library. The fraction of inhibitors that were bases was highly enriched for OCT2 selective (p < 1×10−14) and dual inhibitors (p < 1×10−7) in comparison to the ICONIX library. Bases were also over-represented among the MATE1 selective inhibitors, but at lower significance levels (p < 0.05). Interestingly, zwitterions (e.g., famotidine, telmisartan) were over represented in the OCT2 selective inhibitor group (p < 0.01), but not in the other groups. As expected, acids were overrepresented in the non-inhibitor groups to a highly statistically significant extent (p < 1×10−16), and the same held true for neutral compounds though the significance level was much lower (p < 0.05).
Validation of HTS Screen by Follow-up IC50 Determination
To test the quality of the screening assay as well as to enhance and validate the in silico model development, we determined the IC50 values of various promising drugs against MATE1, MATE2-K, and OCT2 (Table 1). These drugs were selected based on their predicted MATE1 IC50 values (prIC50), their pharmacological interest (e.g., novelty or how commonly they are used) and/or their potential to inhibit other cation transporters (based on published data). Interestingly, for the library used in this study, we identified more compounds that exhibited MATE1 selectivity over MATE2-K than over OCT2 (e.g., gabaxate, granisetron, rimantadine, vecuronium bromide). In Table 1, only topotecan seems to have a slight preference for both MATEs compared to OCT2. Furthermore, ethinyl estradiol proved to be a better OCT2 than MATE inhibitor and chlorhexidine as well as bithionol were pan-inhibitors, i.e., inhibiting MATE1, MATE2-K, and OCT2. Moreover, the IC50 values for OCT1 and OCT3 of chlorhexidine were 0.21 and 0.41 and for bithionol 3.9 and 5.5 μM, respectively (data not shown). Nevertheless, selective inhibitors for MATE1 were identified and include prazosine, buspirone, leucomycine, and the drugs inhibiting at clinically relevant concentrations discussed below. Many of the compounds in Table 1 have not, to the best of our knowledge, been described as inhibitors of organic cation transporters (e.g., domperidone, camostat, dihydroergotamine, epinastine, gabaxate) or have only been shown to interact with the OCTs but not the MATEs (e.g., prazosin, granisetron). It is noteworthy that some compounds in Table 1 were close to clinical significance. For example, tubocurarine has a Cmax,unbound /IC50 of 0.07, which is near the threshold of 0.1.
Development and Utilization of Computational Model (RF-Model-I) to Ratify and Complement HTS Results
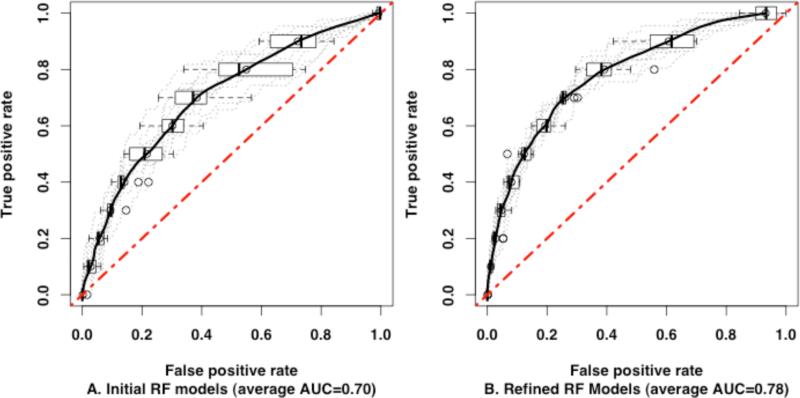
To aid in the targeted validation of the screening hits, we developed an RF classification model of MATE1 inhibitors and non-inhibitors as identified in the HTS. We estimated the performance of the RF algorithm by means of internal cross-validation, using out-of-bag samples, and external validation using ten test sets (Methods). The Receiver Operating Characteristic (ROC) curves for external validation tests are shown in Figure 4A. The average area under the ROC curve (AUC) for the ten tests was 0.70 ±0.05 (permutation test p-value < 0.0001), indicating good quality models. For comparison, we built MATE1 classification models using a k-nearest neighbor (kNN) algorithm and a partial least-squares discriminant analysis (PLS-DA) algorithm. The kNN algorithm is a simple but effective method for similarity clustering and classification. The PLS-DA technique was evaluated because it was successfully used in QSAR modeling of other transporters.15,20 The average AUCs for the kNN (AUC=0.62 ±0.05) and PLS-DA (AUC=0.66 ±0.04) models were significantly lower than for the RF models (Supplementary Figure 2 and Supplementary Table 3).
Figure 4.
Receiver Operating Characteristic curves for ten external data sets. ROC curves of each test are plotted in dotted gray lines and the average ROC curves for the ten tests, in solid black lines. The ROC curve of a random classification is plotted as a red dotted diagonal line. A: ROC curves of the initial RF models. The average AUC is 0.70. B: ROC curves of the refined RF models. The average AUC is 0.78.
Next, we used the entire HTS data set to train an RF model (RF-Model-I), which we employed in several ways. First, we tested the model's ability to rescue false negative compounds with respect to MATE1 inhibition in the screen. As Table 2A illustrates, the model performed well at this task over a broad range of screening-based inhibition values (5-49% inhibition). Remarkable examples were ranitidine and irinotecan, which in the follow-up experiments inhibited MATE1 in the low micromolar range but did not inhibit ASP+ uptake by more than 50% in the screen. The only incorrectly predicted false negative compound was methyl ergonovine, which was surprising because it belongs to the ergot alkaloid-family, which includes several inhibitors of MATE1 (e.g., dihydroergotamine). The rescued negative hits were tested for inhibition of other transporters and several were OCT2 selective. Both, carvedilol and noscapine had IC50 values against OCT2 of 7.5 and 2.6 μM, respectively, and did not inhibit MATE2-K up to 500 μM (data not shown). Although we did not find any additional selective and/or clinically relevant inhibitors, this approach proved useful in rescuing otherwise overlooked inhibitors.
Table 2.
Results of Random Forest-Model-I (RF-Model-I) Validation.
| A.Rescued False Negative Compounds Based on QSAR-Model-I Predictions | |||
|---|---|---|---|
| Compound | Score in Computational Model-I [position out of 898] | ICONIX-Screen [% inhibition] | Actual IC50 [μM] |
| Irinotecan | 3 | 49±13 | 7.9 (5.8-10.7) |
| Carvedilol | 10 | 21±8 | 92.4 (62.0-138) |
| Methyl ergonovine* | 16 | 12±27 | >250 |
| Telmisartan | 18 | 38±10 | 17.9 (14.4-22.2) |
| Astemizole | 23 | 34±9 | 26.4 (18.8-37.1) |
| Noscapine | 26 | 13.4±34 | 34.5 (22.3-53.6) |
| Amantadine | 35 | 5±9 | 37.8 (20.4-70.0) |
| Ranitidine | 118 | 29±14 | 8.3 (5.1-13.5) |
| B.Identification of False-Positive Compounds by QSAR-Model-I Predictions | |||
|---|---|---|---|
| Compound | Score in Computational Model-I [position out of 898] | ICONIX-Screen [% inhibition] | Actual IC50 [μM] |
| Azacitidine | 861 | 50±15 | >500 |
| Ciclopirox | 860 | 52±14 | >500 |
| Sulfacetamide | 842 | 80±4 | >500 |
| Phenacetin | 806 | 53±7 | >250 |
| Niacin | 622 | 54±9 | >500 |
| Thiabendazole | 488 | 55±14 | >500 |
| C.True Negative Compounds in Both QSAR-Model-I and ICONIX Library Screen | |||
|---|---|---|---|
| Compound | Score in Computational Model-I [position out of 898] | ICONIX-Screen [% inhibition] | Actual ICS0 [μM] |
| Enalapril | 355 | 23±18 | >500 |
| Cetirizine | 376 | 2±5 | >500 |
| Diclofenac | 445 | 12±5 | >500 |
| Indomethacin | 454 | 6±6 | >500 |
| Lidocaine | 556 | 7±13 | >500 |
| Furosemide | 617 | 7±6 | >500 |
| Erythromycin | 624 | 7±4 | >500 |
| Dexamethasone | 862 | 30±4 | >500 |
| D.Mismatched Compounds Based on QSAR-Model-I Predictions | |||
|---|---|---|---|
| Compound | Computational Model-I [position out of 898] | ICONIX-Screen [% inhibition] | Actual IC50 [μM] |
| Irbesartan | 71 | 15±4 | >500 |
| Losartan | 73 | 12±5 | >250 |
| Spironolactone | 196 | 27±9 | 18.6 (11.8-29.3) |
| Levofloxacin | 312 | 26±8 | 17.9 (11.5-27.9) |
| Quinidine | 399 | 28±6 | 11.2 (7.2-17.6) |
| Clonidine | 485 | 1±25 | 33.3 (20.9-52.9) |
| Cortisone | 528 | 10±12 | 21.4 (10.8-42.4) |
| Sumatriptan | 581 | 7±7 | 6.7 (3.9-11.6) |
Ranks 1 to 120 were considered as likely hits based on their RF-score. A) Compounds that were negative in the screen but have been rescued based on their RF-model-I values (ranked position 1 to 120).
