Abstract
The Janus cassette permits marker-free allelic replacement or knockout in streptomycin-resistant Streptococcus pneumoniae (pneumococcus) through sequential positive and negative selection. Spontaneous revertants of Janus can lead to high level of false-positives during negative selection, which necessitate a time-consuming post-selection screening process. We hypothesized that an additional counter-selectable marker in Janus would decrease the revertant frequency and reduce false-positives, since simultaneous reversion of both counter-selectable makers is much less likely. Here we report a modified cassette, Sweet Janus (SJ), in which the sacB gene from Bacillus subtilis conferring sucrose sensitivity is added to Janus. By using streptomycin and sucrose simultaneously as selective agents, the frequency of SJ double revertants was about 105-fold lower than the frequency of Janus revertants. Accordingly, the frequency of false-positives in the SJ-mediated negative selection was about 100-fold lower than what was seen for Janus. Thus, SJ enhances negative selection stringency and can accelerate allelic replacement in pneumococcus, especially when transformation frequency is low due to strain background or suboptimal transformation conditions. Results also suggested the sacB gene alone can function as a counter-selectable marker in the Gram-positive pneumococcus, which will have the advantage of not requiring a streptomycin-resistant strain for allelic replacement.
Introduction
Genetic manipulations of Streptococcus pneumoniae (pneumococcus) have been a powerful approach to understand virulence factors and drug targets in this important human pathogen. In particular, the Janus cassette [1] permits marker-free allelic replacement or knockout in Streptococcus pneumoniae through sequential positive and negative selection [2]–[4]. The Janus comprises a kanamycin-resistance (Kmr) marker and a counter-selectable rpsL + marker that confers dominant streptomycin-sensitivity (Sms) in a streptomycin-resistant (Smr) background [1]. Allelic replacement is accomplished in a Smr parental strain by a 2-step transformation procedure. The first transformation tags the locus of interest with the Janus cassette by homologous recombination while conferring a Kmr phenotype that can be selected (positive selection) and Sms. The resulting Janus-carrying strain (KmrSms) is then transformed by donor DNA to replace Janus with an allele of interest. The loss of Janus and acquisition of the allele of interest after the second transformation result in a Smr phenotype that can be selected (negative selection) and Kms. However, Smr can also result from spontaneous conversion of the rpsL + marker in Janus into a Smr rpsL allele (spontaneous revertants), which occurs at a frequency of 10−5 to 10−3 and leads to false-positives during the negative selection [1]. Unlike true Janus replacement transformants (KmsSmr), spontaneous revertants are KmrSmr since Janus is not replaced. To identify true allelic replacement, Smr clones resulted from the negative selection need to be duplicated on Km and Sm plates to screen for Kms ones. When the transformation frequency is around 10−7, as reported for several pneumococcal strains [5], [6], more than 99% of the Smr clones would be false-positives, making the post-selection screening a time-consuming process.
To improve the efficiency of negative selection, we hypothesized that adding another counter-selectable marker, especially one not subject to gene conversion, to Janus would decrease the revertant frequency and reduce false-positives. Here we show that the sacB gene from Bacillus subtilis can be used in pneumococcus as an additional counter-selectable marker in Janus. The sacB gene encodes the enzyme levansucrase that converts sucrose to high molecular-weight fructose polymers (levans) [7], [8]. sacB is commonly used to mediate negative selection in gram-negative bacteria [9] because expression of sacB leads to cell death in the presence of sucrose (Su) [7]. In a few gram-positive bacteria, sacB was also shown to confer sucrose sensitivity (Sus) and function as a counter-selectable marker [10]–[12]. Because simultaneous reversion of both counter-selectable makers is much less likely, the addition of sacB in Janus reduces the revertant frequency to around 10−9 and greatly enhances negative selection efficiency.
