Abstract
Background
The altered composition of immune cells in peripheral blood has been reported to be associated with cancer patient survival. However, analysis of the composition of peripheral immune cells are often limited in retrospective survival studies employing banked blood specimens with long-term follow-up because the application of flow cytometry to such specimens is problematic. The aim of this study was to demonstrate the feasibility of deconvolving blood-based gene expression profiles (GEPs) to estimate the proportions of immune cells and determine their prognostic values for cancer patients.
Methods and Results
Here, using GEPs from peripheral blood mononuclear cells (PBMC) of 108 non-small cell lung cancer (NSCLC) patients, we deconvolved the immune cell proportions and analyzed their association with patient survival. Univariate Kaplan-Meier analysis showed that a low proportion of T cells was significantly associated with poor patient survival, as was the proportion of T helper cells; however, only the proportion of T cells was independently prognostic for patients by a multivariate Cox regression analysis (hazard ratio = 2.23; 95% CI, 1.01–4.92; p = .048). Considering that altered peripheral blood compositions can reflect altered immune responses within the tumor microenvironment, based on a tissue-based GEPs of NSCLC patients, we demonstrated a significant association between poor patient survival and the low level of antigen presentation, which play a critical role in T cell proliferation.
Conclusions
These results demonstrate that it is feasible to deconvolve GEPs from banked blood specimens for retrospective survival analysis of alterations of immune cell composition, and suggest the proportion of T cells in PBMC which might reflect the antigen presentation level within the tumor microenvironment can be a prognostic marker for NSCLC patients.
Introduction
Immune responses to tumor cells in the tumor microenvironment play critical roles in the determination of tumor progression [1], [2]. A better understanding of these immune responses could reveal immune-related markers that could be used to stratify cancer patients with different risks of recurrence or death [3], [4]. For example, high densities of tumor-infiltrating lymphocytes and dendritic cells are associated with prolonged survival in a variety of malignancies, including non-small cell lung cancer (NSCLC) [5], [6], [7], colorectal cancer [8], [9], and ovarian cancer [10], [11]; attributed to their fundamental roles in anti-tumor immunity. It is known that immune cells respond dynamically to variations in the tumor microenvironment [12], [13], thus continuously monitoring these varying immune responses might facilitate the prevention of cancer recurrence or deterioration by enabling treatment protocols to be modulated in a timely manner. However, the immune-related markers in the tumor microenvironment can only be assessed once, at the time of surgical resection for operable cases. To overcome the defect of tumor tissue analysis, the prognostic value of alterations of the immune cell composition in the peripheral blood could be investigated; changes in the immune cell composition reflect the complicated immune status within the tumor microenvironment [14], [15], [16] and would continue to be measureable after surgical resection. In fact, many studies have found that altered compositions of peripheral immune cells, such as lymphocyte proportion, neutrophil proportion, and neutrophil-to-lymphocyte ratios in the peripheral blood, are potential markers for survival in cancer patients, including those with NSCLC [17], [18], colorectal cancer [19] and early gastric cancer [20].
To identify potential prognostic markers, retrospective studies are widely used [21], [22], [23] because the long-term follow-up of cancer patients is rather expensive and time-consuming [24]. Potential prognostic markers are usually validated using prospective studies for clinical application [25]. Tissue or blood specimens are typically accumulated on a long-term basis until there are clear objectives for study. For example, when recent investigations suggest that changes in gene expression or epigenetic may carry information that is predictive of patient outcomes, previously tissue or blood banked specimens with long-term follow-up can be used for microarray analysis to explore prognostic gene or methylation panels [16], [26], [27]. However, it is problematic to measure the proportions of immune cells in such long-term banked blood specimens by flow cytometry which is routinely applied to fresh blood specimens [28], because the specific surface proteins of immune cells may be unstable during long-term cryopreservation [29]. Therefore, other methods for evaluating the proportions of various immune cells in such valuable accumulated blood specimens must be explored for retrospective survival analysis.
Several groups have recently proposed deconvolution methods to estimate the proportions of immune cells in peripheral blood based on gene expression profiles and marker genes that are specifically expressed in immune cells [30], [31]. The proportions estimated by the deconvolution methods are highly correlated with actual proportions in heterozygous blood specimens. For example, the deconvolution method proposed by Gaujoux et al. was previously validated on a microarray dataset of heterogeneous specimens with known proportions of the four cell types (Jurkat, IM-9, Raji, THP-1) [31]. It was found that the mean absolute difference and the pearson correlation coefficient between the actual and estimated proportions were 0.05 and 0.91, respectively. Advantageously, gene expression in mRNA from peripheral whole blood or peripheral blood mononuclear cells (PBMC) is relatively stable under cryopreservation conditions because low temperatures inhibit RNA degradation [32], making deconvolution methods applicable to banked blood specimens for retrospective survival analysis. Deconvolution methods have been applied to the study of some immune diseases, such as systemic lupus erythematosus [30], but not to any cancer research.
