Abstract
Cloning of mammals by somatic cell nuclear transfer (SCNT) is still plagued by low efficiency. The epigenetic modifications established during cellular differentiation are a major factor determining this low efficiency as they act as epigenetic barriers restricting reprogramming of somatic nuclei. In this regard, most factors that promote chromatin decondensation, including histone deacetylase inhibitors (HDACis), have been found to increase nuclear reprogramming efficiency, making their use common to improve SCNT rates. Herein we used valproic acid (VPA) in SCNT to test whether the treatment of nuclear donor cells with this HDACi improves pre- and post-implantation development of cloned cattle. We found that the treatment of fibroblasts with VPA increased histone acetylation without affecting DNA methylation. Moreover, the treatment with VPA resulted in increased expression of IGF2R and PPARGC1A, but not of POU5F1. However, when treated cells were used as nuclear donors no difference of histone acetylation was found after oocyte reconstruction compared to the use of untreated cells. Moreover, shortly after artificial activation the histone acetylation levels were decreased in the embryos produced with VPA-treated cells. With respect to developmental rates, the use of treated cells as donors resulted in no difference during pre- and post-implantation development. In total, five clones developed to term; three produced with untreated cells and two with VPA-treated cells. Among the calves from treated group, one stillborn calf was delivered at day 270 of gestation whereas the other one was delivered at term but died shortly after birth. Among the calves from the control group, one died seven days after birth whereas the other two are still alive and healthy. Altogether, these results show that in spite of the alterations in fibroblasts resulting from the treatment with VPA, their use as donor cells in SCNT did not improve pre- and post-implantation development of cloned cattle.
Introduction
In 1997, cloning of mammals by somatic cell nuclear transfer (SCNT) was shown feasible, becoming a promising technology because of its applications in medicine (e.g. derivation of patient-specific embryonic stem cells) and transgenesis [1]. However, SCNT is still plagued by low efficiency, as a multitude of successful steps are needed for a cloned animal to be born healthy [2]. These include oocyte-mediated reprogramming, pregnancy establishment, development of a functional placenta with adequate maternal–fetal interaction, finally successful delivery and adaptation to the extra uterine life [3]. Problems such as persistence of somatic cell methylation [4] and acetylation patterns in SCNT embryos [5], aberrant expression of imprinted genes [6], failure to produce a functional placenta and consequently poor fetal nutrition are frequently observed in animal clones [7]. These observations suggest that the nuclear reprogramming process is incomplete in most SCNT embryos representing an important target for strategies aiming to improve SCNT efficiency [8].
There are several epigenetic modifications that accompany cell differentiation, including DNA methylation, histone modifications (e.g. H3K9me2/3 methylation; deacetylation), incorporation of histone variants (e.g. macroH2A) and chromatin compaction. All these modifications act as epigenetic barriers restricting reprogramming of somatic nuclei [9]. Since histone deacetylation, followed by DNA compaction, is commonly associated with gene repression in differentiated cells, decondensation of chromatin is required to enable access of transcriptional regulators to genomic targets essential for successful reprogramming of somatic nuclei [10], [11]. Corroborating this hypothesis, most factors that promote chromatin decondensation, including histone deacetylase inhibitors (HDACis), have been found to increase nuclear reprogramming efficiency, making their use common to improve SCNT rates [12], [13].
The use of HDACis, including Valproic Acid (VPA) [14], Sodium Butyrate (NaBu) [15], Trichostatin A (TSA) [16] and Scriptaid [17] is associated with increased rates of successful nuclear reprogramming. An increase of up to five fold in the success rate of mouse cloning was reported when HDACi was used in SCNT [13]. For example, treatment of donor cells with TSA, markedly improved in vitro development of SCNT embryos in rabbits [18] and cattle [19]. In addition, the use of NaBu in cattle improved SCNT [15] whereas the use of VPA enhanced gene reactivation and reprogramming efficiency in pig SCNT [20] and mouse induced pluripotent stem cells (iPSCs) experiments [21]. VPA, a short-chain fatty acid, is widely used in humans as an anticonvulsant and mood stabilizer [22]. Recently, VPA was reported to be a powerful HDACi with low toxicity to the cells, both in vitro and in vivo [23]. VPA relieves HDAC-dependent transcriptional repression and causes hyperacetylation of histones in cultured cells and in vivo [24], [25]. Since histone hyperacetylation promotes neutralization of positive charges of lysine residues, this reduces their DNA binding and may contribute directly to chromatin opening and indirectly to transcriptional upregulation [26]. For this reason, VPA has been used in cattle to investigate its effect on nuclear reprogramming. Treatment of cloned embryos with VPA greatly improved blastocyst formation rate, the number of cells in the inner cell mass (ICM) and trophectoderm (TE), and cell survival in cattle [27]. Recently, Selokar et al. demonstrated in cattle that the use of donor cells treated with VPA was beneficial for efficient reprogramming using handmade cloning procedures [28]. Nonetheless, the use VPA has been limited to experiments involving pre-implantation development as end-points, with no study having evaluated its effect on development of cattle clones to term.
Herein VPA was used in SCNT to test whether the treatment of nuclear donor cells with this HDACi affects pre- and post-implantation development of cattle clones. We hypothesized that the treatment of donor cells with VPA might relax the chromatin structure turning them more amenable cells for reprogramming. As a result, based on previous studies and the low- toxicity of VPA, an increase of the global levels of histone acetylation favoring nuclear reprogramming towards higher developmental rates was expected.
Materials and Methods
All chemicals and reagents used were purchased from Sigma-Aldrich Chemical Company (St. Louis, MO, USA) unless otherwise stated. In vitro experimental procedures were carried out in humidified incubators maintained at 38.5°C in air with 5% CO2. Each experiment consisted of at least three replicates. The experiments were conducted in accordance with the International Guiding Principles for Biomedical Research Involving Animals (Society for the Study of Reproduction).
Ethics Statement
The present study was approved by the “Ethic Committee in the use of animals” of the School of Veterinary Medicine and Animal Science of University of São Paulo, protocol number 2546/2012, which complies with the ethical principles in animal research. We adopted the International Guiding Principles for Biomedical Research Involving Animals (Society for the Study of Reproduction) as well.
Treatment of nuclear donor cells with VPA
Nuclear donor cells were derived from a 14-year old Gir (Bos indicus) bull as described previously [29]. Fibroblasts at third passage were plated at a density of 7×104 cells per 35-mm Petri dish in Alpha Minimum Essential Medium (α-MEM; GIBCO BRL, Grand Island, NY, USA) supplemented with 10% (v/v) fetal calf serum (FCS) and 50 µg/ml gentamicin sulfate. After 48 h post plating, the medium was changed to α-MEM supplemented with 0.5% FCS and 50 µg/ml gentamicin sulfate, and the cells were cultured for another three days to arrest the cell cycle at the G1/G0 stages. The treatment with VPA was performed by adding 0, 1, 2 or 5 mM of VPA to the culture medium 24 h before the end of the culture. Based on the results from an MTT analysis (see below), only 0 mM (control) and 2 mM (treated) of VPA were used in the following experiments.