Materials and Methods
Bacterial strains, DNA, and growth conditions
S. pneumoniae strains used in this study are described in Table 1. All strains were maintained in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY) or on blood agar base no. 2 medium (Becton Dickinson, Sparks, MD) supplemented with 5% defibrinated sheep blood (Northeast Laboratory, Waterville, ME) (SBA). Antibiotic concentrations in SBA selective media were 500 mg/L for kanamycin and 200 mg/L for streptomycin. Genomic DNA was prepared by DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA). Growth media and culture methods for genetic transformations have been described by Pozzi et al. [13]. The transformation of TIGR4 derivatives was induced by competence-stimulating peptide variant 2 (CSP-2) and the transformation of SpnYL002 was induced by CSP-1. The concentrations of donor DNA used in transformation steps were 200 ng/ml for chromosomal DNA and 100 ng/ml for PCR products.
Table 1. Bacterial strains and primers used in this study.
| Strain or primer | Description | Source or reference |
| S. pneumoniae strains | ||
| TIGR4S | Smr TIGR4 (serotype 4 isolate) by selection of mutations | [2] |
| TIGR4J | TIGR4S but cps::kan-rpsL +, KmrSmsSur | [2] |
| SpnYL001 | TIGR4S but cps::sacB-kan-rpsL +, KmrSmsSus | This study |
| 603 | Serotype 6B clinical isolate, KmsSms | [14] |
| SpnYL002 | Smr BR1014 (serotype 23B isolate) by selection of mutations | This study |
| SpnYL003 | SpnYL002 but cps::kan-rpsL +, KmrSmsSur | This study |
| SpnYL004 | SpnYL002 but cps::sacB-kan-rpsL +, KmrSmsSus | This study |
| Primers | ||
| YL229 | GATTGCGGCTATTTTTGGAA | This study |
| YL236 | ATCCTTCCATTCATCCCCATAAGTGACC | This study |
| YL257 | CCCTTATAAGCACGGGCACAAAAGTATGGCTAAAATGAGAATATCACCGGAATTG | This study |
| YL264 | TATTATTTCCCTCCTCTTTTCTACAG | This study |
| YL265 | CTGTAGAAAAGAGGAGGGAAATAATAAATGAACATCAAAAAGTTTGCAAAACAAGCAAC | This study |
| YL266 | ACTTTTGTGCCCGTGCTTATAAGGG CGGCCATCGGCATTTTCTTTTGCG | This study |
Construction of the SJ cassette
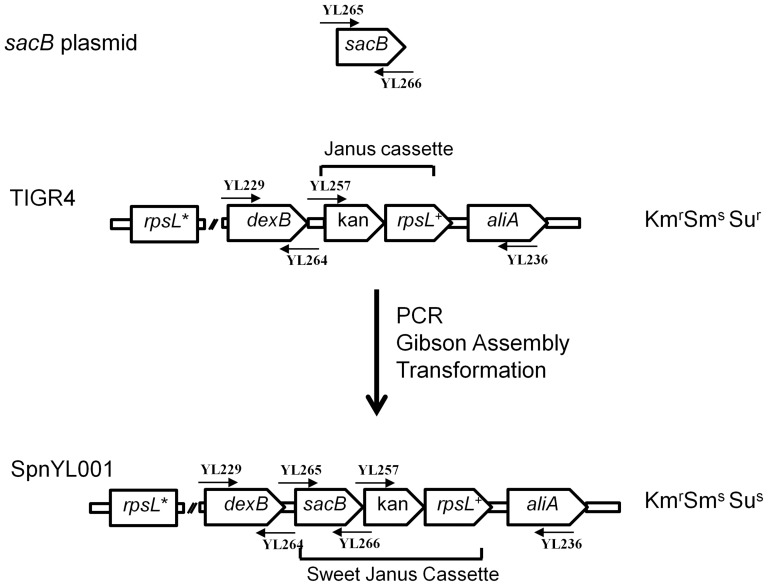
The sacB coding region was amplified from the SacB plasmid (pCre-SacB-Zeo, kindly provided by Kenan Murphy of University of Massachusetts Medical School) by PCR using the primer pair YL265 and YL266. To construct a SJ cassette between DNA sequences corresponding to the genes that flank the capsular polysaccharide biosynthesis locus (cps locus), two DNA fragments, YL229-YL264 and YL257-YL236, were prepared by PCR using chromosomal DNA of TIGR4J as template. DNA fragment YL229-YL264 contains 1422-bp of the cps upstream sequence (dexB) and 81-bp of the constitutive promoter sequence in Janus. DNA fragment YL257-YL236 contains Kan-rpsL + sequence of the Janus and 1363-bp of the cps downstream sequence (aliA). To assemble DNA fragments YL229-YL264, YL265-YL266 and YL257-YL236 into SJ, primers were designed to have 25-bp overlap at the junctions regions and the PCR products were ligated by using the Gibson Assembly Kit (New England BioLabs, Ipswich, MA) according to manufacturer's instructions. The assembly product was used as the template for PCR with primers YL229 and YL236. Product of the PCR was used to transform 403S and SpnYL002 with selection for resistance to kanamycin to create SpnYL001 and SpnYL004, respectively.