In this study, we used the PBMC gene expression profiles of 108 NSCLC patients to demonstrate the feasibility of a deconvolution method for studying the prognostic value of altered peripheral blood composition in cancer patients with banked blood specimens. First, we evaluated the proportions of various immune cells using a deconvolution method and found that the proportion of T cells was an independent prognostic marker for NSCLC patients by a multivariate Cox regression analysis, after adjusting for the other potential prognostic factors that were significant in the univariate analysis. We then obtained a tentative evidence supporting the assumption that the low level of antigen presentation in the tumor microenvironment might be a major cause of the decreased T cell proportion in the peripheral blood by demonstrating a consistent association between the low level of antigen presentation and poor patient outcomes.
Materials and Methods
Microarray data
The dataset of gene expression profiles taken pre-surgery from the PBMC of 108 NSCLC patients with survival information was downloaded from the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/; series accession number GSE13255) [33]. The dataset was generated by the Genome Illumina human-6 v2.0 expression beadchip; the arrays in the dataset were quantile normalized, and the background was subtracted from the expression values, as previously described [34]. The probe sets were annotated using the GPL6102 data file. Probe sets that did not match any gene ID and those that matched multiple gene IDs were deleted. For each sample, the expression intensities of the probe sets that matched the same gene ID were averaged as the expression intensity of that gene ID. The clinical characteristics of patients were summarized in Table 1. As it has been suggested that NSCLC patients are largely insensitive to adjuvant chemotherapy [35], [36], all of the patients were considered together for the following analysis.
Table 1. Clinical characteristics of the NSCLC patients in the PBMC dataset.
| Variable | Patients [N] | [%] |
| All | 108 | 100 |
| Histology | ||
| AD | 67 | 62 |
| SCC | 34 | 31 |
| NSCLC | 7 | 7 |
| Gender | ||
| Male | 53 | 49 |
| Female | 55 | 51 |
| Stage | ||
| I | 66 | 61 |
| II & III | 42 | 39 |
| Age (years) | ||
| <68 | 49 | 45 |
| ≥68 | 59 | 55 |
| Race | ||
| C | 99 | 92 |
| AA | 9 | 8 |
| Smoke | ||
| Never | 6 | 5 |
| Formerly | 87 | 81 |
| Currently | 15 | 14 |
| Chemotherapy | ||
| No | 52 | 48 |
| Yes | 34 | 32 |
| Not sure | 22 | 20 |
| COPD | ||
| Yes | 54 | 50 |
| No | 50 | 46 |
| Not sure | 4 | 4 |
Notes: “AD” and “SCC” represent adenocarcinoma and squamous cell carcinoma, respectively; “C” and “AA” represent Caucasian and African American; “COPD” represents chronic obstructive pulmonary disease.
A tissue-based gene expression dataset of NSCLC (GSE11969) with comprehensive clinical characteristics was selected to study the association between the antigen presentation level in tumor tissues and patient survival. The methods used to generate and normalize the dataset were described previously [37]. Table S1 summarized the clinical characteristics of the patients in the tissue-based dataset.
Evaluating the proportions of immune cells in PBMC
Based on the PBMC gene expression profiles of the 108 NSCLC patients and the marker genes specifically expressed on B lymphocytes, T lymphocytes, natural killer (NK) lymphocytes, dendritic cells (DCs) and monocytes (excluding monocyte-derived DCs) documented in the Immune Response in Silico (IRIS) database [38], we evaluated the proportions of the five immune cells, which together make up 100% of PBMC, using a modified semi-supervised nonnegative matrix factorization method for gene expression deconvolution [31].
The algorithm assumes that the expression intensity of a gene in a sample can be modeled as a linear combination of the expression intensities of that gene in all cell types comprising that sample. Briefly, the expression intensity of the ith gene in the jth sample is the sum of the ith gene expression intensities in all r cell types present in the sample:
| (1) |
where wit is the expression intensity of the ith gene in the tth cell type and htj is the proportion of the tth cell type in the jth sample; εij is a random error.
Given a nonnegative global gene expression matrix X of n genes in p samples, the ssKL algorithm aims to find an approximate matrix decomposition equation:
| (2) |
where W is the n×r matrix representing the gene expression profiles of all r cell types and H is the r×p matrix representing the proportion profiles of all the r cell types in the heterogeneous samples.
Similarly, we estimated the proportions of various immune cells in PBMC based on the marker genes of immune cells characterized by HaemAtlas [39], which classifies T cells into T helper lymphocytes (Th) and cytotoxic T lymphocytes (CTL) and also includes B cells, NK cells, and monocytes (including DCs and other monocytes) using the deconvolution method.
All calculations were performed using the CellMix package in R 2.15.3 software [40].
Survival analysis
Overall survival (OS) was defined as the time from the date of initial surgical resection to the date of death or last contact (censored). For the PBMC dataset, we classified the patients into two groups (Low vs. High) based on the median proportion of each immune cell among all samples. OS was estimated by a univariate analysis using the Kaplan-Meier method, and the OS difference between groups was determined using the log-rank test [41]. The associations between clinical factors and OS were also analyzed using the univariate Kaplan-Meier analysis; the examined factors included histological subtype (adenocarcinoma vs. squamous cell carcinoma), gender (male vs. female), tumor stage (II–III vs. I), age (≥68 years vs. <68), Race (Caucasian vs. African American), smoking status (formerly vs. currently), adjuvant chemotherapy (yes vs. no), and COPD status (present vs. absent). For the prognostic factors that were found to be significant in the univariate analysis, multivariate Cox regression analysis [42] was performed to determine the independent prognostic factors. Significance was defined as a p value<.05.
