Abstract
Rheumatoid arthritis (RA) is the most frequent autoimmune chronic inflammatory disease of the joints and it is characterized by the inflammation of the synovial membrane and the subsequent destruction of the joints. In RA, CD4+ T cells are the main drivers of disease initiation and the perpetuation of the damaging inflammatory process. To date, however, the genetic regulatory mechanisms of CD4+ T cells associated with RA etiology are poorly understood. The genome-wide analysis of expression quantitative trait loci (eQTL) in disease-relevant cell types is a recent genomic integration approach that is providing significant insights into the genetic regulatory mechanisms of many human pathologies. The objective of the present study was to analyze, for the first time, the genome-wide genetic regulatory mechanisms associated with the gene expression of CD4+ T cells in RA. Whole genome gene expression profiling of CD4+ T cells and the genome-wide genotyping (598,258 SNPs) of 29 RA patients with an active disease were performed. In order to avoid the excessive burden of multiple testing associated with genome-wide trans-eQTL analysis, we developed and implemented a novel systems genetics approach. Finally, we compared the genomic regulation pattern of CD4+ T cells in RA with the genomic regulation observed in reference lymphoblastoid cell lines (LCLs). We identified a genome-wide significant cis-eQTL associated with the expression of FAM66C gene (P = 6.51e−9). Using our new systems genetics approach we identified six statistically significant trans-eQTLs associated with the expression of KIAA0101 (P<7.4e−8) and BIRC5 (P = 5.35e−8) genes. Finally, comparing the genomic regulation profiles between RA CD4+ T cells and control LCLs we found 20 genes showing differential regulatory patterns between both cell types. The present genome-wide eQTL analysis has identified new genetic regulatory elements that are key to the activity of CD4+ T cells in RA.
Introduction
Rheumatoid arthritis (RA) is the most frequent autoimmune chronic inflammatory disease of the joints and affects up to 1% of the World population. RA is characterized by synovial membrane hyperplasia, increased vascularity and chronic immune cell infiltration that lead to joint destruction and pain [1]. The predominant immune cell type that infiltrates RA synovial joints is the CD4+ T lymphocyte and, for many years, it has been considered a T-cell driven disease [2]. Consequently, treatments targeting the activation of CD4+ T cells have proven successful in the control of disease activity in RA [3], [4]. This evidence, together with the strong genetic association of the molecules that mediate antigen presentation to CD4+ T cells in RA, clearly indicate that the characterization of the regulatory elements of this cell type will be key to completely understand the disease pathogenesis [5]. To date, however, a global analysis of the regulatory mechanisms of CD4+ T cells in RA has not been performed.
Genome-wide association studies (GWAS), in which common genetic variants are tested for association with complex traits, have revolutionized the identification of genetic risk factors for many common diseases [6]. More recently, the integration of GWAS with gene expression data to identify quantitative trait loci (eQTLs) is starting to provide significant insights into the genetic architecture of human diseases [7]. The number of transcripts expressed by a gene is modified by the variation in genetic regulatory elements. RNA levels can therefore be considered as a quantitative trait and used to map these crucial regulatory elements in the genome [8]. Gene expression microarrays and more recently ultra-high throughput RNA sequencing systems coupled with genome-wide genotyping assays are allowing to scan the whole genome variation to identify trait-specific eQTLs [9]. eQTL studies are leading to the characterization of functional sequence variation as well as the understanding of basic genetic regulatory mechanisms [10]. So far, one of the most important discoveries of genome-wide eQTL mapping has been the finding that a substantial fraction of the gene expression regulation is cell type-specific [11]. Consequently, the understanding of the genomic regulatory basis that underlies a complex disease like RA will only be possible if it is performed at the cell type level, in particular analyzing those cell types that play a crucial role in the disease onset and progression [12], [13].
Recently, studies have been reported that characterize regulatory variants that operate in a cell type-specific manner [11], [14], [15]. However, most of these studies focused only on cis-acting elements rather than identifying trans-acting elements. While cis-eQTLs are likely to influence molecular mechanisms involved in transcription, splicing or mRNA decay [16], trans-eQTLs are more likely to perturb entire pathways and mediate complex epistatic and gene-environment interactions and, therefore, are of particular interest in the study of prevalent diseases with a complex genetic basis like RA [17], [18]. Importantly, while cis-eQTLs are often conserved among different cell types, trans-eQTLs tend to be cell type-specific [19]. The genome-wide analysis of trans-eQTLs, however, requires the analysis of an exponential number of transcript-SNP pairs, resulting in a prohibitive multiple testing problem. As a consequence, very few studies have explored the presence of trans-eQTLs in human traits [20], and new strategies to reduce the burden of multiple testing in these studies must be devised.
In this study, we have analyzed, for the first time, the regulatory variation associated with the gene expression of CD4+ T cells in RA. To do so, we have performed a genome-wide cis-eQTL analysis and we have implemented a robust systems genetic approach to perform the trans-eQTL analysis. In this approach we exploit the presence of cell-specific gene expression networks, together with the power of biological knowledge and the statistical analysis of networks, to efficiently reduce the high dimensionality associated with the global analysis of trans-eQTLs. Finally, in order to identify additional specific gene expression regulation, we have compared the regulatory patterns observed in CD4+ T cells to those of lymphoblastoid cell lines (LCLs), a well-characterized reference cell type. Taken together, the results of this study provide new insights into the key regulatory elements of CD4+ T cells in RA.
Materials and Methods
Patients and samples
A total of 29 patients with rheumatoid arthritis from the Rheumatology Unit of the Vall d'Hebron University Hospital (Barcelona, Spain) were recruited. All patients had been diagnosed as RA following the 1987 American College Rheumatology criteria [21]. In order to obtain a gene expression profile more representative of the disease, all patients had to have a high disease activity at the moment of sample collection. In this study, high disease activity was defined as an European League against Rheumatoid Arthritis (EULAR) Disease Activity Score (DAS28) [22] higher than 3.2. The DAS28 score efficiently reflects the disease activity of the RA patient by combining the evidence of tenderness and swelling in 28 joints together with the patient's global assessment and a systemic marker of inflammation (erythrocyte sedimentation rate or C-reactive protein levels). In order to avoid the influence of treatment over the gene expression patterns in RA, all patients were receiving the same treatment (≤20 mg/wk metothrexate) and were all naïve to biological immunomodulating agents like anti-TNF agents. Patients suspected to have a concomitant infection or were positive for hepatitis B or C viruses (active or inactive) were also not included in this study. The main features of the RA patient cohort used in this study are shown in Table S1. From each patient, 30 mL of venous blood was obtained, from which 5 mL were used for genomic DNA isolation and 25 mL for CD4+ T cell RNA isolation. Genomic DNA was isolated using the Chemagic Magnetic Separation Module I (PerkinElmer, USA). In order to obtain the total RNA from CD4+ T cells, we first isolated the CD4+ lymphocytes from whole blood using the RossetteSep negative selection kit (Stem Cell Technologies, Canada). Isolated cells were immediately preserved in RNA stabilization reagent RNAlater (Qiagen, Hilden, Germany) and frozen at −80°C. In order to determine the level of cell purity, FACS flow cytometry analysis was performed on CD4+ T cells. The CD45+, CD4+, CD3+ and CD8+ T cells were stained by direct immune fluorescence using monoclonal antibodies conjugated with fluorochromes fluorescein isothiocyanate, phycoerythrin, pycoerythrin-cyanin-5 and Phycoerythrin-Texas (all antibodies from Beckman Coulter, FL, USA), respectively. Isotype-matched immunoglobulins with no reactivity against surface markers and the fluorochrome combination were used as negative controls to determine fluorescence background. Acquisition of flow data was performed using an EPICS-XL-MCL cytometer and Expo32 software (Beckman-Coulter, FL, USA) after antibody incubation followed by erythrocyte lysis. This analysis was carried out on the same day of blood extraction in all RA patients and >95% CD4+ T cell purity from all samples was confirmed. RNA extraction from the isolated CD4+ T cells was performed with the RNeasy extraction kit (Qiagen, Hilden, Germany) and its quality determined using the 2100 BioAnalyzer system (Agilent technologies, Waldbronn, Germany).
All the procedures followed were in compliance with the principles of the Declaration of Helsinki. All patients provided written informed consent. The study and the consent procedure were approved by the Institut de Recerca Hospital Universitari Vall d'Hebron ethics committee.
Gene expression profiling
Whole genome transcript abundance from the CD4+ T cells of patients with RA was performed using the Illumina Human-6 v1 Beadchip array system (Illumina, San Diego, CA, USA). This microarray platform measures the gene expression levels of more than 47,000 different transcripts. In order to update the probe annotation for this microarray, we used the NCBI RefSeq database [23]. Matching the microarray probe sequence to the latest RefSeq database version (2nd May 2013) we found that 13,555 probes perfectly mapped to unique transcripts, 26,729 probes that did not map any transcript and 8,013 probes that mapped more than one transcript from which 7,565 mapped to the same gene. Consequently, the updated microarray probe annotation was composed by 21,120 probes matching to known transcripts. Data preprocessing was conducted using the R statistical software [24]. The raw expression intensities of the 29 microarrays were processed using background adjustment. One sample showed intensity dependent biases and it was finally removed. The gene expression intensities were normalized on the log2-scale using the quantile normalization method [25]. In order to remove the potential variability introduced by the presence of different microarray processing batches, we used the ComBat empirical Bayes method [26]. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus [27] and are accessible through GEO Series accession number GSE55468.
