Abstract
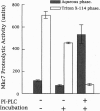
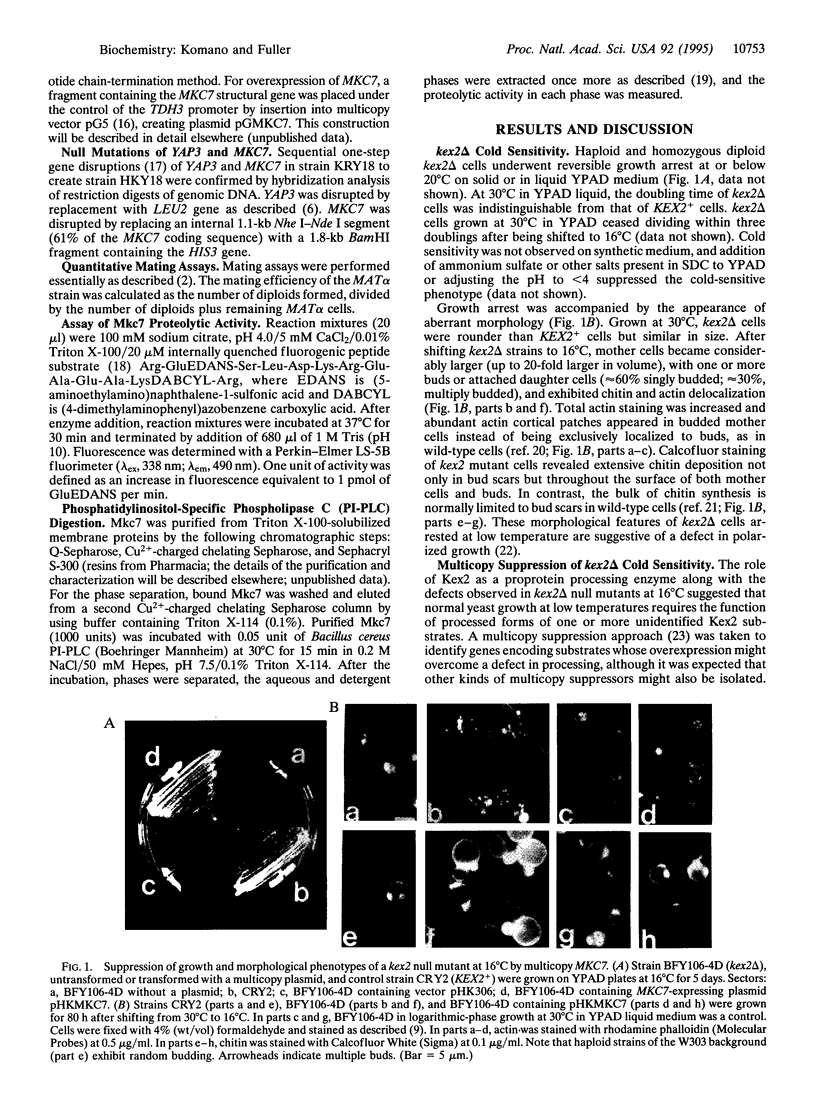
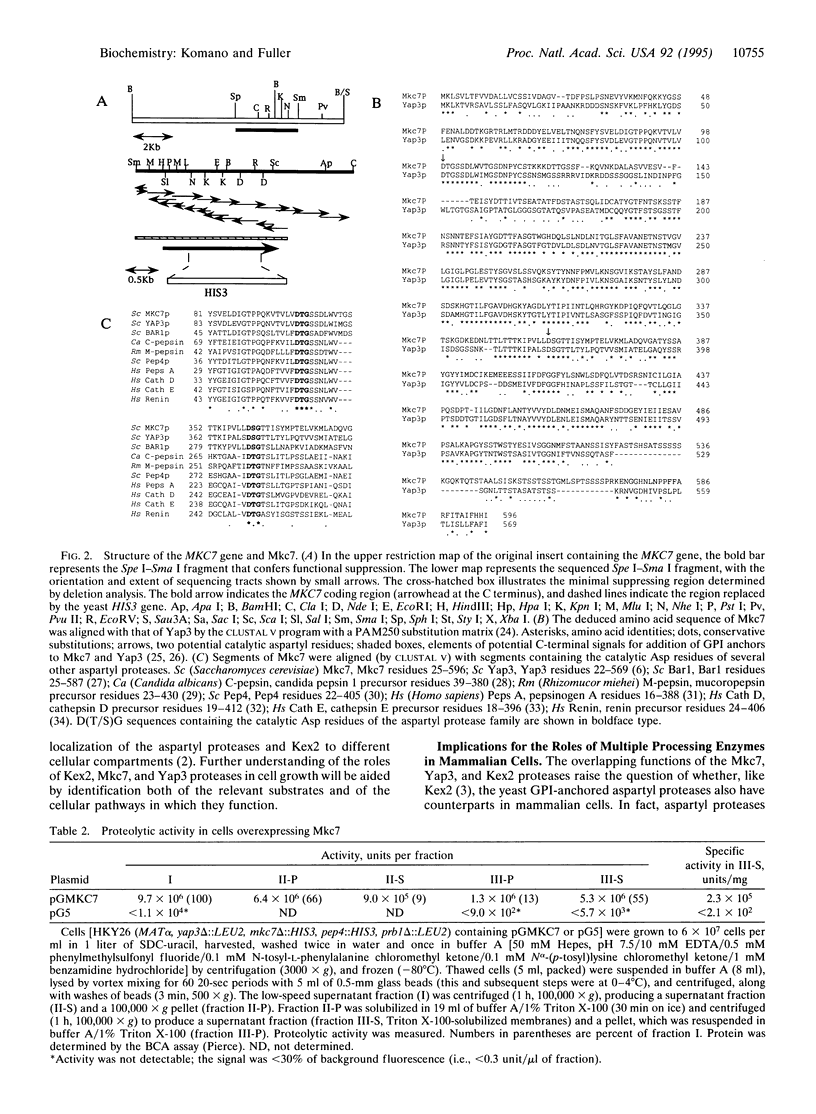
The MKC7 gene was isolated as a multicopy suppressor of the cold-sensitive growth phenotype of a yeast kex2 mutant, which lacks the protease that cleaves pro-alpha-factor and other secretory proproteins at pairs of basic residues in a late Golgi compartment in yeast. MKC7 encodes an aspartyl protease most closely related to product of the YAP3 gene, a previously isolated multicopy suppressor of the pro-alpha-factor processing defect of a kex2 null. Multicopy MKC7 suppressed the alpha-specific mating defect of a kex2 null as well as multicopy YAP3 did, but multicopy YAP3 was a relatively weak suppressor of kex2 cold sensitivity. Overexpression of MKC7 resulted in production of a membrane-associated proteolytic activity that cleaved an internally quenched fluorogenic peptide substrate on the carboxyl side of a Lys-Arg site. Treatment with phosphatidylinositol-specific phospholipase C shifted Mkc7 activity from the detergent to the aqueous phase in a Triton X-114 phase separation, indicating that membrane attachment of Mkc7 is mediated by a glycosyl-phosphatidylinositol anchor. Although disruption of MKC7 or YAP3 alone resulted in no observable phenotype, mkc7 yap3 double disruptants exhibited impaired growth at 37 degrees C. Disruption of MKC7 and YAP3 in a kex2 null mutant resulted in profound temperature sensitivity and more generalized cold sensitivity. The synergism of mkc7, yap3, and kex2 null mutations argues that Mkc7 and Yap3 are authentic processing enzymes whose functions overlap those of Kex2 in vivo.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Azaryan A. V., Wong M., Friedman T. C., Cawley N. X., Estivariz F. E., Chen H. C., Loh Y. P. Purification and characterization of a paired basic residue-specific yeast aspartic protease encoded by the YAP3 gene. Similarity to the mammalian pro-opiomelanocortin-converting enzyme. J Biol Chem. 1993 Jun 5;268(16):11968–11975. [PubMed] [Google Scholar]
- Azuma T., Pals G., Mohandas T. K., Couvreur J. M., Taggart R. T. Human gastric cathepsin E. Predicted sequence, localization to chromosome 1, and sequence homology with other aspartic proteinases. J Biol Chem. 1989 Oct 5;264(28):16748–16753. [PubMed] [Google Scholar]
- Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981 Feb 25;256(4):1604–1607. [PubMed] [Google Scholar]
