Abstract
Gata5 is a transcription factor expressed in the lung, but its physiological role is unknown. To test whether and how Gata5 regulates airway constrictor responsiveness, we studied Gata5−/−, Gata5+/−, and wild-type mice on the C57BL/6J background. Cholinergic airway constrictor responsiveness was assessed invasively in mice without and with induction of allergic airway inflammation through ovalbumin sensitization and aerosol exposure. Gata5-deficient mice displayed native airway constrictor hyperresponsiveness (AHR) in the absence of allergen-induced inflammation. Gata5-deficient mice retained their relatively greater constrictor responsiveness even in ovalbumin-induced experimental asthma. Gata5 deficiency did not alter the distribution of cell types in bronchoalveolar lavage fluid, but bronchial epithelial mucus metaplasia was more prominent in Gata5−/− mice after allergen challenge. Gene expression profiles revealed that apolipoprotein E (apoE) was the fifth most down-regulated transcript in Gata5-deficient lungs, and quantitative RT-PCR and immunostaining confirmed reduced apoE expression in Gata5−/− mice. Quantitative RT-PCR also revealed increased IL-13 mRNA in the lungs of Gata5-deficient mice. These findings for the first time show that Gata5 regulates apoE and IL-13 expression in vivo and that its deletion causes AHR. Gata5-deficient mice exhibit an airway phenotype that closely resembles that previously reported for apoE−/− mice: both exhibit cholinergic AHR in native and experimental asthma states, and there is excessive goblet cell metaplasia after allergen sensitization and challenge. The Gata5-deficient phenotype also shares features that were previously reported for IL-13–treated mice. Together, these results indicate that Gata5 deficiency induces AHR, at least in part, by blunting apoE and increasing IL-13 expression.
Keywords: asthma, transcription factor, apolipoprotein E, IL-13, methacholine
Clinical Relevance
This study demonstrates that the Gata5 transcription factor controls airway constrictor responsiveness and regulates apoE and IL-13 expression in mice. These findings raise the possibility that similar mechanisms might contribute to human asthma.
Asthma is a complex disease syndrome characterized by intermittent airflow obstruction, inflammatory cell infiltrates, increased mucus production, airway remodeling, and airway hyperresponsiveness. Environmental factors and genetic variation influence asthma susceptibility and severity (1–5). To understand the cellular and molecular mechanisms of asthma, identification of the genes responsible for biological variability in airway responsiveness is critical.
Gata5 is a member of the GATA transcription factor family that plays an important role in regulating gene expression during embryonic morphogenesis and cellular differentiation. Gata5 is expressed initially in various mesoderm- and endoderm-derived tissues, such as pulmonary mesenchyme and bronchial (but not vascular) smooth muscle cells, the urogenital ridge, and epithelial cells lining the urogenital sinus, the bladder, and the gut epithelium (6–8). Postnatally, Gata5 expression becomes markedly up-regulated in the lungs, intestine, stomach, bladder, and endocardium (7, 9), and here we show that Gata5 is also expressed in bronchial epithelium. Its tissue-restricted expression and functions during embryogenesis and cell differentiation strongly suggest that Gata5 might modify gene expression in airway, leading to abnormal responses to environmental stimuli.
Apolipoprotein E (apoE) is a glycoprotein encoded by the apoE gene on chromosome 19. In peripheral tissues, apoE is primarily synthesized by the hepatic parenchymal cells and macrophages, and in the brain apoE is mainly produced by astrocytes. Other cell types, such as smooth muscle cells (10) and alveolar type I cells in the rat (11), have been reported to express apoE. The primary biological function of apoE is to facilitate lipid transport into cells by endocytosis mediated by the low-density lipoprotein receptor (LDLR). An additional function of apoE is to modulate adaptive immune responses and host susceptibility to infection. A recent study reveals that apoE functions as an endogenous negative regulator of airway hyperreactivity in experimental house dust mite (HDM)-induced asthma (12–14).
IL-13 is a central mediator of experimental allergic asthma in that its exogenous administration recapitulates many features of allergen induced asthma (15), whereas its genetic deletion (16) or functional blockade (15, 17) ameliorate the allergen-induced asthma phenotype. IL-13 can be produced by migratory inflammatory and resident airway cells, including bronchial epithelial cells (18).
In this study, we evaluated the role of Gata5 in airway constrictor responsiveness in the absence or presence of allergic airway inflammation and analyzed the differential gene expression due to Gata5 gene deletion. We found that Gata5 gene deficiency significantly increases airway responses to intravenous methacholine (MCh) and reduces apoE expression and increases IL-13 expression in mice. Together, the data reported here reveal a role of Gata5 in the pathogenesis of airway hyperresponsiveness, a cardinal feature of asthma, and identify a possible mechanism via regulation of apoE and/or IL-13 expression.
Materials and Methods
Animals
Gata5−/− mice were bred from heterozygotes (19) and maintained on a C57BL/6J background. Animal care and use was IACUC approved.
Laser Capture Microdissection
Lungs and intestine from 10-week-old mice were snap frozen and stored in Optimal Cutting Temperature compound. Sections (6–8 μm) on foil membranes were laser microdissected. Contiguous areas of airway epithelium and smooth muscle, or of intestinal epithelium, were collected. Total RNA was isolated, and cDNAs were transcribed. Segments of full-length Gata5, short Gata5, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) cDNAs were amplified by PCR.
Genotyping of Gata5-Null Mice
Male Gata5 heterozygous mice harboring the exon 2/3-deleted Gata5 allele (19) were bred with C57BL/6 wild-type (WT) female mice in six backcrosses, resulting in > 95% C57BL/6 background. Gata5 genotype was determined by PCR with primers from intron 1, exon 2, and β-galactosidase. We also performed Northern analysis of stomach and small intestinal mucosa, where Gata5 is highly expressed, to confirm Gata5 mRNA deletion (20).
Allergen Sensitization and Airway Challenge and Determination of Airway Constrictor Responsiveness
Allergic airway inflammation was induced with ovalbumin (OVA) in approximately 8-week-old Gata5−/− and Gata5+/− mice essentially as described (21) but using 2 weeks of aerosol OVA challenge; age- and sex-matched controls were injected with PBS instead of OVA and received no aerosol. Airway constrictor responsiveness to intravenous MCh was assessed invasively at the time corresponding to 48 hours after the last challenge as described previously (22, 23), and tissues and cells were obtained for further analysis.
