Abstract
Non-small cell lung cancers (NSCLC) harboring anaplastic lymphoma kinase (ALK) gene rearrangements invariably develop resistance to the ALK tyrosine kinase inhibitor (TKI) crizotinib. Herein, we report the first preclinical evaluation of the next-generation ALK TKI, ceritinib (LDK378) in the setting of crizotinib resistance. Interrogation of in vitro and in vivo models of acquired resistance to crizotinib, including cell lines established from biopsies of crizotinib-resistant NSCLC patients revealed that ceritinib potently overcomes crizotinib resistance mutations. In particular, ceritinib effectively inhibits ALK harboring L1196M, G1269A, I1171T and S1206Y mutations, and a co-crystal of ceritinib bound to ALK provides structural bases for this increased potency. However, we observed that ceritinib did not overcome two crizotinib-resistant ALK mutations, G1202R and F1174C, and one of these mutations were identified in 5 out of 11 biopsies from patients with acquired resistance to ceritinib. Altogether our results demonstrate that ceritinib can overcome crizotinib resistance, consistent with clinical data showing marked efficacy of ceritinib in patients with crizotinib-resistant disease.
Keywords: NSCLC, ALK, ceritinib, LDK378, resistance mechanisms
INTRODUCTION
Chromosomal rearrangements of ALK (anaplastic lymphoma kinase) are detected in 3–7% of NSCLCs (1, 2). These rearrangements result in constitutively active ALK fusion proteins with potent transforming activity (2, 3). Lung cancers with ALK rearrangements are highly sensitive to ALK tyrosine kinase inhibition, underscoring the notion that such cancers are addicted to ALK kinase activity. Based on early phase studies, the multi-targeted tyrosine kinase inhibitor (TKI) crizotinib was approved by the FDA in 2011 to treat patients with advanced NSCLC harboring ALK rearrangements (1). However, despite a high response rate of 60% in ALK-rearranged NSCLC, most patients develop resistance to crizotinib, typically within one to two years.
Studies of ALK-rearranged lung cancers with acquired resistance to crizotinib have identified ALK fusion gene amplification and secondary ALK tyrosine kinase (TK) domain mutations in about one-third of cases (4-6). To date, seven different acquired resistance mutations have been identified among crizotinib-resistant patients. The most frequently identified secondary mutations are L1196M and G1269A. In addition to these mutations, the 1151Tins, L1152R, C1156Y, G1202R, and S1206Y mutations have also been detected in crizotinib-resistant cancers (4, 6-10). In approximately one-third of crizotinib-resistant tumors, there is evidence of activation of bypass signaling tracts such as EGFR or c-KIT (6, 9). In the remaining one-third of crizotinib-resistant tumors, the resistance mechanisms remain to be identified.
Next-generation ALK inhibitors with improved potency and selectivity compared to crizotinib have been developed in order to overcome crizotinib resistance in the clinic. We previously evaluated the ability of several ALK TKIs (TAE684, AP26113, ASP3026 and CH5424802) to inhibit ALK activity in models harboring different ALK secondary mutations (6, 11). These studies revealed variable sensitivity to these ALK inhibitors depending on the specific resistance mutation present. For example, the gatekeeper L1196M mutation was sensitive to TAE684, AP26113 and ASP3026, whereas 1151T-ins conferred resistance to all next generation ALK TKIs. Ceritinib is an ATP-competitive, potent and selective next-generation ALK inhibitor (12). The kinase selectivity has been tested in a cellular proliferation assay against 16 different kinases, and aside from ALK, no inhibition below 100 nM was observed (12). In the phase I study of ceritinib in ALK-positive NSCLC, marked antitumor activity has been observed in both crizotinib-relapsed and crizotinib-naïve patients (13, 14). Based on this impressive clinical activity, ceritinib received FDA approval on April 29, 2014.
Herein, we present the first report examining the activity of ceritinib in preclinical models of ALK-positive lung cancer with acquired resistance to crizotinib, as well as early biological insight into mechanisms of resistance to ceritinib arising in patients.
