Abstract
Protein α-helical coiled coil structures that elicit antibody responses, which block critical functions of medically important microorganisms, represent a means for vaccine development. By using bioinformatics algorithms, a total of 50 antigens with α-helical coiled coil motifs orthologous to Plasmodium falciparum were identified in the P. vivax genome. The peptides identified in silico were chemically synthesized; circular dichroism studies indicated partial or high α-helical content. Antigenicity was evaluated using human sera samples from malaria-endemic areas of Colombia and Papua New Guinea. Eight of these fragments were selected and used to assess immunogenicity in BALB/c mice. ELISA assays indicated strong reactivity of serum samples from individuals residing in malaria-endemic regions and sera of immunized mice, with the α-helical coiled coil structures. In addition, ex vivo production of IFN-γ by murine mononuclear cells confirmed the immunogenicity of these structures and the presence of T-cell epitopes in the peptide sequences. Moreover, sera of mice immunized with four of the eight antigens recognized native proteins on blood-stage P. vivax parasites, and antigenic cross-reactivity with three of the peptides was observed when reacted with both the P. falciparum orthologous fragments and whole parasites. Results here point to the α-helical coiled coil peptides as possible P. vivax malaria vaccine candidates as were observed for P. falciparum. Fragments selected here warrant further study in humans and non-human primate models to assess their protective efficacy as single components or assembled as hybrid linear epitopes.
Introduction
Despite the important reduction in reported malaria incidence during the last decade in a number of countries worldwide, malaria infection still represents one of the major global public health threats. The World Health Organization (WHO) estimated an annual global burden of 207 million malaria cases and 627,000 deaths in 2012 [1].
Of at least six different malaria parasite species which can be transmitted to humans, Plasmodium vivax is the second most parasite species of epidemiological importance with 70–80 million cases estimated per year worldwide [2]. In most malaria-endemic areas, it coexists with P. falciparum, thus making its control more difficult.
Due to the limited impact and cyclical loss of effectiveness of some of the classical malaria control measures, and based on multiple evidence on the feasibility of malaria vaccines, significant efforts have been invested in the development of malaria subunit vaccines over the past 2 to 3 decades [3]–[5]. Significant progress has been achieved with P. falciparum where several vaccine candidates are currently in clinical development [6]; with one now being considered for licensure [7]. In contrast, development of P. vivax vaccines has been significantly neglected and only a few candidates have been selected for clinical testing [8].
Most P. vivax antigens considered to have vaccine potential have been tested in in vitro studies as well as in preliminary preclinical studies in mice and primates [9]–[13]. Only a few of these antigens further selected by classical immuno-serological methods have undergone phase I clinical trials [14]–[16]. In the past, the number of parasite antigens available for vaccine studies has been quite limited. Presently, advances in the establishment of Plasmodium genomes and proteomes [17]–[19] together with high throughout laboratory techniques [20], can potentially accelerate the development of malaria vaccines. Additionally, the use of bioinformatics tools to explore the malaria genome/proteome databases has allowed new approaches for identification of parasite proteins containing α-helical coiled coil domains [21].
Such domains readily fold into stable structures that are capable of eliciting antibodies reactive with structurally native epitopes, and are generally monomorphic [22]; these structures have the capacity to block critical functions of medically important microorganisms [23], [24]. Specifically in P falciparum some antigens containing these domains have been involved in antibody-dependent inhibition of malaria parasite growth [25], [26], and therefore represent targets for vaccine development, thus drastically reducing the time required for antigen selection and preclinical testing [21].
In the past few years, approximately 170 P. falciparum α-helical coiled coil protein fragments have been assessed by combining genome-wide bioinformatics analysis, peptide selection, peptide chemical synthesis, immune and biochemical assays, in vitro functional assays, with associated protection analysis [25], [26] (unpublished data). A total of 140 putative α-helical coil-containing proteins of 200 to 10,000 amino acids in length were identified as new target proteins in P. falciparum asexual blood stages. Here we describe studies carried out using the same technology and approach with P. vivax antigens orthologous to P. falciparum, which have been evaluated for their antigenicity using human sera and immunogenicity in mice.
Materials and Methods
P. vivax genome bioinformatics analysis
Orthologues are good candidates for multi-species vaccines as they have the potential to elicit antigenic reactions against all the species included in the search parameters. A P. vivax Salvador I genome database (PlasmoDB) was used for the selection of P. vivax orthologous to P. falciparum protein sequences from asexual blood stages containing α-helical coiled coil structures, analyzed by COILS software [27]. Fifty P. vivax orthologues were found to have at least 30% homology with the 170 P. falciparum α-helical coiled-coil proteins previously identified. Sequences were of the maximal length possible in order to maximize the stability of the α-helical conformations and to increase the array of conformational epitopes that could be yielded. Selected α-helical coiled coil-containing proteins were further characterized as to possible surface location and GPI anchoring, using the following software: identification of potential signal peptides by SecretomeP and SignalP (http://www.cbs.dtu.dk/services/) [28]; transmembrane spanning region- (TMPRED http://www.ch.embnet.org/ software/TMPRED_rm.html and TMHMM http://www.cbs.dtu.dk/services/TMHMM; [29], and GPI-anchored proteins (http://mendel.imp.univie.ac.at/sat/gpi/gpi_server.html [30]; and prediction of sub-cellular localization (pTARGET) [31]. Additionally, major histocompatibility complex protein (MHC-II) binding predictions were made using the IEDB analysis resource Consensus tool [32], [33] which combines predictions from ANN aka NetMHC [34], [35], SMM [36] and Comblib [37] within the sequence of preselected peptides used in murine immunogenicity studies.
Peptide synthesis
Fifty P. vivax polypeptides 25 to 57 amino acids long were synthesized by fluorenylmethoxycarbonyl (F-moc) solid-phase chemistry [38] using an Intavis AG Bioanalytical synthesizer (Germany) (Table S1). The resulting construct was HPLC-purified; purity was confirmed by analytic C18 HPLC and mass spectrometry (MALDI-TOF; Applied Biosystem, Foster City, CA). All reagents were purchased from Fluka (Buchs, Switzerland) and Novabiochem (Laufelfingen, Switzerland). Additionally, five P. falciparum polypeptides (Pf-P27, Pf-P43, Pf-P45, Pf-P82 and Pf-P96) described previously [26] were used to test cross-reactivity between P. vivax and P. falciparum species.
Circular dichroism studies
Spectra of peptides were recorded on a JASCO J-810 spectrometer (JASCO corporation, Tokyo, Japan) equipped with a temperature controller and a 0.1 cm path length cuvette. The measurements were made in water at pH 7.3 and 22°C and at a peptide concentration of 0.15 mg/mL.
Human sera
Human serum samples from adults living in malaria-endemic areas of Colombia and Papua New Guinea (PNG) as well as from a non-endemic area (Switzerland) were used to assess peptide antigenicity. Sera (n = 42) were collected from Maprik District of the East Sepik Province, a malaria-endemic region of PNG, during a cross-sectional survey described previously [39], whereas the Colombian samples (n = 90) were obtained from two geographically distant and epidemiologically different malaria-endemic sites: Tumaco (Nariño state, n = 51) and Tierralta (Córdoba state, n = 39). Previous infection with P. vivax was confirmed based on a positive P. vivax blood-stage immunofluorescent antibody test (IFAT) result. Ethical clearances for this study were obtained from the PNG Medical Research Advisory Committee as well as from the Institutional Review Boards (IRB) of the Malaria Vaccine and Drug Development Center–MVDC (CECIV) in Cali, Colombia. Written informed consent (IC) was obtained from each volunteer. Negative control samples were obtained from Swiss adult donors with no history of malaria and no previous travel to malaria-endemic areas. Human antibodies specific to Pf-P27 and Pf-P45 [26], were affinity-purified from a pool of human serum samples from adults living in Burkina Faso, and used to test cross-reactivity to the respective P. vivax orthologues.