Bronchoalveolar Lavage and Flow Cytometry Analysis
Bronchoalveolar lavage (BAL) was collected using 0.7 ml of cold PBS twice and pooling the collected fluid. Cells were pelleted and resuspended in PBS, and approximately 2 × 105 BAL cells were immunolabeled for CD3 (clone 17A2), Gr-1 (clone RB6–8C5) (Becton Dickinson, Franklin Lakes, NJ), and CCR3 (clone 83,101.111) (R&D Systems, Minneapolis, MN). DAPI was used to discriminate live cells before flow cytometry on a FACS LSR-II (Becton Dickinson). Eosinophils were identified by CCR3-positive, GR-1–intermediate expression; neutrophils were identified by Gr-1–high, CCR3-negative expression; and T cells were identified by CD3-positive expression.
Histology and Immunostaining
Lungs and livers were fixed with 10% buffered formalin (lungs under 20 cm H2O). Paraffin (5-μm) sections were stained with hematoxylin and eosin, periodic acid Schiff (PAS), and trichrome stains or with goat anti-mouse apoE primary antibody (Santa Cruz Biotechnology, Santa Cruz, CA), HRP-conjugated anti-goat secondary, and 3,3′-DAB (Dako, Glostrup, Denmark). Frozen sections were stained with the same anti-apoE antibody and Alexa 594-conjugated secondary antibody and counterstained with DAPI. Fluorescence micrographs were taken on an SP5 II laser scanning confocal microscope (Leica Microsystems, Buffalo Grove, IL).
Lung sections from five Gata5−/− mice and 5 Gata5+/− mice after OVA challenge were used for analysis of mucus-containing cells as described previously (22). Scoring was performed by examining at least 20 airways per each section. Numerical scores for the abundance of PAS-positive goblet cells in each airway were determined as follows: 0, < 5% goblet cells; 1, 5 to 25% goblet cells; 2, 25 to 50% goblet cells; 3, 50 to 75% goblet cells; and 4, > 75% goblet cells, with 0 being negative and 1 to 4 being positive for PAS-staining bronchi.
Microarray Preparation and Data Analysis
Total RNA from Gata5−/− and WT mouse lungs was extracted and profiled on Affymetrix Mouse Genome 430 2.0 Arrays (Affymetrix, Santa Clara, CA). Raw expression data were processed by AEC Build (v1.3.0.187; Affymetrix, Santa Clara, CA) using RMA and MAS5 and analyzed by Partek Genomics suite (v6.6) (Partek, St. Louis, MO). Gene expression was considered significantly altered when fold change was ≥ 1.5 and false discovery rate (FDR)-corrected P ≤ 0.05. Microarray data were deposited in GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=zjshbumkaaiigfgandacc=GSE47425).
Quantitative Real-Time PCR
Quantitative real-time PCR (qRT-PCR) was performed on cleaned and DNase I–treated RNA using SYBR green (Molecular Probes, Eugene, OR) and gene-specific primers for apoE, Gata6, IL-13, and GAPDH. Relative abundances of apoE/GAPDH and Gata6/GAPDH mRNA ratios were determined by the delta-delta-Ct method; absolute ratios of IL-13/GAPDH were determined using standard curves. Reactions were run in triplicate on the same 384-well plate, and triplicates were averaged to provide the datum.
Non–Microarray-Related Statistical Analysis
ANOVA or unpaired t test was used to evaluate differences in airway response to MCh among groups of mice. Unpaired t test was used to compare differences in log2 mRNA abundances (normalized to GAPDH) determined by qRT-PCR. P < 0.05 was considered statistically significant.
Results
Gata5 Gene Expression in Mouse Airway Epithelium and Smooth Muscle
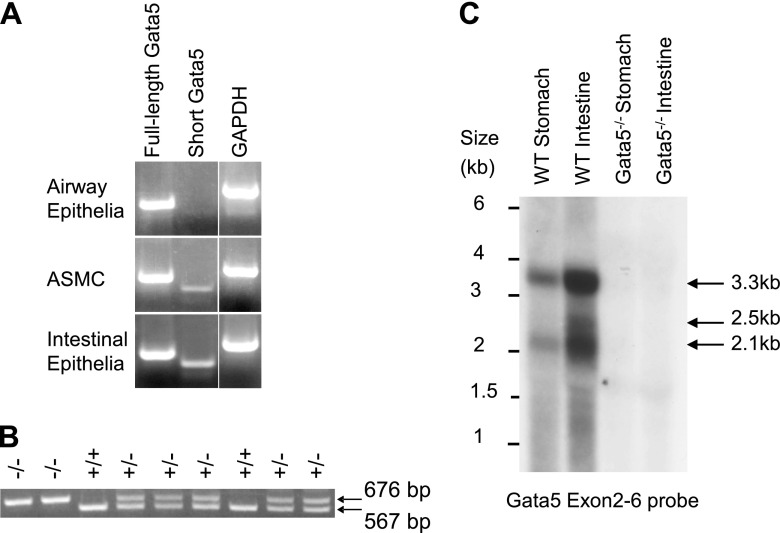
Previous studies have suggested that the Gata5 gene in lung is restricted to bronchial smooth muscle cells during postnatal development (7). We assessed the airway distribution of Gata5 mRNA in adult lungs by laser capture microdissection and qRT-PCR. mRNAs encoding full-length and short Gata5 (20) were present not only in airway smooth muscle cells but also in bronchial epithelium (Figure 1A). However, short Gata5 transcript is expressed at extremely low levels in airway epithelial cells.
Figure 1.