RESULTS
Ceritinib exhibits potent activity in crizotinib-naive ALK-positive NSCLC models
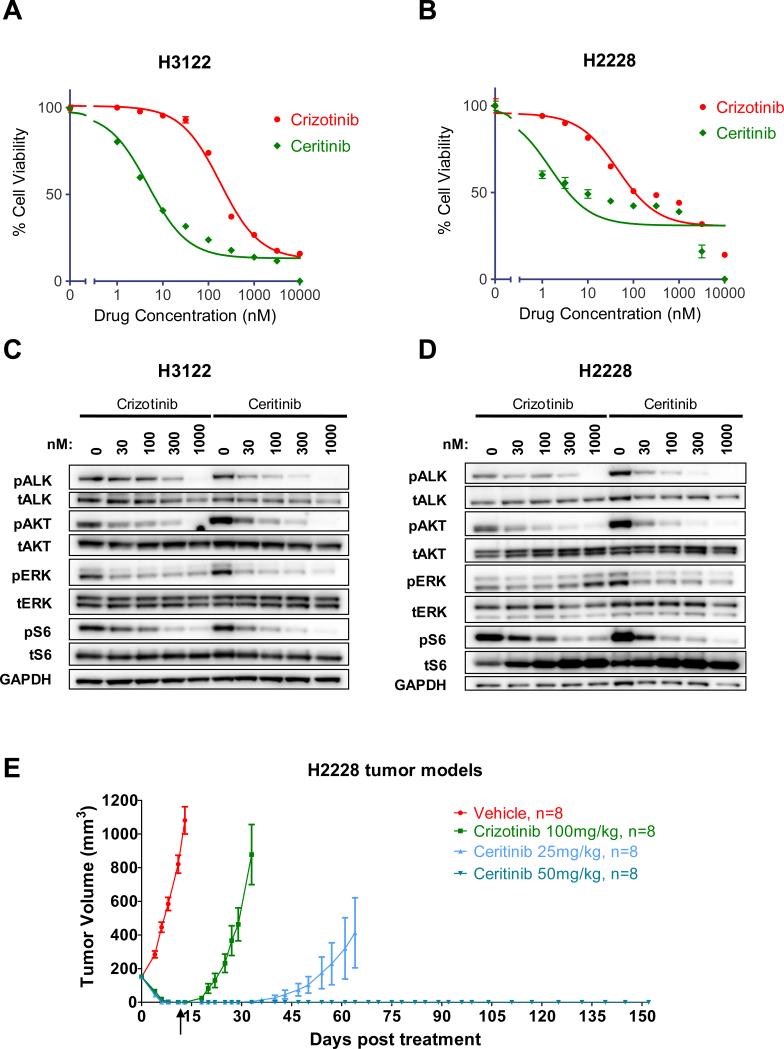
In vitro enzymatic studies revealed that ceritinib was ~20 fold more potent against ALK than crizotinib (Table 1). Similarly, ceritinib was more potent than crizotinib against two ALK-rearranged lung cancer cell lines, H3122 and H2228 (Fig.1A and B, Table 1). Accordingly, ceritinib led to suppression of ALK phosphorylation as well as the downstream PI3K/AKT, MEK/ERK and mTOR signaling pathways at lower doses than crizotinib (Fig.1C and D).
Table 1.
Ceritinib is a potent ALK inhibitor.
| GI50 (nM) | |||
|---|---|---|---|
| Crizotinib | Ceritinib | Fold | |
| ALK enzymatic assay | 3 | 0.15 | 20 |
| H2228 | 107 | 3.8 | 28 |
| H3122 | 245 | 6.3 | 39 |
GI50 values for in vitro ALK enzymatic assay or H3122 and H2228 cell survival assay for crizotinib and ceritinib.
Figure 1. Ceritinib is a potent ALK inhibitor on crizotinib naïve models.
A-B) Cell survival assay of H3122 (A) and H2228 (B) cells treated with the indicated doses of crizotinib or ceritinib for 72 hours. The cell survival was assayed by Cell-Titer-Glo. C-D) H3122 (C) and H2228 (D) cells were treated with the indicated concentrations of crizotinib or ceritinib for 6 hours. Lysates were probed with antibodies directed against the specified proteins. E) SCID beige bearing H2228 cells were administered Crizotinib or ceritinib orally once daily for 14 days. The arrow indicated when treatments were stopped and tumor growth was monitored in animals up to 4 months. Tumor volumes are presented as mean +- SD (n=8).
To further assess the cellular specificity of ceritinib, we determined the GI50 of ceritinib against a panel of tumor cell lines bearing different oncogenic drivers. Whereas ceritinib was potent against the two lung cancer cell lines with ALK-rearrangements, it was not potent against NSCLC or breast cancer cell lines driven by KRAS, EGFR, PI3K or HER2, with GI50s > 1μM (Fig.S1A).
We next compared the efficacy of ceritinib and crizotinib in vivo using treatment-naïve H2228 xenograft models (Fig.1E). Tumor-bearing animals were treated with either high-dose crizotinib (100mg/kg) or ceritinib (25 mg/kg or 50 mg/kg) once daily for 14 days. Both crizotinib (100 mg/kg) and LDK (25 and 50 mg/kg) were well tolerated in this study (Fig.S1B). As expected, marked tumor regression was observed in all groups during the treatment. After treatment was stopped, the animals were monitored for tumor progression. While recurrent tumors were detected within 11 days of drug withdrawal in mice treated with crizotinib, mice treated with ceritinib at 50 mg/kg remained in complete remission with no discernible tumor growth for 4 months. In the mice treated with ceritinib at 25 mg/kg, tumor re-growth was observed in 4 out of 8 animals after 1 month, whereas complete remission was maintained in the other 4 animals for 4 months. Thus LDK had more durable anti-tumor activity than crizotinib, even after the drugs were discontinued. It is also worth noting that the exposure of crizotinib at 100 mg/kg is ~ 3-5 fold greater than the exposures achieved at the human MTD (250 mg, BID)(15) and that ceritinib at 25-50 mg/kg is predicted to be achievable at the human MTD (750mg QD). We also evaluated the efficacy of ceritinib in a primary explant model derived from a crizotinib-naïve NSCLC tumor MGH006 (6). Treatment of these mice with 25 mg/kg ceritinib also led to tumor regressions (Fig.S1C). Altogether, these data demonstrate that ceritinib is potent against crizotinib-naïve ALK-rearranged cell lines and tumor models in vivo and in vitro.