Methyl ergonovine was the only wrongly predicted of the tested rescue compounds.
B) Compounds that were identified as false positive by RF-Model-I (ranked lower than position 120). C) True negative-compounds that were negative in the screen and correctly predicted negative by RF-Model-I. D) Compounds that were mispredicted by the model as either positive (i.e., both sartans) or negative. Note that in the latter case the compounds were not hits in the screen either but were selected based on published data. The results shown in this Table were used to train RF-Model-I and to achieve an improved model (RF-Model-II).
Second, we tested the model's ability to predict false positive screening hits. To do so, we studied compounds that had been assigned a very low score by RF model-I despite >50% inhibition in the initial screen. As Table 2B shows, the model performed well at this task. It is noteworthy that compounds with IC50 values above 100 μM (e.g., phenacetin) might still be substrates, because the screening methodology used in this study is primarily powered to identify inhibitors. For example, when we assessed the inhibition of ASP+ uptake by metformin, we found an IC50 value of 250 μM (data not shown).
Third, we assessed the model's performance in identifying true negative compounds. Table 2C illustrates that the model is also suited well for this task, over a wide array of compounds including cations (e.g., lidocaine) and zwitterions (e.g., furosemide).
Finally, we also identified mismatched compounds based on the RF-Model-I predictions, i.e., compounds that were either predicted to be inhibitors by the model but could not be confirmed experimentally (e.g., irbesartan) or inhibitors confirmed by experiments that were not detected by the model (e.g., propranolol). Table 2D gives an overview of this compound category.
We used the feedback from the experiments to refine the RF models and to generate an improved version (i.e., RF-Model-II; see next paragraph) for further use. Upon refinement with experimental results, the performance of the RF models significantly improved, as assessed in ten external tests (Figure 4A-B). The average AUC of the refined RF models increased to 0.78 ±0.02 (Student's t-test p-value = 0.0003812). Again, we compared the performance of the RF algorithm to those for the kNN and PLS methods. Although the AUCs improved for all three methods, our analysis shows that the RF models performed better in external validation tests (Supplementary Table 3 and Supplementary Figure 2).
Use of RF-Model-II to Screen a Large in silico Library
After improving the initial model based on feedback from experimental testing, we used the new QSAR model (RF-Model-II) for screening the DrugBank in silico library21–23 to explore a larger chemical space than represented by the ICONIX library. Because the applicability domain of the RF model is limited to molecules with physicochemical properties similar to those for the training data set,24–27 we projected the DrugBank compounds onto the principal components’ space of the ICONIX library. We then removed DrugBank molecules with physicochemical properties outside of the 95% confidence interval of the first two principal components computed for the ICONIX library (Supplementary Figure 3).
Next, we classified the remaining 6,122 DrugBank compounds using our RF-Model-II. Five in silico hits, selected based on their diversity from ICONIX-compounds, their novelty, and on their therapeutic interest, were tested successfully as inhibitors (Table 3). Four of the five selected compounds were MATE1 selective inhibitors, consistent with our model. Only pimozide was a false positive. Risperidone and maraviroc were potent and selective MATE1 inhibitors. Even though not fulfilling our criteria for clinical significance, maraviroc gets close (Cmax,unbound /IC50 = 0.02 with 300 mg b.i.d.). In addition, concentrations in the liver (where MATE1 is also expressed) might be even higher due to the elevated concentrations in the portal vein. Maraviroc seems especially intriguing because it is a novel drug from a new therapeutic class (CCR5 receptor-antagonist/entry inhibitor) used to treat HIV-infections.23,24 To confirm that none of the five in silico hits could be found using 2D similarity and substructure searches, we used DrugBank's web interface to retrieve all compounds similar to MATE1 inhibitors. Each MATE1 inhibitor was used as a query for the search. We used 0.6 as a threshold for the Tanimoto similarity coefficient (Tc), which is the default search option in DrugBank. Only pimozide was found using 2D similarity and substructure searching. The pairwise similarity coefficients of in silico hits ranged from below 0.3 (maraviroc and dapiprazole) to 0.724 (pimozide and droperidol) (Table 3). In conclusion, the in silico screening approach using RF-Model-II against the DrugBank-library proved to be helpful and yielded several interesting hits that were confirmed experimentally.
Table 3.
Selected Hits of the DrugBank Library-in silico Screening with RF-Model-II and Their Experimental IC50 Values.
| Compound | Score in Computational Model-II [position out of 6551] | Maximum Tc similarity to ICONIX compounds | MATE1 [IC50 μM, (95% CI)] | MATE2K [IC50 μM, (95% CI)] | OCT2 [IC50 μM, (95% CI)] |
|---|---|---|---|---|---|
| Dabigatran | 102 | 0.528 (irinotecan) | 8.1 (5.7-11.5) | 25.3 (12.0-53.1) | 4.7 (2.8-8.0) |
| Pimozide | 126 | 0.724 (droperidol) | >500 | >500 | 34.2 (23.3-52.2) |
| Aripiprazole | 160 | 0.43 (mosapride) | 130 (77.5-219) | >500 | 239 (70.1-815) |
| Risperidone | 250 | 0.543 (ondansetron) | 1.6 (0.98-1.9) | 291 (117-719) | 11.0 (7.4-16.4) |
| Maraviroc | 579 | <0.3 (dapiprazole) | 17.3 (9.6-31.2) | 297 (166-530) | 255 (155-433) |
IC50 values of compounds that were not contained in the ICONIX library but were identified by in silico screening of the DrugBank library. Compounds were chosen based on their pharmacological interest and novelty compared to previously identified inhibitors. Tadalafil and itraconazole could not be tested due to low solubility.
Dragon Descriptors Important for RF Models of MATE1 Inhibitors
The final set of twenty-one descriptors of RF-Model-II was selected using a backward feature elimination algorithm (Table 4). The importance of each descriptor was evaluated by the decrease in the mean accuracy of the model in the descriptors’ permutation tests (Supplementary Figure 4, Methods).30 Two topological descriptors, the Balaban distance connectivity index and the Balaban V index, as well as molecular weight were the most important for the performance of the model (Supplementary Figure 4). Topological and constitutional descriptors dominated the final set of descriptors. Interestingly, none of the 3D descriptors was present among the optimal selection and only one substructure fragment was selected. Ten topological and seven constitutional descriptors differed significantly between MATE1 inhibitors and non-inhibitors, as assessed using the nonparametric Mann-Whitney U test (Table 4). Unfortunately, it is not possible to interpret topological indices in terms of physicochemical properties. The analysis of constitutional descriptors, which alone were not as predictive as topological plus physicochemical properties, showed that MATE1 inhibitors tend to be larger (higher molecular weight, higher number of bonds, rings and longer circuits) and less electronegative than noninhbitors. The importance of lipophilicity for MATE1 inhibitors is again evident from higher MLOGP coefficients. Additionally, the number of nine-membered nitrogen-containing rings (nR09 descriptor) is greater among MATE1 inhibitors. The nR09 descriptor also had higher values among MATE1 selective inhibitors versus OCT2 selective (p-value = 0.0003082) and dual (p-value = 0.004386) inhibitors.
Table 4.
Overview of the 21 most important descriptors used in the RF-Model-II (mean descriptor values of MATE1 inhibitors and non-inhibitors are presented).
| Descriptor | MATE1 inhibitors | Non-inhbitors | p-value | Description | Category |
|---|---|---|---|---|---|
| Vindex | 0.23 | 0.35 | 3.46681E-13 | Balaban V index | topological |
| Wap | 67766.85 | 86064.57 | 4.89801E-12 | all-path Wiener index | topological |
| J | 1.57 | 1.93 | 8.7592E-12 | Balaban distance connectivity index | topological |
| D/Dr06 | 236.12 | 143.60 | 5.99116E-11 | distance/detour ring index of order 6 | topological |
| Jhetm | 1.95 | 2.62 | 6.09E-11 | Balaban-type index from mass weighted distance matrix | topological |
| MW | 403.64 | 325.35 | 1.47811E-10 | molecular weight | constitutional |
| MPC10 | 191.48 | 112.08 | 3.68301E-10 | molecular path count of order 10 | path and walk count |
| D/Dr09 | 88.25 | 23.44 | 6.80182E-10 | distance/detour ring index of order 9 | topological |
| nCIC | 3.67 | 2.51 | 7.5614E-10 | number of rings | constitutional |
| nR09 | 0.78 | 0.26 | 3.84421E-09 | number of 9-membered rings | constitutional |
| nCIR | 7.04 | 4.58 | 2.33749E-07 | number of circuits | constitutional |
| TI2 | 4.31 | 3.32 | 2.86258E-06 | second Mohar index | topological |
| Ms | 2.28 | 2.52 | 7.97109E-06 | mean electrotopological state | constitutional |
| nN | 2.80 | 1.94 | 4.40216E-05 | number of Nitrogen atoms | constitutional |
| MLOGP | 2.69 | 1.82 | 0.000109292 | Moriguchi octanol-water partition coefficient | molecular |
| C-025 | 1.86 | 1.17 | 0.000154955 | R--CR--R atom centered fragments | atom-centered fragments |
| MSD | 0.24 | 0.26 | 0.00380264 | mean square distance index (Balaban) | topological |
| Me | 1.00 | 1.01 | ns | mean atomic Sanderson electronegativity (scaled on Carbon atom) | constitutional |
| Mv | 0.62 | 0.62 | ns | molecular volume | constitutional |
| ARR | 0.35 | 0.34 | ns | aromatic ratio | constitutional |
| nO | 3.15 | 3.36 | ns | number of Oxygen atoms | constitutional |
Overview of descriptors selected using backward feature elimination algorithm. The differences between descriptor values of MATE1 inhibitors and non-inhibitors are assessed using the nonparametric Mann-Whitney U test.