Detection of Gata5 gene expression. (A) Laser capture microdissection and RT-PCR demonstrated that mRNAs encoding full-length and short Gata5 were present in airway epithelial and smooth muscle cells (ASMCs), but short Gata5 transcript is expressed at extremely low levels in airway epithelial cells. Expected amplicon size is 374 bp for full-length Gata5 mRNA and 303 bp for short Gata5 mRNA. (B) The Gata5 mutant allele was identified by a 676-bp PCR product with Gata5 forward and reverse primer and wild-type (WT) allele by a 567-bp product with Gata5 forward and LacZ primer. Gata5+/− showed both products. (C) Using Gata5 exons 2–6 cDNA probe, Northern analysis was performed on total RNA from stomach and intestinal mucosa of mice that were genotyped by PCR. Gata5 transcripts (3.3, 2.5, and 2.1 kb) are expressed in the WT mouse, whereas there are no transcripts detected in the Gata5−/− mouse. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Gata5 Mutant Mice and PCR Genotyping
Genotyping was performed by PCR using a forward primer from Gata5 intron 1 and reverse primers that map to Gata5 exon 2 or to the LacZ gene in the targeting construct. The WT allele and Gata5 exon 2/3-deleted alleles yielded products of 567 and 676 bp, respectively (Figure 1B). To confirm whether deletion of exon 2/3 allele completely eliminated expression of Gata5, we randomly picked a pair of WT and Gata5−/− mice that had been genotyped by PCR, extracted total RNA from stomach and small intestinal mucosa where Gata5 is highly expressed, and performed Northern analysis using a Gata5 exon 2–6 cDNA probe (Figure 1C). Three Gata5 transcripts of 3.3, 2.5, and 2.1 kb were found in the WT mouse, as we have previously reported (20), but no Gata5-hybridizing transcripts were detected in the Gata5−/− mouse. This confirms that Gata5 exon 2/3–deficient mice are devoid of Gata5. Gata5 heterozygous and homozygous mutants grew and bred normally, as noted previously (19).
Gata5 Deficiency Causes Native Airway Constrictor Hyperresponsiveness
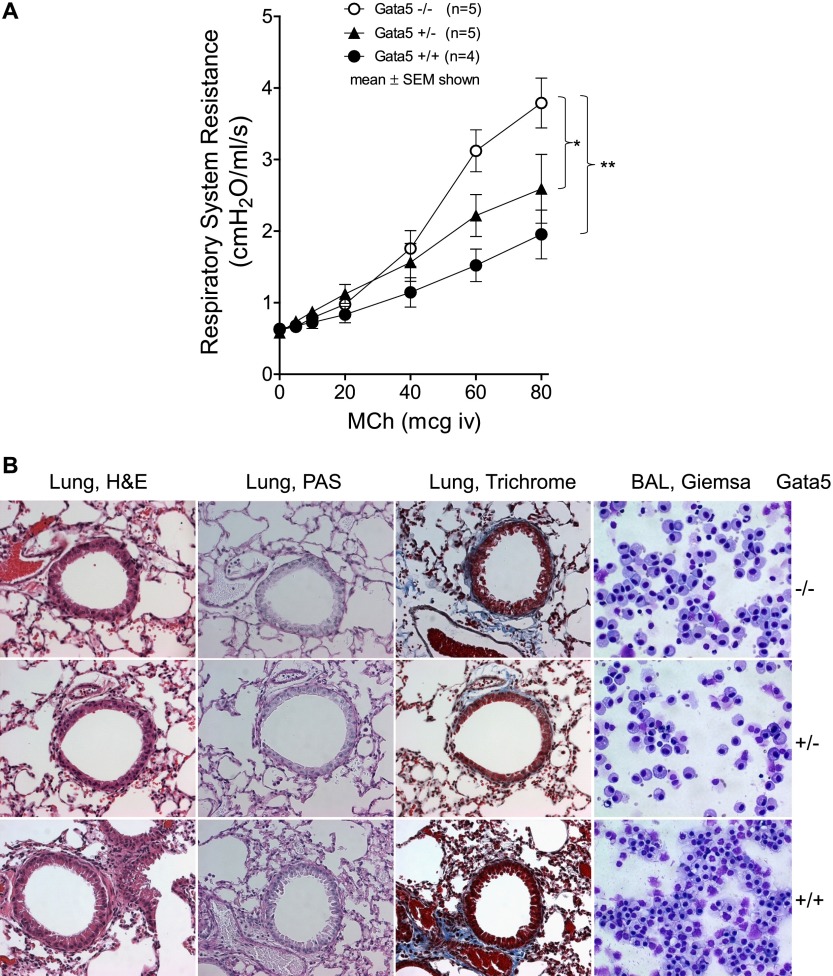
We compared respiratory system resistance responses to intravenous MCh in Gata5 exon 2/3-deleted mice with those of Gata5+/− or WT mice during mechanical ventilation at 10 ml/kg at 150 breaths/min. Gata5−/− mice had substantially greater (more than double) bronchoconstrictor responses to intravenous MCh than did WT mice (P < 0.001), whereas Gata5+/− mice had intermediate responsiveness (Figure 2A).
Figure 2.
(A) Airway constrictor responsiveness to intravenous methacholine (MCh), as reflected in increases in respiratory system resistance, was measured using the Flexivent ventilator (Scireq, Tempe, AZ). Gata5−/− mice had significantly higher bronchoconstrictor responses to intravenous MCh than did WT mice (P < 0.001), whereas Gata5+/− mice had intermediate responsiveness. (B) Representative histological appearance of lungs and bronchoalveolar lavage (BAL) cells in Gata5−/−, Gata5+/−, and WT mice (original magnification, ×40). The airway walls, pulmonary blood vessels, lung parenchyma, and BAL cells appear normal, as judged by qualitative examination of hematoxylin and eosin (H&E), periodic acid Schiff (PAS) (mucus would have stained deep purple), trichrome (fibrotic regions stain blue), and Giemsa stains. There is no evidence of inflammatory cell infiltration, lung edema, mucus metaplasia, excessive fibrosis, or changes of BAL cell composition that might alter airway responsiveness.
The histological appearance of lungs from Gata5-deficient mice was qualitatively normal (Figure 2B). There was no evidence of inflammatory cell infiltration, lung edema, mucus metaplasia, or excessive fibrosis that might alter airway responsiveness. The airway walls, pulmonary blood vessels, and lung parenchyma appeared grossly normal. The composition of BAL cells collected from Gata5−/−, Gata5+/−, and WT mice was predominantly macrophages with a few epithelial cells, which did not show morphological difference among groups. There were few inflammatory cells present in the BAL of Gata5−/−, Gata5+/−, or WT mice. Thus, Gata5 deficiency did not alter the histological appearance of lung or the cell types in BAL fluid. These results indicate that Gata5 deficiency causes native airway constrictor hyperresponsiveness (AHR) without airway inflammation.