- Bourbonnais Y., Ash J., Daigle M., Thomas D. Y. Isolation and characterization of S. cerevisiae mutants defective in somatostatin expression: cloning and functional role of a yeast gene encoding an aspartyl protease in precursor processing at monobasic cleavage sites. EMBO J. 1993 Jan;12(1):285–294. doi: 10.1002/j.1460-2075.1993.tb05655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner C., Fuller R. S. Structural and enzymatic characterization of a purified prohormone-processing enzyme: secreted, soluble Kex2 protease. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):922–926. doi: 10.1073/pnas.89.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelmann A., Riezman H., Desponds C., Bron C. A major 125-kd membrane glycoprotein of Saccharomyces cerevisiae is attached to the lipid bilayer through an inositol-containing phospholipid. EMBO J. 1988 Jul;7(7):2233–2240. doi: 10.1002/j.1460-2075.1988.tb03063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel-Mitani M., Flygenring H. P., Hansen M. T. A novel aspartyl protease allowing KEX2-independent MF alpha propheromone processing in yeast. Yeast. 1990 Mar-Apr;6(2):127–137. doi: 10.1002/yea.320060206. [DOI] [PubMed] [Google Scholar]
- Englund P. T. The structure and biosynthesis of glycosyl phosphatidylinositol protein anchors. Annu Rev Biochem. 1993;62:121–138. doi: 10.1146/annurev.bi.62.070193.001005. [DOI] [PubMed] [Google Scholar]
- Estivariz F. E., Birch N. P., Loh Y. P. Generation of Lys-gamma 3-melanotropin from pro-opiomelanocortin 1-77 by a bovine intermediate lobe secretory vesicle membrane-associated aspartic protease and purified pro-opiomelanocortin converting enzyme. J Biol Chem. 1989 Oct 25;264(30):17796–17801. [PubMed] [Google Scholar]
- Faust P. L., Kornfeld S., Chirgwin J. M. Cloning and sequence analysis of cDNA for human cathepsin D. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4910–4914. doi: 10.1073/pnas.82.15.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller R. S., Sterne R. E., Thorner J. Enzymes required for yeast prohormone processing. Annu Rev Physiol. 1988;50:345–362. doi: 10.1146/annurev.ph.50.030188.002021. [DOI] [PubMed] [Google Scholar]