Revertant frequency
Strains with the cps locus replaced by Janus (TIGR4J and SpnYL003) or SJ (SpnYL001 and SpnYL004) were grown to mid-log phase in THY and subjected to titer determination on SBA plates supplemented with Km (500 µg/ml), Sm (200 µg/ml), Su (10% w/v), or a mix of Sm and Su (Sm+Su, final concentrations 200 µg/ml and 10% w/v, respectively). For this and all subsequent titer determination steps, the result was denoted as one-half the detection limit when no colony was observed on a plate. Revertant frequency was calculated as the ratio of colonies growing on selection plates (Sm, Su, or Sm+Su) to the number growing on the Km plate.
False-positive frequency and screen size
Genomic DNA from a serotype 6B strain (603) or no DNA control (None) was used to transform recipient strains TIGR4J and SpnYL001. Cultures of transformation were subjected to titer determination on SBA plates supplemented with Km, Sm, Su, or Sm+Su. The false-positive frequency was calculated as (TS/TKm)None/(TS/TKm)603, where TS and TKm are the concentrations of colony-forming units on a selection plate (Sm, Su, or Sm+Su) and on the Km plate, respectively. If the observed ratio was greater than 1 then the false-positive frequency was denoted as 0.99. To calculate the screen size (number of colonies on the selection plate to be examined after transformation) that would allow 95% probability of finding at least one true transformant (Kms) among revertants (Kmr), the observed revertant frequency during transformation was used (3.34×10−5 for TIGR4J selection by Sm and 2.06×10−9 for SpnYL001 selection by Sm+Su). For a given transformation frequency, the screen size was calculated as maximum of (1, log(1–0.95)/log(1-f)), where f = transformation frequency/(transformation frequency + revertant frequency).
Results
SJ was constructed by inserting the sacB coding sequence between the promoter region and the start codon of the Km resistance gene in the Janus cassette (Figure 1). The SJ was used to replace the cps loci in the TIGR4S and SpnYL002 strains to construct the SpnYL001 and SpnYL004 strains, respectively (Table 1). The SJ-carrying strains (KmrSmsSus) were subjected to titer determination on SBA plates supplemented with Km, Sm, Su, or a mix of Sm and Su (Sm+Su) to determine sensitivity to selective agents. Two Janus-carrying strains (KmrSmsSur), TIGR4J and SpnYL003, were also subjected to titer determination on the same set of SBA plates for comparison. Both SJ- and Janus-carrying strains showed spontaneous Smr revertant frequencies ranging from 10−6 to 10−5 (Table 2). For Janus-carrying strains, the Sur titer was similar to the Kmr titer. In contrast, the Sur titer was approximately 105-fold lower than the Kmr titer for the SJ-carrying strains (Table 2). Moreover, growth of SJ-carrying strains on plates containing both Sm and Su (Sm+Su) was about 10−9-fold lower than the growth on Km plate (Table 2), which represented about 104-fold additional growth reduction compared to when Sm or Su alone was used as selective agent. The results indicated that SacB in the SJ conferred Sus in pneumococcus and functioned in synergy with rpsL + to reduce the spontaneous revertant frequency.
Figure 1. Construction of Sweet Janus (SJ) cassette in the cps locus of pneumococcus.