Ceritinib is active against patient-derived cell lines from crizotinib-resistant cancers with and without resistant mutations
To investigate the activity of ceritinib against crizotinib-resistance mutations, we used crizotinib-resistant cell line models harboring the two most common EML4-ALK mutations L1196M and G1269A. We have previously described the H3122 CR1 crizotinib-resistant cell line, which developed resistance in vitro by chronic exposure to crizotinib. This cell line harbors both the L1196M EML4-ALK gatekeeper mutation and amplification of the EML4-ALK allele (11). In addition, we also examined two novel cell lines established from biopsies of patients whose ALK-rearranged lung cancers had become resistant to crizotinib in the clinic. These two patient-derived resistant lines, MGH045 and MGH021-4, harbor the L1196M and G1269A mutations, respectively. The MGH021-4 line is a clonal cell line established from MGH021, a tumor harboring both 1151Tins and G1269A mutations; MGH021-4 cells harbor only the G1269A mutation (5). This clone therefore represents an early generation of the patient derived cell line. The GI50 values of ceritinib against all of these resistant cell lines were decreased 6- to 36-fold compared to crizotinib (Fig.2A and S2A-C). Accordingly, phosphorylation of ALK and downstream ERK and AKT were more effectively suppressed by lower doses of ceritinib compared to crizotinib (Fig.2B, C and D).
Figure 2. EML4-ALK L1196M and G1269A mutations are sensitive to ceritinib in vitro.
A) GI50 values of cell survival assay for crizotinib or ceritinib in cell lines harboring L1196M or G1269A crizotinib resistant-mutations. B-D) H3122 CR1 (B), MGH021-4 (C) and MGH045 (D) cells were treated with the indicated concentrations of crizotinib or ceritinib for 6 hours. Lysates were probed with antibodies directed against the indicated proteins. E) Nude mice bearing MGH045 cell line were treated with 100 mg/kg crizotinib or ceritinib 25 mg/kg. Tumor volumes are presented as mean +- SD (n=6).
To further assess the activity of ceritinib against crizotinib-resistant ALK-positive tumors in vivo, we examined the efficacy of ceritinib against xenografts derived from MGH045 cells that harbor the L1196M resistance mutation. As shown in Figure 2E, treatment of MGH045 tumor bearing mice with low-dose ceritinib (25 mg/kg) was more effective than high-dose crizotinib in controlling tumor growth. These data demonstrate that ceritinib is active against cancers derived from patients with acquired resistance to crizotinib and is more potent than crizotinib against ALK-rearranged cancers harboring the L1196M and G1269A resistance mutations.
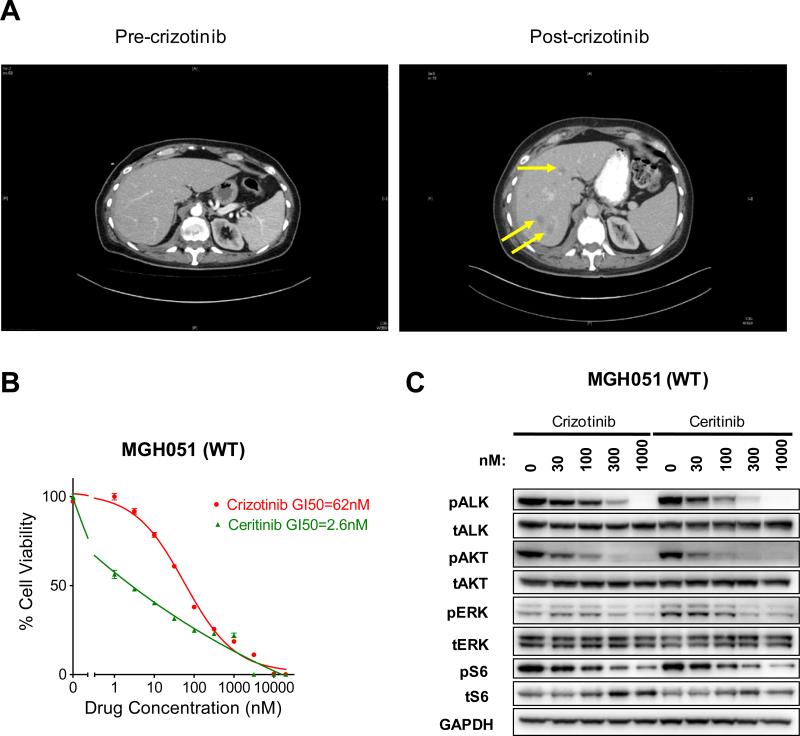
The ongoing clinical trial of ceritinib demonstrates that crizotinib-resistant ALK-positive tumors including tumors without ALK mutation or gene amplification are responsive to ceritinib treatment (13). This raises the possibility that many of these resistant tumors may develop because of inadequate target suppression. We investigated the efficacy of crizotinib and ceritinib against a crizotinib-resistant ALK-positive cell line without ALK resistance mutations, MGH051. As shown in Figure 3A, this cell line was derived from a biopsy of a liver lesion that developed in a patient on crizotinib. Assessment of the biopsy sample revealed no ALK mutations or gene amplification. The cell line derived from the biopsy also did not harbor any ALK resistance mutations. This resistant cell line was highly sensitive to ceritinib in vitro, and surprisingly the MGH051 cell line was also sensitive to crizotinib (Fig. 3B). Accordingly, phosphorylation of ALK and downstream AKT and ERK were efficiently suppressed by crizotinib and ceritinib (Fig. 3C). This data suggests that cancers with acquired resistance to crizotinib without ALK resistant mutations may remain sensitive to ALK inhibition (please see Discussion).