Animals and immunization procedures
Five-week old female BALB/c mice, maintained at the facility of MVDC and handled according to institutional guidelines, were divided into eight groups of four animals each. Mice were injected three times with the selected antigens formulated in Montanide ISA 720 adjuvant (Seppic Inc., Paris, France). Each mouse was injected subcutaneously at the base of the tail with 20 µg of the peptide formulation in a final volume of 50 µL on days 0, 20 and 40. Approximately 150 µL of whole blood were collected eight days before the first immunization, and ten days after second and third immunizations, under anesthesia from the orbital sinus; antibody responses were measured by ELISA as described previously [40]. Twenty days after the final immunization, mice were euthanized by anesthetic inhalation and spleens and lymph nodes were aseptically removed. Mononuclear cells were obtained by lymph node and spleen maceration followed by separation using Ficoll-hystopaque gradients; cells were assayed immediately. IFN-γ production by mononuclear cells was determined using a specific ELIspot assay as described below.
Ethics Statement
This study was carried out in strict accordance with institutional guidelines. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Universidad del Valle (Permit Number: 004-08). All surgery was performed under anesthesia, and all efforts were made to minimize suffering.
ELISA test
Antibody responses to the tested antigens were measured in human and murine sera by ELISA as described previously [40]. Briefly, ELISA plates (Nunc-Immuno Plate, Thermo, USA) were coated with 5 µg/mL of the respective polypeptides overnight. Plates were then blocked with 5% skim milk in PBS+0.05% tween-20 (PBST) pH 7.4 for 2 h at room temperature. After washing, plates were incubated 1 h at room temperature with sera samples prepared in PBST/2.5% skim-milk as follows: human sera were tested at a 1∶200 dilution, whereas murine sera were tested at three-fold serial dilutions starting at 1∶100. IgG antibodies were detected using alkaline phosphatase-conjugated anti-human or anti-mouse immunoglobulin (Sigma Chemical Co., St Louis, MO) at a 1∶1000 dilution. Enzymatic activity was developed after incubation for 30 min at room temperature with para-nitrophenyl phosphate substrate. The final reaction was read at 405 nm in a microplate reader (MRX, Dynex Technologies, Inc., Chantilly, VA). Cut-off points were calculated as three SD above the mean absorbance value of sera from healthy malaria- naïve Swiss volunteers or naïve mice, respectively. Positive responders were classified according to the OD ratio (OD values of tested sample divided by the cut-off value). Results were considered positive when absorbance of the test sera was higher than or equal to the cut-off points. All ELISA experiments were performed in duplicates in two independent experiments.
Since all fragments were orthologous to P. falciparum, we tested the cross-reactivity to this species using P. falciparum antigens and sera from mice immunized with P. vivax α-helical coiled coil fragments (PvPep27, PvPep43, PvPep45, PvPep82 and PvPep96). Likewise, we tested the P. vivax fragments with affinity-purified human IgG specific to Pf-P27 and Pf-P45, two P. falciparum fragments which had previously shown capacity to induce strong monocyte-dependent parasite killing [26]. As negative control, a different α-helical coiled coil non-related antigen was used.
IFA test
Parasite recognition by anti-peptide antibodies was determined by IFAT, using as antigen, P. vivax blood stages obtained from Colombian patients, and mouse sera collected 10 days after last peptide immunization. Briefly, parasites were incubated with sera diluted 1∶20. This reaction was developed with fluorescein-conjugated goat anti-mouse IgG (Jackson Immunoresearch Laboratories, Inc., Baltimore, MD) diluted 1∶1000. Slides were mounted in 30% glycerol and examined under a Nikon Eclipse microscope by epifluorescence. P. falciparum parasite cross-reactivity was also determined by IFAT using as antigen, Pf-FCB-1 blood-stage parasites derived from in vitro cultures [41].
Cellular immune responses in mice
To determine the potential of eight selected peptides to stimulate T-cell responses, in vitro IFN-γ-production by lymph nodes and splenocytes obtained from immunized mice was quantified. For this purpose a commercial mouse anti-IFN-γ ELISpot kit (Mabtech AB, Stockholm, Sweden) was used; the test carried out according to the manufacturer's instructions. Multiscreen 96-well plates (Millipore, Bedford, MA) were coated overnight at room temperature with 5 µg/mL anti-mouse IFN-γ antibodies. RPMI 1640 medium containing 10% fetal bovine serum (FBS, GIBCO) was used as a blocking solution. Freshly isolated mononuclear cells were plated into duplicate wells at 5×105 cells in RPMI 1640 medium supplemented with 10% FBS (100 µL/well). Culture medium alone, Conconavalin A or 10 µg of each synthetic peptide/mL medium (100 µL/well) was added and plates were cultured for 40 h at 37°C in a 5% CO2 humidified atmosphere. After washing, biotinylated antibody at 1 µg/mL was added and incubated for 2 h at room temperature. Plates were washed and alkaline phosphatase-streptavidin (Mabtech AB, Stockholm, Sweden) was added (1∶1000). Spots were visualized by adding 50 µL/well of BCIP/NBT (Sigma), scanned and counted using the AID ELISpot reader (AID Autoimmun Diagnostika GmbH, Germany) to determine the number of spots/well. Results were expressed as the mean number of IFN-γ spot-forming cells (SFC) per 106 cells.
Statistical analysis
Fisher's exact test (2×2 contingency tables) was used to compare differences in seroprevalence between the PNG and Colombian groups; the ANOVA test was used to compare groups. Dunnett's Multiple Comparison Test was used as post-hoc analysis and p value<0.05 was considered statistically significant. Statistical analyses were performed using GraphPad Prism software (version 5.01; GraphPad Software Inc., San Diego, CA, USA.
Results
P. vivax genome bioinformatic analysis
A total of 50 P. vivax fragments, 25–57 residues long and containing the α-helical coiled coil motifs, were selected based on proteome and transcriptome data of P. falciparum orthologues present in erythrocytic parasite stages (Tables 1 and S1). Variable homology (29 to 100% identity) was observed between P. falciparum and the corresponding P. vivax fragments (Table S1), most of which (32 antigens) were greater than 60% homologous. Identification of potential signal peptides, transmembrane (TM) regions, and GPI-anchored or sub-cellular localization prediction revealed five proteins containing TM domains (PvPep39, PvPep101, PvPep122, PvPep123 and PvPep131) and another three involved in secretory pathways (PvPep52, PvPep60, PvPep96.01). These latter peptides also contained a signal peptide. One of the proteins is predicted to be located in the mitochondria (PvPep39); none contained a GPI anchor.