Pentagons represent cassette elements and the dexB and aliA genes flanking the cps locus. Arrows represent oligonucleotides used to amplify fragments of the SJ cassette by PCR (see Table 1 for primer sequences). rpsL* represents the Smr mutant rpsL copy in the genomic region outside the Janus or SJ cassette. The PCR products were assembled into the SJ by using Gibson Assembly and the SJ was used to transform the TIGR4S strain to construct the SpnYL001 strain.
Table 2. Spontaneous reversion of SJ and Janus cassettes in two backgrounds.
| Strain | Growth (CFU/ml) with selection on | Revertant frequency | |||||
| Km | Sm | Su | Sm+Su | Smr | Sur | (Sm+Su)r | |
| TIGR4J | 1.8×109 | 3.6×103 | 1.6×109 | 5.6×103 | 2.0×10−6 | NA | 3.1×10−6 |
| SpnYL001 | 6.3×108 | 6.3×103 | 9.9×103 | 1# | 1.0×10−5 | 1.6×10−5 | 1.6×10−9 |
| SpnYL003 | 1.76×108 | 6.36×103 | 1.30×108 | 5.76×103 | 3.61×10−5 | NA | 3.27×10−5 |
| SpnYL004 | 3.67×108 | 7.88×103 | 9.09×103 | 1# | 2.14×10−5 | 2.48×10−5 | 2.72×10−9 |
Titer below the limit of detection was denoted as one-half the detection limit
NA: Revertant frequency not calculated because strains showed no Su sensitivity.
To test whether the low revertant frequency of SJ could enhance negative selection stringency, the TIGR4J and SpnYL001 strains were transformed with genomic DNA from a type 6B strain (603). The transformation cultures were subjected to titer determination on SBA plates with different selective agents (Table 3). Control transformations with no DNA added (None) were also performed. The false-positive frequency of negative selection was calculated as the ratio between the frequency of selected clones in the control transformation and frequency of selected clones in the DNA transformation. As shown in Table 3, when Sm alone was used as the selective agent, both Janus and SJ showed a high false-positive frequency close to 1. When Su alone was used as the selection agent, the false-positive frequency for SJ was also high (0.50, Table 3). However, when Sm and Su were used together (Sm+Su), the false-positive frequency for SJ decreased to 0.01 (Table 3). Further examination of 10 SmrSur clones in the DNA transformation of SpnYL001 showed that all were Kms and typed as serotype 6B. Thus, nearly all SmrSur clones after DNA transformation of SpnYL001 resulted from replacement of SJ by the cps operon of the donor DNA. In fact, the frequency of the true transformants (SmrSur) was only 5.24×10−7, which was much lower than the spontaneous revertant frequency for either Sm or Su. The low transformation frequency could explain the high false-positive frequency observed when Sm or Su alone was used as the selective agent.
Table 3. Comparison of false positive frequency in the Janus- and SJ-mediated negative selection.
| Recipient Strain | Donor DNA | Growth (CFU/ml) with selection on | False positive frequency | |||||
| Km | Sm | Su | Sm+Su | Selection by Sm only | Selection by Su only | Selection by Sm+Su | ||
| TIGR4J | None | 1.98×107 | 6.61×102 | NA | NA | 0.99 | NA | NA |
| 603(SmS) | 2.28×107 | 7.21×102 | NA | NA | ||||
| SpnYL001 | None | 4.85×107 | 1.80×102 | 2.70×102 | 0.3# | 0.99 | 0.50 | 0.01 |
| 603(SmS) | 5.15×107 | 1.83×102 | 5.71×102 | 27 | ||||
Titer below the limit of detection was denoted as one-half the detection limit.
NA: No data was collected for the TIGR4J strain when Su was used in selection because TIGR4J showed no Su sensitivity.