Clinically Relevant MATE1 Inhibitors and Their Implications for Metformin DDIs
A major goal of this study was to identify selective, potentially clinically relevant MATE1 inhibitors. Specificity was assessed by determining IC50 values for MATE1 as well as MATE2-K, OCT1, OCT2, and OCT3.
We identified four drugs that, at clinically relevant plasma concentrations, selectively inhibited MATE1 (i.e., the IC50 value was at least five-fold lower than that obtained for the other cation transporters). Of these four drugs (indinavir, famotidine, ritonavir, and imatinib), indinavir has so far not been shown to interact with MATE1 and thus is a potential new model inhibitor at clinically relevant plasma concentrations. Of six additional drugs studied, three (i.e., irinotecan, mitoxantrone, ondansetron) inhibited MATE1 at clinical plasma concentrations but were not selective towards MATE1 (Figure 5 and Table 5). Interestingly, mitoxantrone and ondansetron proved to be selective for both MATE1 and MATE2-K over the OCTs, and irinotecan was a dual inhibitor for OCT2 and MATE1. The other three compounds were clinically significant inhibitors for either MATE2-K (i.e., nifekalant) or OCT2 (i.e., pantoprazole, pentamidine). Pantoprazole was truly selective for OCT2 over the MATEs, which is of interest since known DDIs in the kidney now appear to involve MATEs and questions about clinically relevant DDIs involving OCT2 have been raised.18 Though clinical studies are needed, our data suggest that an OCT2 mediated DDI may occur between pantoprazole and metformin.
Figure 5.
Selectivity of putative clinically relevant inhibitors of MATE1, MATE2-K or OCT2. IC50 values were determined for selected drugs in HEK293 cells stably over-expressing MATE1, MATE2-K, OCT1, OCT2, or OCT3. Values are presented as mean±SEM (N=3). Clinical relevancy is defined as Cmax,unbound /IC50 ≥ 0.1.
Table 5.
Summary of IC50 Values for Putative Clinically Relevant Inhibitors of MATE1, MATE2-K or OCT2 in ASP+ Uptake Assay.
| Compound | MATE1 ASP+ [IC50 μM, (95% CI)] | MATE2K ASP+ [IC50 μM, (95% CI)] | OCT1 ASP+ [IC50 μM, (95% CI)] | OCT1 ASP+ [IC50 μM, (95% CI)] | OCT3 ASP+ [IC50 μM, (95% CI)] |
|---|---|---|---|---|---|
| Ritonavir | 4.4 (3.3-6.0) | 23.7 (7.6-74.2) | 33.9 (15.9-72.2) | 24.8 (12.0-51.0) | >300 |
| Indinavir | 7.8 (6.1-9.8) | >500 | 208 (97.6-446) | 142 (59.7-335) | >500 |
| Irinotecan | 7.9 (5.8-10.7) | 78.6 (48.3-127) | 20.8 (14.4-30.2) | 2.7 (1.9-4.0) | 74.6 (47.5-117) |
| Imatinib | 0.35 (0.27-0.44) | 2.9 (2.1-3.8) | 107 (36.4-315) | 4.2 (1.8-9.7) | 25.5 (14.0-46.3) |
| Mitoxantrone | 0.53 (0.46-0.59) | 0.83 (0.73-0.93) | 43.9 (33.6-57.4) | 73.3 (46.1-116) | 60.5 (39.9-91.7) |
| Famotidine | 0.76 (0.66-0.87) | 36.2 (27.3-47.9) | >300 | 36.1 (27.2-47.9) | 10.7 (8.3-13.9) |
| Ondansetron | 0.16 (0.12-0.21) | 0.30 (0.24-0.38) | >300 | 1.7 (1.1-2.6) | 17.4 (13.1-23.2) |
| Nifekalant* | 6.5 (5.3-8.0) | 2.7 (2.0-3.5) | >300 | 10.3 (8.1-13.0) | 146 (96.9-220) |
| Pantoprazole** | 43.2 (29.9-62.4) | >500 | >500 | 1.5 (1.0-2.1) | 137 (87.0-217) |
| Pentamidine** | 2.7 (2.1-3.5) | 10.4 (7.9-13.7) | 22.1 (17.7-27.6) | 1.2 (0.9-1.5) | 9.5 (7.8-11.5) |
Overview of putative clinically relevant inhibitors and their IC50 values against MATE1, MATE2-K, OCT1, OCT2, and OCT3.
Nifekalant does not reach a clinically significant concentration for MATE1 inhibition but does so for MATE2-K.
Similarly, pantoprazole and pentamidine do not reach a clinically significant concentration for MATE1 inhibition but do so for OCT2.
Because the above results were achieved with the probe-fluorophore ASP+, which is not relevant clinically, we used 14C-metformin to more appropriately identify the potential for DDIs. IC50 values were assessed in two cellular models, HEK293 cells and polarized MDCK monolayers in which MATE1 is localized on the apical membranes (data not shown). The latter model may more accurately represent MATE1 activity in renal proximal tubule cells in vivo since it is based on polarized cells.31,32 The Km values of 14C-metformin obtained with the HEK293 cells and with the polarized MDCK-II model were 227 μM33 and 114μM (data not shown), respectively. In general, the IC50 values for inhibition of 14C-metformin uptake were similar to or lower than the ones obtained with ASP+, suggesting that the identified interactions are indeed clinically relevant (Table 6). In general, the two cell models yielded results that were in good agreement. Notable exceptions were ritonavir for which the IC50 was greater in the MDCK-II model compared with the HEK293 cells and pentamidine for which the IC50 was greater in the HEK293 cells compared with the MDCK-II model. Reasons for these differences may be related to differences in access of the compounds to the MATE1 binding site in the two cell lines.
Table 6.
Follow-up IC50 Experiments with Metformin for Putative Clinically Relevant MATE1 Inhibitors in Different Assays.
| Compound | IC50 AgainstMATE1 [μM, (95% CI)] | Cmax [μM] | Protein Binding [% bound] | Cmax,unbound/ IC50 of MATE1 | ||||
|---|---|---|---|---|---|---|---|---|
| ASP+ | Metformin | Metformin Polarized Cells | ASP+ | Metformin | Metformin Polarized Cells | |||
| Ritonavir | 4.4 (3.3-6.0) | 0.08 (0.02- 0.26) | 2.1 (1.55-2.71) | 15.26a | 98 | 0.07 | 3.82 | 0.15 |
| Indinavir | 7.8 (6.1-9.8) | 1.7 (1.2-2.6) | 1.9 (1.03-2.73) | 9.8a | 61 | 0.49 | 2.25 | 2.01 |
| Irinotecan | 7.9 (5.8-10.7) | 2.1 (1.1-4.2) | 4.3 (2.14-6.36) | 4.6b | 50 | 0.29 | 1.10 | 0.54 |
| Imatinib | 0.35 (0.27-0.44) | 0.05 (0.03-0.08) | 0.04 (0.03-0.06) | 3.65c | 95 | 0.52 | 3.65 | 4.56 |
| Mitoxantrone | 0.53 (0.46-0.59) | 0.4 (0.33-0.52) | 0.19 (0.04-0.34) | 0.63b | 78 | 0.26 | 0.35 | 0.73 |
| Famotidine | 0.76 (0.66-0.87) | 0.09 (0.06-0.13) | 0.16 (0.11-0.21) | 0.46b | 15 | 0.51 | 4.34 | 2.44 |
| Ondansetron | 0.16 (0.12-0.21) | 0.03 (0.02-0.05) | <0.01 | 0.44b | 73 | 0.74 | 3.96 | >11.88 |
| Nifekalant* | 6.5 (5.3-8.0) | 1.1 (0.4-2.6) | 0.65 (0.30-1.01) | 3.43d | 90 | 0.05 | 0.31 | 0.53 |
| Pantoprazole** | 43.2 (29.9-62.4) | 2.8 (1.8-4.3) | 2.8 (0.08-5.41) | 5.45e | 98 | 0.003 | 0.12 | 0.12 |
| Pentamidine** | 2.7 (2.1-3.5) | 2.0 (1.1-3.7) | 0.41 (0.33-0.49) | 0.73b | 69 | 0.08 | 0.11 | 0.55 |
Inhibitors tested against MATE1 with either ASP+ or 14C-metformin as substrate. Polarized cells are MDCK-hMATE1 in a transwell setting (see methods). The other values were generated in HEK293-cells stably expressing MATE1.