Airway Constrictor Responsiveness Remains Greater in Gata5−/− than in Gata5+/− Mice Even in the Presence of Allergic Airway Inflammation
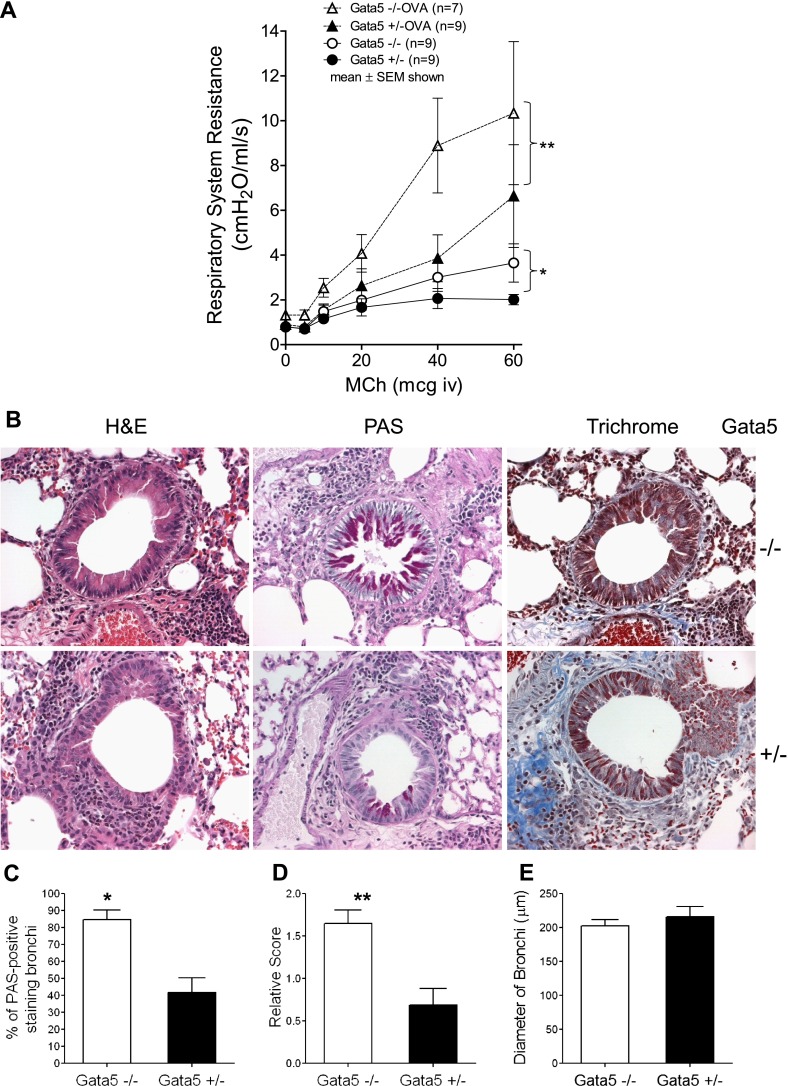
To determine whether Gata5 deficiency–induced airway hyperresponsiveness is manifest in the presence of allergic airway inflammation, Gata5−/− and Gata5+/− mice were sensitized by intraperitoneal injection of OVA and alum and exposed to OVA aerosol 3 d/wk for 2 weeks; control animals received sham sensitization. OVA sensitization and challenge significantly increased MCh responsiveness in Gata5−/− and Gata5+/− mice, but Gata5−/− mice retained their relatively greater responsiveness in allergen challenged cohorts compared with Gata5+/− mice (P = 0.007) (Figure 3A).
Figure 3.
(A) Respiratory system resistance in ovalbumin (OVA)-sensitized and challenged mice. Allergic airway inflammation induced by OVA significantly increased constrictor responsiveness to intravenous MCh in Gata5−/− and Gata5+/− mice. However, Gata5−/− mice retained their relatively greater responsiveness in allergen-challenged cohorts compared with Gata5+/− mice (P < 0.007). (B) Lung histology from OVA-sensitized or OVA-challenged Gata5−/− and Gata5+/− mice (original magnification, ×40). Formalin-fixed tissue was sectioned and stained with H&E, PAS (mucus would have stained deep purple), and trichrome staining (fibrotic regions stain blue). The abundance of inflammatory cells was infiltrated around airways, particularly in regions the adjacent to blood vessels. (C) Quantification of percent of bronchi demonstrating mucus metaplasia (> 5% of cells staining PAS positive). Gata5−/− mice exhibited considerably more bronchi with mucus metaplasia than did Gata5+/− mice. *P < 0.01. (D) The abundance of PAS-positive mucus-containing epithelial cells in the airways of Gata5−/− mice was more than twice that found in Gata5+/− mice. **P < 0.005. (E) The size of bronchi evaluated for PAS-positive scoring was similar between groups.
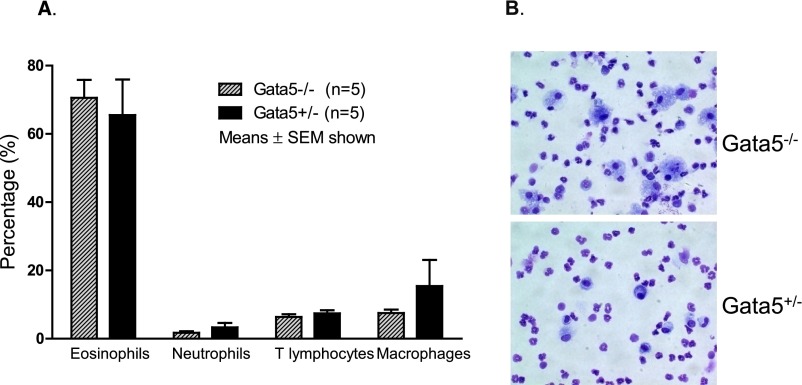
Histology demonstrated that OVA sensitization and challenge induced airway inflammation and hypertrophy of bronchial epithelial cells in Gata5−/− and Gata5+/− mice. The abundance of inflammatory cells was observed around airways, particularly in regions adjacent to blood vessels. PAS-positive mucus-containing cells were present among the hypertrophied airway epithelial cells in Gata5−/− and Gata5+/− mice. However, the increase in PAS-positive cells was clearly more marked in Gata5−/− mice (Figures 3B–3D). OVA sensitization and challenge significantly elevated eosinophils in BAL fluid in Gata5−/− and Gata5+/− mice. Flow cytometry analysis revealed that 70.58% of BAL cells were eosinophils after OVA sensitization and challenge in Gata5−/− mice and 65.51% in Gata5+/− mice (Figure 4). There was no significant difference in BAL cell composition between OVA-challenged Gata5−/− mice and Gata5+/− mice.