Figure 3. Ceritinib is active on ALK wild-type crizotinib resistant cell line.
A) Abdominal computed tomography (CT) images of patient MGH051 prior to treatment with crizotinib and after 11 weeks of crizotinib. Several new hepatic metastases (yellow arrows) were detectable after crizotinib treatment consistent with disease progression. A repeat biopsy of a hepatic metastasis was performed within 2 weeks of crizotinib discontinuation. B) MGH051 cells were treated with the indicated doses of crizotinib or ceritinib for 7 days. After the incubation, the cell survival was assayed by Cell-Titer-Glo. C) MGH051 cells were treated with the indicated concentrations of crizotinib or ceritinib for 24 hours. Lysates were probed with antibodies directed against the indicated proteins.
Assessment of ceritinib activity against a panel of ALK mutations
To systematically assess the potency of ceritinib against ALK resistance mutations, we utilized Ba/F3 cells engineered to express wild type EML4-ALK or one of the 9 different resistance mutations. In this system, ceritinib was approximately 10-fold more potent against wild-type EML4-ALK than crizotinib. Whereas all these secondary mutations induced crizotinib resistance, ceritinib was potent in inhibiting the growth of Ba/F3 cells expressing four of the resistance mutations, including L1196M, G1269A, S1206Y and I1171T (Fig.4A, S3 and Table S1). However, C1156Y, G1202R, 1151T-ins, L1152R and F1174C mutations also conferred resistance to ceritinib, although ceritinib was still more potent than crizotinib against these mutations. Thus, the most common crizotinib-resistance mutations were substantially more sensitive to ceritinib than crizotinib, while less common resistance mutations conferred resistance to both crizotinib and ceritinib.
Figure 4. Ba/F3 models of ALK-crizotinib resistant mutations.
A) IC50 of ceritinib across different Ba/F3 cell lines expressing wild-type or mutated ALK TK and including parental, IL-3–dependent Ba/F3 cells are shown. B-C) ALK resistant mutations mapped onto ALK/ceritinib (PDB 4MKC) (B) and ALK/crizotinib (PDB 2XP2) (C) co-crystal structures. Beta strand secondary structural elements of N-terminal lobe and the αC-helix of the N-terminal lobe are shown in orange and purple, respectively. Helical structural elements of the C-terminal lobe are shown in blue. Residues of the activation loop (A-loop) and catalytic loop are shown in red and orange, respectively. Residues involved in resistant mutations are depicted as green spheres. Inhibitor molecules are depicted as stick representations with carbons colored yellow and cyan for crizotinib and ceritinib, respectively. Nitrogens are colored dark blue, oxygens colored red, and chlorine green for both inhibitors. Fluoride is colored white (crizotinib) and sulfur atoms are colored yellow (ceritinib). Transparent surfaces for the inhibitors are displayed. Zoomed in view boxes for G1269 and L1196 residues are shown. Figures were rendered with MacPymol (The PyMOL Molecular Graphics System, Version 1.4 Schrödinger, LLC).
Structural basis for increased potency of ceritinib against ALK crizotinib-resistance mutations
To glean insights into the structural basis for the ability of ceritinib to maintain activity toward select crizotinib-resistant mutants, the structure of the ALK catalytic domain complexed with ceritinib was determined (Fig. 4B) (PDB 4MKC) and compared to the structure of the ALK catalytic domain bound to crizotinib (Fig. 4C) (PDB 2XP2)(16). As shown in Figure 4A, ceritinib retains potency towards the most common G1269A and L1196M crizotinib-resistant mutants. The co-crystal structure reveals that G1269 is situated just proximal to D1270 of the activation loop DFG-motif. While mutation to Ala in the G1269A mutant would not be predicted to present any steric obstruction to ceritinib binding, it would be predicted to introduce a steric clash to crizotinib binding due to the proximity of the phenyl ring of crizotinib. The Cl-moiety of the pyrimidine hinge-binding core of ceritinib is juxtaposed to the L1196 side chain and participates in a hydrophobic interaction with the Leu side chain. In L1196M mutant, the Cl-moiety of ceritinib can interact with Met, which may compensate the loss of interaction between Cl- and Leu side chain in the wild type ALK. In contrast, introduction of a Met at the gatekeeper position 1196 likely adversely affects crizotinib binding through both steric interference and unfavorable interactions with the 2-amino substituent of the pyridinyl hinge-binding core and the methyl substituent of the alkoxy moiety of crizotinib. These structural findings are in agreement with the increased potency of ceritinib versus crizotinib against these resistance mutations.