Nifekalant does not reach a clinically significant concentration for MATE1 inhibition but does so for MATE2-K.
DISCUSSION
To date, approximately 30 drugs have been shown to interact with MATE transporters and some of them are substrates (e.g., metformin, oxaliplatin, acyclovir).3 A strong overlap of inhibitors (and substrates) among the organic cation transporters in the OCT family and the MATEs is inferred by many authors based on inhibitors and substrates identified so far.3,9 However, little is known about structural features that favor inhibition of a specific human cation transporter and lead to selectivity for MATEs over OCTs. The data presented complements two recent publications that address inhibitor selectivity for OCT2 over other cation transporters15 and for MATE1 over MATE2-K,12 respectively. In the kidney, predicting drugs that may inhibit MATEs or OCTs is important for predicting DDIs that may potentially lead to nephrotoxic events. In particular, inhibitors of MATE1 would lead to drug accumulation in the kidney and possibly enhance nephrotoxicity, whereas inhibitors of OCT2 would be nephroprotective. The purpose of this study was to screen a prescription drug library to (i) identify novel potent inhibitors of MATE1, (ii) to compare MATE1 inhibitors with inhibitors of another important drug transporter in the kidney (OCT2), (iii) to elucidate the physicochemical properties of MATE1 inhibitors and develop a predictive model to identify MATE1-inhibitors, and (iv) to identify MATE1 selective inhibitors that may potentially cause DDIs at clinically relevant concentrations.
Only 84 compounds were identified as hits in this large-scale library screen of MATE1 compared with 244 hits identified in our previous screen of OCT2.15 These findings are in agreement with publications, in which inhibitors of OCT2 and MATE1 were summarized and compared, showing that more drugs strongly inhibit OCT2 than MATE1 in in vitro assays.3,9,19 Furthermore charge distribution was also consistent with previously observed patterns and showed that OCT2 inhibitors were significantly enriched within both the basic compounds and the zwitterions groups, whereas among MATE1 inhibitors zwitterions were not enriched and basic drugs were much less enriched compared to OCT2 (Figure 3). Paradoxically, some physicochemical properties of the MATE1 selective inhibitors, such as molecular weight, number of heavy atoms, and molecular volume, were more variable compared to those for the OCT2 selective inhibitors. For example, the median and the 25 - 75% range of molecular weights for OCT2 inhibitors were 309.33 and 269.88 - 369.41, respectively, whereas for MATE1 inhibitors they were 348.48 and 284.40 to 557.83, respectively (Figure 3). These data suggest that MATE1 can be inhibited by a larger variety of compounds with more diverse chemical properties, yet fewer compounds inhibited MATE1 than OCT2. One possible explanation for this apparent inconsistency is that the structural and molecular features required for OCT2 inhibition are more common in the chemical space typically occupied by prescription drugs. This means that prescription drugs might be biased towards structural features that favor OCT2 inhibition. Another possibility is the occurrence of multiple binding sites in MATE1 and considerably fewer in OCT2. Indeed, a recent study by Astorga et al. indicates that this is a likely assumption since the pharmacophores developed for inhibitors identified with different probes (MPP+ vs. ASP+) were markedly different.12
Several of the important physicochemical parameters identified in this study have been found in earlier studies with the rat and rabbit homologues of MATE1. For example, using rabbit brush border membrane vesicles (BBMV) various publications identified positive charge and, to a lesser extent, lipophilicity as important contributors to MATE1-binding.34–36 Furthermore, studies performed in rats also identified hydrophobicity and charge as major factors for MATE1-binding.37–40 Nevertheless, it seems unlikely that a single molecular descriptor is sufficient to appropriately describe binding to MATE1, as also noted by Astorga et al.,12 and QSAR-models offer the advantage of combining different weighted descriptors to yield a more accurate description of the binding process.
The finding that positive charge, lipophilicity, and molecular weight are important contributors to MATE1-binding is also supported by recently developed structural models of MATE141 based on the crystal structure of the NorM-homologue protein found in Vibrio cholerae.42 From the proposed structural model, it becomes evident that the pore, containing the supposed binding sites, is relatively large and predominantly contains negatively charged as well as hydrophobic patches,41 thus favoring the binding of large, lipophilic, and positively charged compounds as suggested by the results of this study and by previously published QSAR-models based on rodent MATE1-homologues as well as by the pharmacophore-model.
Several studies demonstrated the potential for drug-drug interactions (DDIs) involving MATE1. Indeed, MATE1 knockout mice had significantly increased plasma levels of metformin after intravenous administration, and the urinary excretion of the drug was markedly decreased.43In vitro, cimetidine has been reported to interact with metformin19 and fexofenadine44 by inhibiting MATE1 mediated transport. In human volunteers, pyrimethamine, a MATE inhibitor, was reported to significantly increase the plasma concentrations of metformin and alter its renal elimination.45 With respect to in vivo studies, model inhibitors must exert their effect at clinically relevant concentrations. In this study, four drugs have been identified that selectively inhibit MATE1 at clinically relevant unbound concentrations (i.e., Cmax,unbound /IC50 ≥ 0.1), namely, famotidine, imatinib, ritonavir, and indinavir (Tables 5 and 6 as well as Figure 5). To the best of our knowledge, indinavir has so far not been described as a MATE1 inhibitor. The other three compounds were all found to be MATE1 selective in previous studies. Imatinib has been previously described as potentially clinically relevant by our group.46 This study confirms MATE1 selectivity over OCT3 in addition to MATE2-K, OCT1, and OCT2 that have been reported previously . Interestingly, while both famotidine and cimetidine have been associated with MATE1 selectivity,19 only the latter was examined with respect to clinical significance and was shown to probably cause the clinically observed DDI with metformin.19 As Tables 5 and 6 show, three more compounds were predicted to inhibit MATE1 at clinically relevant concentrations but are not selective over other cation transporters. Nonetheless, they might be of clinical use as model inhibitors and because of their effects on multiple renal transporters may result in clinically significant DDIs. For example, mitoxantrone and ondansetron selectively inhibit both MATEs over the OCTs and irinotecan might act as a dual inhibitor for MATE1 and OCT2. Of these three drugs, ondansetron is to our best knowledge the most potent inhibitor of MATEs (IC50 values in nanomolar range, Table 6), is least toxic, orally available, and may be a specific in vivo probe for studying MATE1 and MATE2-K mediated renal DDIs with basic drugs. Furthermore, three additional compounds are not clinically relevant MATE1 inhibitors, but are otherwise of great interest as in vivo tools in preclinical studies. First, nifekalant is predicted to be a clinically significant MATE2-K inhibitor and (if dosed slightly higher) could act as a pan-inhibitor of all three renal cation transporters. Unfortunately, given its use as a class III-antiarrhythmic drug, such a higher dose might practically not be feasible due to the risk of adverse drug reactions. Second, pantoprazole selectively inhibited OCT2 at clinical concentrations. Whereas previous studies indicate that pantoprazole is selective for OCT2 over OCT1 and OCT3,47 our data extend these findings to the MATEs and suggest that pantoprazole might thus be a useful OCT2 selective model inhibitor based on the IC50 values determined with ASP+. Finally, pentamidine inhibited OCT2 at clinically significant levels and its Cmax,unbound /IC50 ratio for MATE1 (i.e., 0.06) was close to clinical significance. Pentamidine might thus be another dual inhibitor for MATE1 and OCT2 besides irinotecan. Its nephrotoxicity, however, might be a serious limiting factor for its possible use as clinical model inhibitor.
To validate the risk of DDIs associated with the inhibitors described above with ASP+ as a substrate, we determined IC50 values against the clinically relevant model substrate, metformin, which is eliminated exclusively in the kidney, using two distinct models. Metformin is among the most highly prescribed drugs in the world, and DDIs in the kidney might potentially increase the risk of its serious adverse drug reaction, lactic acidosis. OCT2, on the basolateral membrane, and MATE1 and MATE2-K on the apical membrane, appear to be the primary transporters responsible for its secretory clearance.10,48 All drugs showed similar or even lower IC50 values with metformin as a substrate compared to ASP+, underscoring the potential clinical significance of these interactions. Astorga et al. recently showed that distinct pharmacophore models of MATE1 were obtained when different substrates, i.e., ASP+ and MPP+, were used in inhibition assays.12 The finding that the IC50 values for metformin were different from those of ASP+ for several compounds (Table 6) is in agreement with the idea of substrate-dependent pharmacophore models.