Figure 4.
BAL was collected from OVA-sensitized and OVA-challenged Gata5−/− and Gata5+/− mice and resuspended in PBS for cytospin and flow cytometry analysis. (A) OVA sensitization and challenge significantly elevated eosinophils in BAL fluid in Gata5−/− and Gata5+/− mice. Flow cytometry analysis revealed that 70.58% of BAL cells were eosinophils after OVA sensitization and challenge in Gata5−/− mice and that 65.51% were eosinophils in Gata5+/− mice. There is no significant difference of BAL cell composition between OVA-challenged GATA5−/− mice and Gata5 +/− mice. (B) Cytospins showing typical BAL cells from Gata5−/− and Gata5+/− mice (40×).
Together, these data indicate that Gata5 regulates cholinergic airway responsiveness in the absence or presence of allergic airway inflammation in a mechanism that does not involve induction or worsening of airway inflammation, although Gata5 deficiency does potentiate allergic airway inflammation–induced mucus metaplasia.
Identification of Differentially Expressed Genes Affected by Gata5 Gene Deletion
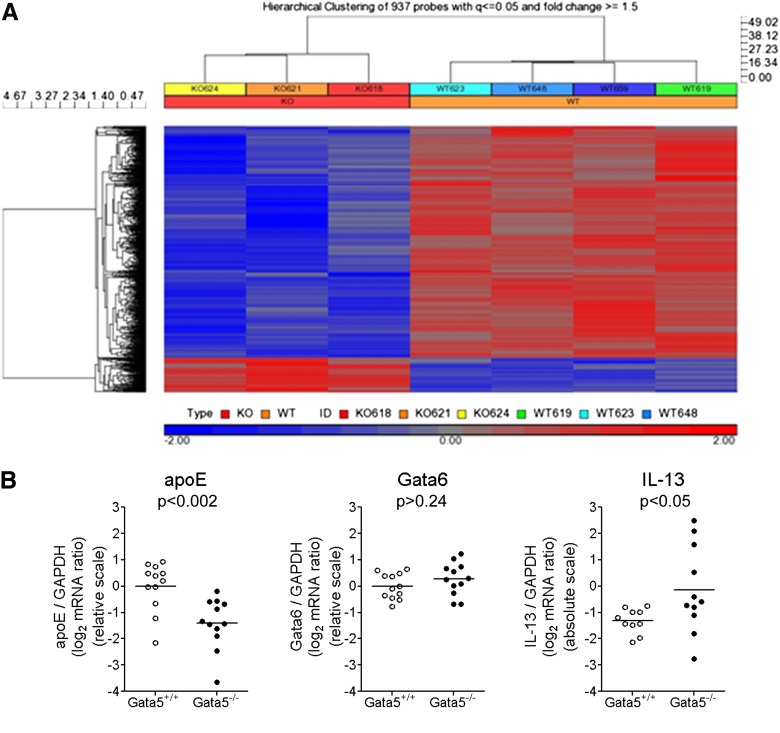
To understand the molecular mechanisms by which Gata5 deficiency leads to AHR, we compared gene expression profiles of lungs from Gata5−/− and WT mice using the Affymetrix Mouse Genome 430 2.0 Array. Total RNA from five Gata5−/− and five WT mouse left lungs was extracted with TRIzol reagent and further cleaned using RNeasy kit. Microarray data were collected following the Affymetrix GeneChip standard protocol and profiling by the University of Chicago Functional Genomics Core. The samples from three Gata5−/− and four WT mice passed quality criteria and were further processed with the Affymetrix Expression Console Build v1.3.0.187 using the RMA and MAS5 algorithms. Expression data were filtered by detection P value ≤ 0.05 in at least three samples, and ANOVA was used to detect differentially expressed genes between Gata5−/− and WT mice. Gata5 gene deletion significantly altered the expression of 843 unique genes (937 probe sets), with 731 genes being down-regulated and 112 genes being up-regulated (fold change ≥ 1.5; P ≤ 0.05; FDR q ≤ 0.05) (Figure 5A).
Figure 5.
ApoE expression is significantly down-regulated due to Gata5 gene deletion. (A) Hierarchical clustering of 834 differentially expressed genes from lungs of three Gata5−/− and four WT mice. Of these, 731 were down-regulated, and 112 were up-regulated due to Gata5 gene deletion. The heat map represents the expression level for each sample, with blue signifying low expression and red signifying high expression. (B) qRT-PCR using lung mRNA from Gata5+/+ and Gata5−/− mice demonstrated significant reduction apolipoprotein E (apoE) mRNA (P < 0.002) and elevation of IL-13 mRNA (P < 0.05) when normalized to GAPDH expression. However, there was no significant change of Gata6 transcript after Gata5 gene deletion (P > 0.24).
ApoE Expression Was Significantly Down-Regulated in Gata5−/− Mice
ApoE was previously reported as an endogenous negative regulator of airway hyperresponsiveness and goblet cell hyperplasia in HDM-induced experimental asthma (12, 13). Our microarray analysis revealed that apoE is the fifth most highly down-regulated transcript caused by Gata5 deletion (fold change = 3.07; P = 0.0019; FDR q = 0.0145). qRT-PCR using RNA from 12 Gata5−/− and 12 WT mouse lungs confirmed significant reduction of apoE transcript (P < 0.002) (Figure 5B). However, Gata6 expression in mouse lungs was not altered by Gata5 gene deletion (P > 0.24).