Similarly, for the tissue-based dataset, we used the univariate Kaplan-Meier analysis and multivariate Cox regression analysis to evaluate the association between OS and the antigen presentation level (Low vs. High) as well as clinical factors, including tumor stage (II–III vs. I), age (≥62 years vs. <62 years), histological subtype (adenocarcinoma vs. squamous cell carcinoma vs. large cell carcinoma) and gender (male vs. female). The antigen presentation level in the tumor microenvironment was characterized by the expression intensities of the major histocompatibility complex (MHC) genes through which DCs present the tumor antigen to T cell receptors [43]. We stratified the patients into two groups (Low vs. High) based on the expression intensities of the MHC genes using the K-means clustering algorithm with Euclidean distance between two samples, which was calculated as follows:
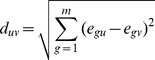 |
(3) |
where m is the number of MHC genes; egu and egv are the expression intensities of the gth gene in the uth and vth samples, respectively.
Results
Association between the proportions of immune cells in PBMC and OS
First, based on the marker genes specifically expressed on B cells, T cells, NK cells, DCs, and monocytes documented in the IRIS database, we adopted the modified semi-supervised nonnegative matrix factorization method to estimate the proportions of the immune cell types in each of the 108 NSCLC patients for which PBMC expression profiles were available (see Materials and Methods). For each type of immune cell, we stratified the patients into two groups, Low (below the median) and High (equal to or greater than the median), according to the median of the cell proportion among the patients and then tested the difference in OS between the groups using the univariate Kaplan-Meier analysis. The OS of the patients in the T cells Low group was significantly worse than that of the patients in the T cells High group (log-rank p = .003; Fig. 1). The proportions of B cells, NK cells, DCs, and monocytes were not observed to be associated with the OS of NSCLC patients (log-rank p>.05; Table 2).
Figure 1. Kaplan-Meier curves for patients stratified by the proportion of T cells.
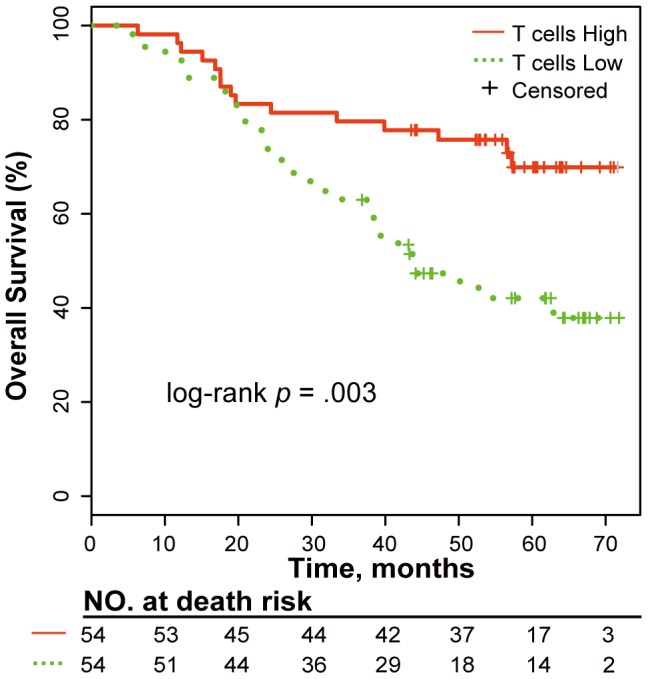
The patients were stratified into two groups based on the median of the T cell proportion among the NSCLC patients: Low (less than the median) and High (greater than or equal to the median). The median overall survival (OS) was assessed using the Kaplan-Meier analysis. p was calculated using the log-rank test.
Table 2. Survival analysis of patients with NSCLC based on the PBMC dataset.
| Univariate | Multivariate | |||
| Variable | log-rank p a | HR | 95% | p b |
| B group (L vs. H) * | .127 | – | – | – |
| T group (L vs. H) * | .003 | 2.23 | 1.01–4.92 | 0.048 |
| NK group (L vs. H) * | .251 | – | – | – |
| DC group (L vs. H) * | .088 | – | – | – |
| Monocyte group (L vs. H) * | .185 | – | – | – |
| Th group (L vs. H) ** | .009 | 1.04 | 0.48–2.28 | 0.92 |
| CTL group (L vs. H) ** | .061 | – | – | – |
| Histology (LCC vs. AD) | .371 | – | – | – |
| Gender (male vs. female) | .738 | – | – | – |
| Stage (II–III vs. I) | .002 | 1.95 | 1.06–3.57 | 0.031 |
| Age (≥68 vs. <68) | .012 | 2.01 | 1.05–3.86 | 0.036 |
| Race (C vs. AA) | .309 | – | – | – |
| Smoking (formerly vs. currently) | .81 | – | – | – |
| Adjuvant chemotherapy (yes vs. no) | .186 | – | – | – |
| COPD (present vs. absent) | .2 | – | – | – |
Note:
The log-rank p value was derived from the Kaplan-Meier method using the log-rank test;
the p value was derived from the Cox regression model; HR, hazard ratio; CI, confidence interval;
* The cell proportions estimated using marker genes in the IRIS, L, group with a lower cell proportion (<median), H, group with a higher cell proportion (≥median).