Furthermore, some of the 84 HTS-hits are strong and selective MATE1 inhibitors, but are not expected to inhibit MATE1 at clinical concentrations (Table 1). For example, leucomycine selectively inhibits MATE1 without affecting MATE2-K or OCT2 while topotecan inhibits both MATE1 and MATE2-K and only to a lesser extent OCT2. In addition, chlorhexidine strongly inhibits all tested cation transporters. These compounds provide valuable in vitro tools for predicting the contribution of the various renal transporters to overall renal excretion. Reviewing the HTS-data, it became apparent that MATE1 inhibitors were not necessarily also MATE2-K inhibitors and that the constellation of physicochemical properties that determines inhibitors of MATE2-K is likely distinct from those of MATE1 and OCT2. Interestingly, MATE1 inhibitors tested in follow-up studies more frequently inhibited OCT2 mediated transport rather than affecting MATE2-K, as illustrated by gabaxate, vecuronium bromide or rimantadine. Indeed, in this study we only identified a few MATE-selective inhibitors over OCT2 (e.g., mitoxantrone, ondansetron, topotecan) when at least a five-fold difference in IC50-values was used as the criterion. Previous publications show the same trend.11,12,19 Although the study was focused on the identification of MATE1 selective inhibitors, two compounds that seem to be predominantly OCT2 selective were found. As Table 1 shows, ethinyl estradiol exhibits a certain selectivity for OCT2 over the MATEs.49 The other compound, pantoprazole, has been discussed above. Additionally, we also demonstrate the usefulness of a combined in silico and experimental approach with iterative model improvement (i.e., development of different RF-Models based on experimental feedback). This approach proved helpful in identifying false positive and false negative screening hits and to identify additional MATE1-inhibitors from a large in silico library (e.g., maraviroc). None of the inhibitors identified in the current study were tested as substrates of the transporters; therefore, direct experiments testing the uptake of the inhibitors by the relevant transporters as well as their mechanism (e.g., competitive or noncompetitive) are needed.
In summary, we tested 910 prescription drugs for their potential to inhibit MATE1 and identified 84 hits. We identified several MATE1 selective inhibitors (e.g., indinavir) as well as general inhibitors of renal transporters with less selectivity. Compounds expected to cause clinically relevant DDIs were identified including compounds that can be used as selective model inhibitors of the individual transporters. Through the development of predictive models, distinct constellations of physicochemical properties were identified that differentiated OCT2 from MATE1 inhibitors. Several interesting MATE1 inhibiting drugs, not contained in the original screening library, were predicted and then experimentally validated, including maraviroc and risperidone. This study adds new in vitro probes as well as possible in vivo model inhibitors to clarify drug excretory pathways and enhances the understanding of both OCTs and MATEs.
EXPERIMENTAL SECTION
Reagents
The ICONIX compound screening library was obtained through the Small Molecule Discovery Center (University of California, San Francisco, CA) and contains 910 FDA-approved drugs.15 4-(4-(Dimethylamino)styryl)-N-methylpyridinium iodide (ASP+) was purchased from Molecular Probes (Grand Island, NY). All other reagents were obtained from Sigma-Aldrich (St. Louis, MI) and Santa Cruz Biotechnology (Santa Cruz, CA) and were of analytical grade with at least 95% purity, unless otherwise stated.
Cell Culture
Flp-In human embryonic kidney (HEK-293-Flp-In) cell lines stably expressing human MATE1, MATE2-K, OCT1, OCT2 and OCT3 were previously established in our laboratory.15,50–52 Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 200 μg/mL hygromycine at 37 °C in a humidified atmosphere with 5% CO2.
Madin Darby canine kidney type II (MDCK-II) cells were cultured in modified DMEM with 1 g/L glucose and1 g/L NaHCO3 (Caisson Labs, North Logan, UT) supplemented with 50 U/mL penicillin, 50 μg /mL streptomycin and 10% fetal bovine serum, at 37 °C in a humidified atmosphere with 5% CO2.
Fluorescent-Probe Uptake Assay
MATE1-overexpressing cells were seeded in black poly-D-lysine-coated 96-well plates (Greiner Bio-One, Monroe, NC) and grown for 48 hours until approximately 90% confluent. After washing twice with Hank's Buffered Saline Solution (HBSS), the cells were pre-incubated for 30 minutes in a 30 mM NH4Cl solution in HBSS at pH 6.5. Uptake was initiated by application of HBSS at pH 7.4 containing the serially diluted fluorescent substrate ASP+ (4-(4-(dimethylamino)styryl)-N-methylpyridinium iodide). After incubating for 1.5 min at room temperature, substrate uptake was stopped by aspirating the incubation mixture and washing the cells twice with ice-cold HBSS containing 500 μM cimetidine to stop the reaction. Similar experiments were performed using 5 μM of ASP+ and stopping the reaction at 1, 2, 3, 5, 7, 10, 20 and 30 min. Each test condition was analyzed in triplicate. The intensity of accumulated ASP+ fluorescence was measured using an Analyst AD plate reader (Molecular Devices, Sunnyvale, CA) with excitation and emission filters tuned at 485 and 585 nm wavelength, respectively. Transport kinetics studies for MATE2-K and OCT1, 2, and 3 were performed similarly with the following modifications: No pre-loading of cells with NH4Cl was required for ASP+ uptake by the OCT-transfected cells and the HBSS stop solution contained 200 μM spironolactone and 200 μM corticosterone for OCT1 and OCT3, respectively. Transport kinetics and Z’-values were calculated as described elsewhere.15
High throughput Screen (HTS) of Transporter Inhibition
The high throughput screen was performed at the Small Molecule Discovery Center at the University of California, San Francisco. Assay buffers were prepared by diluting 1 mM inhibitor stock solutions in DMSO with HBSS (pH 7.4) containing ASP+ (5 μM) to a final inhibitor concentration of 20 μM (2% DMSO). Nonspecific transport was determined in separate wells on each assay plate using 500 μM cimetidine as inhibitor of MATEs. After subtraction of the nonspecific transport, residual transport rates were used for further calculations. Predicted half-maximum inhibitory concentrations (prIC50) were estimated from the screening inhibition measurements as V = (V0/(1+ [I] *prIC50), where V and V0 are the activity with and without inhibitor, respectively, and I is the inhibitor concentration of 20 μM. The estimated prIC50 values were considered representative for 20%<V<80%, where inhibition plots are linear.
Experimental IC50 Determination
Selected compounds, which reduced ASP+ uptake by 50% or more in the HTS, were subjected to experimental IC50 determination. The selection was based on literature-reported transporter selectivity (IC50 values versus MATE2-K, OCT1, OCT2, and OCT3), clinical relevance, chemical structure, and novelty. To determine IC50 values, after pre-incubation as described above, the stably transfected HEK293 cells were simultaneously exposed to ASP+ (2 μM) and the compound of interest at various concentrations (ranging from 0 to 500 μM) in HBSS at pH 7.4. The dilutions were prepared from 25 mM DMSO stock solutions and the DMSO concentration was kept constant at 2% in all samples. After 1.5 minutes transport was stopped by removing the uptake media and washing twice with ice-cold stop solution as described above. Then, fluorescence within the cells was determined and IC50 values were calculated using the Prism Software. Values were accepted if the R2 value of the resulting sigmoidal curve was higher than 0.98 and an acceptable standard deviation of the data-points was achieved. The uptake of ASP+ was measured in triplicate at each inhibitor-concentration. IC50 determinations for selected compounds as inhibitors of MATE2-K, OCT1, OCT2, and OCT3 were performed similarly with the following exceptions: No pre-incubation with NH4Cl is required for ASP+ uptake by OCT-transfected cells, the incubation time for OCT-transfected cells was 3 minutes, and the HBSS stop solution in the OCT1 and OCT3 experiments contained 200 μM spironolactone and 200 μM corticosterone, respectively.
MATE1 IC50 Determinations With 14C-Labelled Metformin in HEK293 cells
The determinations were performed as described elsewhere,46 with the modification that 28μM 14C-metformin (1 μCi/ml) were used.