MTT cell proliferation assay
To evaluate fibroblast viability and proliferation after VPA treatment, we used the 3-(4,5-dimethylthiazol-2-yl)-2, 4-diphenyl-tetrazolium bromide assay [30] (MTT) as previously described [29], with a few modifications. Briefly, fibroblasts were seeded in 96-well plates at 7×103 cells per well and treated with 0, 1, 2 and 5 mM of VPA as described above. Thereafter, 100 µl of the MTT solution (0.5 mg/ml final concentration) were added into each well of the 96-well plates and cultured in a humidified incubator with 5% CO2 at 38.5°C. After 3 h, the MTT solution was removed and 200 µl of acidic isopropanol (0.04 M HCl in absolute isopropanol) were added into each well to dissolve the formed formazan crystals. Each plate was read using a spectrophotometer Thermo Scientific Multiskan FC (Thermo Scientific, Wilmington, DE, USA) at 570 nm wavelength. Absorbance values are expressed as percentages in relation to a control group (without VPA).
Somatic cell nuclear transfer
Somatic cell nuclear transfer was performed as described by Sangalli et al. [29]. Briefly, cumulus-oocyte complexes (COCs) were aspirated from ovaries obtained from local abattoirs and selected based on morphological characteristics. COCs were then subjected to in vitro maturation (IVM) and the oocytes denuded of cumulus cells by gentle pipetting. The oocytes with visible first polar body (PB) were enucleated by removing the 1st PB and metaphase-II (MII) plate by gentle aspiration using a 15-µm (internal diameter) glass pipette (ES transferTip; Eppendorf, Hamburg, Germany). Enucleated oocytes were reconstructed by injection of a single fibroblast (previously untreated or treated with VPA) into the perivitelline space. The resulting couplet was fused in electrofusion solution (0.28 M mannitol, 0.1 mM MgSO4, 0.5 mM HEPES and 0.05% BSA in ultra-pure water) by applying one pulse of alternating current (0.05 kV/cm for 5 s) and two pulses of continuous current (1.75 kV/cm for 45 µs). Successfully fused couplets were fixed immediately before activation (see below) or artificially activated 26 h post-IVM. Artificial activation was performed by treatment with 5 µM ionomycin for 5 min, washing in 3% BSA solution and treatment with 2 mM of 6-dimethylaminopurine (6-DMAP) for 3 h [29]. Presumptive zygotes were cultured for an additional period of 2 h post-activation (h.p.a.) and fixed for immunocytochemistry analysis (5 h.p.a.) or cultured for 7 days in SOF supplemented with 2.5% v/v FCS, 0.5% bovine serum albumin (BSA), 0.2 mM sodium pyruvate and 50 µg/ml gentamicin sulfate. Cleavage and blastocyst rates were evaluated at 48 and 168 h.p.a., respectively. In all experiments, parallel parthenogenetic embryos were generated and used as controls.
Embryo transfer and pregnancy evaluation
Blastocysts at day 7 of development were individually transferred non-surgically into the uterus of previously synchronized recipient cows [31]. Recipients were evaluated for pregnancy by transrectal palpation/ultrasonography at 30, 60, 90 and 270 days after embryo transfer. Abortion rate was evaluated on a monthly basis.
Immunodetection of H3K9ac and 5-methylcytosine
Immunocytochemistry for histone 3 lysine 9 acetylation (H3K9ac) was performed using fused couplets (immediately before the activation) or presumptive zygotes (5 h.p.a.). Nuclear donor cells were also evaluated by immunocytochemistry for H3K9ac and global levels of DNA methylation (5-mC) as previously reported [32]–[34], with a few modifications. Briefly, samples were fixed for 12 min in 4% (w/v) paraformaldehyde diluted in phosphate buffer solution (PBS) with 0.1% (w/v) polyvinylpyrrolidone (PBS-PVP) and permeabilized for 20 min in PBS-PVP with 1% (v/v) Triton X-100, washed in PBS-PVP containing 0.1% (v/v) Tween 20 (TW-PBS) and incubated for 15 min in 4 N HCl. Afterwards, samples were extensively washed in TW-PBS and non-specific binding sites were blocked by incubation overnight at 4°C in a blocking buffer consisting of PBS-PVP containing 0.4% (w/v) BSA. Samples were incubated for 3 h at 37°C with primary antibodies diluted at 1∶1000. DNA methylation staining (fibroblasts) was performed using the anti-mouse 5-mC (Santa Cruz Biotechnology, Santa Cruz, CA, USA) whereas the anti-rabbit H3K9ac (Millipore, Bedford, MA, USA) was used for histone acetylation staining (fibroblasts, fused couplets and presumptive zygotes). Next, samples were washed in TW-PBS and incubated for 1 h at room temperature with secondary antibodies diluted at 1∶500. The goat anti-rabbit conjugated with fluorescein isothiocyanate-FITC (Vector Laboratories, Burlingame, CA, USA) was used for histone acetylation staining and the rabbit anti-mouse conjugated with rhodamine (Santa Cruz Biotechnology) for DNA methylation staining. After several washes in TW-PBS, fibroblasts, fused couplets and presumptive zygotes were mounted on glass slides with coverslips using Prolong Antifade reagent (Invitrogen). An epifluorescent microscope (Axioplan; Carl Zeiss, Zeppelingstrasse, Germany) was used for evaluation of both DNA methylation and histone acetylation. As negative control, PBS was used in place of primary antibody.
Images were captured using a digital camera (Zeiss MC 80 DX; Carl Zeiss) and the same settings for all samples subject to the same staining (e.g. methylation or acetylation) procedure. The acetylation status was analyzed using at least 15 images of individual couplets for each experimental group. With regards to donor cells, 5 images of a monolayer of fibroblasts were evaluated for each experimental group to evaluate both acetylation and methylation status. Individual nuclei were outlined (excluding overlapping and folded nuclei) using the Adobe Photoshop v. 7 (Adobe Systems, San Jose, CA, USA). About 30 nuclei were outlined in each fibroblast image. The background was subtracted from pixel intensity in all analysis and the averaged signal intensity was calculated using the ImageJ software (ImageJ, National Institute of Health, Bethesda, MD, USA). Here, intensity refers to the degree of brightness of colored pixels (in a gray scale: 0–255; 0 = black and 255 = white) where brighter intensity indicates greater immunocytochemistry reactivity.
Gene expression analysis
RNA was extracted from control and treated fibroblasts using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's recommendations, with a few modifications. In brief, a mix containing 100 µl of TRIzol reagent, 5 µg of linear acrylamide (Ambion Inc., Austin, TX, USA) and 5 µl of diethylene pyrocarbonate-treated H2O was added to each sample. The extracted RNA was directly dissolved in 10 µl of DNase I solution (Invitrogen) plus 1 unit/µl RNase OUT for DNA degradation, as suggested by the manufacturer. Then, the RNA was immediately reverse transcribed into cDNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to manufacturer's protocol and stored at −20°C until use.