Whole genome genotyping
The genome-wide genotyping of the 29 RA patients was performed using the Illumina Quad610 Beadchips (Illumina, San Diego, California, USA). The Quad610 genotyping arrays scan 618,150 polymorphisms (598,258 SNPs and 19,892 CNV probes). Genotype calling was performed using the GenomeStudio data analysis software (v2011.1, Illumina, San Diego, California, USA). The quality control evaluation was performed using PLINK software [28]. All the autosomal SNPs were initially selected (n = 582,539). A total of 7,886 markers showing >10% missing data were excluded, as well as 1,724 SNPs with P<1e-5 for test of deviance from Hardy-Weinberg equilibrium. After all quality control steps, a final number of 572,980 SNPs were used for the eQTL analysis in CD4+ T cells. In order to compare the exact same polymorphisms in the CD4+ T cell and LCL eQTL analyses, 6,962 SNPs had to be excluded from the initial dataset. In this case, a final number of 566,018 SNPs were used for the comparative eQTL analysis between both cell types. The presence of population stratification in the study samples was estimated using principal component analysis (PCA) implemented in EIGENSOFT (v4.2) software [29]. Using the top ten principal components over ten iterations and using a threshold of six standard deviations, we excluded two samples showing an outlier genetic background.
Statistical association analysis
Cis- and trans-eQTL analyses were performed using Matrix eQTL software [30]. Matrix eQTL efficiently performs large numbers of eQTL analyses by the use of large matrix multiplications. Most gene regulatory elements that act in cis have been previously reported to be located in close proximity to the gene [31]. However, there is clear evidence that cis regulatory elements like enhancers can be located as far as 1 Mb from the gene they regulate [32]. Consequently, we used 1 Mb as the maximal distance at which cis regulation can occur. This distance is in accordance to most recent studies on genome-wide eQTLs [10]. For each transcript-SNP pair, we fitted the following linear model assuming additive effect of genotype on gene expression:
The genotype Xi of individual i at given SNP is encoded by 0, 1 and 2 according to the number of minor alleles present in the genotype of the individual. The gene expression Yi is the normalized log-expression level of the probe for individual i. The εi captures all other factors which influence the gene expression. The null hypothesis of the statistical test was that there is no association between the genotype and the gene expression (β = 0). In the present study, we included gender as a covariate. In order to avoid false positives due to low allele frequency, we filtered those SNPs with a minor allele frequency <10%. Multiple testing correction was performed using the False Discovery Rate (FDR) method [33]. A total of 21,120 transcripts and 572,980 SNPs measured in 26 individuals were finally used for the cis-eQTL analysis in CD4+ T cells.
Novel systems genetics approach for trans-eQTL identification
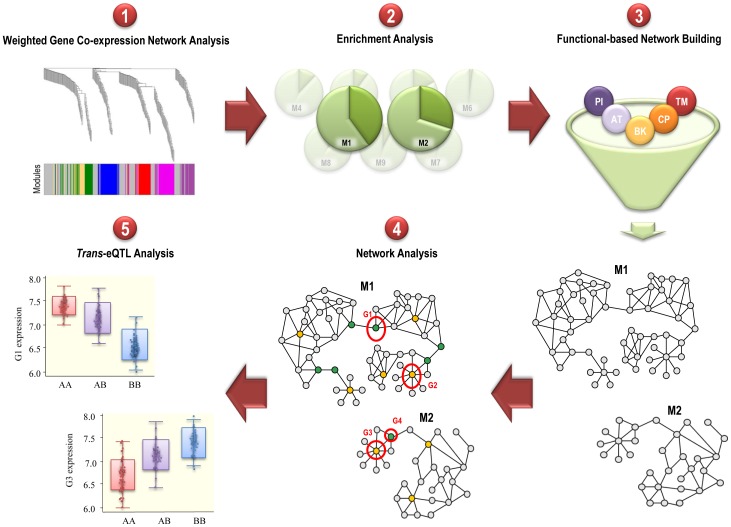
We present a novel systems genetics approach to address the dimensionality problem present in trans-eQTL analyses. The pipeline of this approach is composed by five consecutive steps (Figure 1).
Figure 1. Workflow of the systems genetics approach for trans-eQTL identification.
The new systems genetics approach is based on four steps that are performed before the trans-eQTL analysis in order to efficiently reduce the number of analyzed genes: 1) Identification of the gene expression modules (M) that characterize CD4+ T cell gene expression 2) Enrichment analysis of a specific biological process that is related to the trait of interest 3) Construction of the functional-based networks using biological knowledge within the significantly enriched modules 4) Network analysis to identify those genes that are likely to play a central role in the functional-based networks 5) Trans-eQTL analysis using the subset of genes that show the highest centrality in each module. Abbreviations: AT, association transfer between organisms; BK, curated biological pathway knowledge; CP, computational predictions; G, gene; M, module; PI, physical interactions; TM, text-mining.
In the first step, the gene expression modules that characterize the cell type of interest are identified using the genome-wide expression data. There are several methods that can be used for this objective. In our study, we used the weighted correlation network analysis implemented in the WGCNA R software package [34]. In this method, the correlation between genes is used to compute a network adjacency matrix, which fully determines the gene co-expression networks. From this network adjacency matrix, WGCNA then uses an unsupervised clustering approach to identify the gene expression modules that best characterize the gene expression of the cell or tissue of interest.
In the second step, the gene expression modules that characterize the cell type of interest are analyzed for enrichment of a specific biological process that is related to the disease or trait of interest (i.e. cell cycle in cancer studies). For this objective, a biological annotation database like the gene ontology (GO) database [35] is used. The enrichment can then be quantitatively measured using well-known statistical methods. In our study, we assessed the statistical significance of the GO functional enrichment using the Fisher's exact test as described previously [36]. Only those gene expression modules that are significantly enriched by the biological process of interest are selected for the next step.
In the third step, the biological knowledge is used to build functional-based networks from the gene expression modules of interest. In these functional-based networks, the nodes are the genes from the selected gene expression modules. Then, according to the presence or absence of biological evidence between each gene pair, the edges of the network are established. For this objective, a complete and updated database of functional associations is required. In our study, we used STRING (v9.1, 2013) software tool [37]. STRING is a powerful bioinformatics database that integrates five different sources of functional associations between genes in more than 1,100 organisms. These sources are i) physical interactions, ii) curated biological pathway knowledge, iii) computational predictions, iv) text-mining and v) association transfer between organisms. The association transfer between organisms is based on the principle of interaction conservation, which means that a pair of proteins binding in one organism is expected to be binding in another organism if both genes are present in both genomes. Therefore, the functional associations in one organism can be transferred to another organism using comparative genomics.
In the fourth step of this approach, network analysis methods are used to identify those genes that are likely to play a central role in the previously identified functional-based networks. In network analysis, the most influential genes are those that show either many connections to other genes and/or that exert an essential connection between gene (node) subnetworks. These two features are commonly known as degree centrality (DC) and betweenness centrality (BC), respectively. Genes with high DC are defined as hubs and may also be more evolutionary conserved than non-hubs [38]. Bottlenecks, that is genes with high BC, are more likely to be essential than proteins with low BC [39], [40]. In our study, the network analysis was performed using the Cytoscape v3.0.1 software [41].
In the last step of this systems genetics method, only those genes that are more likely to play a central role in the cell-specific network are selected and analyzed for the trans-eQTL analysis. These genes are selected according to their DC and BC values obtained in the previous step and, therefore, will have a higher probability to be associated to an influential genetic variation in the cell type of interest.
With this systems genetics approach, biological and network information is efficiently used to objectively reduce the number of genes included in the trans-eQTL analysis to those with highest influence on the cell type of interest and, therefore, increase the likelihood of identifying relevant trans-eQTL associations.
Analysis of differential genomic regulation profiles
In order to compare the genomic regulation profiles of CD4+ T cells and LCL cells, we focused on the study of cis- and trans-eQTL associations of two different groups of genes. In the first group, we analyzed those genes previously associated with RA risk and belonging to known biological pathways [42]. In the second group, we analyzed those genes associated with the T cell differentiation into different CD4+ T cell subtypes, including T helper 1 (Th1), T helper 2 (Th2), T helper 17 (Th17) and T regulatory (Treg) subtypes. This last set of genes was extracted from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [43]. In total, the comparative eQTL analysis between CD4+ T cells and LCL cells was performed using 145 transcripts, corresponding to 113 different genes (Table S2). The LCL gene expression profiles and the corresponding whole genome genotype data were obtained from 26 unrelated Caucasian European (CEU) individuals from the HapMap reference project [44].