Table 1. Bioinformatics analysis of coiled coil fragments.
| Peptide | MW | P. vivax | P. falciparum | aa sequence | Position | Cell localization/function | Coiled coil Domain |
| PvPep5 | 2936 | PVX_003585 | PFB0145c | IADIKISLEKLKYEVKDKKDCLENV | 203–227 | Hypothetical protein | 84–380 |
| PvPep12 | 5039 | PVX_003585 | PFB0145c | YKKELEEKAKIIEDLKDKICTLTNEVMDLKNVKNELAERDSSL | 1023–1065 | Hypothetical protein | 1016–1244 |
| PvPep27 | 3169 | PVX_113335 | MAL6P1.37 | KKQNAEKELSVLKKNYDAMSEEIEEIT | 654–680 | Hypothetical protein | 636–743 |
| PvPep40 | 3634 | PVX_119385 | PFC0235w | NETIQRMSNSLLKYEQDIETYQNEVSTLTGK | 675–705 | Hypothetical protein | 594–744 |
| PvPep42 | 3348 | PVX_087730 | PF07_0014 | NTPDYYKKITTKLQNNINNVEEYINNITNDINILKSSID | 154–192 | Hypothetical protein | 164–259 |
| PvPep43 | 4583 | PVX_089660 | PFD0685c | SVDINALNEQVKKLREELNKVTNEYDDFKNKLELLYQK | 779–816 | Chromosome associated protein | 726–917 |
| PvPep45 | 4333 | PVX_123385 | PF11_0207 | KEVKVEVNEVGEEVNEVKEEVNEAKEEVIEKKEEMTE | 650–686 | Hypothetical protein | 557–781 |
| PvPep52 | 3617 | PVX_123480 | PFL0770w | VEQVKKEINQINEQININETKITHLRNKIE | 176–205 | Secretory pathway | 166–207 |
| PvPep63 | 3658 | PVX_118160 | PF07_0086 | NNEMDETLSKLKKDINKLNEKIQKYDNYVK | 207–236 | Hypothetical protein | 162–244 |
| PvPep82.02 | 6721 | PVX_122740 | MAL13P1.96 | ETINQIDQKMEEIENNINLALEELKNLDQKILELQASFTCYENEIKQVIKKIEGLEK | 862–918 | Structural maintenance of chromosome 2 | 980–1045 |
| PvPep82.03 | 6574 | PVX_091910 | MAL13P1.96 | IEQLNTKMKNINENSNDSEHVNLAEFELKIAELKEDVNNINNMMKTFEMKFSALEK | 471–526 | Kelch domain-containing protein | 462–551 |
| PvPep83 | 4536 | PVX_087730 | PFC0345w | LQNNINNVEEYINNITNDINILKSSIDDERNERIIYNN | 166–203 | Hypothetical protein | 164–259 |
| PvPep90 | 4164 | PVX_00072 | PFD0520c | TRRMHSELSDGNKELKKLKKNIVQSDVLNAQLELNI | 63–98 | Hypothetical protein | 64–98 |
| PvPep95 | 3512 | PVX_117455 | PF14_0574 | EKGLKDLNDKIRNYDSIIENQKKELEHLK | 145–173 | Hypothetical protein | 145–245 |
| PvPep96.01 | 6595 | PVX_124060 | PF13_0107 | VEAVPENAEAAPENADPVHENAEAAPENAEPVHENAE | 773–809 | Secretory pathway | 773–809 |
| PvPep96.03 | 4482 | PVX_084385 | PF13_0107 | DVQRIDTINKNISTINDDVDHINSNINNINDNLHKINSH | 2051–2089 | Hypothetical protein | 2049–2088 |
| PvPep101 | 3554 | PVX_085155 | PF14_0255 | NKLTEMRRKLKIIDEKVQSVYKAIHAVLNN | 314–343 | CorA-like Mg2+ transporter protein | 313–343 |
| PvPep106 | 3441 | PVX_114430 | MAL6P1.163 | KTIDQLDFEINDLNSKLKNYEKSVSQNKK | 673–701 | Hypothetical protein | 430–799 |
| PvPep123 | 3455 | PVX_117855 | PF14_0500 | EKYSLIKEEIKYLNEDLDDLDNSVNVVKK | 43–71 | Hypothetical protein | 40–86 |
| PvPep125 | 3092 | PVX_099410 | PFI0975c | ILRKIEHSLKGWEADYNELKGKYNSV | 1990–2015 | Hypothetical protein | 1924–2082 |
Circular dichroism studies
Circular dichroism (CD) studies of 16 randomly selected peptides indicate that they assume a total or partial α-helical conformation in water. Peptides 40-43, 55, 60 and 65 exhibit a CD pattern characteristic of a high α-helical content as indicate for PvPep40 (Figure 1A), whereas the remaining peptides (2, 5, 12, 27, 41, 45, 48 59 and 63) show CD profiles similar to that shown for peptide PvPep63 (Figure 1B) or intermediate between those shown in Figures 1A and 1B, all characteristic of a partial α-helical organization.
Figure 1. Representative CD spectra of peptides (A) PvPep40 and (B) PvPep63.
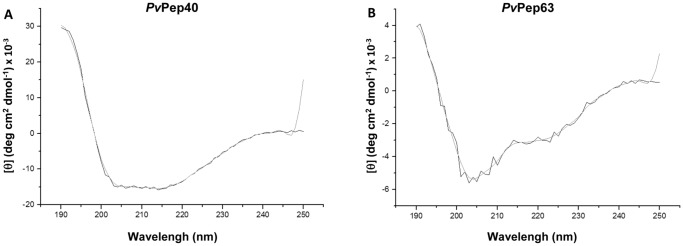
The CD's were done at room temperature on 150 µg/mL samples in aqueous solutions. Spectrums came from the averages of duplicates in the far UVs, from 190 to 250 nm and were smoothed with a 5 points filter.
Recognition of α-helical coiled coil peptides by human sera
Out of the 50 α-helical coiled coil peptides tested by ELISA using human sera, 43 were recognized by PNG (n = 42) sera at variable prevalence, however in all cases prevalence was >10%; 20 antigens displayed reactivity >29% (see Table S1). In addition, 17 peptides, which showed more than 30% of prevalence with PNG samples, were further tested with Colombian sera; all these peptides were antigenic with variable prevalence (Table 2). Ten peptides (PvPep27, PvPep42, PvPep43, PvPep45, PvPep82.02, PvPep82.03, PvPep83, PvPep95, PvPep96.01 and PvPep96.03) tested with the 90 human Colombian sera samples displayed a high degree of recognition, ranging from 30% to 86%. Recognition of the 17 peptides by PNG sera ranged between 29-71%, whereas recognition by Colombian sera for the same 17 peptides varied between 2–86%.