The target genes of interest were POU5F1, IGF2R and PPARGC1A whereas ACTB was used as a reference gene. Primers used for real-time reverse transcription PCR (RT-PCR) were designed using the Primer Express software v.3.1 (Applied Biosystems) based upon sequences available in GenBank (Table 1). Relative quantification of gene-specific mRNA transcripts was performed in 20-µl reactions containing 0.2 µM of primers (POU5F1, IGF2R, PPARGC1A and ACTB) plus 1× Power SYBR Green PCR Master Mix (Applied Biosystems) and 2 µl of a 8-fold diluted cDNA. All gene-specific cDNAs amplified for a particular sample were run in triplicate in the same PCR plate. A non-template control was always run in parallel with samples using 2 µl of water instead of cDNA. The following cycling conditions were applied for amplification: initial denaturation at 95°C for 10 min followed by 40 cycles consisting of 95°C for 15 sec and 60°C for 1 min. The SYBR Green fluorescence was read at the end of each extension step (60°C). Pilot experiments using five different concentrations of cDNA (spanning a 60-fold range) were run to set up real time RT-PCR conditions. The specificity of PCR products was confirmed by analysis of melting curves. Target transcript amounts were determined using the following formula: E(target) −ΔCt(target)/E(ref) −ΔCt(ref), in which E corresponds to the amplification efficiency and ΔCt to the difference of cycle threshold (Ct) between control and treated samples. Values of Ct were averaged from sample duplicates whereas E referred to the mean efficiency estimated for each primer set (which varied from 93.6% to 95.2%) using LinRegPCR program [35]–[37]. Gene expression data from seven biological replicates are presented.
Table 1. Primer sequences used in real time RT-PCR.
| Gene Symbol | Gene Name | Accession number | Primer Sequences (5′-3′) | Product (bp) | Annealing Temp. (°C) |
| POU5F1 | POU class 5 homeobox 1 | NM_174580.2 | F: CAGGCCCGAAAGAGAAAGC | 78 | 60 |
| R: CGGGCACTGCAGGAACA | |||||
| IGF2R | Insulin-like growth factor 2 receptor | NM_174352.2 | F: CAGGTCTTGCAACTGGTGTATGA | 83 | 60 |
| R: ACGAAGCTGATGACGCTCTTG | |||||
| PPARGC1A | Peroxisome proliferator-activated receptor gamma, coactivator 1 alpha | NM_177945.3 | F: ACCATGCAAACCATAATCACAGGAT | 82 | 60 |
| R: CTCTTCGCTTTATTGCTCCATGAAT | |||||
| ACTB | Actin beta | NM_173979.3 | F: CAGCAGATGTGGATCAGCAAGC | 91 | 60 |
| R: AACGCAGCTAACAGTCCGCC |
Statistical analysis
All experiments were repeated at least three times. Statistical analysis was performed using the SAS System v. 9.3 (SAS/STAT, SAS Institute Inc., Cary, NC). Developmental rates were analyzed using chi-square test. Remaining data were tested for normality of residuals and homogeneity of variances, and analysed as follows. MTT and immunocytochemistry data were analysed by regression analysis and t-Student's test, respectively. Gene expression data were analyzed by one-way ANOVA followed by Tukey post-hoc test. Gene expression data are presented in natural log (Ln) scale because of the log normal distribution considered for analysis. Differences with probabilities (P)<0.05 were considered significant. In the text, values are presented as means ± the standard error of the mean (SEM).
Results
Valproic Acid increases proliferation/viability of donor cells
In order to ascertain whether VPA-treated cells were suitable as nuclear donors and monitor the changes caused by VPA treatment, bovine fibroblasts were evaluated by the MTT assay after the treatment with 0, 1, 2 and 5 mM VPA for 24 h. The rate of proliferation/viability of these cells fitted (P = 0.0001) a second grade polynomial regression (Figure 1). The cells treated with 1, 2 and 5 mM VPA increased proliferation/viability by 134±5,32%, 135±5,53% and 127±4,97%, respectively, compared to untreated cells (100±5,60%). Thus it was concluded that since the treatment did not impair cell viability the cells treated with VPA were suitable as nuclear donors.
Figure 1. Valproic Acid increases proliferation/viability of donor cells.
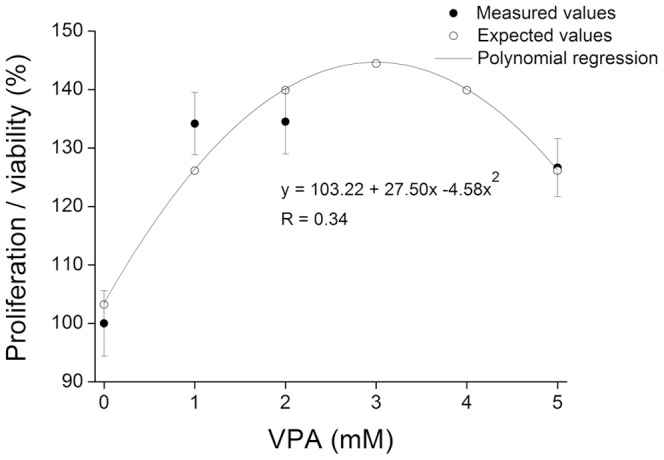
Proliferation/viability rates of donor cells treated with 0, 1, 2 and 5 mM VPA measured by the MTT assay. Values fitted a second grade polynomial regression (P = 0.0001).
VPA treatment increases H3K9 acetylation without affecting DNA methylation of donor cells
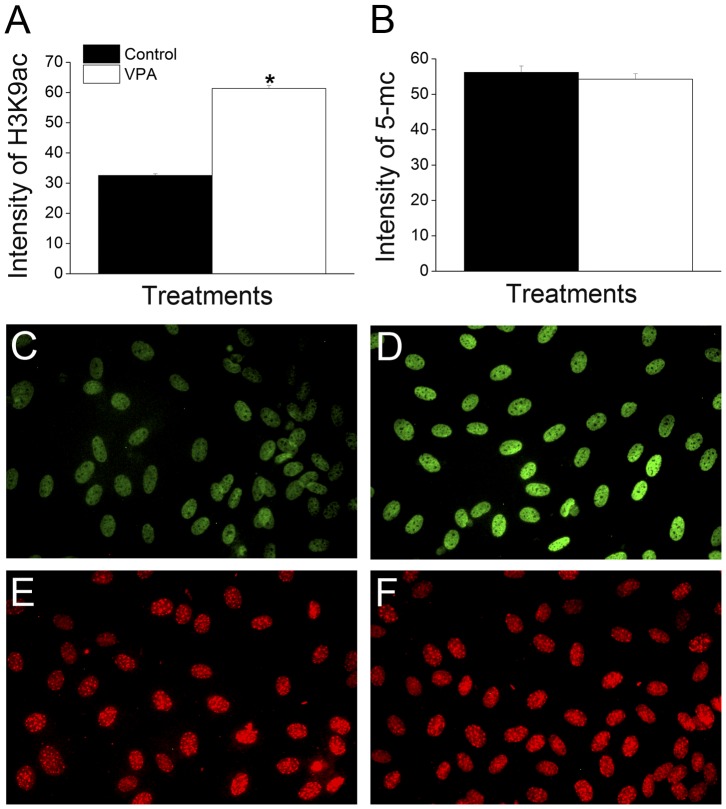
In order to confirm the effect of VPA treatment on histone acetylation, global histone acetylation levels of fibroblasts treated with 2 mM VPA were evaluated by immunocytochemistry (Figure 2). Compared to control cells, the treatment with VPA increased H3K9ac by about 1.9 fold (P<0.0001). Moreover, since VPA has been shown to decrease DNA methylation in cultured cells [38], we evaluated the global DNA methylation levels, but no differences were found between control and treated cells (Figure 2).