MATE1 IC50 Determinations With 14C-Labelled Metformin in polarized MDCK-II cells
MDCK-II cells were seeded on 96-well porous membrane insert plates (Millipore, Carrigtwohill, Ireland) coated with rat tail type I collagen (Millipore, Temecula, CA), at a seeding density of 60K per well 24 hours prior to transfection. Fully confluent MDCK cell monolayers were transfected with pCI-MATE-1 (accession number NM_018242) or pCI-GFP control plasmid DNA at a final concentration of 30ng/μl, using a proprietary Opti-Expression transfection technology (Optivia Biotechnology, Menlo Park, CA). The transiently transfected MDCK cells were incubated for 48 hours to allow the cell monolayers to become polarized. The plates were washed apically and basally three times with warm HBSS (Mediatech, Manassas, VA). After the final wash, the apical sides of the MDCK monolayers were incubated in 150 μl of HBSS-30 mM NH4Cl buffer for 20 minutes at 37C on a 60 rpm shaking platform to allow for intracellular acidification. The basolateral side of the insert plate was blotted and left dry. After pre-incubation, the cells were washed once with warm HBSS and replaced with 10 μM 14C-metformin dosing solution (Moravek Biochemicals, Brea, CA) containing varying concentrations of test inhibitor or 0.5% DMSO as vehicle control. As a inhibition control, 10 μM cimetidine was added as a reference inhibitor in the presence of 10 μM 14C-metformin. The plates were placed on a 60 rpm shaking platform for 5 minutes at 37Cfor 5 minutes. After this time, the cells were immediately washed both apically and basally with three washes of ice cold PBS. To assess 14C-metformin intracellular accumulation, 60 μl of 50:50 Acetonitrile:H2O was added to the inserts and shaken at 37C for 15 minutes. A 30 μl sample of the solubilized cell monolayers was taken for liquid scintillation counting (MicroBeta, Perkin-Elmer, Santa Clara, CA).
In silico Modeling
Three-dimensional structures of 910 compounds were generated using the Corina version 3.20 package (Molecular Networks GmbH, Erlangen, Germany). SMILES strings were used as input, hydrogen atoms were added, small fragments were removed, charges were neutralized, and 100 ring conformations were generated for each compound. The lowest energy conformations of each molecule were used to compute Dragon (Talete s.r.l., Milano, Italy) molecular descriptors. Ten compounds were removed due to computational failure in the Corina or Dragon software. Descriptors with variance near zero were removed and pair-wise correlation coefficients were computed for the remaining descriptors. One randomly chosen descriptor was removed from highly correlated descriptor pairs (correlation coefficient greater than 0.90). All descriptor values were centered and scaled. The random forest (RF) algorithm17 was used to develop a binary classification model that differentiates between MATE1 inhibitors and non-inhibitors. Class labels were assigned to all compounds. Compounds were assigned either a class label 1 if they showed inhibition greater than 50% in the HTS (inhibitor) or a class label 0 otherwise (non-inhibitor). A training and a test set were constructed by partitioning the ICONIX data set into two non-overlapping subsets (training and test sets) each consisting of 450 compounds. The ratio of inhibitors to non-inhibitors in the subsets was kept the same as in the whole data set (approximately 1:10). Next, a RF model was tuned by the means of out-of-bag validation of the training set.30 Random forest parameters were tuned to maximize the area under the receiver operating characteristic curve (AUC). The best model from the inner loop was tested on the withheld test data set. The entire validation was repeated ten times for different partitioning of data. Descriptor importance was computed and a feature elimination algorithm was used to select the 21 most important molecular descriptors. The algorithm iteratively removes molecular descriptors in the inner loop if the AUC does not decrease. A permutation test was performed to access the statistical significance of the final models. This test studies the null hypothesis that the molecular descriptors and the class labels are independent, that is, that there is no difference between the classes (i.e., inhibitors and non-inhibitors). The null distribution under this hypothesis was estimated by permuting the labels of the dataset 1000 times. The results of the permutation test showed that models achieve significantly higher AUCs than expected by chance (p < 0.0001). RF models were used to compute a classification score in the range [0,1] for each of the 900 compounds. The score cutoff was calibrated against the classifier error in the outer validation loop and was set to 0.2 for subsequent analyses. The compounds with a score greater than or equal to 0.2 were classified as inhibitors, and compounds with a score less than 0.2 were classified as non-inhibitors. All computational modeling was performed using the caret library in R.
The kNN and PLS-DA models were constructed following the same protocol. The Student's t-test was used to assess the differences between kNN and RF as well as between PLS-DA and RF algorithms.
In silico Model Refinement
The first RF model (RF-Model-I) was refined by incorporating the validation results to yield RF-Model-II. During the refinement, compound labels were updated to reflect new experimental results and new models were developed and tested as described above. To enable comparisons between the initial and refined models, data partitioning was done with the same seed for the random number generator. A paired Student's t-test was used to assess the model improvement.
DrugBank Ligand Screening
Three-dimensional structures of 6,363 compounds were retrieved from the DrugBank database and 21 Dragon molecular descriptors (Table 4) were computed for each compound. The final RF model (RF-Model-II) was used to predict the MATE1 inhibition status for each of the DrugBank compounds. Compounds with a score greater than or equal to 0.2 were classified as inhibitors, and compounds with a score less than 0.2 were classified as non-inhibitors.
DrugBank similarity and substructure searches were performed using DrugBank's web interface with default options. Tanimoto coefficients of similarity were computed using the Drugbank interface.
Principal Component Analysis of Physicochemical Properties
Three-dimensional structures of 910 compounds were generated as described above (In Silico Modeling). The lowest energy conformations of each molecule were used to compute molecular weight, molecular volume, number of heavy atoms, number of rotatable bonds, number of hydrogen bond donors and acceptors, SLogP, topological polar surface area and charge. ChemAxon Calculator (cxcalc) version 5.11.1 was used to compute the formal charge, ApKa and BpKa at pH 7.4. The compounds were binned into acids (ApKa < 9.4), bases (BpKa > 5.4), zwitterions (ApKa < 9.4 and BpKa > 5.4), and neutral (ApKa > 9.4 and BpKa < 5.4). Compounds for which ChemAxon Calculator failed to compute ApKa and BpKa values were denoted as unknown. Principal component analysis was performed with the printcomp function in the stats library in R.
Statistical Analysis
Statistical differences in physicochemical properties of OCT2 inhibitors, MATE1 inhibitors, dual inhibitors and non-inhibitors were assessed with the Dunnett-Tukey-Kramer pairwise multiple comparison test adjusted for unequal variances and unequal sample sizes. Over- or under-representation of bases, acids, zwitterions, and neutral compounds among OCT2 inhibitors, MATE1 inhibitors, dual inhibitors and non-inhibitors was determined with the hypergeometric test. Differences between AUC values of ten independent test sets of two different models were assessed with the Student's t-test. Differences between properties of Dragon descriptors of MATE1 inhibitors and non-inhibitors were assessed using nonparametric Mann-Whitney U test.
Supplementary Material
ACKNOWLEDGEMENT
We gratefully acknowledge Dr. Anton Rosenbaum for fluorescent reader advice, Steven Chen from the UCSF Small Molecule Discovery Center for help with the assay automation, and Drs. Avner Schlessinger and Pär Matsson as well as Eugene Chen for helpful discussions. This work is generously supported by the National Institute of General Medical Sciences (NIGMS) under grants R44 GM086970 (YH and KG), U01 GM61390 (KG and AS), and U54 GM074929 (KG and AS). The authors would also like to gratefully acknowledge funding for Matthias Wittwer (The Swiss National Science Foundation's grant for prospective researchers PBBSP3-133384), Arik Zur (FDA-CDER/ORISE) and Kari Morrissey (NIH Training Grant T32 GM007175, FDA-CDER/ORISE).