Table 2. Prevalence of antibodies reactive to P. vivax coiled coil fragments in volunteers from PNG and Colombia.
| PNG (n = 42)a | Colombia (n = 90)a | ||||||||
| Seroprevalenceb | Ratio>2 | Seroprevalence | Ratio>2 | ||||||
| Peptide | Protein | n | Percentage | Percentagec | n | Percentage | Percentage | p valued seroprevalence | p valued ratio>2 |
| PvPep5 | PVX_003585 | 15 | 36 | 24 | 8 | 9 | 5 | 0.0004 | 0.0016 |
| PvPep27 | PVX_113335 | 25 | 60 | 43 | 66 | 73 | 24 | ns | 0.0421 |
| PvPep40 | PVX_119385 | 13 | 31 | 17 | 8 | 9 | 1 | 0.0021 | 0.0014 |
| PvPep42 | PVX_087730 | 12 | 29 | 26 | 69 | 77 | 24 | 0.0001 | ns |
| PvPep43 | PVX_089660 | 23 | 55 | 43 | 27 | 30 | 13 | 0.0075 | 0.0002 |
| PvPep45 | PVX_123385 | 24 | 57 | 33 | 39 | 43 | 15 | ns | 0.0194 |
| PvPep52 | PVX_123480 | 18 | 43 | 19 | 25 | 27 | 16 | ns | ns |
| PvPep63 | PVX_118160 | 23 | 55 | 26 | 12 | 13 | 2 | 0.0001 | 0.0001 |
| PvPep82.02 | PVX_122740 | 15 | 36 | 24 | 40 | 44 | 15 | ns | ns |
| PvPep82.03 | PVX_091910 | 20 | 48 | 26 | 77 | 86 | 36 | 0.0001 | ns |
| PvPep83 | PVX_087730 | 22 | 52 | 33 | 48 | 53 | 17 | ns | 0.0421 |
| PvPep90 | PVX_000725 | 13 | 31 | 19 | 18 | 20 | 7 | ns | ns |
| PvPep95 | PVX_117455 | 17 | 40 | 29 | 39 | 43 | 11 | ns | 0.0220 |
| PvPep96.01 | PVX_124060 | 27 | 64 | 43 | 25 | 28 | 20 | 0.0001 | 0.0110 |
| PvPep96.03 | PVX_084385 | 30 | 71 | 60 | 36 | 40 | 12 | 0.0013 | 0.0001 |
| PvPep101 | PVX_085155 | 16 | 38 | 10 | 2 | 2 | 1 | 0.0001 | 0.0353 |
| PvPep106 | PVX_114430 | 17 | 40 | 36 | 25 | 28 | 9 | ns | 0.0004 |
Human sera sample were tested at 1∶200 dilution. bCorresponds to number and percentage of positive volunteers; Percentage of positive responses evaluated as OD values above the negative control mean + 3SD. Sera samples obtained from Swiss adult donors with no malaria history and no previous travel to malaria-endemic areas were used as negative control. cPercentage of OD ratio higher than 2 between the mean of the experimental and the mean of the control sera OD. dp value calculated by Fisher's exact test between PNG and Colombia. NS = not significant (p>0.05).
Interestingly, seven of the 17 selected peptides were the most antigenic (>50% of responders) in PNG (PvPep27, PvPep43, PvPep45, PvPep63, PvPep83, PvPep96.01, PvPep96.03), four peptides (PvPep27, PvPep42, PvPep82.03 and PvPep83) were the most reactive with Colombian sera (Table 2). Differences in reactivity were also observed between the two malaria-endemic sites in Colombia, Tierralta and Tumaco (data not shown). Responses against 16/17 peptides were stronger with PNG as compared with Colombian sera, presenting with OD ratios >2 (Table 2).
Immunogenicity of α-helical coiled coil peptides in mice
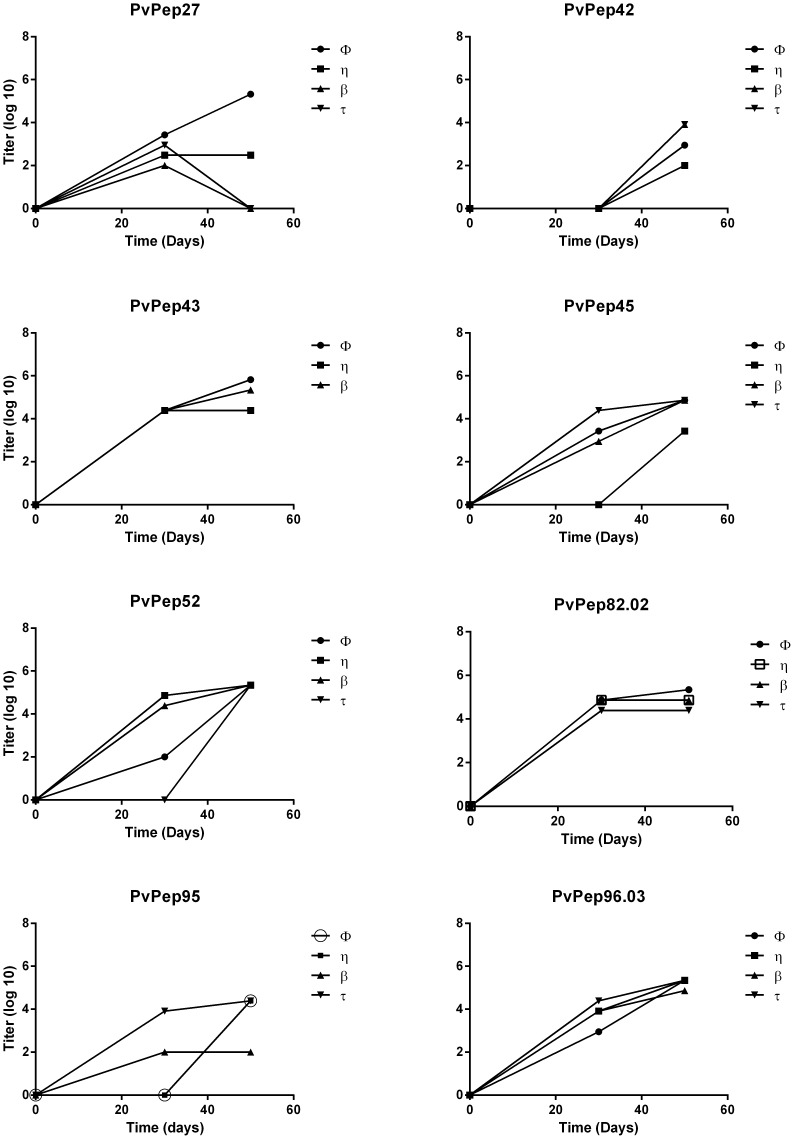
Eight peptides that showed prevalence >50% either with PNG or Colombian sera were further tested for their immunogenicity in BALB/c mice. Immunized mice developed specific IgG antibodies to the α-helical coiled coil fragments after the second immunization dose as determined by ELISA with the exception of those immunized with PvPep42; three immunization doses were needed to produce detectable antibody levels (Figure 2). Antibody titers increased steadily with titers ranging from 9×102 to 2×106 after the third immunization (Table 3). Mice immunized with PvPep27 and PvPep95 showed variable responses that were not uniform in all animals; two animals in each group failed to develop the typical boosting response after third dose. Antibody titers decreased and became negative (PvPep27) or remained stable (PvPep95); neither recognized the native protein in the IFAT (Table 3).
Figure 2. Immunogenicity of coiled coil peptides in BALB/c mice.
Titration of IgG antibody responses to coiled coil peptides in immunized mice. Evaluation on days 0, 30 and 50. Titers shown are according to a Log10 scale. “φ, η, β and τ corresponds to an identification mark for each one of the animals per group. ELISA experiments were performed by duplicated in two independent experiments.
Table 3. Immunogenicity of P. vivax coiled coil fragments in BALB/c mice.
| Antigen | Protein IDa | ELISA titer range | ELISA responders | IFAb | |
| n | Percentage | 1∶20 | |||
| PvPep27 | PVX_113335 | 3.0×102–2.4×104 | 2 | 50% | − |
| PvPep42 | PVX_087730 | 9×102–8×103 | 3 | 75% | − |
| PvPep43 | PVX_089660 | 6.6×105–2.0×106 | 3 | 100% | + |
| PvPep45 | PVX_123385 | 2.7×103–2.2×105 | 3 | 75% | ++ |
| PvPep52 | PVX_123480 | 7.2×104–2.0×106 | 4 | 100% | − |
| PvPep82.02 | PVX_122740 | 2.4×104–2.2×105 | 4 | 100% | ++ |
| PvPep95 | PVX_117455 | 2.4×104–7.2×104 | 3 | 75% | − |
| PvPep96.03 | PVX_084385 | 7.2×104–2.2×105 | 4 | 100% | + |
ID from PlasmoDB; b(-) negative, (+) positive with 1-10, and (++) positive between 10 to 20 fluorescent parasites per well, respectively.