** The cell proportions estimated using marker genes in the HaemAtlas.
Then, based on the marker genes of the immune cells characterized by HaemAtlas, we estimated the proportions of Th and CTL cells, B cells, NK cells, and monocytes in PBMC and evaluated the associations of these proportions with OS using the univariate Kaplan-Meier analysis. The patients with a low proportion of Th cells (< median) had significant worse OS than those with a high proportion of Th cells (≥ median; log-rank p = .009; Fig. 2A), and patients with low proportion of CTL cells was marginally associated with worse OS (log-rank p = .061; Fig. 2B). The proportions of B cells, NK cells, and monocytes were not found to be significantly or marginally associated with the OS of NSCLC patients (log-rank p>.05; data not shown).
Figure 2. Kaplan-Meier curves for patients stratified by the proportion of T cell subsets.
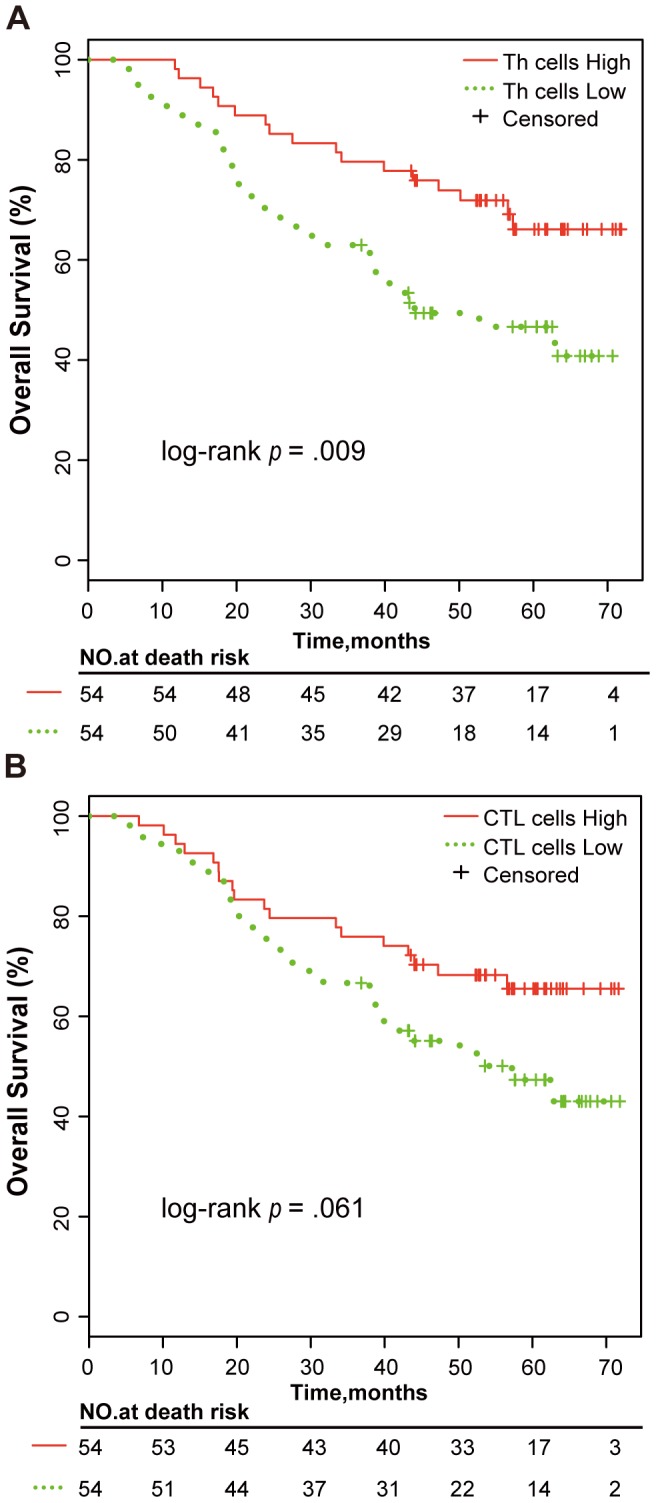
The patients were stratified into two groups according to the median of (A) Th cell proportion and (B) CTL cell proportion in PBMC among the NSCLC patients.
Taken together, these results suggest that the proportions of T cells and its subset Th cells are potential prognostic factors for NSCLC patients.
The proportion of T cells in PBMC is an independent prognostic marker of OS
Multivariate Cox regression analysis was performed of the T cell proportion, Th cell proportion and the clinical factors that were found to be significant in the univariate analysis, including the age and tumor stage of patients. The proportion of T cells remained significantly associated with OS as an independent prognostic factor for NSCLC patients (T cells Low vs. T cells High groups: hazard ratio [HR] = 2.23, 95% CI, 1.07–4.21; p = .048). The age and tumor stage of patients were also independently prognostic for NSCLC patients in the multivariate model (Table 2).