ABBREVIATIONS
- ASP+
4-(4-(dimethylamino)styryl)-N-methylpyridinium iodide
- HBSS
Hanks’ balanced salt solution
- HEK293
Human embryonic kidney cell line
- kNN
k-nearest neighbor
- MATE1
Multidrug and toxin extrusion transporter 1
- MATE2-K
Multidrug and toxin extrusion transporter 2 - kidney specific splicing variant
- OCT
Organic cation transporter
- PCA
Principal component analysis
- PLS-DA
partial least squares discriminant analysis
- QSAR
Quantitative structure activity relationship
- RF
Random forest
- AUC
Area under the receiver operating curve
- SD
Standard deviation
- SEM
Standard error of the mean
- SLC
solute carrier
Footnotes
Author Contributions
M.B.W. and A.A.Z. designed the research, conducted experiments, analyzed the data, interpreted the results, generated the figures and tables, and together with K.M.G. wrote the paper. N.K. developed the in silico models, performed statistical data analysis, , made figures and tables, and contributed to writing the paper. A.S. participated in results interpretation and discussion and proof-read the manuscript. Y.K. conducted the HTS screen. A.K. and X.Z. conducted the polarized MDCK inhibition studies, and together with Y.H., contributed to the discussion regarding clinical DDIs. K.M.M helped with finding optimal assay conditions, gave inputs for the writing, and proof-read the manuscript. K.M.G. provided conceptual guidance, contributed to the writing of the paper, and led the study. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Supporting Information. HTS data (% inhibition at 20 μM and SD) as well as names and SMILES codes of all ICONIX library drugs (in .xls format); Physicochemical properties of compounds in the ICONIX library; Comparison of the performance of three computational methods, kNN, PLS-DA and RF; non-significant descriptors of MATE1 inhibitors and of the other groups; development and improvement of kNN and PLS-DA models; PCA plot of the DrugBank compounds; Depiction of the importance of Dragon descriptors in RF-Model-II. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.You G, Morris ME, editors. Wiley Series in Drug Discovery and Development. 1st ed. Wiley-Interscience, Inc.; Hoboken, NJ: 2007. Drug Transporters: Molecular Characterization and Role in Drug Disposition. [Google Scholar]
- 2.Giacomini KM, Huang S-M, Tweedie DJ, Benet LZ, Brouwer KLR, Chu X, Dahlin A, Evers R, Fischer V, Hillgren KM, Hoffmaster KA, Ishikawa T, Keppler D, Kim RB, Lee CA, Niemi M, Polli JW, Sugiyama Y, Swaan PW, Ware JA, Wright SH, Yee SW, Zamek-Gliszczynski MJ, Zhang L. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9:215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nies AT, Koepsell H, Damme K, Schwab M. Organic Cation Transporters (OCTs, MATEs), In Vitro and In Vivo Evidence for the Importance in Drug Therapy. In: Fromm MF, Kim RB, editors. Drug Transporters. Springer Berlin Heidelberg. Vol. 201. Heidelberg; Berlin: 2011. pp. 105–167. [DOI] [PubMed] [Google Scholar]
- 4.Burckhardt BC, Burckhardt G. Transport of organic anions across the basolateral membrane of proximal tubule cells. Reviews of Physiology, Biochemistry and Pharmacology. 2003;146146:95–158. doi: 10.1007/s10254-002-0003-8. [DOI] [PubMed] [Google Scholar]
- 5.Masereeuw R, Russel FGM. Therapeutic implications of renal anionic drug transporters. Pharmacology & Therapeutics. 2010;126:200–216. doi: 10.1016/j.pharmthera.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Otsuka M, Matsumoto T, Morimoto R, Arioka S, Omote H, Moriyama Y. A human transporter protein that mediates the final excretion step for toxic organic cations. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17923–17928. doi: 10.1073/pnas.0506483102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masuda S, Terada T, Yonezawa A, Tanihara Y, Kishimoto K, Katsura T, Ogawa O, Inui K. Identification and functional characterization of a new human kidney-specific H+/organic cation antiporter, kidney-specific multidrug and toxin extrusion 2. J. Am. Soc. Nephrol. 2006;17:2127–2135. doi: 10.1681/ASN.2006030205. [DOI] [PubMed] [Google Scholar]
- 8.Komatsu T, Hiasa M, Miyaji T, Kanamoto T, Matsumoto T, Otsuka M, Moriyama Y, Omote H. Characterization of the human MATE2 proton-coupled polyspecific organic cation exporter. Int. J. Biochem. Cell Biol. 2011;43:913–918. doi: 10.1016/j.biocel.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Meyer zu Schwabedissen HE, Verstuyft C, Kroemer HK, Becquemont L, Kim RB. Human multidrug and toxin extrusion 1 (MATE1/SLC47A1) transporter: functional characterization, interaction with OCT2 (SLC22A2), and single nucleotide polymorphisms. Am. J. Physiol. Renal Physiol. 2010;298:F997–F1005. doi: 10.1152/ajprenal.00431.2009. [DOI] [PubMed] [Google Scholar]
- 10.König J, Zolk O, Singer K, Hoffmann C, Fromm MF. Double-transfected MDCK cells expressing human OCT1/MATE1 or OCT2/MATE1: determinants of uptake and transcellular translocation of organic cations. Br. J. Pharmacol. 2011;163:546–555. doi: 10.1111/j.1476-5381.2010.01052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanihara Y, Masuda S, Sato T, Katsura T, Ogawa O, Inui K-I. Substrate specificity of MATE1 and MATE2-K, human multidrug and toxin extrusions/H(+)-organic cation antiporters. Biochem. Pharmacol. 2007;74:359–371. doi: 10.1016/j.bcp.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Astorga B, Ekins S, Morales M, Wright SH. Molecular determinants of ligand selectivity for the human multidrug and toxin extruder proteins MATE1 and MATE2-K. J. Pharmacol. Exp. Ther. 2012;341:743–755. doi: 10.1124/jpet.112.191577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahlin G, Karlsson J, Pedersen JM, Gustavsson L, Larsson R, Matsson P, Norinder U, Bergström CAS, Artursson P. Structural requirements for drug inhibition of the liver specific human organic cation transport protein 1. J. Med. Chem. 2008;51:5932–5942. doi: 10.1021/jm8003152. [DOI] [PubMed] [Google Scholar]
- 14.Badolo L, Rasmussen LM, Hansen HR, Sveigaard C. Screening of OATP1B1/3 and OCT1 inhibitors in cryopreserved hepatocytes in suspension. Eur J Pharm Sci. 2010;40:282–288. doi: 10.1016/j.ejps.2010.03.023. [DOI] [PubMed] [Google Scholar]
- 15.Kido Y, Matsson P, Giacomini KM. Profiling of a prescription drug library for potential renal drug-drug interactions mediated by the organic cation transporter 2. J. Med. Chem. 2011;54:4548–4558. doi: 10.1021/jm2001629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yonezawa A, Inui K. Importance of the multidrug and toxin extrusion MATE/SLC47A family to pharmacokinetics, pharmacodynamics/toxicodynamics and pharmacogenomics. Br. J. Pharmacol. 2011;164:1817–1825. doi: 10.1111/j.1476-5381.2011.01394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breiman L. Random forests. Machine learning. 2001;45:5–32. [Google Scholar]
- 18.Ito S, Kusuhara H, Yokochi M, Toyoshima J, Inoue K, Yuasa H, Sugiyama Y. Competitive inhibition of the luminal efflux by multidrug and toxin extrusions, but not basolateral uptake by organic cation transporter 2, is the likely mechanism underlying the pharmacokinetic drug-drug interactions caused by cimetidine in the kidney. J. Pharmacol. Exp. Ther. 2012;340:393–403. doi: 10.1124/jpet.111.184986. [DOI] [PubMed] [Google Scholar]
- 19.Tsuda M, Terada T, Ueba M, Sato T, Masuda S, Katsura T, Inui K. Involvement of human multidrug and toxin extrusion 1 in the drug interaction between cimetidine and metformin in renal epithelial cells. J. Pharmacol. Exp. Ther. 2009;329:185–191. doi: 10.1124/jpet.108.147918. [DOI] [PubMed] [Google Scholar]
- 20.Karlgren M, Vildhede A, Norinder U, Wisniewski JR, Kimoto E, Lai Y, Haglund U, Artursson P. Classification of Inhibitors of Hepatic Organic Anion Transporting Polypeptides (OATPs): Influence of Protein Expression on Drug–Drug Interactions. J Med Chem. 2012;55:4740–4763. doi: 10.1021/jm300212s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V, Djoumbou Y, Eisner R, Guo AC, Wishart DS. DrugBank 3.0: a comprehensive resource for “omics” research on drugs. Nucleic Acids Res. 2011;39:D1035–1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheridan RP. Three useful dimensions for domain applicability in QSAR models using random forest. J Chem Inf Model. 2012;52:814–823. doi: 10.1021/ci300004n. [DOI] [PubMed] [Google Scholar]
- 25.Dimitrov S, Dimitrova G, Pavlov T, Dimitrova N, Patlewicz G, Niemela J, Mekenyan O. A stepwise approach for defining the applicability domain of SAR and QSAR models. J Chem Inf Model. 2005;45:839–849. doi: 10.1021/ci0500381. [DOI] [PubMed] [Google Scholar]