However, sera from four of the eight immunized groups were able to recognize native protein on P. vivax asexual blood stages in IFAT assays at a 1∶20 dilution; two showed strong reactivity (Table 3). Control mice, which received only adjuvant in saline solution, were non-responsive as indicated by ELISA and IFAT (data not shown).
Cross-reactivity tests
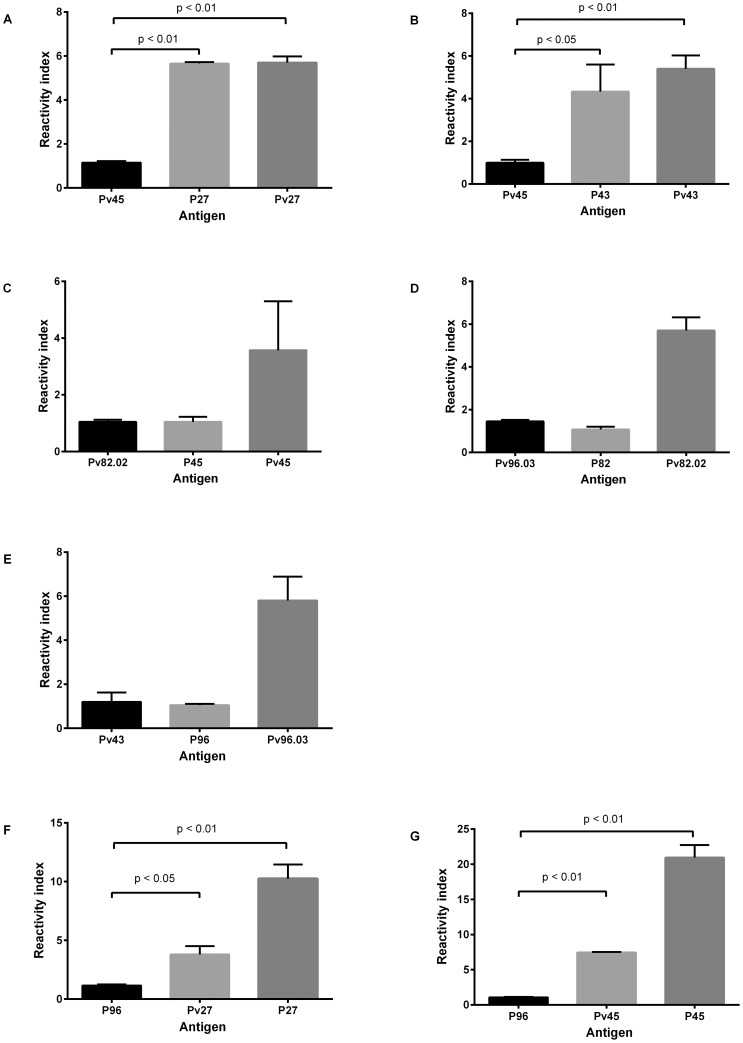
Sera from mice immunized with PvPep27 and PvPep43 were reactive with the corresponding orthologues Pf-P27 and Pf-P43 with similar reactivity indices as compared to a control sample (Figure 3). None of the other antigens (Pf-P45, Pf-P82 or Pf-P96) showed significant cross-reactivity. Moreover, cross-reactivity was also observed when specific affinity-purified human IgG to Pf-P27 and Pf-P45 were tested with the corresponding P. vivax orthologue; three-fold less reactivity was observed in both cases as compared with the positive control. Additionally, cross-reactivity with whole P. falciparum parasites was observed by IFAT (Table 4). No relationship was observed between homology and reactivity since fragments with low identity, such as PvPep45, were highly reactive with both the P. vivax fragment and the P. falciparum parasite, whereas PvPep82.02 with greater than 60% homology was not reactive (Table 4).
Figure 3. Cross-reactivity of P. vivax and P. falciparum antigens.
Antigenic cross-reactivity between P. vivax and P. falciparum was tested by ELISA, testing mice sera samples A. anti-PvPep27; B. anti-PvPepP43; C. anti-PvPep 45; D. anti-PvPep82 and E. anti-PvPep96; at 1∶200 dilution with the corresponding P. vivax and P. falciparum antigens. Additionally, affinity-purified human antibodies specific to F. PfP27 and G. PfP45 were used to test the reactivity of homologous P. falciparum and P. vivax antigens. Human IgG was tested at a 1∶200 dilution. In all cases, a non-related antigen was used as a negative control. Reactivity index defined as OD values of tested sample divided by the cut-off value, are reported as mean ± SEM for each mouse serum. Cross reactivity experiments were performed in duplicate in two independent experiments.
Table 4. Reactivity of IgG tested with different parasite antigen fragments and whole parasites.
| Origin | Antibody | Identitya (%) | P. vivax fragmentsb | P. falciparum fragmentsc | P. vivax parasited | P. falciparum parasitee |
| Mouse | anti PvPep27 | 63 | + | + | - | - |
| Mouse | anti-PvPep43 | 82 | + | + | + | - |
| Mouse | anti-PvPep45 | 44 | + | - | + | - |
| Mouse | anti-PvPep82 | 61 | + | - | + | - |
| Mouse | anti-PvPep96 | 43 | + | - | + | - |
| Human | anti Pf-P27 | NAd | + | + | + | + |
| Human | anti Pf-P45 | NA | + | + | + | + |
Identity between P. vivax and P. falciparum orthologous antigens; bReactivity tested by ELISA test using P. vivax antigens; cReactivity tested by ELISA test using P. falciparum antigens; dReactivity tested by IFA test with P. vivax blood stages; eReactivity tested by IFA test with P. falciparum blood stages; dDoes not apply.
Cellular immune responses in mice
T-cell IFN-γ production was induced by six (PvPep27, PvPep42, PvPep43, PvPep45, PvPep52 and PvPep82.02) of the eight peptides tested by ELIspot; PvPep95 and PvPep96.03 were not recognized by murine lymphocytes (Table 3). The greatest IFN-γ production was induced by PvPep43 and PvPep52 (mean SFC 344.7±15.33 and 304±60.8, respectively) followed by PvPep45, PvPep27 and PvPep42 (mean SFC 176.7±98.1, 127.3±47.6 and 68.3±47.6, respectively) (Figure 4).
Figure 4. Production of IFN-γ by mononuclear cells from immunized mice.