Figure 2. VPA treatment increases H3K9 acetylation without affecting DNA methylation of donor cells.
(A) Relative average intensity of H3K9 acetylation (H3K9ac) of control and VPA-treated cells. (B) Relative average intensity of 5-methylcytosine (5-mC) of control and VPA-treated cells. (C–D) Immunofluorescence labeling for H3K9ac of control (C) and VPA-treated cells (D). (E–F) Immunofluorescence labeling for 5-mC of control (E) and VPA-treated cells (F). The (*) denotes a significant difference between experimental groups (P<0.0001). All images were taken in the same magnification (200×).
Expression of IGF2R and PPARGC1A is increased in donor cells treated with VPA
In order to further understand the effect of VPA on the cells, we evaluated expression of POU5F1, IGF2R and PPARGC1A by real-time RT-PCR (Figure 3). The genes evaluated are known for their role in regulating fetal growth (IGF2R) [39], promoting pluripotency (POU5F1) [40] and metabolism regulation (PPARGC1A) [41]. With respect to IGF2R, it was found that the treatment with VPA increased (P<0.0001) transcript abundance by about two fold compared to control cells (Figure 3a). No difference in expression of POU5F1 was found (Figure 3b). Finally, expression of PPARGC1A was increased (P<0.0001) over five fold in the cells treated with VPA (Figure 3c). In summary, treatment of cells with VPA increased expression of IGF2R and PPARGC1A, but not of POU5F1.
Figure 3. Expression of IGF2R and PPARGC1A is increased in donor cells treated with VPA.
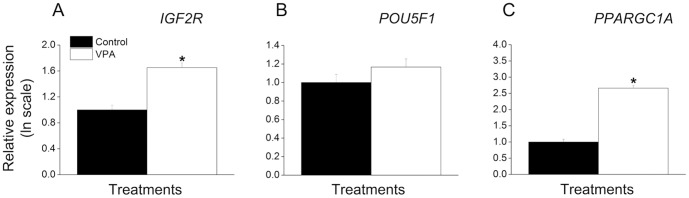
The amounts of IGF2R (A), POU5F1 (B) and PPARGC1A (C) transcripts are expressed in relation to the control group. The asterisk (*) denotes difference between control and VPA-treated groups (P<0.0001).
The higher levels of histone acetylation in donor cells are not maintained after nuclear transfer
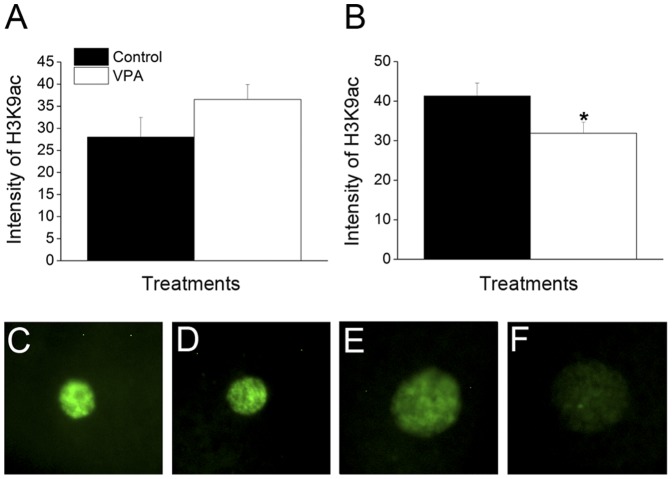
Fibroblasts with increased levels of histone acetylation after VPA treatment were used as SCNT donors cells. To confirm whether the higher acetylation levels were maintained after nuclear transfer, global histone acetylation levels were measured in fused couplets before and after artificial activation concerning their global histone acetylation levels (Figure 4). Although H3K9ac was marginally superior in the VPA group (28.0±4.44 versus 36.5±3.37, respectively for control and VPA), no significant difference was found between groups before activation (P = 0.12). Unexpectedly, when H3K9ac was evaluated after artificial activation (5 h.p.a.), a significant decrease of about 1.3 fold was verified in the VPA group (P = 0.03). Taken together, it appears that the higher levels of histone acetylation of donor cells treated with VPA are not maintained after nuclear transfer.
Figure 4. The higher levels of histone acetylation in donor cells are not maintained after nuclear transfer.
(A) Relative average intensity of H3K9ac of fused couplets from control and VPA-treated groups. (B) Relative average intensity of H3K9ac of presumptive zygotes 5 h.p.a from control and VPA-treated groups. (C–D) Immunofluorescence labelling for H3K9ac of fused couplets from (C) control and (D) VPA-treated groups. (E–F) Immunofluorescence labelling for H3K9ac of presumptive zygotes 5 h.p.a. from (E) control and (F) VPA-treated groups. The asterisk (*) denotes difference between control and VPA-treated groups (P = 0.03). All images were taken in the same magnification (200×).
Higher levels of histone acetylation in donor cells has no effect on pre- and post-implantation development of clones
Since the cells treated with VPA showed higher levels of histone acetylation, they were used as donors in SCNT to evaluate the effect of donor cell acetylation levels on developmental rates (Table 2). A total of 254 cloned embryos were produced in four experimental replicates, 180 controls and 168 treated, but no difference in overall rates of fusion, cleavage or blastocyst rates were found between groups. Although this finding indicates that the treatment of donor cells with VPA does not affect the development of SCNT embryos, blastocysts produced from both groups were transferred to recipients to evaluate whether the treatment affected post-implantation development (Table 3). Sixty-four blastocysts, 34 controls and 30 treated, were transferred individually into synchronized recipients. Although fewer full term gestations were obtained from VPA-treated donor cells, no significant difference was found between control and treated groups at any stage during pregnancy, further indicating that the treatment of donor cells with VPA does not affect developmental rates of cattle clones.
Table 2. Effect of treating nuclear-donor cells with Valproic Acid on pre-implantation developmental rates of cloned embryos.
| Groups | Oocytes N | Fused embryos N (%) | Cleavage N (%) | Blastocysts N (%) |
| Control | 180 | 139 (77.22) | 112 (80.57) | 44 (31.65) |
| VPA | 168 | 115 (68.45) | 90 (78.26) | 38 (33.04) |
Values within the same column did not differ statistically.
Fusion rate: number of embryos fused/number embryos reconstructed.
Cleavage rate: number of cleaved embryos/number of fused embryos.
Blastocyst rate: number of blastocysts/number of fused embryos.
Table 3. Effect of treating nuclear-donor cells with VPA on post-implantation developmental of clones.
| Groups | Transferred embryos N | Pregnancies N (%) | Live born offspring | |||
| Day 30 | Day 60 | Day 90 | Day 270 | Day 289 | ||
| Control | 34 | 7 (20.58) | 3 (8.82) | 3 (8.82) | 3 (8.82) | 3 (8.82) |
| VPA | 30 | 6 (20.00) | 2 (6.66) | 2 (6.66) | 2 (6.66) | 1 (3.33) |
Values within the same column did not differ statistically.