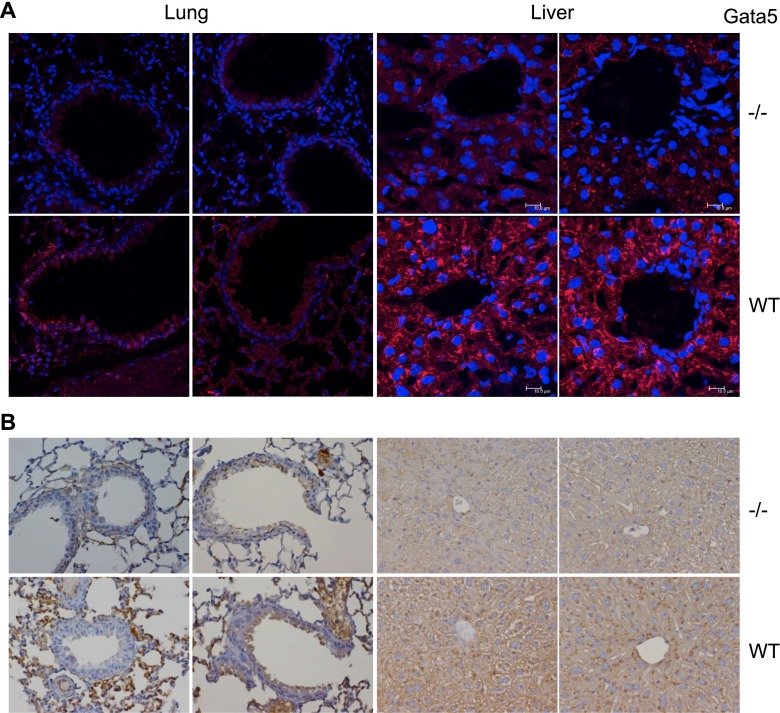
Approximately three fourths of plasma apoE is normally synthesized by hepatocytes (10). Therefore, we evaluated apoE protein expression in Gata5−/− and WT mouse liver and lung using immunofluorescence and immunohistochemical staining. Both methods revealed a marked reduction of apoE protein in Gata5−/− mouse liver and lung compared with WT controls (Figure 6). Taken together, these data demonstrate that Gata5 deficiency significantly reduced apoE expression in mice.
Figure 6.
Immunofluorescence (A, red) and immunohistochemical (B, brown) staining for apoE demonstrated that Gata5 deficiency reduced apoE protein expression of liver and lung tissues in Gata5−/− mouse compared with WT controls.
IL-13 Expression Was Significantly Up-Regulated in Gata5−/− Mice
Exogenous administration of IL-13 is sufficient to cause airway hyperresponsiveness and increased bronchial mucus in mice (15). In light of the abnormally increased cholinergic constrictor responsiveness demonstrated in the absence (Figure 2) or presence of allergic airway inflammation in Gata5-deficient mice and in light of their exaggerated bronchial mucus metaplasia after allergen sensitization and challenge (Figure 3), we wondered whether Gata5 deletion might have altered IL-13 expression. To test this possibility, we prepared RNA from whole lung homogenates of 11 Gata5−/− and 10 Gata5+/+ mice (these mice were not allergen exposed) and quantified IL-13 mRNA in these specimens using qRT-PCR. Copy numbers were interpolated from a simultaneously amplified standard curve generated with known quantities of the amplicon, and IL-13 mRNA abundance was normalized to that of GAPDH mRNA in the same sample. We found that lungs from Gata5-deficient mice contain substantially more IL-13 mRNA than do those of Gata5-replete mice (P < 0.05) (Figure 5B).
Discussion
AHR is characteristic of atopic and nonatopic asthmatic subjects, and AHR has a strong genetic component (1, 2, 24). Our data indicate that Gata5 is expressed airway epithelial and smooth muscle cells (Figure 1) and that Gata5−/− mice have substantially greater bronchoconstrictor responses to intravenous MCh than do Gata5+/− or WT mice in the absence of allergen-induced airway inflammation (Figure 2A). Gata5 deficiency alone did not alter the histological appearance of lung or of cell types in BAL fluid (Figure 2B). These results demonstrate that Gata5 gene deficiency causes non–inflammation-induced (i.e., native) AHR. Furthermore, Gata5−/− mice retain their relatively greater responsiveness in the presence of allergen-induced airway inflammation (Figure 3A). Gata5 deficiency does not change the intensity or nature of such inflammation (Figures 3B and 4) but does potentiate airway inflammation–induced mucus metaplasia of the bronchial epithelium (Figures 3C and 3D). Therefore, Gata5 is a previously unrecognized candidate gene whose expression or silencing might alter the genetic predisposition to AHR, a cardinal feature of asthma.
Gata5 is a transcription factor that plays an important role in regulating gene expression during embryonic morphogenesis and cellular differentiation. To appreciate the molecular mechanisms by which Gata5 deficiency induces native AHR in mice, we compared gene expression profiles of lungs from Gata5−/− and WT mice and found that 731 genes were down-regulated and 112 genes up-regulated due to Gata5 gene deletion (Figure 5A). One of the most highly down-regulated genes is apoE, which was previously identified as an endogenous negative regulator of airway responsiveness and goblet cell hyperplasia in experimental allergic asthma (12–14) (discussed further below). We confirmed that apoE expression is significantly reduced in Gata5−/− mice at the mRNA (Figure 5B) and protein (Figure 6) levels. We therefore infer that Gata5 deficiency induces AHR at least in part by reducing apoE expression. The human apoE promoter is known to be regulated by GATA factors, which bind at a polymorphic site at −219 bp relative to the transcription start site (25). A search of the mouse apoE promoter region against the TRANSFAC database reveals possible GATA binding sites at −413 and −496 bp. In addition, Gata5 is highly expressed in liver (20), the site of most apoE production. Thus, regulation of mouse apoE expression by Gata5 binding to its gene promoter seems plausible. Further study is required to test this directly. It is conceivable that other mechanisms independent of apoE contribute to the phenotype induced by Gata5 gene deletion.