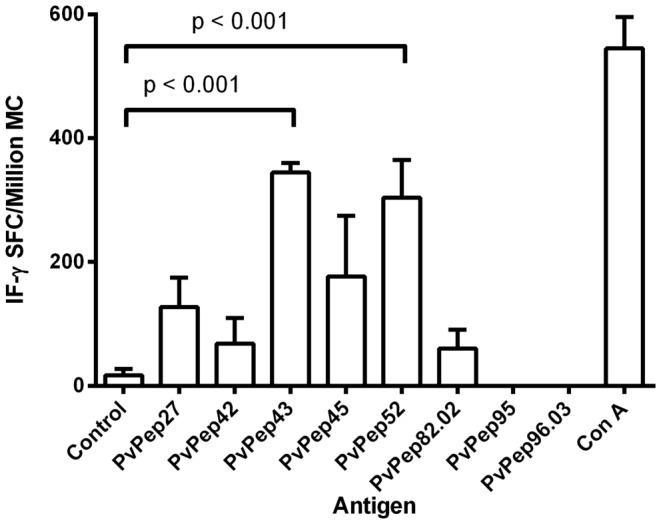
Proliferative responses of mononuclear cells obtained from mice immunized with eight synthetic peptides, and further in vitro-stimulated with 10 µg/mL of corresponding coiled coil peptides. Conconavalin A (Con A) mitogen was used as a positive control. RPMI 1640 medium was used as a negative control. Data of SFC were reported as mean ± SEM for each peptide. P value was calculated by the ANOVA test. MN: mononuclear cells.
Additionally, the selected peptides presented potential CD4+ epitopes in their amino acid sequences as confirmed by bioinformatics analysis (Table 5). No apparent relation was observed between the affinity of the predicted epitope and the IFN-γ results obtained, when mouse epitopes were described (Table 5). Higher affinity, defined as the lower percentile rank, were observed for PvPep27 and PvPep82.02 epitopes. When the alleles from human were tested, higher affinity was observed in all cases compared with mouse epitopes, although differences were observed in the main epitopes found. Same epitopes predicted for mouse alleles could be present in human alleles but with lower affinity.
Table 5. Cell immune response and association with HLA II epitopes prediction.
| Antigen | IFN-γ production SFCa | Mouse | Human | |||||
| Mean | range | Peptide | Allele | Percentile rank | Peptide | Allele | Percentile rank | |
| PvPep27 | 164 | 125–228 | KKQNAEKELSVLKKN | H2-Iad | 9.8 | VLKKNYDAMSEEIEE | HLA-DQA1*0401/DQB1*0402 | 1.02 |
| KKQNAEKELSVLKKN | HLA-DPA1*0201/DPB1*0501 | 16.09 | ||||||
| PvPep42 | 133 | 104–169 | PDYYKKITTKLQNNI | H2-Iad | 20.72 | INNITNDINILKSSI | HLA-DRB1*0301 | 0.31 |
| PDYYKKITTKLQNNI | HLA-DRB5*0101 | 1.4 | ||||||
| PvPep43 | 344 | 314–360 | VKKLREELNKVTNEY | H2-Iad | 43.01 | TNEYDDFKNKLELLY | HLA-DRB1*0801 | 2.37 |
| VKKLREELNKVTNEY | HLA-DPA1*0201/DPB1*0501 | 9.14 | ||||||
| PvPep45 | 254 | 191–339 | NEAKEEVIEKKEEMT | H2-Ied | 48.85 | INKNISTINDDVDHI | HLA-DRB3*0101 | 0.01 |
| PvPep52 | 277 | 122–375 | NINETKITHLRNKIE | H2-Ied | 32.44 | INEQININETKITHL | HLA-DRB1*0701 | 0.35 |
| NINETKITHLRNKIE | HLA-DRB1*0827 | 4.29 | ||||||
| PvPep82.02 | 69 | 36–142 | NLDQKILELQASFTC | H2-Iad | 7.93 | NEIKQVIKKIEGLEK | HLA-DRB5*0101 | 0.39 |
| NLDQKILELQASFTC | HLA-DRB1*0102 | 0.39 | ||||||
| PvPep95 | NRb | NR | LKDLNDKIRNYDSII | H2-Ied | 47.22 | EKGLKDLNDKIRNYD | HLA-DRB3*0101 | 0.26 |
| LKDLNDKIRNYDSII | HLA-DRB5*0101 | 17.04 | ||||||
| PvPep96.03 | NR | NR | NDDVDHINSNINNIN | H2-Iab | 42.28 | INKNISTINDDVDHI | HLA-DRB3*0101 | 0.01 |
| NDDVDHINSNINNIN | HLA-DRB1*0102 | 12.61 | ||||||
SFC: spot-forming colonies×106; bNo response observed.
Discussion
In an attempt to identify new target parasite antigens for malaria vaccine development, bioinformatics tools have been previously used to select proteins containing α-helical coiled coil motifs in P. falciparum proteins. In this study, similar algorithms were used in a pilot search of P. falciparum orthologous antigens in the P. vivax genome, and 50 α-helical coiled coil P. vivax segments showing a high degree of homology to the previously identified orthologous P. falciparum fragments were selected, and were further assess in antigenicity and immunogenicity studies; at the end four antigens were identified as potential targets for additional testing as vaccine candidates (Figure 5).
Figure 5. Schematic representation of the antigen selection process.
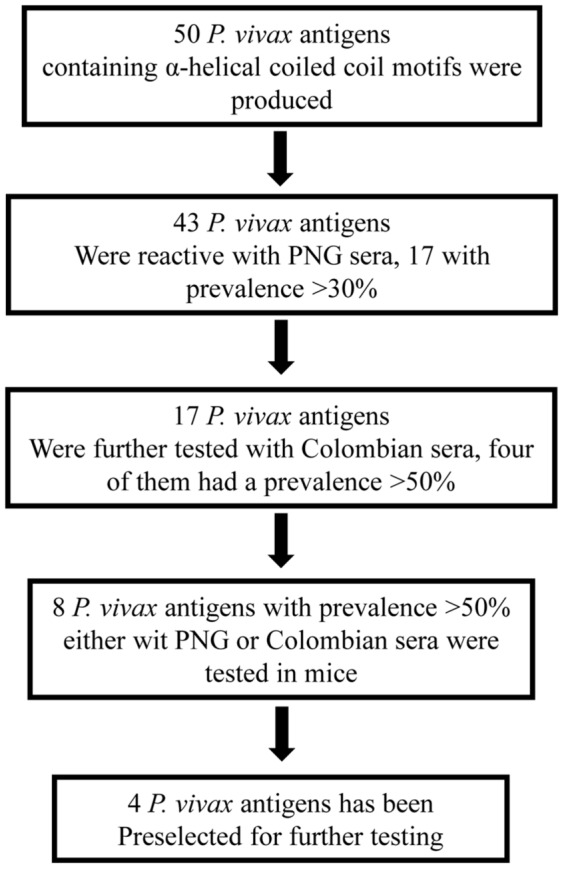
Download selection is represented: first, 50 P. vivax antigens containing α-helical coiled coil motifs, selected from orthologues of P. falciparum, were chemically synthesized and tested with PNG sera. Seventeen reactive with prevalence >30% were tested with Colombian sera. Eight antigens with prevalence >50% either with PNG or Colombian sera were used for mice immunization and immune response testing. Four antigens were finally selected for further pre-clinical testing.
It is interesting to note that of the 50 fragments tested containing α-helical coils, 19 were recognized by sera of individuals living in P. vivax endemic areas of PNG and Colombia. Most of the fragments were antigenic with variable prevalence depending on the origin of the serum samples. Variation in reactivity among sera appeared to be associated mainly with the distinct malaria transmission conditions in these two regions [42], [43]. Whereas PNG is highly endemic for P. vivax and accounts for a large proportion of the malaria cases, Colombia is a low- to moderate malaria-endemic region where P. vivax is the prevalent Plasmodium parasite. However, other factors such as differences in the genetic background of the host and parasites, and transmission rate may also explain the differences observed in the recognition frequency. These results are similar to those found in previous studies where different reactivity was observed when antigens were tested with sera from different endemic areas [26], [44].