Association of tissue antigen presentation level with OS
One of the major sources leading to reduced proportion of T cells in PBMC might be the low antigen presentation level by DCs in the tumor microenvironment; DCs play a critical role in T cell proliferation in secondary lymphoid organ, from which the proliferated T cells migrate to peripheral blood [44]. Thus, we next determined whether the level of antigen presentation by DCs in tissues could predict patient outcome, consisting with T cell proportion in PBMC.
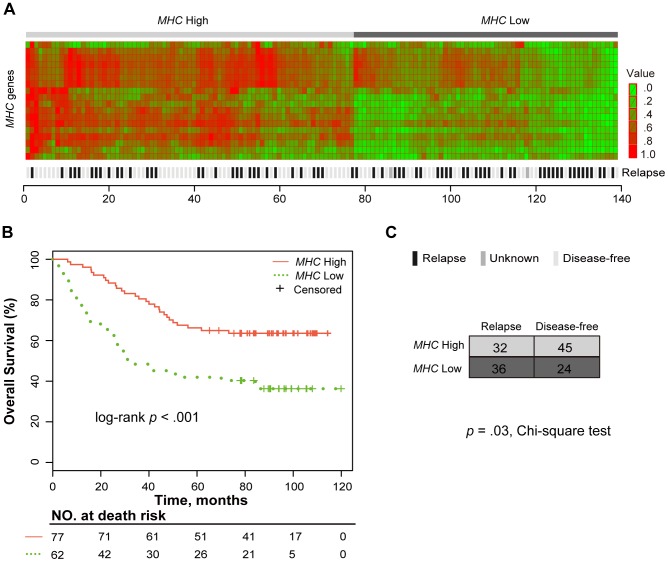
For the tissue-based expression profile dataset of 139 NSCLC patients, we applied the two-means clustering algorithm (see Materials and Methods) to stratify patients into two MHC-related groups based on the expression intensities of the MHC genes, which characterize the level of antigen presentation by DCs in the tumor microenvironment. The patients with a low expression pattern of MHC genes comprised the MHC Low group, while the remaining patients comprised the MHC High group, as shown in Fig. 3A. In the univariate Kaplan-Meier analysis, OS of the patients in the MHC Low group was significantly worse than that of the patients in the MHC High group (log-rank p<.001, Fig. 3B).
Figure 3. The MHC genes signature identifies two groups with different death and relapse risks.
(A) Heat map of the MHC genes in the NSCLC patients. For the tissue expression dataset for NSCLC, the patients were classified into two MHC-related groups (MHC High and MHC Low) based on the expression intensities of 18 detected MHC genes listed in Table S2, using two-means clustering. The expression intensities of the MHC genes characterize the level of antigen presentation by DCs in the tumor microenvironment. (B) Kaplan-Meier curve for patients stratified into two MHC-related groups. The p value was calculated using the log-rank test. (C) The distribution of relapsing patients in the MHC-related groups. The chi-square test was used to compare the correlation between the level of MHC gene expression and relapse. Two patients were excluded because information about their relapse was unknown.
Next, we performed a multivariate Cox regression analysis of the level of antigen presentation, tumor stage and age, which were found to be significant in the univariate analysis. We found that the expression level of MHC genes was an independent prognostic factor for OS after adjusting for the tumor stage and age of patients (MHC Low vs. MHC High: HR = 2.45, 95% CI, 1.51–3.99; p<.001; Table 3). In addition, we found that the relapsing patients were significantly overrepresented in the MHC Low group (p = .03; chi-squared test; Fig. 3C).
Table 3. Survival analysis of NSCLC patients based on the tissue dataset.
| Univariate | Multivariate | |||
| Variable | log-rank p a | HR | 95% CI | p b |
| MHC group (H vs. L) * | 3.0E-04 | 2.45 | 1.51–3.99 | 3.0E-04 |
| Stage (II–III vs. I) | 1.4E-08 | 2.15 | 1.63–2.84 | 5.4E-08 |
| Age (≥63 years vs. <62 years) | .021 | 1.94 | 1.18–3.19 | 8.8E-03 |
| Histology (AD vs. SCC vs. LCC) | .125 | – | – | – |
| Gender (male vs. female) | .159 | – | – | – |
Note:
The log-rank p value was derived from the Kaplan–Meier method using the log-rank test;
the p value was derived from the Cox regression model; HR, hazard ratio; CI, confidence interval;
*L, the group with low MHC gene expression pattern; H, the group with high MHC gene expression pattern.
Discussion
In this study, using the gene expression profiles of NSCLC patients, we demonstrated the feasibility of deconvolving proportions of peripheral immune cells to study their prognostic values for cancer patients using long-term banked blood specimens, which are important materials for retrospective survival analysis. The results suggested that the proportion of T cells in PBMC is a promising prognostic biomarker for NSCLC patients. Our analysis revealed that the low proportion of T cells was significantly associated with poor survival of NSCLC patients. In addition, the deconvolution method could be used to further explore the prognostic values of more refined immune cell subsets in cancer patients based on the same gene expression profiles. For instance, our analysis further demonstrated that the low proportion of one T cell subset, Th cells, was also significantly associated with survival of NSCLC patients. While the proportion of T cells was the only independent prognostic immune marker for overall survival after adjusting for other potential prognostic variables. As both of Th cells and CTL cells play major roles in anti-tumor immunity by specifically identifying tumor cells as “non-self” [45], the degree to which overall proportion of T cells decrease in peripheral blood may effectively reflect the degree of the deterioration of the immune response to tumor cells and thus may correlate closely with patient survival. Compared with the prognostic gene panels identified by gene expression profiles [16], [46], the T cell proportion in peripheral blood may be more acceptable as a prognostic marker for clinical application because of its easy detection and explicable biological principle.