In order to avoid the study of redundant eQTLs (i.e. neighboring SNPs in high linkage disequilibrium associated to one single transcript), we divided the autosomal chromosomes into 32,962 independent loci according to the localization of high recombination sites (i.e. hotspots) [45]. The whole set of SNPs were mapped to these independent loci in order to be able to determine the Transcript Complexity Value (TCV). The TCV represents the number of independent loci that are associated to the expression of one particular gene. Using the cis- (P<0.05) and trans-eQTLs (P<1e−5) identified in the CD4+ T cells and LCLs analyses, we computed the TCV for each of the 113 selected genes. The statistical significance of the differences in TCVs between both cell types was assessed using the Fisher's exact test.
Results
Genome-wide cis-eQTL analysis in RA CD4+ T cells
After performing the cis-eQTL analysis of the gene expression of CD4+ T cell from RA patients having an active disease (n = 8,747,394 tests), we detected two genome-wide significant associations with FAM66C gene: SNP rs7976243 (chromosome 12p13.3, PFDR = 2.85e−2) and SNP rs2244822 (chromosome 12p13.3, PFDR = 2.85e−2). Both genome-wide significant cis-eQTLs are only 2.14 Kb apart and, as expected, they are in high linkage disequilibrium (r2 = 0.95, HapMap Caucasian European samples) and consequently, they represent the same association signal. A complete list of cis-eQTLs from RA CD4+ T cells having a PFDR<0.5 is shown in Table S3.
Trans-eQTL analysis in RA CD4+ T cells using the novel systems genetics approach
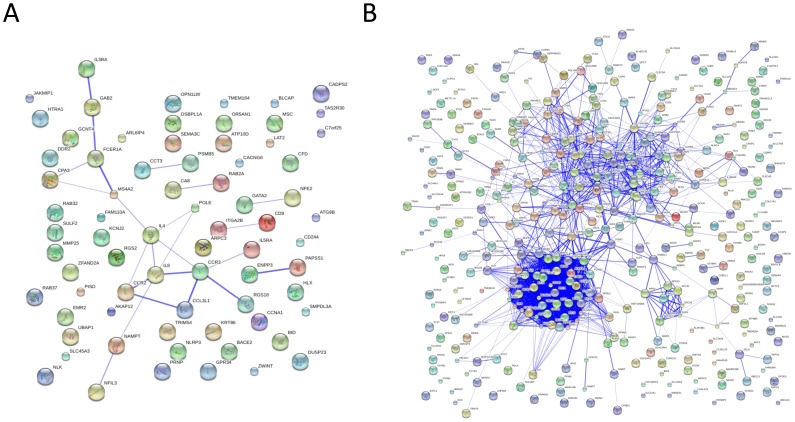
A total of 16 gene expression modules were found to characterize the CD4+ T cell gene expression of RA patients with active disease (Table 1). Given that RA is an autoimmune disease characterized by the chronic activity of inflammatory cells, we performed the functional enrichment analysis of the CD4+ T cell modules over GO terms related to the immune system. From the 16 gene expression modules identified in RA CD4+ T cells, we found two modules to be highly significantly enriched in genes related to the immune system (P = 9.23e−21 and P = 9.14e−5 for modules 9 and 12, respectively). Using these two immune-related gene expression modules, we built their corresponding functional-based networks. The M9 CD4+ T cell module functional analysis revealed a network of 15 interconnected genes (Figure 2 A). The analysis of the M12 gene-expression module showed larger and more complex network involving 247 genes. The topological structure of the M12 functional network (Figure 2 B) suggests the existence of different subnetworks that are connected by a few genes (i.e. genes showing high BC).
Table 1. Gene expression modules identified in RA CD4+ T cells.
| Module | Number of Transcripts | Immune System Process | P-value |
| M1 | 288 | NS | 3.69e-01 |
| M2 | 2326 | Underrepresented | 5.00e-03* |
| M3 | 1682 | Underrepresented | 3.41e-06* |
| M4 | 83 | Underrepresented | 1.26e-02* |
| M5 | 755 | NS | 2.64e-01 |
| M6 | 155 | NS | 3.69e-01 |
| M7 | 10003 | NS | 7.71e-01 |
| M8 | 199 | NS | 4.94e-01 |
| M9 | 75 | Overrepresented | 9.23e-21* |
| M10 | 253 | NS | 1.36e-01 |
| M11 | 157 | NS | 7.50e-01 |
| M12 | 402 | Overrepresented | 9.14e-05* |
| M13 | 124 | Underrepresented | 5.34e-23* |
| M14 | 136 | Underrepresented | 2.82e-06* |
| M15 | 2904 | NS | 7.90e-02 |
| M16 | 1578 | Underrepresented | 3.50e-05* |
Applying the new systems genetics approach for trans-eQTL identification in RA CD4+ T cells and using WGCNA, 16 genes expression modules (M) that characterize the CD4+ T cells were identified. For each gene expression module, the number of gene transcripts representing each module is shown, as well as the results of the immunological enrichment analysis.
Abbreviations: NS, No Significant.
* Significant (P<0.05).
Figure 2. Building of functional-based networks for M9 and M12 CD4+ gene expression modules using biological knowledge.
A: Functional-based network obtained from the immunologically enriched M9 gene expression module where 15 genes display a connected network. B: Functional-based network obtained from the immunologically enriched M12 gene expression module, which shows a complex network structure with different subnetworks involving 247 interconnected genes.
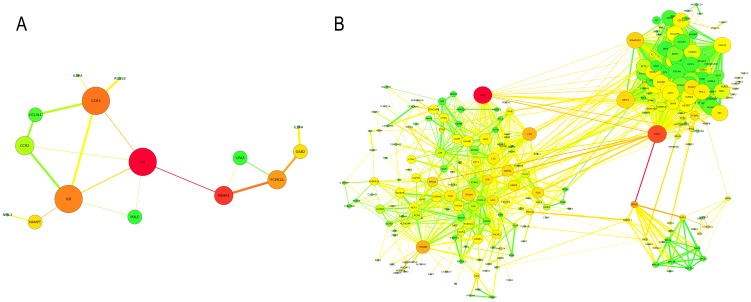
After computing the network properties of each gene in the identified M9 and M12 functional networks (Table 2), we selected those genes showing the highest centrality measures. A total of 13 genes showing either high connectivity with other genes (DCM9≥5, DCM12≥51) or high connectivity between node subnetworks (BCM9≥0.26, BCM12≥0.05) were selected. These selected genes were IL4, MS4A2, CCR3, IL8 and FCER1A genes from M9 module and TSPO, CDK1, RPS3, TYROBP, CD4, BIRC5, CDC45 and KIAA0101 genes from M12 module (Figure 3). Using this set of genes that have a very high probability of being relevant in the CD4+ T cell pathophysiology in RA, we finally performed the trans-eQTL analysis (NM9 = 3,435,556 tests, NM12 = 5,725,245 tests). After multiple test correction, we identified six statistically significant trans-eQTLs (Table 3). A complete list of the trans-eQTLs having a nominal P-value<1e-5 in modules M9 and M12 is shown in Tables S4 and S5, respectively.
Table 2. Network statistics of M9 and M12 RA CD4+ gene expression modules.
| Module | Gene Symbol | BC | DC | Module | Gene Symbol | DC | BC |
| M9 | IL4 | 0.5329 | 5 | M9 | IL4 | 5 | 0.5329 |
| M9 | MS4A2 | 0.4395 | 3 | M9 | CCR3 | 5 | 0.3297 |
| M9 | CCR3 | 0.3297 | 5 | M9 | IL8 | 5 | 0.3021 |
| M9 | IL8 | 0.3021 | 5 | M9 | MS4A2 | 3 | 0.4395 |
| M9 | FCER1A | 0.2637 | 3 | M9 | FCER1A | 3 | 0.2637 |
| M9 | NAMPT | 0.1429 | 2 | M9 | CCR2 | 3 | 0.0549 |
| M9 | GAB2 | 0.1429 | 2 | M9 | NAMPT | 2 | 0.1429 |
| M9 | CCR2 | 0.0549 | 3 | M9 | GAB2 | 2 | 0.1429 |
| M9 | CCL3L1 | 0.0110 | 2 | M9 | CCL3L1 | 2 | 0.0110 |
| M9 | RGS18 | 0.0000 | 1 | M9 | POLE | 2 | 0.0000 |
| M12 | TSPO | 0.2436 | 55 | M12 | CDK1 | 57 | 0.1717 |
| M12 | CDK1 | 0.1717 | 57 | M12 | TSPO | 55 | 0.2436 |
| M12 | RPS3 | 0.0905 | 11 | M12 | BIRC5 | 53 | 0.0432 |
| M12 | TYROBP | 0.0836 | 35 | M12 | CDC45 | 51 | 0.0138 |
| M12 | CD4 | 0.0738 | 36 | M12 | KIAA0101 | 50 | 0.0515 |
| M12 | KIAA0101 | 0.0515 | 50 | M12 | TOP2A | 50 | 0.0156 |
| M12 | MNDA | 0.0472 | 20 | M12 | CCNA2 | 50 | 0.0036 |
| M12 | MAPK1 | 0.0455 | 25 | M12 | TK1 | 49 | 0.0246 |
| M12 | BIRC5 | 0.0432 | 53 | M12 | CDT1 | 49 | 0.0156 |
| M12 | ITGAM | 0.0425 | 34 | M12 | DLGAP5 | 49 | 0.0103 |
Applying the new systems genetics approach for trans-eQTL identification in RA CD4+ T cells, the BC and DC values were computed for each particular gene of M9 and M12 immunity-enriched modules. On the left side of the table, the top 10 genes showing the highest BC values in each module are sorted by BC in decreasing order. On the right side of the table, the top 10 genes showing the highest DC values in each module are sorted by DC in decreasing order.