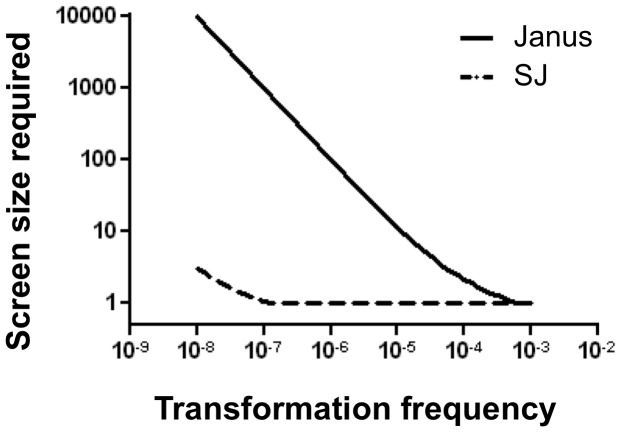
For both Janus- and SJ-mediated allelic exchange, an additional screening step is usually needed after the negative selection in order to identify true transformants (Kms) among spontaneous revertants (Kmr). The screen size (number of clones to be examined) is determined by transformation frequency and spontaneous-revertant frequency. Using the observed revertant frequency in the presence of CSP-2, we estimated the screen size required to ensure 95% probability of finding at least one true transformant (Figure 2). The screen size required by SJ would be more than 100-fold lower than the screen size required by Janus when the transformation frequency is below 10-6 (Figure 2).
Figure 2. SJ allows more efficient identification of true transformants.
Screen size (number of colonies on the selection plate to be examined after transformation) that would allow 95% probability of finding at least one true transformant (Kms) among revertants (Kmr) at indicated transformation frequency was calculated for Janus (solid line) and SJ (dashed line). The observed revertant frequency during transformation of Janus and SJ (Table 3) was used in screen size calculation (see Materials and Methods for calculation details).
We observed that growing the SJ-carrying strains on a Su plate resulted in very small colonies. To test whether these colonies are true Sur revertants, we duplicated 21 clones of SpnYL004 found on Su plate to another Su plate and a plate with no selective agent. 20 of these clones showed no growth on either plates, and 1 clone showed growth on both plates. For the one stable Sur revertant of SpnYL004, we sequenced the SacB coding region and found a C to T substitution at position 1078, which will cause an arginine-to-cysteine change in the encoded protein.
Discussion
In this study we constructed a modified Janus cassette, SJ, by adding an additional counter selectable marker, sacB, to Janus. Introduction of SJ to pneumococcus resulted in sensitivity to both Sm and Su. The frequency of SJ spontaneous double revertants (SmrSur) was about 104-fold lower than the frequency of Janus spontaneous revertants (Smr). Accordingly, the frequency of false-positives in the SJ-mediated negative selection was about 100-fold lower than what was seen for the Janus-mediated negative selection. We propose that the SJ cassette can greatly accelerate constructing marker-free allelic replacement or knockout in Streptococcus pneumoniae by reducing the amount of work required for post-selection screening.
The false-positive frequency during negative selection is determined by the relative frequency between transformation and spontaneous reversion since both events generate the selectable Smr phenotype. Transformation frequency was reported to vary among pneumococcal strains and a frequency below 10−6 was not unusual [5], [6]. Transformation conditions, including the choice of competence-stimulating peptide variants [13], can also influence transformation frequency. When transformation frequency is lower than 10−6 due to, for example, strain background or suboptimal transformation conditions, a Janus-mediated negative selection would require screening hundreds of Smr clones after selection (Figure 2) in order to identify a true transformant because vast majority of the Smr clones would be spontaneous revertants. In contrast, a screen size less than 10 would usually be sufficient for the SJ-mediate negative selection (Figure 2) due to its extremely low double revertant frequency.
Growth of the SJ-carrying strains, and not the Janus-carrying strains, was severely inhibited in the presence of sucrose (Table 2). The results suggested the sacB gene can confer sucrose-sensitivity, making it possible to use sacB alone as a counter-selectable marker in the Gram-positive pneumococcus. Compared to using rpsL+, using sacB alone to mediate negative selection in pneumococcus will have the advantage of not requiring a Smr background to perform allelic replacement, such that the strain after allelic replacement would not remain Smr and be truly maker-free. Use of SacB alone would work best when transformation frequency is high relative to the frequency of Sur revertants; otherwise high false-positive frequency may result similar to the situation when rpsL + is used alone. In addition, the sucrose sensitivity resulting from expression of sacB has been shown in many Gram-negative bacteria but only in a few Gram-positive bacteria [9]. The basis of the toxicity is still unclear. It will be interesting to understand the mechanism by which sacB expression causes cell death of the Gram-positive pneumococcus in the presence of sucrose.