In contrast to G1269A and L1196M mutations, ceritinib is not potent against the G1202R crizotinib-resistance mutation (Fig. 4A). The crystal structure reveals that mutation of G1202 to a larger, bulky, and charged side chain would be incompatible with ceritinib or crizotinib ALK binding due to steric hindrance (6). This steric obstruction leads to a loss in potency as reflected by the shift in IC50 values observed for ceritinib and crizotinib. In contrast to the G1202R mutation, the T1151 insertion, L1152P, C1156Y, and F1174C inhibitor resistant mutants all map to the N-terminal lobe of the ALK catalytic domain and flank opposing ends of the αC-helix. The locations of these mutants do not directly contribute to inhibitor binding in co-crystal structures. Interestingly, positions T1151 and F1174 in ALK have been previously identified as sites of activating gain-of-function mutations in neuroblastoma (17). Though difficult to predict without structural and biochemical analyses of these mutants, T1151 is adjacent to the catalytically important K1150, and insertion at this position, along with the F1174C, L1152P, and C1156Y inhibitor resistance mutants likely influence αC-helix mobility and conformational dynamics of the catalytic domain. Previously reported structures of non-phosphorylated ALK in the apo, ADP, and inhibitor bound forms suggest the ALK catalytic domain structure possesses a “DFG-in” conformation (18) with a unique activation loop conformation. It is conceivable that these mutations destabilize the ALK conformation and shift the conformational equilibrium towards those that no longer are able to bind inhibitor. It is also possible that these mutations could decrease the Km for ATP, rendering ceritinib/crizotinib a less effective ATP competitive inhibitor.
Crizotinib resistant tumors harboring EML4-ALK wild-type, I1171T or C1156Y mutations are sensitive to ceritinib in vivo
To evaluate the activity of ceritinib against crizotinib resistant tumors in vivo, crizotinib-resistant H2228 xenograft tumors were generated by treatment with escalating doses of crizotinib (from 50 to 100 mg/kg). Tumors that progressed during treatment with 100 mg/kg crizotinib were analyzed for resistance mechanisms. Typical tumor responses and resistance are shown for 3 animals in Figure S4, and are representative of the 80 animals used in this study. To determine mechanisms of resistance to crizotinib, we sequenced the ALK kinase domain of all 80 tumors and identified 3 distinct resistance mutations in 6 tumors. The G1202R, C1156Y, and I1171T mutations were detected in 3, 2, and 1 resistant tumors, respectively. Of these 3 mutations, G1202R and C1156Y have been previously reported in NSCLC patients who relapsed on crizotinib (6, 7). Interestingly, I1171T has not yet been reported from crizotinib-resistant patients but was identified in an in vitro mutagenesis screen for resistance mutations (19).
The efficacy of ceritinib was tested against these crizotinib-resistant H2228 xenograft tumor models as well as one of the resistance models that did not harbor a resistance mutation nor ALK amplification (data not shown). While each was resistant to crizotinib at 100 mg/kg, ceritinib suppressed tumor growth in multiple resistance models (Fig.5A, B, C and D). In the wild-type and I1171T resistant models, ceritinib demonstrated impressive anti-tumor activity, while it was less active in the C1156Y resistant model and was inactive against the G1202R resistant model. This data is consistent with the Ba/F3 models in which ceritinib was more potent against I1171T than the C1156Y and G1202R mutants (Fig. 4A). The studies shown herein provide evidence that ceritinib can overcome resistance in vivo, especially in tumors harboring wild-type, L1196M or I1171T ALK fusion genes at a dose that is predicted to be achievable in humans. Of note, it is rather interesting that ceritinib overcame crizotinib resistance in the tumor that did not harbor an ALK resistance mutation as this recapitulates observations in the clinic and with the patient-derived cell line shown in Figure 3 (please see Discussion).
Figure 5. EML4-ALK C1156Y, I1171T, G1202R mutations sensitivity to ceritinib.
A-D) SCID beige mice bearing H2228 crizotinib resistant tumors EML4-ALK wild-type (A), I1171T (B), C1156Y (C) or G1202R (D) were treated with 100 mg/kg crizotinib or 50 mg/kg ceritinib, once daily for 12 to 22 days. Tumor volumes are presented as mean +- SD (n=5-8).
Acquired resistance to ceritinib in patients
Ceritinib has demonstrated impressive activity in the clinic in crizotinib-resistance patients (13). However, similar to other targeted therapy successes, despite initial and durable responses, tumors do develop resistance. We have now biopsied 11 cancers with acquired resistance to ceritinib (2 of which were from different sites from the same patient). As shown in Figure 6A, 5 of these biopsies revealed the development of mutations at either G1202 or F1174 in the ceritinib resistant cancers. In the patient, JFCR021, who had 2 sites of disease biopsied, two different ceritinib resistant mutations were identified, underscoring the heterogeneity of resistance mechanisms that can be identified in a single patient (6). Of note, 2 of the patients had crizotinib-resistance mutations before enrolling on ceritinib (MGH011 –Fig.6B - and JFCR021) that our laboratory studies suggested would be sensitive to ceritinib. In the ceritinib resistant cancers, those mutations were no longer detected, but the G1202R mutation emerged (Fig. 6B). These findings are consistent with preclinical studies presented in this manuscript demonstrating the activity of ceritinib against G1269A and S1206Y crizotinib-resistance mutations, and its lack of potency against the G1202R mutation.