Abbreviations: BC, Betweenness centrality; DC, Degree centrality.
Figure 3. Functional-based networks analyzed in each enriched CD4+ T cell gene expression module.
A: Functional-based network obtained from the immunologically enriched M9 gene expression module. B: Functional-based network obtained from the immunologically enriched M12 gene expression module. The dimensions of each node (i.e. gene) are proportional to its DC value and its color is based on its BC value, ranging from green (lowest BC values) to red (highest BC values). The edge width is proportional to the strength of the functional association evidences between two genes. The edge betweenness determines the edge color, ranging from green (edges connecting nodes with the lowest BC values) to red (edges connecting nodes with the highest BC values).
Table 3. Trans-eQTL associations revealed by the application of the systems genetics approach.
| Module | SNP | Gene | SNP Coordinates (bp) | Gene Coordinates (bp) | β | P-value | PFDR |
| M9 | rs9293162 | FCER1A | chr5:25065470 | chr1:159253678–159278014 | 1,851 | 1.1927e-07 | 0.077 |
| M9 | rs978897 | FCER1A | chr7:15598156 | chr1:159253678–159278014 | 1,670 | 3.2865e-07 | 0.094 |
| M12 | rs3862556 | KIAA0101 | chr10:76540987 | chr15:64657210–64673702 | −0,924 | 2.4332e-08 | 0.046* |
| M12 | rs711114 | KIAA0101 | chr12:78003541 | chr15:64657210–64673702 | −0,848 | 5.0807e-08 | 0.048* |
| M12 | rs10283761 | BIRC5 | chr9:26755930 | chr17:76210276–76221716 | −0,345 | 5.3493e-08 | 0.048* |
| M12 | rs9561023 | KIAA0101 | chr13:93022401 | chr15:64657210–64673702 | −0,999 | 6.0208e-08 | 0.048* |
| M12 | rs2513046 | KIAA0101 | chr11:62236094 | chr15:64657210–64673702 | −0,999 | 6.0208e-08 | 0.048* |
| M12 | rs17009383 | KIAA0101 | chr3:21771769 | chr15:64657210–64673702 | −0,573 | 7.3813e-08 | 0.048* |
| M12 | rs4806933 | KIAA0101 | chr19:3414020 | chr15:64657210–64673702 | −0,727 | 1.6853e-07 | 0.079 |
| M12 | rs4745758 | KIAA0101 | chr10:76568257 | chr15:64657210–64673702 | −0,814 | 4.7989e-07 | 0.095 |
| M12 | rs11001178 | KIAA0101 | chr10:76601805 | chr15:64657210–64673702 | −0,814 | 4.7989e-07 | 0.095 |
| M12 | rs10824245 | KIAA0101 | chr10:76666799 | chr15:64657210–64673702 | −0,814 | 4.7989e-07 | 0.095 |
| M12 | rs736086 | KIAA0101 | chr10:76683785 | chr15:64657210–64673702 | −0,814 | 4.7989e-07 | 0.095 |
| M12 | rs4746248 | KIAA0101 | chr10:76705881 | chr15:64657210–64673702 | −0,814 | 4.7989e-07 | 0.095 |
| M12 | rs7046685 | KIAA0101 | chr9:28659143 | chr15:64657210–64673702 | −0,873 | 5.0499e-07 | 0.095 |
| M12 | rs12341535 | KIAA0101 | chr9:28663042 | chr15:64657210–64673702 | −0,873 | 5.0499e-07 | 0.095 |
| M12 | rs17101861 | KIAA0101 | chr14:34232772 | chr15:64657210–64673702 | −0,873 | 5.0499e-07 | 0.095 |
| M12 | rs6756606 | CDK1 | chr2:178700057 | chr10:62538211–62554610 | −0,688 | 5.2009e-07 | 0.095 |
| M12 | rs10497482 | CDK1 | chr2:178805428 | chr10:62538211–62554610 | −0,688 | 5.2009e-07 | 0.095 |
| M12 | rs6756606 | KIAA0101 | chr2:178700057 | chr15:64657210–64673702 | −0,955 | 5.5269e-07 | 0.095 |
| M12 | rs10497482 | KIAA0101 | chr2:178805428 | chr15:64657210–64673702 | −0,955 | 5.5269e-07 | 0.095 |
| M12 | rs2714384 | BIRC5 | chr18:25302948 | chr17:76210276–76221716 | −0,348 | 6.4318e-07 | 0.098 |
| M12 | rs2617950 | BIRC5 | chr18:25309414 | chr17:76210276–76221716 | −0,348 | 6.4318e-07 | 0.098 |
| M12 | rs2851757 | BIRC5 | chr18:25313101 | chr17:76210276–76221716 | −0,348 | 6.4318e-07 | 0.098 |
Applying the new systems genetics approach for trans-eQTL identification in RA CD4+ T cells, the associations obtained after performing the trans-eQTL analysis are displayed for each immunity-enriched module (PFDR<0.01).
Abbreviations: Bp, Base pair; β, Slope coefficient; PFDR, P-value False Discovery Rate.
* Significant (PFDR<0.05).
The statistically significant trans-eQTLs identified with our novel systems genetics approach were associated to the expression of KIAA0101 and BIRC5 genes, both central genes of the M12 CD4+ T cell network. Five of the significant trans-eQTLs were associated to the expression of KIAA0101 and are located in different chromosomic regions: SNP rs3862556 (intergenic variant, chromosome 10q22.2, PFDR = 4.6e−2), SNPS rs711114 (intergenic variant, chromosome 12q21.1, PFDR = 4.8e−2), SNP rs9561023 (GPC5 locus, chromosome 13q31.3, PFDR = 4.8e−2), SNP rs2513046 (AHNAK locus, chromosome 11q12.1, PFDR = 4.8e−2) and SNP rs17009383 (ZNF385D locus, chromosome 3p24.3, PFDR = 4.8e−2). The remaining trans-eQTL was established between the genetic variation at SNP rs10283761 (intergenic variant, chromosome 9p21.2, PFDR = 4.8e-2) and the expression levels of the BIRC5 gene.
Analysis of differential genomic regulation profiles
The comparison of the genomic regulation profiles of CD4+ T cells and LCL cells revealed several genes showing significantly different genetic regulatory mechanisms between both cell types.
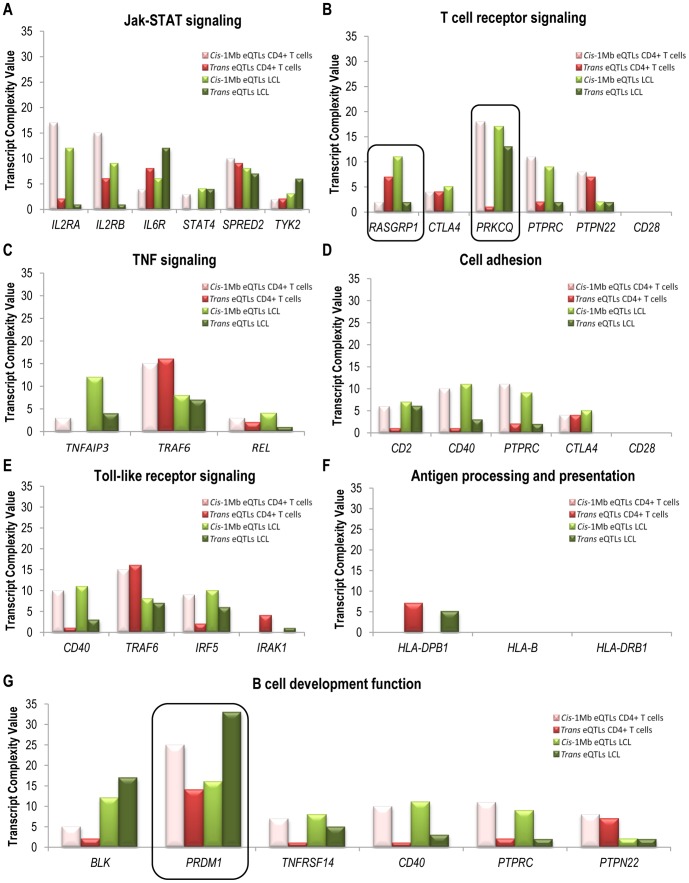
In the RA risk gene group, three genes showing a highly differential genomic regulation between CD4+ T cells and LCLs were identified (Figure 4). In the T cell receptor signaling pathway, PRKCQ gene had similar TCVs in LCLs cis- and trans-eQTLs while CD4+ T cells had a high cis-TCV and a practically absent trans-TCV (P = 4.1e−3). In this same pathway, RASGRP1 gene showed an opposite regulatory pattern in both cell types, with a predominant trans-TCV in CD4+ T cells and a predominant cis-TCV in LCLs (P = 7.3e−3). Finally, PRDM1 gene, belonging to the B cell development pathway, was found to be mainly cis-regulated in CD4+ T cells and trans-regulated in LCLs (P = 4.9e−3).