- Gray G. L., Hayenga K., Cullen D., Wilson L. J., Norton S. Primary structure of Mucor miehei aspartyl protease: evidence for a zymogen intermediate. Gene. 1986;48(1):41–53. doi: 10.1016/0378-1119(86)90350-1. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Bleasby A. J., Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992 Apr;8(2):189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Imai T., Miyazaki H., Hirose S., Hori H., Hayashi T., Kageyama R., Ohkubo H., Nakanishi S., Murakami K. Cloning and sequence analysis of cDNA for human renin precursor. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7405–7409. doi: 10.1073/pnas.80.24.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J. V., Adams A. E. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984 Mar;98(3):922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurjan J., Herskowitz I. Structure of a yeast pheromone gene (MF alpha): a putative alpha-factor precursor contains four tandem copies of mature alpha-factor. Cell. 1982 Oct;30(3):933–943. doi: 10.1016/0092-8674(82)90298-7. [DOI] [PubMed] [Google Scholar]
- Lott T. J., Page L. S., Boiron P., Benson J., Reiss E. Nucleotide sequence of the Candida albicans aspartyl proteinase gene. Nucleic Acids Res. 1989 Feb 25;17(4):1779–1779. doi: 10.1093/nar/17.4.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay V. L., Welch S. K., Insley M. Y., Manney T. R., Holly J., Saari G. C., Parker M. L. The Saccharomyces cerevisiae BAR1 gene encodes an exported protein with homology to pepsin. Proc Natl Acad Sci U S A. 1988 Jan;85(1):55–59. doi: 10.1073/pnas.85.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackin R. B., Noe B. D., Spiess J. The anglerfish somatostatin-28-generating propeptide converting enzyme is an aspartyl protease. Endocrinology. 1991 Oct;129(4):1951–1957. doi: 10.1210/endo-129-4-1951. [DOI] [PubMed] [Google Scholar]
- Martin C., Young R. A. KEX2 mutations suppress RNA polymerase II mutants and alter the temperature range of yeast cell growth. Mol Cell Biol. 1989 Jun;9(6):2341–2349. doi: 10.1128/mcb.9.6.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matayoshi E. D., Wang G. T., Krafft G. A., Erickson J. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science. 1990 Feb 23;247(4945):954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- Nuoffer C., Jenö P., Conzelmann A., Riezman H. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol Cell Biol. 1991 Jan;11(1):27–37. doi: 10.1128/mcb.11.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K., Holcomb C., Fuller R. S. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. J Cell Biol. 1991 May;113(3):527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J. Gene overexpression in studies of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:239–251. doi: 10.1016/0076-6879(91)94019-9. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloat B. F., Adams A., Pringle J. R. Roles of the CDC24 gene product in cellular morphogenesis during the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1981 Jun;89(3):395–405. doi: 10.1083/jcb.89.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogawa K., Fujii-Kuriyama Y., Mizukami Y., Ichihara Y., Takahashi K. Primary structure of human pepsinogen gene. J Biol Chem. 1983 Apr 25;258(8):5306–5311. [PubMed] [Google Scholar]
- Steiner D. F., Smeekens S. P., Ohagi S., Chan S. J. The new enzymology of precursor processing endoproteases. J Biol Chem. 1992 Nov 25;267(33):23435–23438. [PubMed] [Google Scholar]
- Struck D. K., Lennarz W. J., Brew K. Primary structural requirements for the enzymatic formation of the N-glycosidic bond in glycoproteins. Studies with alpha-lactalbumin. J Biol Chem. 1978 Aug 25;253(16):5786–5794. [PubMed] [Google Scholar]
- Wilcox C. A., Fuller R. S. Posttranslational processing of the prohormone-cleaving Kex2 protease in the Saccharomyces cerevisiae secretory pathway. J Cell Biol. 1991 Oct;115(2):297–307. doi: 10.1083/jcb.115.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox C. A., Redding K., Wright R., Fuller R. S. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol Biol Cell. 1992 Dec;3(12):1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowicz D., Lu C. F., Kurjan J., Lipke P. N. Cell surface anchorage and ligand-binding domains of the Saccharomyces cerevisiae cell adhesion protein alpha-agglutinin, a member of the immunoglobulin superfamily. Mol Cell Biol. 1993 Apr;13(4):2554–2563. doi: 10.1128/mcb.13.4.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford C. A., Daniels L. B., Park F. J., Jones E. W., Van Arsdell J. N., Innis M. A. The PEP4 gene encodes an aspartyl protease implicated in the posttranslational regulation of Saccharomyces cerevisiae vacuolar hydrolases. Mol Cell Biol. 1986 Jul;6(7):2500–2510. doi: 10.1128/mcb.6.7.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T., Anraku Y. Nucleotide sequence of AMS1, the structure gene of vacuolar alpha-mannosidase of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1989 Sep 15;163(2):908–915. doi: 10.1016/0006-291x(89)92308-5. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]