Additionally, it is very promising to find that eight peptide fragments were able to induce a significant antibody response in immunized mice with concomitant induction of IFN-γ producing T-cells with six of the peptides. Furthermore, specific antibodies to four of the fragments resulted in positive reactions in IFA assays using P. vivax blood- stage parasites; two of these antibodies were also reactive with P. falciparum orthologous antigens, although none was reactive to P. falciparum parasite antigens. On the other hand, affinity purified human antibodies specifics to two P. falciparum antigens were reactive with the P. vivax parasite antigens. All eight preselected antigens induced antibody responses although to a variable degree regarding antibody titers and antibody kinetics. Similar results were obtained in previous studies using P. falciparum orthologous antigens, which elicited variable intermediate-to- high antibody responses [26]. Responses do not seem to be associated with fragment length since strong antibody titers were observed in response to smaller fragments such as PvPep43. However, only four peptides (PvPep43, PvPep45, PvPep82.02, and PvPep96.03) induced antibodies in mice that were able to react with whole P. vivax parasites; these four peptides induced the strongest antibody responses. Peptides PvPep43 and PvPep82.02 are chromosome-associated proteins with the other two being hypothetical proteins.
Most interestingly, antibodies to PvPep43 were cross-reactive with the orthologous P. falciparum antigen, which could represent a clear advantage for multispecies malaria vaccine development provided that cross reactivity will be also observed with the P. falciparum parasite protein. Additionally, considering the interest on Pf-P27, previously described as a promising malaria vaccine candidate [26], we also tested the cross-reactivity to this antigen. Both sera from mice immunized with PvPep27 and specific purified human IgG were reactive with both Pf-P27 and PvPep27. Homology of the two peptides, PvPep27 and PvPep43, is variable (60% and 83%, respectively). Surprisingly, Pv82.02, which shares an identity of 60% with the corresponding orthologue, did not show cross-reactivity; interestingly, PvPep45 was shown to be reactive with purified human IgG anti-Pf-P45, however conversely, the P. falciparum antigen was not reactive with anti-PvPep45 mouse sera. None of the antibodies to P. vivax antigen tested showed cross-reactivity with the native protein in blood stages as detected by IFAT possibly due the lower sensitivity of the test due to a mixture of stages present in the donor's samples or the low protein expression.
Most of the peptides induced strong IFN-γ production as expected because of the presence of MHC-II epitopes predicted by bioinformatics analysis. PvPep43, PvPep45 and PvPep52 induced higher levels of IFN-γ along with strong antibody responses. PvPep95 and PvPep96.03 did not induce detectable IFN-γ production in agreement with the low affinity CD4+ cell epitopes predicted as assessed by the IEDB analysis resource Consensus tool. It is worthy to note that peptides inducing the greatest IFN-γ production also induced the strongest antibody responses, which indicates a great potential for vaccine development. Since not association was observed between mouse and human predicted epitopes, additional experiments should be performed in non-human primates to assess the cell immune response.
Most of the antigens that have trans-membrane segments or are involved in secretory pathways were found to be poorly antigenic, suggesting that these fragments may not be expressed on the parasite surface or are not present in sufficient concentrations to allow recognition. Further investigations are warranted to determine the actual localization of the corresponding antigens. Although most antigenic fragments were not associated with trans-membrane domains with only two (PvPep52 and PvPep96.01) involved in secretory pathways, it has been shown that soluble proteins released at the time of schizont rupture are equally effective at triggering immune responses [45]–[47].
Though desirable, the functional activity of antibodies elicited in mice or humans as measured by a parasite growth inhibition assay could not be performed due to the lack of P. vivax in vitro cultures. Further preclinical studies, including experimental infection in non-human primates, must be carried out to address this question. Taken together, present data, along with that previously published, point to coiled coil peptides as an important potential source of malaria vaccine candidates. Analysis of α-helical coiled coil motifs should be extended to the entire group of erythrocytic parasite antigens. Poly-subunit antigens should be designed, containing both relevant P. vivax and P. falciparum fragments that are capable of inducing effective immune responses. Thus, this study has direct relevance to P. vivax asexual blood- stage vaccine design and suggests that some of the antigens tested could be effective in different malaria settings such as PNG and Colombia.
Supporting Information
Bioinformatics analysis of coiled coil fragments and Antibody response to PNG sera samples of all coiled coil P. vivax tested antigens.
(XLSX)
Acknowledgments
Authors are grateful for the participation of the community from malaria-endemic regions of PNG and Colombia as well as Swiss volunteers. We thank Geraldine Frank and Eliecer Jiménez for their expert technical support. Dr. Alice Koumaré of the Centre National de Transfusion Sanguine in Ouagadougou, Burkina Faso for providing immune plasma. We also thank Seppic Inc, Paris, France for the supply of Montanide adjuvant.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All data are included within the Supporting Information files.
Funding Statement
This work was funded by Colciencias (Contract 278-2008, Contract 360-2011, Contract 458-2012 and Contract 719-2013) and NIH (Grant number 5U19AI089702. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO (2013) World malaria report 2013. World Health Organization.
- 2. Mendis K, Sina BJ, Marchesini P, Carter R (2001) The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg 64: 97–106. [DOI] [PubMed] [Google Scholar]
- 3. Greenwood BM, Targett GA (2011) Malaria vaccines and the new malaria agenda. Clin Microbiol Infect 17: 1600–1607. [DOI] [PubMed] [Google Scholar]
- 4. Hill AV (2011) Vaccines against malaria. Philos Trans R Soc Lond B Biol Sci 366: 2806–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schwartz L, Brown GV, Genton B, Moorthy VS (2012) A review of malaria vaccine clinical projects based on the WHO rainbow table. Malar J 11: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Salvador A, Hernandez RM, Pedraz JL, Igartua M (2012) Plasmodium falciparum malaria vaccines: current status, pitfalls and future directions. Expert Rev Vaccines 11: 1071–1086. [DOI] [PubMed] [Google Scholar]
- 7. Agnandji ST, Lell B, Fernandes JF, Abossolo BP, Methogo BG, et al. (2012) A phase 3 trial of RTS,S/AS01 malaria vaccine in African infants. N Engl J Med 367: 2284–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Valencia SH, Rodriguez DC, Acero DL, Ocampo V, Arevalo-Herrera M (2011) Platform for Plasmodium vivax vaccine discovery and development. Mem Inst Oswaldo Cruz 106 Suppl 1179–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arevalo-Herrera M, Castellanos A, Yazdani SS, Shakri AR, Chitnis CE, et al. (2005) Immunogenicity and protective efficacy of recombinant vaccine based on the receptor-binding domain of the Plasmodium vivax Duffy binding protein in Aotus monkeys. Am J Trop Med Hyg 73: 25–31. [DOI] [PubMed] [Google Scholar]
- 10. Bell BA, Wood JF, Bansal R, Ragab H, Cargo J III, et al. (2009) Process development for the production of an E. coli produced clinical grade recombinant malaria vaccine for Plasmodium vivax. Vaccine 27: 1448–1453. [DOI] [PubMed] [Google Scholar]