Figure 4. Transcript Complexity Value analysis of RA risk genes.
TCV from the RA CD4+ T cell and control LCLs analyzed in RA risk genes. A: JAK-STAT signaling pathway. B: T cell receptor signaling pathway. C: TNF signaling pathway. D: Cell adhesion pathway. E: Toll-like receptor signaling pathway. F: Antigen processing and presentation pathway. G: B cell development function pathway. The genes that are framed represent those genes showing significant genomic regulation profiles between RA CD4+ T cells and LCLs (P<0.05).
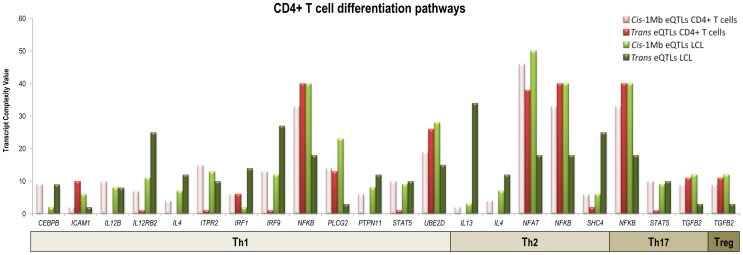
Among those genes involved in T cell differentiation pathways, we found a total of 17 genes showing a differential regulatory profile between both cell types. Taking into account that CD4+ T cell differentiation pathways can share several genes, we found a differential regulation in 13, 5, 3 and 1 genes from the Th1, Th2, Th17 and Treg differentiation pathways, respectively (Figure 5).
Figure 5. Transcript Complexity Value analysis of T cell differentiation genes.
The type and quantity of significant eQTLs in RA CD4+ T cells and LCL cells are compared in the genes from the Th1, Th2, Th17 and Treg differentiation pathways. Only those genes showing a significantly different genomic regulation profile (P<0.05) are shown.
NFKB (P = 8.0e−3), ICAM1 (P = 1.9e−2), UBE2D (P = 3.5e−2) and TGFB2 (P = 4.6e−2) genes showed opposite regulatory profiles between both cell types, with a higher contribution of trans regulation in CD4+ T cells and a higher contribution of cis regulation in LCLs. Conversely, we found that the expression of IRF9 (P = 7.9e−6), IL12RB2 (P = 4.9e−3), SHC4 (P = 5.6e−3), IL12B (P = 9.5e−3), IL13 (P = 1.3e−2), ITPR2 (P = 1.4e−2), PTPN11 (P = 1.7e−2), STAT5 (P = 2.3e−2) and IL4 (P = 3.7e−2) genes were predominantly regulated by cis-acting elements in CD4+ T cells from RA patients.
The IRF1 gene expression, whose cis- and trans- regulation was clearly found to be similar in RA CD4+ T cells, showed a markedly higher trans-eQTL regulation and almost absent cis-eQTL evidence of regulation in LCLs (P = 4.4e−2). PLCG2 gene expression regulation was also similar in CD4+ T cells, but had a markedly high cis regulation and almost absent trans regulation in LCLs (P = 6.2e−3). Finally, the regulatory profile of CEBPB gene expression was characterized by the absence of a trans regulation in RA CD4+ T cells and the clear effect of trans-acting regulatory elements in LCLs (P = 3.3e−4).
The complete list of cis- and trans-eQTLs in CD4+ T cells and LCLs that were used to characterize these regulatory profiles are shown in Table S6, Table S7, Table S8 and Table S9.
Discussion
In the present study we have performed a genome-wide analysis of eQTLs in CD4+ T cells and we have identified new genetic regulatory variants associated with the gene expression of this key cell type in RA. Analyzing the transcriptome of CD4+ T cells from RA patients with active disease we found a genome-wide significant cis-eQTL regulating the expression of FAM66C gene. In order to detect significant trans-eQTLs, we developed a novel systems genetics approach that integrates gene expression and network biology information to reduce the multiple test burden associated with this type of analysis. Using this new approach, we found statistically significant trans-eQTLs regulating the expression of BIRC5 and KIAA0101 genes in CD4+ T cells in RA. Finally, comparing the genetic regulatory patterns of RA CD4+ T cells with control LCLs, we found several differential regulatory patterns. This study represents the first global analysis of the CD4+ T cell regulatory architecture associated with RA.
FAM66C is a long non-coding RNA (lncRNA) gene, whose biological functionality is still unknown. LncRNAs genes are transcribed into non-protein coding transcripts that are longer than 200 nucleotides and have been shown to act as modulators of the gene expression through epigenetic changes, transcriptional regulation and post-transcriptional regulation [46]. To our knowledge, the FAM66C lncRNA gene has not been previously associated with the pathophysiology of RA or any other complex disease. Recently, lncRNAs have emerged as possible contributors to the basis of human diseases by regulating the expression of neighbouring protein-coding genes [46], [47]. Interestingly, FAM66C lncRNA gene maps near C3AR1 gene (<115 Kb), which encodes a complement receptor that has been shown to be crucial in the modulation of the function of CD4+ T cell subtypes [48]. C3AR1 expression promotes the proinflammatory activity of CD4+ T cells by enhancing the survival and function of Th1 and Th17 cells, while its inhibition leads to the induction of Treg CD4+ cells [49]. Additional studies will need to be carried out to determine if this or other biological mechanisms are responsible for the observed FAM66C association in the RA CD4+ T cell transcriptome.
Genes that encode proteins with high DC and BC values are likely to have a high impact in the network functionality. Consequently, the characterization of such central genes in the functional networks of specific cell types can be a powerful strategy to identify regulatory variants that contribute to disease [50], [51]. Based on this assumption we developed a new dimensionality reduction approach that allowed us to determine the most influential genes in the CD4+ T cell specific networks and identify significant trans-eQTL associations with two of these genes, BIRC5 and KIAA0101. These cell type-specific regulatory mechanisms are therefore likely to be of high importance in the activity of CD4+ T cells associated with RA pathology.
BIRC5 gene encodes survivin, an antiapoptotic protein that has been strongly associated to RA pathogenesis [52], [53], [54]. Survivin mRNA levels in peripherial blood mononuclear cells have been significantly associated with disease activity and the extent of joint damage in RA [55]. Importantly, survivin expression has been shown to be a key promoter of T cell proliferation after antigen presentation as well as a powerful antagonizer of apoptosis in activated T cells [56], [57]. Increased proliferation and reduced cell death are two of the main characteristics of the CD4+ T cell population infiltrating the synovial membrane in RA [58], [59]. Survivin-mediated cell survival could therefore have a major role in maintaining this pathogenic CD4+ T cell features. Additionally, there is recent evidence demonstrating that survivin expression in CD4+ T cells is activated by TNF-α cytokine which is the key regulator of the inflammatory and tissue-destructive pathways in RA [56].
KIAA0101 gene encodes for the Proliferating Cell Nuclear Antigen (PCNA) associated factor that acts as a regulator of DNA repair during DNA replication [60]. Importantly, PCNA-associated factor interacts with PCNA which increases the DNA polymerase's processivity during elongation of the leading strand and, therefore, accelerates the cell cycle progression [61]. In RA, an increased rate of cell proliferation has been shown to be associated with high levels of PCNA in synovial fibroblasts [62]. Accordingly, we suggest that the increased proliferation and reduced apoptosis of CD4+ T cells in the synovial membrane in RA is also regulated by the expression of KIAA0101 gene.
The main goal of the new systems genetics approach described in this study is to objectively reduce the multiple test problem associated with the genome-wide analysis of trans-eQTLs. An exhaustive trans-eQTL analysis at a genome-wide scale would have required to perform approximately 12,000e6 association tests. This extremely high number of tests would have lead to an excessive penalization for multiple testing and, consequently, the inability to detect true associations. Using the new systems genetics approach we reduced this number of tests to 3.4e6 and 5.7e6 trans-eQTL analyses for M9 and M12 functional networks, respectively. Clearly, our study demonstrates that this methodological approach can be useful to identify significant trans-eQTLs without the need to explore all the combinatorial space. Importantly, the proposed systems genetics workflow includes several steps that can be easily customized to incorporate new or alternative bioinformatics methodologies. For example, in the functional enrichment analysis we used the gene ontology database but other functional annotation databases like the KEGG database can be used instead. The flexibility of this systems genetics approach workflow also makes this method a powerful strategy to uncover the relevant trans-eQTLs associated with human traits or diseases, including the upcoming studies based on RNA-seq technologies.