Pregnancy rate: number of pregnancies/number transferred embryos.
Analysis of the health status of cloned calves
Five calves were born after SCNT; two from the VPA group and three from the control group (Table 3). The recipient that delivered one of the calves from the VPA group showed signs of severe hydrallantois on day 270 of gestation (Figure 5A). We performed an emergency cesarean section, and delivered a stillborn calf, in spite of intensive care management and attempts of resuscitation. The calf was born with enlarged umbilical cord and ascites (Figure 5B). The other four calves that developed to term were delivered by cesarean section as well, but on day 289 of pregnancy. Among these, the remaining calf from the VPA group presented intestinal atony and died 12 h after birth due to gastrointestinal complications. One calf from the control group, showed poor viability after birth, received intensive care, including intranasal oxygen supplementation and colostrum via an esophageal tube, but died seven days after birth from septicemia secondary to omphalitis. The other two calves from the control group had no apparent abnormality, showed strong suckling reflex and drank colostrum with vigor. Both had clinical parameters monitored (e.g. glycemia, respiratory and heart rate, rectal temperature, blood gas) during the first 24 h. After this period, as they showed no clinical abnormalities they were discharged to a ventilated and clean stall. At the time of writing this manuscript, the surviving clones (both from the control group) were 13 months old, healthy and normal (Figure 5C) compared to their age-matched peers derived from natural reproduction at the time of preparation of this manuscript. It is noteworthy that between the two survivors calves, one is morphologically normal, adjusted all vital parameters and nursed the surrogate cow (Figure 5D) faster than the other calf. Surprisingly, the four calves (two from VPA and two from control) had some degree of brachygnathism (Figure 5B and 5E). Furthermore, the surviving calf that had brachygnathism also had monorchidism (Figure 5F).
Figure 5. Analysis of health status of cloned calves.
(A) Recipient female from the VPA-treated group showing signals of severe hydroallantois on day 270 of gestation. (B) Stillborn calf from VPA-treated group with enlarged umbilical cord and ascites (white arrow) and brachygnatism (red arrow). (C) Viable calves from the control group. (D) Calf from control group nursing, picture highlighting the correct morphology of the mandible (white arrow). (E) Mandible of cloned calf from control group with moderate brachygnatism (white arrow). (F) Picture highlighting the inguinal region of the calf from the control group evidencing monorchidism.
Discussion
Almost two decades have passed since the first mammal was cloned by using donor cells from an adult animal [1]. Yet, SCNT is still an inefficient procedure in which less than 10% of the embryos transferred to recipients results in the birth of viable offspring [2], [42], [43]. Among the factors affecting SCNT efficiency, chromatin compaction is thought to be a challenge for reprogramming donor cells as it acts as an epigenetic barrier to complete nuclear reprogramming [9], [11]. HDAC inhibitors such as VPA are molecules that increase the global levels of histone acetylation [25] and the treatment of donor cells with HDAC inhibitors have been reported to effectively increase in vitro cloning efficiency [15], [19]. Thus, we hypothesized that the treatment of donor cells with VPA should increase the global levels of histone acetylation, leading to an “open chromatin” state. The use of these cells in SCNT might result in increased development rates of SCNT as their nuclei are expected to be more amenable to reprogramming [9], [25]. To address this hypothesis, we investigated the effects of donor cells treatment with VPA on cellular proliferation/viability, epigenetic remodeling, gene expression, and pre- and post-implantational development of cattle clones derived from them. Our results showed that, albeit VPA treatment increased cellular proliferation/viability, expression of PPARGC1A and IGF2R, and the levels of H3K9ac in donor cells, no effect on efficiency of SCNT was found when treated cells were used as nuclear donors.
The viability of the cells used as nucleus donors is a major point in SCNT as their nuclei drive development of SCNT embryos after oocyte reconstruction. Previous studies have shown that development of clones is negatively impacted when donor cells are treated with chromatin modifying agents (CMAs) that compromise cell cycle distribution, morphological characteristics and cellular proliferation [19], [29], [44], [45]. Therefore, donor fibroblasts were treated for 24 h with increasing concentrations of VPA (1, 2 and 5 mM) and evaluated based on an MTT assay. Since all doses resulted in significantly higher proliferation/viability according to the MTT assay, 2 mM of VPA was used in the subsequent experiments as this concentration has been shown to increase the rates of induced pluripotent stem cells (iPSCs) [21] production and SCNT in mice [14]. It is noteworthy that based on the MTT assay, the treatment with VPA increased cell proliferation/viability while these cells were not expected to proliferate because of serum starvation to synchronize the cell cycle [46], [47]. However, this increase might be caused as a side effect of VPA on cell metabolism as the MTT assay is based on activity of metabolic enzymes that reduces tetrazolium salts [30]. According to a recent paper, inhibition of HDAC doubled the activity of enzymes related to intermediate cell metabolism [48]. PPARGC1A is a transcriptional coactivator with a central role in mitochondrial biogenesis and metabolism regulation in the cell [41]. The finding that VPA treatment increased expression of PPARGC1A further supports its effect on cell metabolism as previously reported [49], [50]. Based on these findings, 2 mM of VPA were found to be a suitable concentration to be used in the subsequent experiments.
In a second experiment we confirmed the effect of VPA on histone acetylation by evaluation of fibroblasts treated with 2 mM of VPA for 24 h in comparison to untreated cells. In addition, since VPA has been shown to cause DNA demethylation [38], the global levels of DNA methylation were evaluated, but no difference was found between control and treated cells. Since VPA causes demethylation by stimulating accessibility of a demethylase to DNA [38], we believe that a more prolonged treatment may be needed in order to obtain detectable levels of DNA demethylation. Next, since we hypothesized that the effects of VPA treatment would persist into embryo development, we attempted to confirm that the higher acetylation levels of donor cells were maintained after nuclear transfer. However, similar levels of histone acetylation were found between cloned embryos produced with control and treated cells. Moreover, a significant decrease in acetylation levels was found in SCNT couplets at 5 h.p.a. when fibroblasts treated with VPA were used as nuclear donors. This is in contrast with a report in the mouse, in which histone modifications caused by treatment of 8-cell embryos with VPA were maintained at least until the blastocyst stage [51]. On the other hand, oocytes are known to have special enzymatic activities, such as histone-modifying and DNA demethylating enzymes [11], that are responsible for reprogramming the sperm after fertilization [52]. Gao et al. showed that reprogramming by nuclear transfer uses the developmental program that is normally used after fertilization, termed by the authors as “erase-and-rebuild” process [53]. Furthermore, we speculate that the oocyte might have erased the epigenetic marks brought by the transferred nucleus, to reestablish a new developmental program. According to our data, this erasing process begins immediately after nuclear transfer, as no difference of acetylation was seen between control and treated groups immediately before artificial activation (approximately 1 to 2 h after couplet fusion). The ability of the oocyte to reverse the hyperacetylation caused by VPA, lowering the acetylation levels compared to a control group (5 h.p.a.), raises questions about the relevance of pretreating donor cells with CMAs. Enright et al. also found hypoacetylation in SCNT embryos produced by treatment of donor cells with TSA [19]. The authors suggested this was caused by a rebound effect from drug treatment, because the effect of HDACis is reversible [19]. In rabbits, similar results were found after treatment with NaBu and the authors argued that the aberrant epigenetic marks of clones cannot be corrected by the treatment with HDAC inhibitors [18]. In summary, although the treatment of donor cells with VPA caused histone hyperacetylation, this epigenetic state was reversed after oocyte reconstruction resulting in zygotes from treated group with lower levels of H3K9ac than the control group.