The altered composition of peripheral immune cells might be the result of altered immune responses within the tumor microenvironment. Therefore, we assumed that one of the major sources leading to a reduced T cell proportion in PBMC could be a low level of antigen presentation by DCs in the tumor microenvironment of NSCLC patients, as DCs play a central role in T cell proliferation in secondary lymphoid organ, from which the proliferated T cells migrate to peripheral blood (and then to the tumor site) [44], [47]. Consistent with this assumption, our results revealed that the low expression level of MHC genes through which DCs present the tumor antigen to T cells [43], [48], in tumor tissues was also independently prognostic of poor survival in NSCLC patients. This result also supports a previous report that DCs dysfunction in tumor tissues, which is a critical mechanism for escaping the immune surveillance of tumor cells [48], is associated with poor survival in NSCLC patients [7], [49]. To further verify this assumption, we must simultaneously determine the gene expression profiles in tumor tissue, secondary lymphoid organ, and peripheral blood in the same cohort of NSCLC patients in future studies. This result also suggests that the outcomes of NSCLC patients with the low level of antigen presentation of DCs, could be improved by reinvigorating the immune status of DCs in the tumor microenvironment using immunotherapies such as DCs vaccines [50]. It is known that the tissue-based immune markers are limited and uncertain for clinical application as they could be influenced by the differences in tumor region, such as the center or the invasive margin of the tumor, of sampling from the cancer [51]. Detecting the prognostic markers (such as T cell proportion) in peripheral blood could avoid this problem. Additionally, because the peripheral prognostic markers can be easily detected in post-surgery blood specimens from cancer patients at regular intervals, they could also provide information to help physicians modulate treatment protocols for patients in a timely manner to improve outcomes. Therefore, the T cell proportion in peripheral blood, which might reflect the antigen presentation level in tumor tissues, may become a promising biomarker to predict the outcomes and monitor the progression for NSCLC patients.
Notably, the proportions of immune cells estimated by the deconvolution method are just positively correlated with the actual proportions in complex biological samples [31]. Thus, after using the deconvolution method to estimate immune cell proportions in banked blood specimens of patients and identify prognostic markers, we recommend that prospective studies should utilize flow cytometry to validate the results and determine the optimal cutoff values of the proportions for clinical application. Specifically, flow cytometry in large-scale prospective studies must be used to determine the clinical applicability of the T cell proportion in PBMC as a prognostic marker for NSCLC patients.
Supporting Information
Clinical Characteristics of the NSCLC patients in the tissue dataset. Note: “AD”, “SCC”, and “LCC” represent adenocarcinoma, squamous cell carcinoma, and large cell carcinoma, respectively.
(PDF)
MHC genes list detected in the NSCLC tissue dataset. Note: The MHC genes family, which are also called human leukocyte antigen (HLA), is mainly divided into two subgroups: class I, class II; MHC I genes present antigens to the TCRs of CTL cells and MHC II genes present antigens to the TCRs of Th cells. The genes were clustered using hierarchical clustering by Euclidean distance and the order was consistent with that displayed in the Fig. 3A.
(PDF)
Funding Statement
This work was supported by the National Natural Science Foundation of China (Grant Nos. 91029717, 81071646, 81372213 and 81201822), and Research Fund for the Doctoral Program of Higher Education of China (Grant No. 20112307110011). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. de Visser KE, Eichten A, Coussens LM (2006) Paradoxical roles of the immune system during cancer development. Nat Rev Cancer 6: 24–37. [DOI] [PubMed] [Google Scholar]
- 2. Suzuki K, Kachala SS, Kadota K, Shen R, Mo Q, et al. (2011) Prognostic immune markers in non-small cell lung cancer. Clin Cancer Res 17: 5247–5256. [DOI] [PubMed] [Google Scholar]
- 3. Pautu JL, Kumar L (2003) Intratumoral T cells and survival in epithelial ovarian cancer. Natl Med J India 16: 150–151. [PubMed] [Google Scholar]
- 4. Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, et al. (2005) Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med 353: 2654–2666. [DOI] [PubMed] [Google Scholar]
- 5. Ruffini E, Asioli S, Filosso PL, Lyberis P, Bruna MC, et al. (2009) Clinical significance of tumor-infiltrating lymphocytes in lung neoplasms. Ann Thorac Surg 87: 365–371 discussion 371–362. [DOI] [PubMed] [Google Scholar]
- 6. Johnson SK, Kerr KM, Chapman AD, Kennedy MM, King G, et al. (2000) Immune cell infiltrates and prognosis in primary carcinoma of the lung. Lung Cancer 27: 27–35. [DOI] [PubMed] [Google Scholar]