Trans-eQTL associations present in CD4+ T cells and absent in LCLs could be indicative of cell-specific regulatory processes that are specifically activated in RA. In the group of genes associated with RA risk, we found RASGRP1 and PRDM1 genes to have a differential regulation between both cell types. RASGRP1 has been shown to be a critical regulator of the ERK/MAP signaling pathway which is crucial for T cell development, homeostasis and differentiation. T cells from patients with RA have been shown to have hyperresponsive ERK activity upon TCR stimulation [63]. Consequently, RASGRP1 expression could be a specific modulator of the CD4+ T cell hyperresponsiveness to autoantigens associated with RA. PRDM1 gene, instead, has been shown to drive the maturation of B-lymphocytes into immunoglobulin-secreting cells [64]. Consistently, we found a predominant trans regulation of PRDM1 gene expression in LCLs, which are cell lines originally generated from B cells.
Among those genes involved in the CD4+ T cell differentiation pathways, we found a markedly differential regulatory profile of Nuclear Factor Kappa B (NFKB) and Transforming Growth Factor Beta 2 (TGFB2) genes. NKFB is a well known transcription factor that has been associated with the T cell differentiation into Th1, Th2 and Th17 subtypes [65] and is a pivotal regulator of the inflammatory process present in rheumatoid arthritis [66], [67], [68]. The identification of the genetic variants that control NFKB gene expression in CD4+ T cells could lead to a better understanding of the biological mechanisms that are more relevant in the regulation of this cell type in RA.
TGFB2 is a Transforming Growth Factor family cytokine that has been associated with immunological tolerance and Treg and Th17 pathways [69]. Previous studies have shown that normal Treg/Th17 cell balance is not maintained in RA, with an increase in the differentiation of CD4+ T cells into the proinflammatory CD4+ Th17 phenotype and a decrease in the production of anti-inflammatory CD4+ Tregs [70], [71], [72]. Therefore, the increased trans-eQTLs associated with CD4+ T cells compared to LCLs, could indicate a specific regulatory mechanism associated with the increase of T cell autoreactivity observed in RA pathophysiology.
The different methodologies used in this study for the characterization of the CD4+ T cell-specific genetic regulation in RA have nonetheless some limitations. The CD4+ T cell population is highly heterogeneous with different subtypes exerting sometimes opposing regulatory activities in inflammation. Therefore, a more comprehensive analysis would have required the isolation and separate analysis of each CD4+ subpopulation. However, by using a homogeneous cohort of RA patients with a high level of disease activity, we favored the collection of a highly similar gene expression profile representative of the pathogenic regulation of CD4+ T cells in RA. Also, the comparison of the CD4+ regulatory pattern against LCLs of control individuals could have limited the identification of additional relevant regulatory mechanisms. As more eQTL data on different cell subtypes becomes available, more cell-to-cell comparisons can be performed in order to completely characterize the specific regulatory mechanisms of CD4+ T cells in RA.
In the analysis of differential genomic regulation profiles we focused on genes associated with the susceptibility to develop RA as well as genes associated with T cell differentiation. Another potential limitation of this approach is that other genes that encode proteins associated with RA pathophysiology are not included. From these, the pro-inflammatory cytokines TNF-α [73], IL-1β [74] and IFN-γ [75] and the anti-inflammatory cytokine IL-10 [76] have shown to be key in the development and chronification of RA. However, analyzing the regulatory profiles for these genes only a significant differential regulation for IL-1β is observed (data not shown). This association is due to a predominant cis-regulation in CD4+ T cells compared to an increased trans-regulation in LCLs. This result is consistent with the high level of expression of this cytokine observed in different immune and non-immune cell types like monocytes, tissue macrophages or synovial fibroblasts [77], [78]. Together, the results of our study support the use of this methodology to characterize the functionality of disease risk genes as well as genes annotated to the cell type of interest.
One of the most important challenges ahead in human genetics is to identify the regulatory elements that control the gene expression and how they contribute to disease. This study is the first approach to the characterization of the CD4+ T cell regulatory profile associated with RA. In this comprehensive genetic study we report genetic regulatory variants that are significantly associated with the expression of FAM66C lncRNA, BIRC5 and KIAA0101 genes in RA CD4+ T cells. These results highlight the importance of the cell cycle processes in the pathological activity of RA CD4+ T cells infiltrating the synovial membrane, as well as the potential implication of lncRNA in the genetic regulatory basis of RA. This study represents a significant progress in the characterization of the genetic regulation of the main immune cell type involved in the pathogenesis of RA.
Supporting Information
Main epidemiological and clinical features of the RA patients included in this study. Abbreviations: DAS28, Disease Activity Score.
(DOC)
Total genes used in the comparative eQTL analysis between RA CD4+ T cells and LCLs.
(XLS)
Cis -1Mb associations obtained in the genome-wide cis -eQTL analysis in RA CD4+ T cells (PFDR<0.5). Abbreviations: Gene chr, Chromosomal location of the expressed gene; Gene coord, Coordinates of the expressed gene; Bp, Base pair; SNP chr, Chromosomal location of the SNP; SNP coord, Coordinates of the SNP; FDR, P-value False Discovery Rate.
(XLS)
Trans associations in CD4+ T cells for M9 module genes (P<1e-5). Abbreviations: Gene chr, Chromosomal location of the expressed gene; Gene coord, Coordinates of the expressed gene; Bp, Base pair; SNP chr, Chromosomal location of the SNP; SNP coord, Coordinates of the SNP; Beta, Slope coefficient; FDR, P-value False Discovery Rate.
(XLS)
Trans associations in CD4+ T cells for M12 module genes (P<1e-5). Abbreviations: Gene chr, Chromosomal location of the expressed gene; Gene coord, Coordinates of the expressed gene; Bp, Base pair; SNP chr, Chromosomal location of the SNP; SNP coord, Coordinates of the SNP; Beta, Slope coefficient; FDR, P-value False Discovery Rate.
(XLS)
Cis -1Mb associations in CD4+ T cells (P<5e-2) that were used to characterize the gene regulatory profiles. Abbreviations: Gene chr, Chromosomal location of the expressed gene; Gene coord, Coordinates of the expressed gene; Bp, Base pair; SNP chr, Chromosomal location of the SNP; SNP coord, Coordinates of the SNP; Beta, Slope coefficient; FDR, P-value False Discovery Rate.
(XLS)
Trans associations in CD4+ T cells (P<1e-5) that were used to characterize the gene regulatory profiles. Abbreviations: Gene chr, Chromosomal location of the expressed gene; Gene coord, Coordinates of the expressed gene; Bp, Base pair; SNP chr, Chromosomal location of the SNP; SNP coord, Coordinates of the SNP; Beta, Slope coefficient; FDR, P-value False Discovery Rate.
(XLS)
Cis -1Mb associations in LCLs (P<5e-2) that were used to characterize the gene regulatory profiles. Abbreviations: Gene chr, Chromosomal location of the expressed gene; Gene coord, Coordinates of the expressed gene; Bp, Base pair; SNP chr, Chromosomal location of the SNP; SNP coord, Coordinates of the SNP; Beta, Slope coefficient; FDR, P-value False Discovery Rate.
(XLS)
Trans associations in LCLs (P<1e-5) that were used to characterize the gene regulatory profiles. Abbreviations: Gene chr, Chromosomal location of the expressed gene; Gene coord, Coordinates of the expressed gene; Bp, Base pair; SNP chr, Chromosomal location of the SNP; SNP coord, Coordinates of the SNP; Beta, Slope coefficient; FDR, P-value False Discovery Rate.
(XLS)
Acknowledgments
We thank Joachim L. Schultze and Svenja Debey-Pascher from the LIMES Institute (Bönn, Germany) for technical support in microarray analysis.
We thank Devin Absher from the HudsonAlpha Institute for Biotechnology (Alabama, United States) for technical support in GWAS genotyping.
Data Availability
The authors confirm that, for approved reasons, some access restrictions apply to the data underlying the findings. The data discussed in this publication have been deposited in GEO (FOR REVIEWERS ONLY: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=sbsruqocflkfhcz&acc=GSE55468).