Beyond its primary function in regulating lipid transport and metabolism (10), adaptive immune responses, and host susceptibility to infection (26), apoE is known to regulate airway constrictor responsiveness (12, 13). ApoE-deficient mice exhibit native airway constrictor hyperresponsivess (12, 13). AHR was significantly further increased and goblet cell hyperplasia accentuated in HDM-challenged apoE−/− mice compared with HDM-challenged WT mice, and administration of an apoE mimetic peptide rescued the asthmatic phenotype of apoE−/− mice (13). Furthermore, HDM-challenged LDLR−/− mice displayed a similar phenotype as apoE−/− mice, but the apoE mimetic peptide did not attenuate AHR in sensitized and challenged LDLR−/− mice (13). Thus, apoE exerts its airway effect through an apoE–LDLR pathway (13).
Like apoE-deficient mice, our Gata5-deficient mice exhibited native AHR without spontaneous inflammation or an increase in airway mucus production (Figure 2). Also like apoE−/− mice, our Gata5-deficient mice exhibited greater AHR after allergen sensitization and challenge than did their heterozygous counterparts (Figure 3), and the degree of allergen-induced inflammation was similar in Gata5−/− and Gata5+/− mice (Figure 4). Furthermore, paralleling a similar observation in apoE−/− mice, our Gata5−/− mice exhibited accentuated goblet cell hyperplasia after allergen challenge (Figures 3B–3D). These close phenotypic parallels between Gata5-deficient and apoE-deficient mice, together with the observed reduction of apoE expression in Gata5-deficient mice, imply that apoE deficiency is one mechanism responsible for cholinergic AHR in Gata5 deficiency.
Another mechanism that seems likely to contribute to the Gata5-deficient phenotype is the increased lung expression of IL-13. IL-13 mRNA was increased almost 4-fold on average in the lungs of Gata5−/− mice in the absence of allergen sensitization and challenge. Because exogenous IL-13 administration can induce an asthma-like phenotype (15), it seems likely that the increased expression of IL-13 in Gata5-deficient mice contributes to their asthma-like features as well. However, we have not tested this association causally; experiments to determine whether Gata5−/− phenotype persists in the additional presence of IL-13 deficiency or blockade could address this possibility.
IL-13 and apoE expression have been inversely associated in two different paradigms. First, apoE-peptide replacement reduced IL-13 expression in the lungs of allergen-inflamed apoE-deficient mice (13), although this treatment also blunted inflammation broadly. Second, in a study of plasma biomarkers associated with apoE genotype and Alzheimer’s disease (27), there was an inverse relationship between plasma IL-13 and plasma apoE levels when considered across apoE allele groups. As such, although Gata5 deletion reduces lung apoE expression and increases lung IL-13 expression, it is uncertain whether these are independent consequences of Gata5 deletion or if one consequence of Gata5 deletion leads to the other. Note that Gata5 (Figure 1), apoE (Figure 6), and IL-13 (18) are all expressed in bronchial epithelium.
Our results raise several questions worthy of future study: (1) What is the relationship between apoE and IL-13 expression changes that result from Gata5 deficiency? (2) What are the molecular mechanisms by which Gata5-deficiency alters apoE and/or IL-13 expression? (3) Is the observed AHR specific to cholinergic stimulation? We anticipate that elucidation of these questions will raise attractive hypotheses about parallel mechanisms that might operate in human asthma.
Acknowledgments
Acknowledgments
The authors thank Jason Churchill for assistance in measuring mouse respiratory system resistance.
Footnotes
This work was supported by National Institutes of Health grants K12HL090003, CTSA UL1TR000430, and SCOR P50HL56399.
Author Contributions: Study conception and design: B.C. and J.S. Execution of experiments: B.C., T.V.M., Z.L. Data analysis and interpretation: B.C., A.I.S., C.Z., J.A., A.R., N.B., Y.H., E.E.M., P.J.G., J.S. Writing the manuscript: B.C., J.S.
Originally Published in Press as DOI: 10.1165/rcmb.2013-0294OC on November 7, 2013
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Postma DS, Bleecker ER, Amelung PJ, Holroyd KJ, Xu J, Panhuysen CI, Meyers DA, Levitt RC. Genetic susceptibility to asthma: bronchial hyperresponsiveness coinherited with a major gene for atopy. N Engl J Med. 1995;333:894–900. doi: 10.1056/NEJM199510053331402. [DOI] [PubMed] [Google Scholar]
- 2.Tremblay K, Daley D, Chamberland A, Lemire M, Montpetit A, Laviolette M, Musk AW, James AL, Chan-Yeung M, Becker A, et al. Genetic variation in immune signaling genes differentially expressed in asthmatic lung tissues. J Allergy Clin Immunol. 2008;122:529–536, e517. doi: 10.1016/j.jaci.2008.05.049. [DOI] [PubMed] [Google Scholar]
- 3.Nicolae D, Cox NJ, Lester LA, Schneider D, Tan Z, Billstrand C, Kuldanek S, Donfack J, Kogut P, Patel NM, et al. Fine mapping and positional candidate studies identify HLA-G as an asthma susceptibility gene on chromosome 6p21. Am J Hum Genet. 2005;76:349–357. doi: 10.1086/427763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, Torrey D, Pandit S, McKenny J, Braunschweiger K, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002;418:426–430. doi: 10.1038/nature00878. [DOI] [PubMed] [Google Scholar]
- 5.Xu J, Meyers DA, Ober C, Blumenthal MN, Mellen B, Barnes KC, King RA, Lester LA, Howard TD, Solway J, et al. Collaborative Study on the Genetics of Asthma. Genomewide screen and identification of gene-gene interactions for asthma-susceptibility loci in three U.S. populations: collaborative study on the genetics of asthma. Am J Hum Genet. 2001;68:1437–1446. doi: 10.1086/320589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laverriere AC, MacNeill C, Mueller C, Poelmann RE, Burch JB, Evans T. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J Biol Chem. 1994;269:23177–23184. [PubMed] [Google Scholar]