- 26.Sahigara F, Mansouri K, Ballabio D, Mauri A, Consonni V, Todeschini R. Comparison of different approaches to define the applicability domain of QSAR models. Molecules. 2012;17:4791–4810. doi: 10.3390/molecules17054791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dragos H, Gilles M, Alexandre V. Predicting the predictability: a unified approach to the applicability domain problem of QSAR models. J Chem Inf Model. 2009;49:1762–1776. doi: 10.1021/ci9000579. [DOI] [PubMed] [Google Scholar]
- 28.Abel S, Back DJ, Vourvahis M. Maraviroc: pharmacokinetics and drug interactions. Antivir. Ther. (Lond.) 2009;14:607–618. [PubMed] [Google Scholar]
- 29.Abel S, Russell D, Whitlock LA, Ridgway CE, Nedderman ANR, Walker DK. Assessment of the absorption, metabolism and absolute bioavailability of maraviroc in healthy male subjects. Br J Clin Pharmacol. 2008;65(Suppl 1):60–67. doi: 10.1111/j.1365-2125.2008.03137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and regression trees. Wadsworth International Group; Belmont, CA: 1984. [Google Scholar]
- 31.Steele RE, Preston AS, Johnson JP, Handler JS. Porous-bottom dishes for culture of polarized cells. Am. J. Physiol. 1986;251:C136–139. doi: 10.1152/ajpcell.1986.251.1.C136. [DOI] [PubMed] [Google Scholar]
- 32.Handler JS, Preston AS, Steele RE. Factors affecting the differentiation of epithelial transport and responsiveness to hormones. Fed. Proc. 1984;43:2221–2224. [PubMed] [Google Scholar]
- 33.Chen Y, Teranishi K, Li S, Yee SW, Hesselson S, Stryke D, Johns SJ, Ferrin TE, Kwok P, Giacomini KM. Genetic variants in multidrug and toxic compound extrusion-1, hMATE1, alter transport function. Pharmacogenomics J. 2009;9:127–136. doi: 10.1038/tpj.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright SH, Wunz TM, Wunz TP. Structure and interaction of inhibitors with the TEA/H+ exchanger of rabbit renal brush border membranes. Pflugers Arch. 1995;429:313–324. doi: 10.1007/BF00374145. [DOI] [PubMed] [Google Scholar]
- 35.Wright SH, Wunz TM. Influence of substrate structure on turnover of the organic cation/H+ exchanger of the renal luminal membrane. Pflugers Arch. 1998;436:469–477. doi: 10.1007/s004240050658. [DOI] [PubMed] [Google Scholar]
- 36.Wright SH, Wunz TM. Influence of substrate structure on substrate binding to the renal organic cation/H+ exchanger. Pflugers Arch. 1999;437:603–610. doi: 10.1007/s004240050823. [DOI] [PubMed] [Google Scholar]
- 37.Ullrich KJ, Papavassiliou F, David C, Rumrich G, Fritzsch G. Contraluminal transport of organic cations in the proximal tubule of the rat kidney. I. Kinetics of N1-methylnicotinamide and tetraethylammonium, influence of K+, HCO3-, pH; inhibition by aliphatic primary, secondary and tertiary amines and mono- and bisquaternary compounds. Pflugers Arch. 1991;419:84–92. doi: 10.1007/BF00373751. [DOI] [PubMed] [Google Scholar]
- 38.Ullrich KJ, Rumrich G, Neiteler K, Fritzsch G. Contraluminal transport of organic cations in the proximal tubule of the rat kidney. II. Specificity: anilines, phenylalkylamines (catecholamines), heterocyclic compounds (pyridines, quinolines, acridines). Pflugers Arch. 1992;420:29–38. doi: 10.1007/BF00378638. [DOI] [PubMed] [Google Scholar]
- 39.David C, Rumrich G, Ullrich KJ. Luminal transport system for H+/organic cations in the rat proximal tubule. Kinetics, dependence on pH; specificity as compared with the contraluminal organic cation-transport system. Pflugers Arch. 1995;430:477–492. doi: 10.1007/BF00373884. [DOI] [PubMed] [Google Scholar]
- 40.Ullrich KJ, Rumrich G. Luminal transport system for choline+ in relation to the other organic cation transport systems in the rat proximal tubule. Kinetics, specificity: alkyl/arylamines, alkylamines with OH, O, SH, NH2, ROCO, RSCO and H2PO4-groups, methylaminostyryl, rhodamine, acridine, phenanthrene and cyanine compounds. Pflugers Arch. 1996;432:471–485. doi: 10.1007/s004240050159. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, He X, Baker J, Tama F, Chang G, Wright SH. Twelve Transmembrane Helices Form the Functional Core of Mammalian MATE1 (Multidrug and Toxin Extruder 1) Protein. J. Biol. Chem. 2012;287:27971–27982. doi: 10.1074/jbc.M112.386979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He X, Szewczyk P, Karyakin A, Evin M, Hong W-X, Zhang Q, Chang G. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature. 2010;467:991–994. doi: 10.1038/nature09408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuda M, Terada T, Mizuno T, Katsura T, Shimakura J, Inui K. Targeted disruption of the multidrug and toxin extrusion 1 (mate1) gene in mice reduces renal secretion of metformin. Mol. Pharmacol. 2009;75:1280–1286. doi: 10.1124/mol.109.056242. [DOI] [PubMed] [Google Scholar]
- 44.Matsushima S, Maeda K, Inoue K, Ohta K, Yuasa H, Kondo T, Nakayama H, Horita S, Kusuhara H, Sugiyama Y. The inhibition of human multidrug and toxin extrusion 1 is involved in the drug-drug interaction caused by cimetidine. Drug Metab. Dispos. 2009;37:555–559. doi: 10.1124/dmd.108.023911. [DOI] [PubMed] [Google Scholar]
- 45.Kusuhara H, Ito S, Kumagai Y, Jiang M, Shiroshita T, Moriyama Y, Inoue K, Yuasa H, Sugiyama Y. Effects of a MATE protein inhibitor, pyrimethamine, on the renal elimination of metformin at oral microdose and at therapeutic dose in healthy subjects. Clin. Pharmacol. Ther. 2011;89:837–844. doi: 10.1038/clpt.2011.36. [DOI] [PubMed] [Google Scholar]
- 46.Minematsu T, Giacomini KM. Interactions of Tyrosine Kinase Inhibitors with Organic Cation Transporters and Multidrug and Toxic Compound Extrusion Proteins. Mol Cancer Ther. 2011;10:531–539. doi: 10.1158/1535-7163.MCT-10-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nies AT, Hofmann U, Resch C, Schaeffeler E, Rius M, Schwab M. Proton pump inhibitors inhibit metformin uptake by organic cation transporters (OCTs). PLoS ONE. 2011;6:e22163. doi: 10.1371/journal.pone.0022163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato T, Masuda S, Yonezawa A, Tanihara Y, Katsura T, Inui K-I. Transcellular transport of organic cations in double-transfected MDCK cells expressing human organic cation transporters hOCT1/hMATE1 and hOCT2/hMATE1. Biochem. Pharmacol. 2008;76:894–903. doi: 10.1016/j.bcp.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 49.Han Y-H, Busler D, Hong Y, Tian Y, Chen C, Rodrigues AD. Transporter studies with the 3-O-sulfate conjugate of 17alpha-ethinylestradiol: assessment of human kidney drug transporters. Drug Metab. Dispos. 2010;38:1064–1071. doi: 10.1124/dmd.109.031526. [DOI] [PubMed] [Google Scholar]
- 50.More SS, Li S, Yee SW, Chen L, Xu Z, Jablons DM, Giacomini KM. Organic Cation Transporters Modulate the Uptake and Cytotoxicity of Picoplatin, a Third-Generation Platinum Analogue. Molecular Cancer Therapeutics. 2010;9:1058–1069. doi: 10.1158/1535-7163.MCT-09-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang S, Lovejoy KS, Shima JE, Lagpacan LL, Shu Y, Lapuk A, Chen Y, Komori T, Gray JW, Chen X, Lippard SJ, Giacomini KM. Organic Cation Transporters Are Determinants of Oxaliplatin Cytotoxicity. Cancer Res. 2006;66:8847–8857. doi: 10.1158/0008-5472.CAN-06-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shu Y, Sheardown SA, Brown C, Owen RP, Zhang S, Castro RA, Ianculescu AG, Yue L, Lo JC, Burchard EG, Brett CM, Giacomini KM. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J. Clin. Invest. 2007;117:1422–1431. doi: 10.1172/JCI30558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. [Jun 12, 2012]; Home - MICROMEDEX® 2.0 http://www.thomsonhc.com/micromedex2/librarian/ND_T/evidencexpert/ND_PR/evidenc expert/CS/CDB742/ND_AppProduct/evidencexpert/DUPLICATIONSHIELDSYNC/BFA79B/ND_PG/evidencexpert/ND_B/evidencexpert/ND_P/evidencexpert/PFActionId/pf.HomePage.
- 54.Thummel Kenneth E., Shen Danny D. Appendix II - Design and Optimization of Dosage Regimens: Pharmacokinetic Data. In: Hardman Joel G., Limbird Lee E., Goodman Gilman Alfred., editors. Goodman & Gilman's The Pharmacological Basis of Therapeutics. McGraw-Hill, Inc.; New York, NY: 2001. pp. 1917–2023. [Google Scholar]
- 55.Clarke's Analysis of Drugs and Poisons. Moffat Anthony C., Osselton M. David, Widdop Brian, Galichet Laurent Y. Third ed. Vol. 2. Pharmaceutical Press; London, UK: 2004. [Google Scholar]
- 56.Frye RF, Fitzgerald SM, Lagattuta TF, Hruska MW, Egorin MJ. Effect of St John's wort on imatinib mesylate pharmacokinetics. Clin. Pharmacol. Ther. 2004;76:323–329. doi: 10.1016/j.clpt.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 57.Nakaya H, Uemura H. Electropharmacology of nifekalant, a new class III antiarrhythmic drug. Cardiovasc. Drug Rev. 1998;16:133–144. [Google Scholar]
- 58.Andersson T, Holmberg J, Röhss K, Walan A. Pharmacokinetics and effect on caffeine metabolism of the proton pump inhibitors, omeprazole, lansoprazole, and pantoprazole. Br J Clin Pharmacol. 1998;45:369–375. doi: 10.1046/j.1365-2125.1998.t01-1-00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.