Funding Statement
This work was supported by the Spanish Ministry of Economy and Competitiveness grants [PSE-010000-2006-6 and IPT-010000-2010-36] and the Fondo de Investigación Sanitaria grant [PI040720]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Choy EH, Panayi GS (2001) Cytokine pathways and joint inflammation in rheumatoid arthritis. N Engl J Med 344: 907–916. [DOI] [PubMed] [Google Scholar]
- 2. Firestein GS (2003) Evolving concepts of rheumatoid arthritis. Nature 423: 356–361. [DOI] [PubMed] [Google Scholar]
- 3. Maxwell L, Singh JA (2009) Abatacept for rheumatoid arthritis. Cochrane Database Syst Rev: CD007277 doi: 007210.001002/14651858.CD14007277.pub14651852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pieper J, Herrath J, Raghavan S, Muhammad K, Vollenhoven R, et al. (2013) CTLA4-Ig (abatacept) therapy modulates T cell effector functions in autoantibody-positive rheumatoid arthritis patients. BMC Immunol 14 34: 10.1186/1471–2172-1114-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skapenko A, Leipe J, Lipsky PE, Schulze-Koops H (2005) The role of the T cell in autoimmune inflammation. Arthritis Res Ther 7: S4–14. Epub 2005 Mar 2016. [DOI] [PMC free article] [PubMed]
- 6. Manolio TA (2010) Genomewide association studies and assessment of the risk of disease. N Engl J Med 363: 166–176 doi: 110.1056/NEJMra0905980 [DOI] [PubMed] [Google Scholar]
- 7. Montgomery SB, Dermitzakis ET (2011) From expression QTLs to personalized transcriptomics. Nat Rev Genet 12: 277–282 doi: 210.1038/nrg2969. Epub 2011 Mar 1039 [DOI] [PubMed] [Google Scholar]
- 8. Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M (2009) Mapping complex disease traits with global gene expression. Nat Rev Genet 10: 184–194 doi: 110.1038/nrg2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Majewski J, Pastinen T (2011) The study of eQTL variations by RNA-seq: from SNPs to phenotypes. Trends Genet 27: 72–79 doi: 10.1016/j.tig.2010.1010.1006. Epub 2010 Nov 1029 [DOI] [PubMed] [Google Scholar]
- 10. Nica AC, Dermitzakis ET (2013) Expression quantitative trait loci: present and future. Philos Trans R Soc Lond B Biol Sci 368: 20120362 doi: 20120310.20121098/rstb.20122012.20120362. Print 20122013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, et al. (2009) Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325: 1246–1250 doi: 1210.1126/science.1174148. Epub 1172009 Jul 1174130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, et al. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 doi: 110.1038/nature07829. Epub 02009 Mar 07818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ackermann M, Sikora-Wohlfeld W, Beyer A (2013) Impact of natural genetic variation on gene expression dynamics. PLoS Genet 9: e1003514 doi: 1003510.1001371/journal.pgen.1003514. Epub 1002013 Jun 1003516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, et al. (2008) Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol 6: e1 doi: 10.1371/journal.pbio.1000001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grundberg E, Kwan T, Ge B, Lam KC, Koka V, et al. (2009) Population genomics in a disease targeted primary cell model. Genome Res 19: 1942–1952 doi: 1910.1101/gr.095224.095109. Epub 092009 Aug 095224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doss S, Schadt EE, Drake TA, Lusis AJ (2005) Cis-acting expression quantitative trait loci in mice. Genome Res 15: 681–691. Epub 2005 Apr 2018. [DOI] [PMC free article] [PubMed]
- 17. van Nas A, Ingram-Drake L, Sinsheimer JS, Wang SS, Schadt EE, et al. (2010) Expression quantitative trait loci: replication, tissue- and sex-specificity in mice. Genetics 185: 1059–1068 doi: 1010.1534/genetics.1110.116087. Epub 112010 May 116083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fehrmann RS, Jansen RC, Veldink JH, Westra HJ, Arends D, et al. (2011) Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet 7: e1002197 doi: 1002110.1001371/journal.pgen.1002197. Epub 1002011 Aug 1002194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Civelek M, Lusis AJ (2013) Systems genetics approaches to understand complex traits. Nat Rev Genet 3. [DOI] [PMC free article] [PubMed]
- 20. Small KS, Hedman AK, Grundberg E, Nica AC, Thorleifsson G, et al. (2011) Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Genet 43: 561–564 doi: 510.1038/ng.1833. Epub 2011 May 1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, et al. (1988) The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 31: 315–324. [DOI] [PubMed] [Google Scholar]
- 22. Prevoo ML, van 't Hof MA, Kuper HH, van Leeuwen MA, van de Putte LB, et al. (1995) Modified disease activity scores that include twenty-eight-joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum 38: 44–48. [DOI] [PubMed] [Google Scholar]
- 23. Pruitt KD, Tatusova T, Brown GR, Maglott DR (2012) NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res 40: D130–135 doi: 110.1093/nar/gkr1079. Epub 2011 Nov 1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ihaka R, Gentleman RC (1996) R: A language for data analysis and graphics. Journal of Computational and Graphical Statistics 5: 299–314. [Google Scholar]
- 25. Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193. [DOI] [PubMed] [Google Scholar]
- 26.Johnson WE, Li C, Rabinovic A (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8: 118–127. Epub 2006 Apr 2021. [DOI] [PubMed]
- 27. Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. Epub 2007 Jul 2025. [DOI] [PMC free article] [PubMed]
- 29.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904–909. Epub 2006 Jul 2023. [DOI] [PubMed]
- 30. Shabalin AA (2012) Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28: 1353–1358 doi: 1310.1093/bioinformatics/bts1163. Epub 2012 Apr 1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S, et al. (2005) Genome-wide associations of gene expression variation in humans. PLoS Genet 1: e78. Epub 2005 Dec 2016. [DOI] [PMC free article] [PubMed]
- 32. Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, et al. (2003) A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 12: 1725–1735. [DOI] [PubMed] [Google Scholar]
- 33.Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100: 9440–9445. Epub 2003 Jul 9425. [DOI] [PMC free article] [PubMed]
- 34. Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9 559: 10.1186/1471–2105-1189-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 doi: 10.1038/nprot.2008.1211 [DOI] [PubMed] [Google Scholar]
- 37. Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, et al. (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41: D808–815 doi: 810.1093/nar/gks1094. Epub 2012 Nov 1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vallabhajosyula RR, Chakravarti D, Lutfeali S, Ray A, Raval A (2009) Identifying hubs in protein interaction networks. PLoS One 4: e5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hahn MW, Kern AD (2005) Comparative genomics of centrality and essentiality in three eukaryotic protein-interaction networks. Mol Biol Evol 22: 803–806. Epub 2004 Dec 2022. [DOI] [PubMed]
- 40. Joy MP, Brock A, Ingber DE, Huang S (2005) High-betweenness proteins in the yeast protein interaction network. J Biomed Biotechnol 2005: 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Julia A, Marsal S (2013) The genetic architecture of rheumatoid arthritis: from susceptibility to clinical subphenotype associations. Curr Top Med Chem 13: 720–731. [DOI] [PubMed] [Google Scholar]
- 43. Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, et al. (1999) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 27: 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. The International HapMap Project. Nature 426: 789–796. [DOI] [PubMed] [Google Scholar]
- 45. Myers S, Bottolo L, Freeman C, McVean G, Donnelly P (2005) A fine-scale map of recombination rates and hotspots across the human genome. Science 310: 321–324. [DOI] [PubMed] [Google Scholar]
- 46. Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155–159 doi: 110.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- 47. Esteller M (2011) Non-coding RNAs in human disease. Nat Rev Genet 12: 861–874 doi: 810.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- 48. Kolev M, Le Friec G, Kemper C (2013) The role of complement in CD4(+) T cell homeostasis and effector functions. Semin Immunol 25: 12–19 doi: 10.1016/j.smim.2013.1004.1012. Epub 2013 May 1029 [DOI] [PubMed] [Google Scholar]
- 49. Strainic MG, Shevach EM, An F, Lin F, Medof ME (2013) Absence of signaling into CD4(+) cells via C3aR and C5aR enables autoinductive TGF-beta1 signaling and induction of Foxp3(+) regulatory T cells. Nat Immunol 14: 162–171 doi: 110.1038/ni.2499. Epub 2012 Dec 1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, et al. (2004) Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 430: 88–93. Epub 2004 Jun 2009. [DOI] [PubMed]
- 51. Zhu M, Gao L, Li X, Liu Z, Xu C, et al. (2009) The analysis of the drug-targets based on the topological properties in the human protein-protein interaction network. J Drug Target 17: 524–532 doi: 510.1080/10611860903046610 [DOI] [PubMed] [Google Scholar]