Figure 6. Ceritinib resistant tumors acquired mutations at positions G1202 or F1174.
A) ALK mutational status in ceritinib resistant patient tumors before and after ceritinib treatment. B) Thoracic computed tomography (CT) images of patient MGH011 during crizotinib or ceritinib treatments. Sites of biopsies (red arrows) revealed the presence of different ALK secondary mutations throughout the treatments. Tumor growth observed during ceritinib treatment is consistent with disease progression.
DISCUSSION
Since its recent approval in the United States in 2011, the ALK inhibitor crizotinib has emerged as a standard of care for patients with advanced NSCLC harboring the ALK fusion oncogene. Unfortunately, as has been observed with other targeted therapies, the emergence of resistance has ultimately limited the benefit of this therapy. Next-generation ALK inhibitors (ceritinib, CH5424802, ASP3026, AP26113, and X-396) have been developed with the hope that they may overcome acquired resistance to crizotinib. We previously reported differential activity of some of these ALK inhibitors depending on the resistance mutations present within the ALK TK domain (6, 11). In an ongoing early phase clinical study, ceritinib has exhibited dramatic activity in patients with ALK-rearranged NSCLC (13).
In these studies, we find that ceritinib is a more potent ALK inhibitor than crizotinib, and has marked activity in crizotinib-naïve models of ALK-positive NSCLC, including H2228, H3122, Ba/F3 cell lines in vitro and MGH006 primary explants in vivo. To better characterize the activity of ceritinib in crizotinib resistance, we developed a variety of crizotinib-resistant models, including cell lines derived from biopsies from patients whose cancers had developed resistance to crizotinib in the clinic. These models harbored different resistance mechanisms, including various ALK resistance mutations. The activity of ceritinib varied depending on the specific ALK resistance mutation. For example, in Ba/F3 models, ceritinib was highly active against L1196M, G1269A, S1206Y and I1171T EML4-ALK mutants, and less active against the less common mutations, C1156Y, G1202R, 1151T-ins, L1152P and F1174C. It is notable that in the phase 1 study of ceritinib, 5 of 19 crizotinib-resistant tumors harbored resistance mutations at residues 1196, 1269 and 1206, with one tumor harboring both G1269A and 1151Tins. The patients harboring these resistance mutations all exhibited significant tumor shrinkage (13).
Importantly, as has been observed in the clinic, ceritinib showed potent efficacy in vitro and in vivo against crizotinib-resistant tumor that did not harbor an ALK resistance mutation or gene amplification (Fig 3B). Interestingly, the patient-derived cell line also retained sensitivity to crizotinib in vitro, demonstrating that these cells are still sensitive to ALK inhibition. One potential explanation for this finding is that, in the clinic, crizotinib fails to achieve tumor levels that completely inhibit ALK, and that tumor cells can survive through modest input from activation of bypass tracks such as EGFR. However, these cells remain sensitive to complete ALK inhibition. In the setting of a more potent ALK inhibitor, ALK is inhibited fully, abrogating the functional role of bypass tracks and leading to the elimination of tumor cells. It is also possible that this patient relapsed on crizotinib because of poor adherence to therapy or due to a stromal contribution. Similar findings were also observed in the H2228 xenograft model that developed resistance to crizotinib in vivo, did not develop an ALK mutation, and was sensitive to ceritinib (Fig. 5A). These findings may explain, at least in part, the finding that ceritinib is highly active in crizotinib resistant cancers with or without ALK resistance mutations.
The initial interrogation of ceritinib resistant patient biopsies supports the notion that ceritinib is able to effectively suppress many crizotinib-resistance mutations, but the G1202R and F1174V/C mutants are resistant to ceritinib. It is noteworthy that in two cases the crizotinib resistance mutations, S1206Y and G1269A, were no longer observed in the ceritinib resistant biopsies in which the G1202R mutations were observed (Fig. 6A). This suggests that predominant clones with the S1206Y and G1269A mutations were suppressed by ceritinib, whereas much more rare clones with G1202R mutations were selected by ceritinib. These findings give further support to the notion that there are multiple populations of resistant clones whose emergence is dependent on the selective pressure applied.
Altogether, our in vitro and in vivo data, including cell lines models established from crizotinib-resistant patient samples, demonstrate that the next generation ALK inhibitor ceritinib is active against most crizotinib resistant tumors. This is consistent with the marked clinical activity of ceritinib in ALK-positive NSCLC patients who progressed on crizotinib. As resistance to ceritinib has already been observed in the clinic, future studies will need to identify mechanisms of resistance to ceritinib other than mutations in the G1202 and F1174 residues in order to maximize the clinical benefit afforded by next generation ALK-targeted therapies.
Materials and Methods
Cell lines and reagents
All human lung cancer samples were obtained from patient with informed consent at MGH and JFCR, and all procedures were conducted under an Institutional Review Board–approved protocol. Cells in pleural effusion were collected by centrifugation at 440g for 10 min. After red blood cells were lysed with Red Blood Cell Lysis Solution (Biolegend), cells were grown in ACL-4 (Invitrogen) supplemented with 1% fetal bovine serum (FBS) or RPMI supplemented with 10% FBS and 1× Antibiotic-Antimycotic. After the cells started growing stably, clonal cell lines were also established.