- 7. Dieu-Nosjean MC, Antoine M, Danel C, Heudes D, Wislez M, et al. (2008) Long-term survival for patients with non-small-cell lung cancer with intratumoral lymphoid structures. J Clin Oncol 26: 4410–4417. [DOI] [PubMed] [Google Scholar]
- 8. Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, et al. (2011) Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res 71: 1263–1271. [DOI] [PubMed] [Google Scholar]
- 9. Sandel MH, Dadabayev AR, Menon AG, Morreau H, Melief CJ, et al. (2005) Prognostic value of tumor-infiltrating dendritic cells in colorectal cancer: role of maturation status and intratumoral localization. Clin Cancer Res 11: 2576–2582. [DOI] [PubMed] [Google Scholar]
- 10. Leffers N, Gooden MJ, de Jong RA, Hoogeboom BN, ten Hoor KA, et al. (2009) Prognostic significance of tumor-infiltrating T-lymphocytes in primary and metastatic lesions of advanced stage ovarian cancer. Cancer Immunol Immunother 58: 449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eisenthal A, Polyvkin N, Bramante-Schreiber L, Misonznik F, Hassner A, et al. (2001) Expression of dendritic cells in ovarian tumors correlates with clinical outcome in patients with ovarian cancer. Hum Pathol 32: 803–807. [DOI] [PubMed] [Google Scholar]
- 12. Croci DO, Zacarias Fluck MF, Rico MJ, Matar P, Rabinovich GA, et al. (2007) Dynamic cross-talk between tumor and immune cells in orchestrating the immunosuppressive network at the tumor microenvironment. Cancer Immunol Immunother 56: 1687–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Showe MK, Kossenkov AV, Showe LC (2012) The peripheral immune response and lung cancer prognosis. Oncoimmunology 1: 1414–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mohr S, Liew CC (2007) The peripheral-blood transcriptome: new insights into disease and risk assessment. Trends Mol Med 13: 422–432. [DOI] [PubMed] [Google Scholar]
- 15. Satomi A, Murakami S, Ishida K, Mastuki M, Hashimoto T, et al. (1995) Significance of increased neutrophils in patients with advanced colorectal cancer. Acta Oncol 34: 69–73. [DOI] [PubMed] [Google Scholar]
- 16. Burczynski ME, Twine NC, Dukart G, Marshall B, Hidalgo M, et al. (2005) Transcriptional profiles in peripheral blood mononuclear cells prognostic of clinical outcomes in patients with advanced renal cell carcinoma. Clin Cancer Res 11: 1181–1189. [PubMed] [Google Scholar]
- 17. Sarraf KM, Belcher E, Raevsky E, Nicholson AG, Goldstraw P, et al. (2009) Neutrophil/lymphocyte ratio and its association with survival after complete resection in non-small cell lung cancer. J Thorac Cardiovasc Surg 137: 425–428. [DOI] [PubMed] [Google Scholar]
- 18. Li YX, Chen YJ, Chang LJ, Hendryx M, Luo JH (2012) Clinical Biomarkers and Prognosis in Taiwanese Patients with Non-Small Cell Lung Cancer (NSCLC). Journal of Cancer Therapy 03: 412–423. [Google Scholar]
- 19. Walsh SR, Cook EJ, Goulder F, Justin TA, Keeling NJ (2005) Neutrophil-lymphocyte ratio as a prognostic factor in colorectal cancer. J Surg Oncol 91: 181–184. [DOI] [PubMed] [Google Scholar]
- 20. Hirashima M, Higuchi S, Sakamoto K, Nishiyama T, Okada H (1998) The ratio of neutrophils to lymphocytes and the phenotypes of neutrophils in patients with early gastric cancer. J Cancer Res Clin Oncol 124: 329–334. [DOI] [PubMed] [Google Scholar]
- 21. Devaux BC, O'Fallon JR, Kelly PJ (1993) Resection, biopsy, and survival in malignant glial neoplasms. A retrospective study of clinical parameters, therapy, and outcome. J Neurosurg 78: 767–775. [DOI] [PubMed] [Google Scholar]
- 22. Wronski M, Arbit E, Burt M, Galicich JH (1995) Survival after surgical treatment of brain metastases from lung cancer: a follow-up study of 231 patients treated between 1976 and 1991. J Neurosurg 83: 605–616. [DOI] [PubMed] [Google Scholar]
- 23. Cedres S, Torrejon D, Martinez A, Martinez P, Navarro A, et al. (2012) Neutrophil to lymphocyte ratio (NLR) as an indicator of poor prognosis in stage IV non-small cell lung cancer. Clin Transl Oncol 14: 864–869. [DOI] [PubMed] [Google Scholar]
- 24. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719–748. [PubMed] [Google Scholar]
- 25. Solsona E, Iborra I, Rubio J, Casanova JL, Ricos JV, et al. (2001) Prospective validation of the association of local tumor stage and grade as a predictive factor for occult lymph node micrometastasis in patients with penile carcinoma and clinically negative inguinal lymph nodes. J Urol 165: 1506–1509. [PubMed] [Google Scholar]
- 26. van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, et al. (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530–536. [DOI] [PubMed] [Google Scholar]
- 27. Wei SH, Balch C, Paik HH, Kim YS, Baldwin RL, et al. (2006) Prognostic DNA methylation biomarkers in ovarian cancer. Clin Cancer Res 12: 2788–2794. [DOI] [PubMed] [Google Scholar]
- 28. Jaye DL, Bray RA, Gebel HM, Harris WA, Waller EK (2012) Translational applications of flow cytometry in clinical practice. J Immunol 188: 4715–4719. [DOI] [PubMed] [Google Scholar]