In a third experiment we investigated the effect of VPA treatment on gene expression in fibroblasts. It was reported that the treatment with VPA, even for a short period, had a significant effect on gene expression in cultured cells [25], [54]. Since treatment of donor cells has been reported to improve SCNT rates [15], [28], we hypothesized that this effect might be mediated, among other factors, by an effect of VPA on expression of key genes to SCNT. Since an appropriate expression of POU5F1 [55] and IGF2R [56] during early embryogenesis is critical to the success of SCNT, we evaluated whether the expression of these genes is altered in fibroblasts treated with VPA. We found no effect of the treatment on the expression of POU5F1, but VPA did increase expression of IGF2R compared to untreated cells. POU5F1 plays an essential role in controlling cellular pluripotency and therefore is a key factor in nuclear reprogramming in SCNT [40]. Treatment of myogenic cells with VPA was been shown to increase expression of POU5F1 [57]. Thus, SCNT might benefit from an overexpression of POU5F1 in donor cells as SCNT embryos frequently fail to express this gene [55]. However, as we found that VPA did not affect POU5F1 expression, we believe that other epigenetic modifications than histone acetylation are involved in the control of POU5F1 expression [58]. With respect to IGF2R, this gene plays an important role as a negative effector in fetal growth since it promotes IGF2 arrest into lysosomes followed by degradation [59], [60]. It has been hypothesized that dysregulation of imprinted genes such as IGF2R are the cause of poor developmental rates observed in SCNT [56], and alterations of IGF2R expression has often been associated to common problems in cloned embryos [61]. For instance, in sheep, parthenogenetic embryos that express low levels of IGF2 and high levels of IGF2R and H19, have retarded growth when compared to control embryos [62]–[64]. On the other hand, the reduced IGF2R expression in ovine embryos cultured in vitro is associated with Large Offspring Syndrome [39]. Baqir et al. observed that most imprinted genes have their expression increased following exposure of mouse embryonic stem cells to TSA [65]. Interestingly, they also found that expression of several imprinted genes remained high and in some cases, increased further, after drug removal or even after cell passageing, indicating a long lasting and retarded effect of the treatment on gene expression [65]. Herein we confirmed the effect of VPA on increased expression of an imprinted gene, providing further evidence that acetylation is involved in regulation of imprinting [65], [66]. The higher expression levels of IGF2R induced by the treatment of donor cells with VPA might affect SCNT as the epigenetic modifications induced by VPA on IGF2R have a long lasting effect on development [65]. Hence, the treatment of donor cells with VPA resulted on increased expression of IGF2R without affecting expression of POU5F1.
Although the hyperacetylation caused by VPA on donor cells was reversed after nuclear transfer, we decided to evaluate development of cloned embryos to further characterize the effect of VPA on SCNT. It is possible that some of the modifications induced by VPA on donor cells (e.g. overexpression of IGF2R) remain after nuclear transfer, with a consequent effect on cloning efficiency. Several groups have reported increased rates of SCNT when donor cells were treated with HDAC inhibitors, including VPA [28], TSA (cattle) [19], [32] and NaBu (cattle [15] and rabbits [18]). Yet, here we found no effect of donor cells treated with VPA on pre-implantation development of SCNT embryos. Although surprisingly this finding is in agreement with a previous reports in which NaBu was used to produce pig clones [45]. This report described that the treatment of donor cells with NaBu resulted in histone hyperacetylation, but the use of these cells in SCNT did not affect development of cloned embryos. These contradictory results provide evidence that the effect of CMAs may diverge depending on several factors including species and reprogramming system (e.g. SCNT or iPSCs). This notion is supported by a recent report with cloned mice which showed that several HDAC inhibitors such as TSA, scriptaid, suberoylanilide hydroxamic acid and oxamflatin reduced the rate of apoptosis in blastocysts and improved full term development, whereas VPA had no effect on SCNT efficiency [67]. The authors argued that VPA is an inhibitor of HDAC classes I and IIa, whereas the others are inhibitors of HDAC classes I and IIa/b, suggesting that inhibition of HDAC class IIb is a key step for successful reprogramming in the mouse [67]. In contrast, Costa-Borges et al. found that VPA improved in vitro development, blastocyst quality, and full term development of cloned mice, at comparable level to TSA [14]. Treatment with VPA also improved pre-implantation development of cattle [68] and pigs [69] when the cloned embryos were treated during in vitro culture. The treatment resulted in an increase of blastocyst rate and cell number in the inner cell mass (ICM) [68], [69]. Selokar et al showed that the treatment of donor cells with VPA improved bovine SCNT blastocysts production, reduced the apoptosis and H3K9 methylation levels, similar to those of embryos derived from IVF [28]. While these results differ from our, it should be noted that Selokar et al used handmade cloning, which is characterized by significant differences in the technique from that used in the present study, which might explain the differences observed in pre-implantation development. In addition, VPA was recently found to regulate pluripotency in iPSCs, increasing the efficiency as measured by the number of colonies, and up-regulation of pluripotency genes [21]. In summary, the effect of CMAs such as VPA on SCNT is not clear [70], but here we found no effect of VPA on developmental rates of SCNT embryos.
Taking into account that VPA might have affected SCNT embryos later during development, the blastocysts derived from the previous experiment were transferred to recipients to evaluate post-implantation development. Cloning of cattle by SCNT is typically associated with a high incidence of pregnancy failure throughout gestation [7], [71]. Our data are in accordance with previous reports, showing that the majority of established pregnancies (60–70%) were lost around the time of implantation [56], [72]. In cloned cattle, the losses that occur between days 30 and 60 of pregnancy are frequently associated to morphological (placentomegaly and faulty vascularization) and functional (steroidogenesis) abnormalities in placentas [71]. With respect to VPA, no effect of its treatment was seen on post-implantation development as the rate of pregnancy failure between 60 and 270 days and rate of development to term did not appreciably differ between groups. This unexpected result supports the hypothesis that all the modifications induced by the treatment were either inconsequential for post blastocyst development or reversed after nuclear transfer. Moreover, whereas two out of three clones produced with untreated cells were healthy at birth, the two clones that derived from VPA-treated cells died after caesarian section. This highlights the importance of evaluating post-implantation development and survival rate when CMAs are employed in SCNT. This agrees with the report by Kang et al. with pigs who described that VPA improved in vitro development of cloned embryos but did not improve survival to adulthood [73]. Interestingly, herein four out of five calves produced by SCNT (two from untreated cells and two from treated cells) presented brachygnatism. This trait is widely accepted to be a congenital and heritable abnormality [74]. However, the bull whose donor cells were derived did not present this phenotype. We speculate that the cell culture environment or an incomplete nuclear reprograming might have led to such abnormality. In a previous study, Johnson et al. also reported the occurrence of brachygnatism in foals derived by SCNT, trait that was not present in the horse used to donor cell derivation [74]. Strikingly, one of the viable calves, in addition to brachygnatism, had monorchidism whereas the other calf did not present noticeable abnormalities. After birth, the calf without abnormalities, stood up, drank colostrum and adjusted the physiological parameters faster than the other one with brachygnatism and monorchidism. This suggests that the process of nuclear reprogramming is stochastic, with calves derived from the same cell line presenting variable levels of “reprogramming” and health status. In summary, the treatment of donor cells with VPA did not affect developmental rates resulting in one stillborn and one calf that survived only 12 h after birth.