H3122, H2228, A549, H460, H1299, HCC827 and H522 cell lines were provided by the Center for Molecular Therapeutics (CMT) at Massachusetts General Hospital which performs routine cell line authentication testing by single-nucleotide polymorphism and short-tandem repeat analysis. BT-474, SKBR3 and the ALK positive patient derived cell lines used in this study are from the Engelman laboratory and have been previously tested for mutation status to confirm their authenticity. A549, H460, H1299, HCC827, H522, SKBR3, H2228, H3122, H3122 CR1 and MGH021-4 cell lines were cultured in RPMI 1640 supplemented with 10% FBS. For survival assay, H2228 were cultured in 1% FBS. The MGH045, cell line was cultured in ACL4 supplemented with 1% FBS, MGH051 and BT-474 in DMEM supplemented with 10% FBS.
Mouse myeloma Ba/F3 cells were cultured in DMEM supplemented with 10% FBS with (parental) or without (EML4-ALK) IL-3 (0.5 ng/ml). cDNAs encoding EML4-ALK variant1 or EML4-ALK variant3 containing different point mutations were cloned into retroviral expression vectors, and virus was produced as previously described (11). After retroviral infection, Ba/F3 cells were selected in puromycin (0.5ug/mL) for 2 weeks. IL-3 was withdrawn from the culture medium for more than 2 weeks before experiments.
Crizotinib was purchased from ChemieTek and ceritinib was provided by Novartis. Both were dissolved in DMSO for in vitro experiments. Ceritinib was formulated in 0.5% Methyl cellulose / 0.5% Tween 80 and crizotinib in 0.1N HCl or 0.5% Methyl cellulose / 0.5% Tween 80 for in vivo studies.
Western blot
5×105 cells were treated in 6 well plates for 6 hours with the indicated drugs. Cells protein lysates were prepared as previously described (6, 11). Phospho-ERK (T202/Y204), ERK, S6, phosphor S6, phospho-AKT (S473 and T308), AKT, phospho-ALK (Y1282/1283), and ALK antibodies were obtained from Cell Signaling Technology. GAPDH was purchased from Millipore.
Survival assays
2000 or 5000 cells were plated in triplicates into 96-well plates. 72 hours (48 hours for Ba/F3 cells and 7 days for MGH051) after drug treatments, cells were incubated with CellTiter-Glo assay reagent (Promega) for 15 min, and luminescence was measured with a Centro LB 960 microplate luminometer (BertholdTechnologies).
In vivo efficacy study of ceritinib
SCID beige mice for crizotinib resistant H2228 xenograft tumor models, nude mice for MGH006 primary explants and MGH045 cells were randomized into groups of 5, 6 or 8 mice with an average tumor volume of ~150 mm3 and received Crizotinib or ceritinib daily treatments by oral gavage as indicated in each study. Tumor volumes were determined by using caliper measurements and calculated with the formula (Length × Width × Height)/2.
In vitro enzymatic assay
Enzymatic assay for recombinant ALK kinase domain (1066-1459) was conducted using the Caliper mobility shift methodology, utilizing fluorescently labeled peptides as kinase substrates. The Caliper assay was performed at 30°C for 60 min in a total volume of 9 μL. The reaction was terminated by the addition of 16 μL of stop solution (100 mM Hepes, 5 % (v/v) DMSO, 0.1 % (v/v) Coating reagent, 10 mM EDTA, 0.015 % (v/v) Brij 35). After termination of the reactions, the plates were transferred into the Caliper LabChip 3000 workstation for analysis.
Analysis of ALK/ceritinib and ALK/crizotinib co-structures
The ALK/ceritinib co-structure was determined by soaking of 2mM ceritinib into apo crystals grown in 0.2M sodium acetate trihydrate/20% PEG3350 using protein expressed and purified as previously described (18). The ALK/ceritinib final model determined to 2.0Å (PDB 4MKC- on hold) was superimposed with the coordinates of the ALK/crizotinib co-structure (PDB 2XP2) for analyses.
Patient sample analyses
The ALK-positive NSCLC patients with acquired ceritinib resistance underwent biopsy of their resistant tumors between January 2011 and September 2013. Standard histopathology was performed to confirm the diagnosis of malignancy as previously described (6). The electronic medical record was reviewed retrospectively to obtain clinical information under an Institutional Review Board–approved protocol. This study was approved by the institutional review board of MGH or the Cancer Institute Hospital of JFCR
Supplementary Material
SIGNIFICANCE.
The second-generation ALK inhibitor ceritinib can overcome several crizotinib-resistance mutations and is potent against several in vitro and in vivo laboratory models of acquired resistance to crizotinib. These findings provide the molecular basis for the marked clinical activity of ceritinib in ALK-positive NSCLC patients with crizotinib-resistant disease.