- 7.Morrisey EE, Ip HS, Tang Z, Lu MM, Parmacek MS. GATA-5: a transcriptional activator expressed in a novel temporally and spatially-restricted pattern during embryonic development. Dev Biol. 1997;183:21–36. doi: 10.1006/dbio.1996.8485. [DOI] [PubMed] [Google Scholar]
- 8.Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- 9.Peterkin T, Gibson A, Loose M, Patient R. The roles of GATA-4, -5 and -6 in vertebrate heart development. Semin Cell Dev Biol. 2005;16:83–94. doi: 10.1016/j.semcdb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 11.Caselli RJ, Chen K, Bandy D, Smilovici O, Boeve BF, Osborne D, Alexander GE, Parish JM, Krahn LE, Reiman EM. A preliminary fluorodeoxyglucose positron emission tomography study in healthy adults reporting dream-enactment behavior. Sleep. 2006;29:927–933. doi: 10.1093/sleep/29.7.927. [DOI] [PubMed] [Google Scholar]
- 12.Yao X, Dai C, Fredriksson K, Lam J, Gao M, Keeran KJ, Nugent GZ, Qu X, Yu ZX, Jeffries N, et al. Human apolipoprotein E genotypes differentially modify house dust mite-induced airway disease in mice. Am J Physiol Lung Cell Mol Physiol. 2012;302:L206–L215. doi: 10.1152/ajplung.00110.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao X, Fredriksson K, Yu ZX, Xu X, Raghavachari N, Keeran KJ, Zywicke GJ, Kwak M, Amar MJ, Remaley AT, et al. Apolipoprotein E negatively regulates house dust mite-induced asthma via a low-density lipoprotein receptor-mediated pathway. Am J Respir Crit Care Med. 2010;182:1228–1238. doi: 10.1164/rccm.201002-0308OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao X, Vitek MP, Remaley AT, Levine SJ. Apolipoprotein mimetic peptides: a new approach for the treatment of asthma. Front Pharmacol. 2012;3:37. doi: 10.3389/fphar.2012.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, Donaldson DD. Interleukin-13: central mediator of allergic asthma. Science. 1998;282:2258–2261. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 16.Walter DM, McIntire JJ, Berry G, McKenzie AN, Donaldson DD, DeKruyff RH, Umetsu DT. Critical role for IL-13 in the development of allergen-induced airway hyperreactivity. J Immunol. 2001;167:4668–4675. doi: 10.4049/jimmunol.167.8.4668. [DOI] [PubMed] [Google Scholar]
- 17.Grünig G, Warnock M, Wakil AE, Venkayya R, Brombacher F, Rennick DM, Sheppard D, Mohrs M, Donaldson DD, Locksley RM, et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282:2261–2263. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allahverdian S, Harada N, Singhera GK, Knight DA, Dorscheid DR. Secretion of IL-13 by airway epithelial cells enhances epithelial repair via HB-EGF. Am J Respir Cell Mol Biol. 2008;38:153–160. doi: 10.1165/rcmb.2007-0173OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh MK, Li Y, Li S, Cobb RM, Zhou D, Lu MM, Epstein JA, Morrisey EE, Gruber PJ. Gata4 and Gata5 cooperatively regulate cardiac myocyte proliferation in mice. J Biol Chem. 2010;285:1765–1772. doi: 10.1074/jbc.M109.038539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen B, Yates E, Huang Y, Kogut P, Ma L, Turner JR, Tao Y, Camoretti-Mercado B, Lang D, Svensson EC, et al. Alternative promoter and GATA5 transcripts in mouse. Am J Physiol Gastrointest Liver Physiol. 2009;297:G1214–G1222. doi: 10.1152/ajpgi.00165.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirano A, Kanehiro A, Ono K, Ito W, Yoshida A, Okada C, Nakashima H, Tanimoto Y, Kataoka M, Gelfand EW, et al. Pirfenidone modulates airway responsiveness, inflammation, and remodeling after repeated challenge. Am J Respir Cell Mol Biol. 2006;35:366–377. doi: 10.1165/rcmb.2005-0452OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong J, Bandulwala HS, Clay BS, Anders RA, Shilling RA, Balachandran DD, Chen B, Weinstock JV, Solway J, Hamann KJ, et al. Fas-positive T cells regulate the resolution of airway inflammation in a murine model of asthma. J Exp Med. 2006;203:1173–1184. doi: 10.1084/jem.20051680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen B, Liu G, Shardonofsky F, Dowell M, Lakser O, Mitchell RW, Fredberg JJ, Pinto LH, Solway J. Tidal breathing pattern differentially antagonizes bronchoconstriction in C57BL/6J vs. A/J mice. J Appl Physiol (1985) 2006;101:249–255. doi: 10.1152/japplphysiol.01010.2004. [DOI] [PubMed] [Google Scholar]
- 24.Wills-Karp M, Ewart SL. The genetics of allergen-induced airway hyperresponsiveness in mice. Am J Respir Crit Care Med. 1997;156:S89–S96. doi: 10.1164/ajrccm.156.4.12-tac-3. [DOI] [PubMed] [Google Scholar]
- 25.Maloney B, Ge YW, Petersen RC, Hardy J, Rogers JT, Pérez-Tur J, Lahiri DK. Functional characterization of three single-nucleotide polymorphisms present in the human APOE promoter sequence: differential effects in neuronal cells and on DNA-protein interactions. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:185–201. doi: 10.1002/ajmg.b.30973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allan LL, Hoefl K, Zheng DJ, Chung BK, Kozak FK, Tan R, van den Elzen P. Apolipoprotein-mediated lipid antigen presentation in B cells provides a pathway for innate help by NKT cells. Blood. 2009;114:2411–2416. doi: 10.1182/blood-2009-04-211417. [DOI] [PubMed] [Google Scholar]
- 27.Soares HD, Potter WZ, Pickering E, Kuhn M, Immermann FW, Shera DM, Ferm M, Dean RA, Simon AJ, Swenson F, et al. Biomarkers Consortium Alzheimer’s Disease Plasma Proteomics Project. Plasma biomarkers associated with the apolipoprotein E genotype and Alzheimer disease. Arch Neurol. 2012;69:1310–1317. doi: 10.1001/archneurol.2012.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]