Growing the SJ-carrying strains on a Su plate resulted in very small colonies and most of these small colonies could not be cultured on either a Su plate or a plate with no selective agents. These observations suggested that many of the apparent Sur revertants on the Su plate are phenotypic variants that did not acquire stable resistance to Su. It is possible that the small colony observed is composed of dead or dying bacteria accumulated during a slowing killing kinetics mediated by expression of the SacB gene in pneumococcus. Further investigation is needed to understand the source of the small colonies.
Acknowledgments
The sequence of the SJ cassette, including the sacB sequence, is deposited in GeneBank (Accession number: KJ845726).
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant sequence data are available from the GeneBank database (accession number KJ845726).
Funding Statement
This work was supported by award 5R01AI048935-12 to ML from the United States National Institute of Allergy and Infectious Diseases (http://www.niaid.nih.gov). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sung CK, Li H, Claverys JP, Morrison DA (2001) An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl Environ Microbiol 67: 5190–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Trzcinski K, Thompson CM, Lipsitch M (2003) Construction of otherwise isogenic serotype 6B, 7F, 14, and 19F capsular variants of Streptococcus pneumoniae strain TIGR4. Appl Environ Microbiol 69: 7364–7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li Y, Gierahn T, Thompson CM, Trzcinski K, Ford CB, et al. (2012) Distinct effects on diversifying selection by two mechanisms of immunity against Streptococcus pneumoniae. PLoS Pathog 8: e1002989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li Y, Weinberger DM, Thompson CM, Trzcinski K, Lipsitch M (2013) Surface Charge of Streptococcus pneumoniae Predicts Serotype Distribution. Infect Immun 81: 4519–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Joloba ML, Kidenya BR, Kateete DP, Katabazi FA, Muwanguzi JK, et al. (2010) Comparison of transformation frequencies among selected Streptococcus pneumoniae serotypes. Int J Antimicrob Agents 36: 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Evans BA, Rozen DE (2013) Significant variation in transformation frequency in Streptococcus pneumoniae. ISME J 7: 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Steinmetz M, Le Coq D, Djemia HB, Gay P (1983) [Genetic analysis of sacB, the structural gene of a secreted enzyme, levansucrase of Bacillus subtilis Marburg]. Mol Gen Genet 191: 138–144. [DOI] [PubMed] [Google Scholar]
- 8. Dedonder R (1966) Levansucrase from bacillus subtilis . Methods Enzymol 8: 6. [Google Scholar]
- 9. Reyrat JM, Pelicic V, Gicquel B, Rappuoli R (1998) Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infect Immun 66: 4011–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun X, Yang D, Wang Y, Geng H, He X, et al. (2013) Development of a markerless gene deletion system for Streptococcus zooepidemicus: functional characterization of hyaluronan synthase gene. Appl Microbiol Biotechnol 97: 8629–8636. [DOI] [PubMed] [Google Scholar]
- 11. Jager W, Schafer A, Puhler A, Labes G, Wohlleben W (1992) Expression of the Bacillus subtilis sacB gene leads to sucrose sensitivity in the gram-positive bacterium Corynebacterium glutamicum but not in Streptomyces lividans. J Bacteriol 174: 5462–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pelicic V, Reyrat JM, Gicquel B (1996) Expression of the Bacillus subtilis sacB gene confers sucrose sensitivity on mycobacteria. J Bacteriol 178: 1197–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pozzi G, Masala L, Iannelli F, Manganelli R, Havarstein LS, et al. (1996) Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J Bacteriol 178: 6087–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Trzcinski K, Thompson CM, Srivastava A, Basset A, Malley R, et al. (2008) Protection against nasopharyngeal colonization by Streptococcus pneumoniae is mediated by antigen-specific CD4+ T cells. Infect Immun 76: 2678–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant sequence data are available from the GeneBank database (accession number KJ845726).