In conclusion, accumulated evidence suggests that CMAs are highly important to aid nuclear reprogramming with a consequent increase in SCNT efficiency in the mouse [75]. In cattle the use of CMAs remains largely controversial [70], [76]. Altogether, our results show that in spite of the alterations caused in fibroblasts by the treatment with VPA, their use as donor cells in SCNT does not improve pre- and post-implantation development of cloned cattle. In the future, it will be interesting to dissect the roles that CMAs have in donor cells and cloned embryos, to gain mechanistic insights on their use.
Acknowledgments
The authors gratefully acknowledge the help provided by Dr. Paulo Fantinato Neto and Dr. Flávio José Minieri Marchese.
Funding Statement
JRS is supported by São Paulo Research Foundation (FAPESP), grant number 2013/06673-7, and previously by Coordination for the Improvement of Higher Level Personnel (CAPES/DFAIT). This work was granted by São Paulo Research Foundation (FAPESP; grant numbers 2011/51126-9, 2010/13384-3, 2010/19768-8). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH (1997) Viable offspring derived from fetal and adult mammalian cells. Nature 385: 810–813. [DOI] [PubMed] [Google Scholar]
- 2. Wilmut I, Beaujean N, de Sousa PA, Dinnyes A, King TJ, et al. (2002) Somatic cell nuclear transfer. Nature 419: 583–586. [DOI] [PubMed] [Google Scholar]
- 3. Meirelles FV, Birgel EH, Perecin F, Bertolini M, Traldi AS, et al. (2010) Delivery of cloned offspring: experience in Zebu cattle (Bos indicus). Reprod Fertil Dev 22: 88–97. [DOI] [PubMed] [Google Scholar]
- 4. Santos F, Zakhartchenko V, Stojkovic M, Peters A, Jenuwein T, et al. (2003) Epigenetic marking correlates with developmental potential in cloned preimplantation embryos. Curr Biol 13: 1116–1121. [DOI] [PubMed] [Google Scholar]
- 5. Wee G, Koo DB, Song BS, Kim JS, Kang MJ, et al. (2006) Inheritable histone H4 acetylation of somatic chromatins in cloned embryos. J Biol Chem 281: 6048–6057. [DOI] [PubMed] [Google Scholar]
- 6. Suzuki J, Therrien J, Filion F, Lefebvre R, Goff AK, et al. (2011) Loss of methylation at H19 DMD is associated with biallelic expression and reduced development in cattle derived by somatic cell nuclear transfer. Biol Reprod 84: 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arnold DR, Fortier AL, Lefebvre R, Miglino MA, Pfarrer C, et al. (2008) Placental insufficiencies in cloned animals - a workshop report. Placenta 29 Suppl A: S108–10. [DOI] [PubMed] [Google Scholar]
- 8. Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, et al. (2001) Conservation of methylation reprogramming in mammalian development: Aberrant reprogramming in cloned embryos. Proc Natl Acad Sci U S A 24: 13734–13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pasque V, Jullien J, Miyamoto K, Halley-Stott RP, Gurdon JB (2011) Epigenetic factors influencing resistance to nuclear reprogramming. Trends Genet 27: 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M (2011) Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol 12: 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gurdon JB, Wilmut I (2011) Nuclear transfer to eggs and oocytes. Cold Spring Harb Perspect Biol 3 (6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao J, Hao Y, Ross JW, Spate LD, Walters EM, et al. (2010) Histone deacetylase inhibitors improve in vitro and in vivo developmental competence of somatic cell nuclear transfer porcine embryos. Cell Reprogram 12: 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kishigami S, Mizutani E, Ohta H, Hikichi T, Thuan NVan, et al. (2006) Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem Biophys Res Commun 340: 183–189. [DOI] [PubMed] [Google Scholar]
- 14. Costa-borges N, Santaló J, Ibáñes E (2010) Comparison between the effects of valproic acid and trichostatin A on the in vitro development, blastocyst quality, and full-term development of mouse somatic cell nuclear transfer embryos. Cell Reprogram 12: 437–446. [DOI] [PubMed] [Google Scholar]
- 15. Shi W, Hoeflich A, Flaswinkel H, Stojkovic M, Wolf E, et al. (2003) Induction of a senescent-like phenotype does not confer the ability of bovine immortal cells to support the development of nuclear transfer embryos. Biol Reprod 69: 301–309. [DOI] [PubMed] [Google Scholar]
- 16. Lee MJ, Kim SW, Lee HG, Im GS, Yang BC, et al. (2011) Trichostatin A promotes the development of bovine somatic cell nuclear transfer embryos. J Reprod Dev 57: 34–42. [DOI] [PubMed] [Google Scholar]
- 17. Zhao J, Ross JW, Hao Y, Spate LD, Walters EM, et al. (2009) Significant improvement in cloning efficiency of an inbred miniature pig by histone deacetylase inhibitor treatment after somatic cell nuclear transfer. Biol Reprod 81: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang F, Hao R, Kessler B, Brem G, Wolf E, et al. (2007) Rabbit somatic cell cloning: effects of donor cell type, histone acetylation status and chimeric embryo complementation. Reproduction 133: 219–230. [DOI] [PubMed] [Google Scholar]
- 19. Enright BP, Kubota C, Yang X, Tian XC (2003) Epigenetic characteristics and development of embryos cloned from donor cells treated by trichostatin A or 5-aza-2′-deoxycytidine. Biol Reprod 69: 896–901. [DOI] [PubMed] [Google Scholar]
- 20. Miyoshi K, Mori H, Mizobe Y, Akasaka E, Osawa A, et al. (2010) Valproic acid enhances in vitro development and OCT-3/4 expression of miniature pig somatic cell nuclear transfer embryos. Cell Reprogram 12: 67–74. [DOI] [PubMed] [Google Scholar]
- 21. Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, et al. (2008) Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol 26: 795–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Phiel CJ, Zhang F, Huang EY, Guenther MG, Lazar Ma, et al. (2001) Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J Biol Chem 276: 36734–36741. [DOI] [PubMed] [Google Scholar]
- 23. Göttlicher M, Minucci S, Zhu P, Krämer OH, Schimpf A, et al. (2001) Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J 20: 6969–6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marchion DC, Bicaku E, Daud AI, Sullivan DM, Munster PN (2005) Valproic acid alters chromatin structure by regulation of chromatin modulation proteins. Cancer Res 65: 3815–3822. [DOI] [PubMed] [Google Scholar]