- 52. Svensson B, Hafstrom I, Forslind K, Albertsson K, Tarkowski A, et al. (2010) Increased expression of proto-oncogene survivin predicts Joint destruction and persistent disease activity in early rheumatoid arthritis. Ann Med 42: 45–54 doi: 10.3109/07853890903376280 [DOI] [PubMed] [Google Scholar]
- 53. Andersson SE, Svensson MN, Erlandsson MC, Dehlin M, Andersson KM, et al. (2012) Activation of Fms-like tyrosine kinase 3 signaling enhances survivin expression in a mouse model of rheumatoid arthritis. PLoS One 7: e47668 doi: 47610.41371/journal.pone.0047668. Epub 0042012 Oct 0047617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bokarewa M, Brink M, Erlandsson M, Rantapaa Dahlqvist S (2014) Survivin but not Fms-like tyrosine kinase 3 ligand is up-regulated before the onset of rheumatoid arthritis: a pilot study. Arthritis Res Ther 16: R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ahn JK, Oh JM, Lee J, Bae EK, Ahn KS, et al. (2010) Increased extracellular survivin in the synovial fluid of rheumatoid arthritis patients: fibroblast-like synoviocytes as a potential source of extracellular survivin. Inflammation 33: 381–388 doi: 310.1007/s10753-10010-19196-10751 [DOI] [PubMed] [Google Scholar]
- 56. Song J, So T, Cheng M, Tang X, Croft M (2005) Sustained survivin expression from OX40 costimulatory signals drives T cell clonal expansion. Immunity 22: 621–631. [DOI] [PubMed] [Google Scholar]
- 57.Song J, Salek-Ardakani S, So T, Croft M (2007) The kinases aurora B and mTOR regulate the G1-S cell cycle progression of T lymphocytes. Nat Immunol 8: 64–73. Epub 2006 Nov 2026. [DOI] [PubMed]
- 58. Salmon M, Scheel-Toellner D, Huissoon AP, Pilling D, Shamsadeen N, et al. (1997) Inhibition of T cell apoptosis in the rheumatoid synovium. J Clin Invest 99: 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu H, Pope RM (2003) The role of apoptosis in rheumatoid arthritis. Curr Opin Pharmacol 3: 317–322. [DOI] [PubMed] [Google Scholar]
- 60. Kais Z, Barsky SH, Mathsyaraja H, Zha A, Ransburgh DJ, et al. (2011) KIAA0101 interacts with BRCA1 and regulates centrosome number. Mol Cancer Res 9: 1091–1099 doi: 1010.1158/1541-7786.MCR-1010-0503. Epub 2011 Jun 1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Burkovics P, Hajdu I, Szukacsov V, Unk I, Haracska L (2009) Role of PCNA-dependent stimulation of 3′-phosphodiesterase and 3′-5′ exonuclease activities of human Ape2 in repair of oxidative DNA damage. Nucleic Acids Res 37: 4247–4255 doi: 4210.1093/nar/gkp4357. Epub 2009 May 4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Karouzakis E, Gay RE, Michel BA, Gay S, Neidhart M (2009) DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum 60: 3613–3622 doi: 3610.1002/art.25018 [DOI] [PubMed] [Google Scholar]
- 63. Deshpande P, Cavanagh MM, Le Saux S, Singh K, Weyand CM, et al. (2013) IL-7- and IL-15-mediated TCR sensitization enables T cell responses to self-antigens. J Immunol 190: 1416–1423 doi: 1410.4049/jimmunol.1201620. Epub 1202013 Jan 1201616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, et al. (2003) Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity 19: 607–620. [DOI] [PubMed] [Google Scholar]
- 65. Oh H, Ghosh S (2013) NF-kappaB: roles and regulation in different CD4(+) T-cell subsets. Immunol Rev 252: 41–51 doi: 10.1111/imr.12033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Makarov SS (2001) NF-kappa B in rheumatoid arthritis: a pivotal regulator of inflammation, hyperplasia, and tissue destruction. Arthritis Res 3: 200–206. Epub 2001 Mar 2026. [DOI] [PMC free article] [PubMed]
- 67. Feldmann M, Andreakos E, Smith C, Bondeson J, Yoshimura S, et al. (2002) Is NF-kappaB a useful therapeutic target in rheumatoid arthritis? Ann Rheum Dis 61: ii13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Simmonds RE, Foxwell BM (2008) Signalling, inflammation and arthritis: NF-kappaB and its relevance to arthritis and inflammation. Rheumatology (Oxford) 47: 584–590 doi: 510.1093/rheumatology/kem1298. Epub 2008 Jan 1029 [DOI] [PubMed] [Google Scholar]
- 69. Li MO, Sanjabi S, Flavell RA (2006) Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity 25: 455–471. [DOI] [PubMed] [Google Scholar]
- 70. Eisenstein EM, Williams CB (2009) The T(reg)/Th17 cell balance: a new paradigm for autoimmunity. Pediatr Res 65: 26R–31R doi: 10.1203/PDR.1200b1013e31819e31876c31817 [DOI] [PubMed] [Google Scholar]
- 71. Noack M, Miossec P (2014) Th17 and regulatory T cell balance in autoimmune and inflammatory diseases. Autoimmun Rev 11: 00008–00001. [DOI] [PubMed] [Google Scholar]
- 72. Wang W, Shao S, Jiao Z, Guo M, Xu H, et al. (2012) The Th17/Treg imbalance and cytokine environment in peripheral blood of patients with rheumatoid arthritis. Rheumatol Int 32: 887–893 doi: 810.1007/s00296-00010-01710-00290. Epub 02011 Jan 00211 [DOI] [PubMed] [Google Scholar]
- 73.Vasanthi P, Nalini G, Rajasekhar G Role of tumor necrosis factor-alpha in rheumatoid arthritis: a review.
- 74. Contassot E, Beer HD, French LE (2012) Interleukin-1, inflammasomes, autoinflammation and the skin. Swiss Med Wkly 142 w13590: 10.4414/smw.2012.13590. [DOI] [PubMed] [Google Scholar]
- 75. Page CE, Smale S, Carty SM, Amos N, Lauder SN, et al. (2010) Interferon-gamma inhibits interleukin-1beta-induced matrix metalloproteinase production by synovial fibroblasts and protects articular cartilage in early arthritis. Arthritis Res Ther 12: R49 doi: 10.1186/ar2960. Epub 2010 Mar 1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fu LH, Ma CL, Cong B, Li SJ, Chen HY, et al. (2011) Hypomethylation of proximal CpG motif of interleukin-10 promoter regulates its expression in human rheumatoid arthritis. Acta Pharmacol Sin 32: 1373–1380 doi: 1310.1038/aps.2011.1398. Epub 2011 Oct 1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Smolen JS, Aletaha D, Redlich K (2012) The pathogenesis of rheumatoid arthritis: new insights from old clinical data? Nat Rev Rheumatol 8: 235–243 doi: 210.1038/nrrheum.2012.1023 [DOI] [PubMed] [Google Scholar]
- 78. Pope RM, Shahrara S (2013) Possible roles of IL-12-family cytokines in rheumatoid arthritis. Nat Rev Rheumatol 9: 252–256 doi: 210.1038/nrrheum.2012.1170. Epub 2012 Oct 1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Main epidemiological and clinical features of the RA patients included in this study. Abbreviations: DAS28, Disease Activity Score.
(DOC)
Total genes used in the comparative eQTL analysis between RA CD4+ T cells and LCLs.
(XLS)
Cis -1Mb associations obtained in the genome-wide cis -eQTL analysis in RA CD4+ T cells (PFDR<0.5). Abbreviations: Gene chr, Chromosomal location of the expressed gene; Gene coord, Coordinates of the expressed gene; Bp, Base pair; SNP chr, Chromosomal location of the SNP; SNP coord, Coordinates of the SNP; FDR, P-value False Discovery Rate.
(XLS)
Trans associations in CD4+ T cells for M9 module genes (P<1e-5). Abbreviations: Gene chr, Chromosomal location of the expressed gene; Gene coord, Coordinates of the expressed gene; Bp, Base pair; SNP chr, Chromosomal location of the SNP; SNP coord, Coordinates of the SNP; Beta, Slope coefficient; FDR, P-value False Discovery Rate.
(XLS)
Trans associations in CD4+ T cells for M12 module genes (P<1e-5). Abbreviations: Gene chr, Chromosomal location of the expressed gene; Gene coord, Coordinates of the expressed gene; Bp, Base pair; SNP chr, Chromosomal location of the SNP; SNP coord, Coordinates of the SNP; Beta, Slope coefficient; FDR, P-value False Discovery Rate.
(XLS)
Cis -1Mb associations in CD4+ T cells (P<5e-2) that were used to characterize the gene regulatory profiles. Abbreviations: Gene chr, Chromosomal location of the expressed gene; Gene coord, Coordinates of the expressed gene; Bp, Base pair; SNP chr, Chromosomal location of the SNP; SNP coord, Coordinates of the SNP; Beta, Slope coefficient; FDR, P-value False Discovery Rate.
(XLS)
Trans associations in CD4+ T cells (P<1e-5) that were used to characterize the gene regulatory profiles. Abbreviations: Gene chr, Chromosomal location of the expressed gene; Gene coord, Coordinates of the expressed gene; Bp, Base pair; SNP chr, Chromosomal location of the SNP; SNP coord, Coordinates of the SNP; Beta, Slope coefficient; FDR, P-value False Discovery Rate.
(XLS)
Cis -1Mb associations in LCLs (P<5e-2) that were used to characterize the gene regulatory profiles. Abbreviations: Gene chr, Chromosomal location of the expressed gene; Gene coord, Coordinates of the expressed gene; Bp, Base pair; SNP chr, Chromosomal location of the SNP; SNP coord, Coordinates of the SNP; Beta, Slope coefficient; FDR, P-value False Discovery Rate.
(XLS)
Trans associations in LCLs (P<1e-5) that were used to characterize the gene regulatory profiles. Abbreviations: Gene chr, Chromosomal location of the expressed gene; Gene coord, Coordinates of the expressed gene; Bp, Base pair; SNP chr, Chromosomal location of the SNP; SNP coord, Coordinates of the SNP; Beta, Slope coefficient; FDR, P-value False Discovery Rate.
(XLS)
Data Availability Statement
The authors confirm that, for approved reasons, some access restrictions apply to the data underlying the findings. The data discussed in this publication have been deposited in GEO (FOR REVIEWERS ONLY: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=sbsruqocflkfhcz&acc=GSE55468).