- 29. Tollerud DJ, Brown LM, Clark JW, Neuland CY, Mann DL, et al. (1991) Cryopreservation and long-term liquid nitrogen storage of peripheral blood mononuclear cells for flow cytometry analysis: effects on cell subset proportions and fluorescence intensity. J Clin Lab Anal 5: 255–261. [DOI] [PubMed] [Google Scholar]
- 30. Abbas AR, Wolslegel K, Seshasayee D, Modrusan Z, Clark HF (2009) Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS One 4: e6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gaujoux R, Seoighe C (2012) Semi-supervised Nonnegative Matrix Factorization for gene expression deconvolution: a case study. Infect Genet Evol 12: 913–921. [DOI] [PubMed] [Google Scholar]
- 32. Fan H, Hegde PS (2005) The transcriptome in blood: challenges and solutions for robust expression profiling. Curr Mol Med 5: 3–10. [DOI] [PubMed] [Google Scholar]
- 33. Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, et al. (2007) NCBI GEO: mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res 35: D760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Showe MK, Vachani A, Kossenkov AV, Yousef M, Nichols C, et al. (2009) Gene expression profiles in peripheral blood mononuclear cells can distinguish patients with non-small cell lung cancer from patients with nonmalignant lung disease. Cancer Res 69: 9202–9210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, et al. (2003) Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med 349: 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bonadonna G, Valagussa P (1981) Dose-response effect of adjuvant chemotherapy in breast cancer. N Engl J Med 304: 10–15. [DOI] [PubMed] [Google Scholar]
- 37. Takeuchi T, Tomida S, Yatabe Y, Kosaka T, Osada H, et al. (2006) Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J Clin Oncol 24: 1679–1688. [DOI] [PubMed] [Google Scholar]
- 38. Abbas AR, Baldwin D, Ma Y, Ouyang W, Gurney A, et al. (2005) Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun 6: 319–331. [DOI] [PubMed] [Google Scholar]
- 39. Watkins NA, Gusnanto A, de Bono B, De S, Miranda-Saavedra D, et al. (2009) A HaemAtlas: characterizing gene expression in differentiated human blood cells. Blood 113: e1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gaujoux R, Seoighe C (2013) CellMix: a comprehensive toolbox for gene expression deconvolution. Bioinformatics 29: 2211–2212. [DOI] [PubMed] [Google Scholar]
- 41. Harrington DP, Fleming TR (1982) A class of rank test procedures for censored survival data. Biometrika 69: 553–566. [Google Scholar]
- 42.Fox J (2002) Cox Proportional-Hazards Regression for Survival Data. Thousand Oaks,CA: Sage Publications Inc.
- 43. Banchereau J, Steinman RM (1998) Dendritic cells and the control of immunity. Nature 392: 245–252. [DOI] [PubMed] [Google Scholar]
- 44. Guermonprez P, Valladeau J, Zitvogel L, Thery C, Amigorena S (2002) Antigen presentation and T cell stimulation by dendritic cells. Annu Rev Immunol 20: 621–667. [DOI] [PubMed] [Google Scholar]
- 45. Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ (2011) Natural innate and adaptive immunity to cancer. Annu Rev Immunol 29: 235–271. [DOI] [PubMed] [Google Scholar]
- 46. Kossenkov AV, Dawany N, Evans TL, Kucharczuk JC, Albelda SM, et al. (2012) Peripheral immune cell gene expression predicts survival of patients with non-small cell lung cancer. PLoS One 7: e34392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen DS, Mellman I (2013) Oncology meets immunology: the cancer-immunity cycle. Immunity 39: 1–10. [DOI] [PubMed] [Google Scholar]
- 48. Pinzon-Charry A, Maxwell T, Lopez JA (2005) Dendritic cell dysfunction in cancer: a mechanism for immunosuppression. Immunol Cell Biol 83: 451–461. [DOI] [PubMed] [Google Scholar]
- 49. Kikuchi E, Yamazaki K, Torigoe T, Cho Y, Miyamoto M, et al. (2007) HLA class I antigen expression is associated with a favorable prognosis in early stage non-small cell lung cancer. Cancer Sci 98: 1424–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hirschowitz EA, Foody T, Kryscio R, Dickson L, Sturgill J, et al. (2004) Autologous dendritic cell vaccines for non-small-cell lung cancer. J Clin Oncol 22: 2808–2815. [DOI] [PubMed] [Google Scholar]
- 51. Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, et al. (2006) Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313: 1960–1964. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical Characteristics of the NSCLC patients in the tissue dataset. Note: “AD”, “SCC”, and “LCC” represent adenocarcinoma, squamous cell carcinoma, and large cell carcinoma, respectively.
(PDF)
MHC genes list detected in the NSCLC tissue dataset. Note: The MHC genes family, which are also called human leukocyte antigen (HLA), is mainly divided into two subgroups: class I, class II; MHC I genes present antigens to the TCRs of CTL cells and MHC II genes present antigens to the TCRs of Th cells. The genes were clustered using hierarchical clustering by Euclidean distance and the order was consistent with that displayed in the Fig. 3A.
(PDF)