- 11. Moreno A, Caro-Aguilar I, Yazdani SS, Shakri AR, Lapp S, et al. (2008) Preclinical assessment of the receptor-binding domain of Plasmodium vivax Duffy-binding protein as a vaccine candidate in rhesus macaques. Vaccine 26: 4338–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vicentin EC, Francoso KS, Rocha MV, Iourtov D, Dos Santos FL, et al. (2014) Invasion-inhibitory antibodies elicited by immunization with Plasmodium vivax apical membrane antigen-1 expressed in Pichia pastoris yeast. Infect Immun 82: 1296–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Teixeira LH, Tararam CA, Lasaro MO, Camacho AG, Ersching J, et al. (2014) Immunogenicity of a prime-boost vaccine containing the circumsporozoite proteins of Plasmodium vivax in rodents. Infect Immun 82: 793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Herrera S, Bonelo A, Perlaza BL, Fernandez OL, Victoria L, et al. (2005) Safety and elicitation of humoral and cellular responses in colombian malaria-naive volunteers by a Plasmodium vivax circumsporozoite protein-derived synthetic vaccine. Am J Trop Med Hyg 73: 3–9. [DOI] [PubMed] [Google Scholar]
- 15. Herrera S, Fernandez OL, Vera O, Cardenas W, Ramirez O, et al. (2011) Phase I safety and immunogenicity trial of Plasmodium vivax CS derived long synthetic peptides adjuvanted with montanide ISA 720 or montanide ISA 51. Am J Trop Med Hyg 84: 12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu Y, Ellis RD, Shaffer D, Fontes E, Malkin EM, et al. (2008) Phase 1 trial of malaria transmission blocking vaccine candidates Pfs25 and Pvs25 formulated with montanide ISA 51. PLoS One 3: e2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, et al. (2008) Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455: 757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gardner MJ, Hall N, Fung E, White O, Berriman M, et al. (2002) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419: 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, et al. (2002) A proteomic view of the Plasmodium falciparum life cycle. Nature 419: 520–526. [DOI] [PubMed] [Google Scholar]
- 20. Doolan DL, Southwood S, Freilich DA, Sidney J, Graber NL, et al. (2003) Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci U S A 100: 9952–9957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Corradin G, Villard V, Kajava AV (2007) Protein structure based strategies for antigen discovery and vaccine development against malaria and other pathogens. Endocr Metab Immune Disord Drug Targets 7: 259–265. [DOI] [PubMed] [Google Scholar]
- 22. Kulangara C, Kajava AV, Corradin G, Felger I (2009) Sequence conservation in Plasmodium falciparum alpha-helical coiled coil domains proposed for vaccine development. PLoS One 4: e5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singh S, Soe S, Roussilhon C, Corradin G, Druilhe P (2005) Plasmodium falciparum merozoite surface protein 6 displays multiple targets for naturally occurring antibodies that mediate monocyte-dependent parasite killing. Infect Immun 73: 1235–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tripet B, Kao DJ, Jeffers SA, Holmes KV, Hodges RS (2006) Template-based coiled-coil antigens elicit neutralizing antibodies to the SARS-coronavirus. J Struct Biol 155: 176–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Olugbile S, Villard V, Bertholet S, Jafarshad A, Kulangara C, et al. (2011) Malaria vaccine candidate: design of a multivalent subunit alpha-helical coiled coil poly-epitope. Vaccine 29: 7090–7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Villard V, Agak GW, Frank G, Jafarshad A, Servis C, et al. (2007) Rapid identification of malaria vaccine candidates based on alpha-helical coiled coil protein motif. PLoS One 2: e645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lupas A, Van Dyke M, Stock J (1991) Predicting coiled coils from protein sequences. Science 252: 1162–1164. [DOI] [PubMed] [Google Scholar]
- 28. Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795. [DOI] [PubMed] [Google Scholar]
- 29. Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580. [DOI] [PubMed] [Google Scholar]
- 30. Eisenhaber B, Bork P, Eisenhaber F (1999) Prediction of potential GPI-modification sites in proprotein sequences. J Mol Biol 292: 741–758. [DOI] [PubMed] [Google Scholar]
- 31. Guda C, Subramaniam S (2005) pTARGET [corrected] a new method for predicting protein subcellular localization in eukaryotes. Bioinformatics 21: 3963–3969. [DOI] [PubMed] [Google Scholar]
- 32. Kim Y, Ponomarenko J, Zhu Z, Tamang D, Wang P, et al. (2012) Immune epitope database analysis resource. Nucleic Acids Res 40: W525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang P, Sidney J, Kim Y, Sette A, Lund O, et al. (2010) Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics 11: 568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, et al. (2008) NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res 36: W509–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nielsen M, Lundegaard C, Worning P, Lauemoller SL, Lamberth K, et al. (2003) Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci 12: 1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peters B, Sette A (2005) Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinformatics 6: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sidney J, Assarsson E, Moore C, Ngo S, Pinilla C, et al. (2008) Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Atherton E, Hubscher W, Sheppard RC, Woolley V (1981) Synthesis of a 21-residue fragment of human proinsulin by the polyamide solid phase method. Hoppe Seylers Z Physiol Chem 362: 833–839. [DOI] [PubMed] [Google Scholar]
- 39. Alpers MP, al-Yaman F, Beck HP, Bhatia KK, Hii J, et al. (1992) The Malaria Vaccine Epidemiology and Evaluation Project of Papua New Guinea: rationale and baseline studies. P N G Med J 35: 285–297. [PubMed] [Google Scholar]
- 40. Arevalo-Herrera M, Roggero MA, Gonzalez JM, Vergara J, Corradin G, et al. (1998) Mapping and comparison of the B-cell epitopes recognized on the Plasmodium vivax circumsporozoite protein by immune Colombians and immunized Aotus monkeys. Ann Trop Med Parasitol 92: 539–551. [PubMed] [Google Scholar]
- 41. Trager W, Jensen JB (1976) Human malaria parasites in continuous culture. Science 193: 673–675. [DOI] [PubMed] [Google Scholar]
- 42. Koepfli C, Ross A, Kiniboro B, Smith TA, Zimmerman PA, et al. (2011) Multiplicity and diversity of Plasmodium vivax infections in a highly endemic region in Papua New Guinea. PLoS Negl Trop Dis 5: e1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arevalo-Herrera M, Quinones ML, Guerra C, Cespedes N, Giron S, et al. (2012) Malaria in selected non-Amazonian countries of Latin America. Acta Trop 121: 303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cespedes N, Arevalo-Herrera M, Felger I, Reed S, Kajava AV, et al. (2013) Antigenicity and immunogenicity of a novel chimeric peptide antigen based on the P. vivax circumsporozoite protein. Vaccine 31: 4923–4930. [DOI] [PubMed] [Google Scholar]
- 45. Jafarshad A, Dziegiel MH, Lundquist R, Nielsen LK, Singh S, et al. (2007) A novel antibody-dependent cellular cytotoxicity mechanism involved in defense against malaria requires costimulation of monocytes FcgammaRII and FcgammaRIII. J Immunol 178: 3099–3106. [DOI] [PubMed] [Google Scholar]
- 46. Kulangara C, Luedin S, Dietz O, Rusch S, Frank G, et al. (2012) Cell biological characterization of the malaria vaccine candidate trophozoite exported protein 1. PLoS One 7: e46112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Olugbile S, Kulangara C, Bang G, Bertholet S, Suzarte E, et al. (2009) Vaccine potentials of an intrinsically unstructured fragment derived from the blood stage-associated Plasmodium falciparum protein PFF0165c. Infect Immun 77: 5701–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bioinformatics analysis of coiled coil fragments and Antibody response to PNG sera samples of all coiled coil P. vivax tested antigens.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All data are included within the Supporting Information files.