Acknowledgements
We thank Thomas Marsilje, Celin Tompkins, Auzon Steffy for expert technical assistance and input on the studies described in the manuscript. We thank Atsushi Horiike for helping to obtain repeat biopsy samples. We thank Be a Piece of the Solution and the Evan Spirito Memorial Foundation for support of lung cancer research at MGH.
Financial support: This work was supported by a grant from the US National Institutes of Health (5R01CA164273-02 to A.T.S and J.A.E.), by a V Foundation Translational Research Grant (to A.T.S. and J.A.E.) and by the NIH/National Cancer Institute (R01CA137008 to J.A.E.). The study was also supported by a grant from JSPS KAKENHI (25710015 to R.K.).
Footnotes
Competing interests: JAE is a consultant for Novartis and receives research support from Novartis. ATS is a consultant for Novartis. NL, CCL, P-Y M, SK, ACP, JL, SK, FS, XS, SH, PM, and JLH are employees and stockholders of Novartis.
REFERENCES
- 1.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki R, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. The New England journal of medicine. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 3.Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, Holmes AJ, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doebele RC, Pilling AB, Aisner DL, Kutateladze TG, Le AT, Weickhardt AJ, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res. 2012;18:1472–1482. doi: 10.1158/1078-0432.CCR-11-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gainor JF, Varghese AM, Ou SH, Kabraji S, Awad MM, Katayama R, et al. ALK Rearrangements Are Mutually Exclusive with Mutations in EGFR or KRAS: An Analysis of 1,683 Patients with Non-Small Cell Lung Cancer. Clin Cancer Res. 2013;19:4273–4281. doi: 10.1158/1078-0432.CCR-13-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katayama R, Shaw AT, Khan TM, Mino-Kenudson M, Solomon BJ, Halmos B, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Science translational medicine. 2012;4:120ra117. doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi YL, Soda M, Yamashita Y, Ueno T, Takashima J, Nakajima T, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. The New England journal of medicine. 2010;363:1734–1739. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 8.Lovly CM, Pao W. Escaping ALK inhibition: mechanisms of and strategies to overcome resistance. Science translational medicine. 2012;4:120ps122. doi: 10.1126/scitranslmed.3003728. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki T, Koivunen J, Ogino A, Yanagita M, Nikiforow S, Zheng W, et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer research. 2011;71:6051–6060. doi: 10.1158/0008-5472.CAN-11-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasaki T, Okuda K, Zheng W, Butrynski J, Capelletti M, Wang L, et al. The neuroblastoma-associated F1174L ALK mutation causes resistance to an ALK kinase inhibitor in ALK-translocated cancers. Cancer research. 2010;70:10038–10043. doi: 10.1158/0008-5472.CAN-10-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katayama R, Khan TM, Benes C, Lifshits E, Ebi H, Rivera VM, et al. Therapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4-ALK. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7535–7540. doi: 10.1073/pnas.1019559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marsilje TH, Pei W, Chen B, Lu W, Uno T, Jin Y, et al. Synthesis, Structure-Activity Relationships, and in Vivo Efficacy of the Novel Potent and Selective Anaplastic Lymphoma Kinase (ALK) Inhibitor 5-Chloro-N2-(2-isopropoxy-5-methyl-4-(piperidin-4-yl)phenyl)-N4-(2-(isopropylsulf onyl)phenyl)pyrimidine-2,4-diamine (LDK378) Currently in Phase 1 and Phase 2 Clinical Trials. Journal of medicinal chemistry. 2013 doi: 10.1021/jm400402q. [DOI] [PubMed] [Google Scholar]
- 13.Shaw AT, Kim DW, Mehra R, Tan DS, Felip E, Chow LQ, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014;370(13):1189–97. doi: 10.1056/NEJMoa1311107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Early Results Promising for LKD378 in ALK-positive NSCLC. Cancer discovery. 2013;3:OF5. [Google Scholar]
- 15.FDA, CENTER FOR DRUG EVALUATION AND RESEARCH 2011 APPLICATION NUMBER: 202570Orig1s000 Reference ID: 3006911. [Google Scholar]
- 16.Cui JJ, Tran-Dube M, Shen H, Nambu M, Kung PP, Pairish M, et al. Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). Journal of medicinal chemistry. 2010;54:6342–6363. doi: 10.1021/jm2007613. [DOI] [PubMed] [Google Scholar]
- 17.Chand D, Yamazaki Y, Ruuth K, Schonherr C, Martinsson T, Kogner P, et al. Cell culture and Drosophila model systems define three classes of anaplastic lymphoma kinase mutations in neuroblastoma. Disease models & mechanisms. 2013;6:373–382. doi: 10.1242/dmm.010348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee CC, Jia Y, Li N, Sun X, Ng K, Ambing E, et al. Crystal structure of the ALK (anaplastic lymphoma kinase) catalytic domain. The Biochemical journal. 2010;430:425–437. doi: 10.1042/BJ20100609. [DOI] [PubMed] [Google Scholar]
- 19.Zhang S, Wang F, Keats J, Zhu X, Ning Y, Wardwell SD, et al. Crizotinib-resistant mutants of EML4-ALK identified through an accelerated mutagenesis screen. Chemical biology & drug design. 2011;78:999–1005. doi: 10.1111/j.1747-0285.2011.01239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.