- 25. Hezroni H, Sailaja BS, Meshorer E (2011) Pluripotency-related, valproic acid (VPA)-induced genome-wide histone H3 lysine 9 (H3K9) acetylation patterns in embryonic stem cells. J Biol Chem 286: 35977–35988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Turner BM (2007) Defining an epigenetic code. Nat Cell Biol 9: 2–6. [DOI] [PubMed] [Google Scholar]
- 27. Xu W, Wang Y, Li Y, Wang L, Xiong X, et al. (2012) Valproic acid improves the in vitro development competence of bovine somatic cell nuclear transfer embryos. Cell Reprogram 14: 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Selokar NL, St John L, Revay T, King WA, Singla SK, et al. (2013) Effect of histone deacetylase inhibitor valproic acid treatment on donor cell growth characteristics, cell cycle arrest, apoptosis, and handmade cloned bovine embryo production efficiency. Cell Reprogram 15: 531–542. [DOI] [PubMed] [Google Scholar]
- 29. Sangalli JR, De Bem THC, Perecin F, Chiaratti MR, Oliveira LDJ, et al. (2012) Treatment of nuclear-donor cells or cloned zygotes with chromatin-modifying agents increases histone acetylation but does not improve full-term development of cloned cattle. Cell Reprogram 14: 235–247. [DOI] [PubMed] [Google Scholar]
- 30. Mosmann T (1983) Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J Immunol Methods 65: 55–63. [DOI] [PubMed] [Google Scholar]
- 31. Nasser LF, Reis EL, Oliveira MA, Bó GA, Baruselli PS (2004) Comparison of four synchronization protocols for fixed-time bovine embryo transfer in Bos indicus x Bos taurus recipients. Theriogenology 62: 1577–1584. [DOI] [PubMed] [Google Scholar]
- 32. Ding X, Wang Y, Zhang D, Guo Z, Zhang Y (2008) Increased pre-implantation development of cloned bovine embryos treated with 5-aza-2′-deoxycytidine and trichostatin A. Theriogenology 70: 622–630. [DOI] [PubMed] [Google Scholar]
- 33. Giraldo AM, Lynn JW, Purpera MN, Godke RA (2007) DNA methylation and histone acetylation patterns in cultured bovine fibroblasts for nuclear transfer. Mol Reprod Dev 74: 1514–1524. [DOI] [PubMed] [Google Scholar]
- 34. Martinez-Diaz MA, Che L, Albornoz M, Seneda MM, Collis D, et al. (2010) Pre- and postimplantation development of swine-cloned embryos derived from fibroblasts and bone marrow cells after inhibition of histone deacetylases. Cell Reprogram 12: 85–94. [DOI] [PubMed] [Google Scholar]
- 35. Ramakers C, Ruijter JM, Deprez RHL, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66. [DOI] [PubMed] [Google Scholar]
- 36. Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 38. Detich N, Bovenzi V, Szyf M (2003) Valproate induces replication-independent active DNA demethylation. J Biol Chem 278: 27586–27592. [DOI] [PubMed] [Google Scholar]
- 39. Young LE, Fernandes K, McEvoy TG, Butterwith SC, Gutierrez CG, et al. (2001) Epigenetic change in IGF2R is associated with fetal overgrowth after sheep embryo culture. Nat Genet 27: 153–154. [DOI] [PubMed] [Google Scholar]
- 40. Boiani M, Eckardt S, Schöler HR, McLaughlin KJ (2002) Oct4 distribution and level in mouse clones: consequences for pluripotency. Genes Dev 16: 1209–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Austin S, St-Pierre J (2012) PGC1α and mitochondrial metabolism–emerging concepts and relevance in ageing and neurodegenerative disorders. J Cell Sci 125: 4963–4971. [DOI] [PubMed] [Google Scholar]
- 42. Gurdon JB, Melton DA (2008) Nuclear reprogramming in cells. Science 322: 1811–1815. [DOI] [PubMed] [Google Scholar]
- 43. Yang X, Smith SL, Tian XC, Lewin HA, Renard JP, et al. (2007) Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat Genet 39: 295–302. [DOI] [PubMed] [Google Scholar]
- 44. Enright BP, Sung LY, Chang CC, Yang X, Tian XC (2005) Methylation and acetylation characteristics of cloned bovine embryos from donor cells treated with 5-aza-2′-deoxycytidine. Biol Reprod 72: 944–948. [DOI] [PubMed] [Google Scholar]
- 45. Das ZC, Gupta MK, Uhm SJ, Lee HT (2010) Increasing histone acetylation of cloned embryos, but not donor cells, by sodium butyrate improves their in vitro development in pigs. Cell Reprogram 12: 95–104. [DOI] [PubMed] [Google Scholar]
- 46. Campbell KH (1999) Nuclear equivalence, nuclear transfer, and the cell cycle. Cloning 1: 3–15. [DOI] [PubMed] [Google Scholar]
- 47. Miranda MDS, Bressan FF, Zecchin KG, Vercesi AE, Mesquita LG, et al. (2009) Serum-starved apoptotic fibroblasts reduce blastocyst production but enable development to term after SCNT in cattle. Cloning Stem Cells 11: 565–573. [DOI] [PubMed] [Google Scholar]
- 48. Zhao S, Xu W, Jiang W, Yu W, Lin Y, et al. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327: 1000–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chiang MC, Cheng YC, Lin KH, Yen CH (2013) PPARγ regulates the mitochondrial dysfunction in human neural stem cells with tumor necrosis factor alpha. Neuroscience 229: 118–129. [DOI] [PubMed] [Google Scholar]
- 50. Aouali N, Palissot V, El-Khoury V, Moussay E, Janji B, et al. (2009) Peroxisome proliferator-activated receptor gamma agonists potentiate the cytotoxic effect of valproic acid in multiple myeloma cells. Br J Haematol 147: 662–671. [DOI] [PubMed] [Google Scholar]
- 51. VerMilyea MD, O'Neill LP, Turner BM (2009) Transcription-independent heritability of induced histone modifications in the mouse preimplantation embryo. PLoS One 4: e6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jullien J, Pasque V, Halley-Stott RP, Miyamoto K, Gurdon JB (2011) Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process? Nat Rev Mol Cell Biol 12: 453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gao T, Zheng J, Xing F, Fang H, Sun F, et al. (2007) Nuclear reprogramming: the strategy used in normal development is also used in somatic cell nuclear transfer and parthenogenesis. Cell Res 17: 135–150. [DOI] [PubMed] [Google Scholar]
- 54. Felisbino MB, Tamashiro WMSC, Mello MLS (2011) Chromatin remodeling, cell proliferation and cell death in valproic acid-treated HeLa cells. PLoS One 6: e29144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bortvin A, Eggan K, Skaletsky H, Akutsu H, Berry DL, et al. (2003) Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development 130: 1673–1680. [DOI] [PubMed] [Google Scholar]
- 56. Smith LC, Suzuki J, Goff AK, Filion F, Therrien J, et al. (2012) Developmental and epigenetic anomalies in cloned cattle. Reprod Domest Anim 47 Suppl 4: 107–114. [DOI] [PubMed] [Google Scholar]
- 57. Teng HF, Kuo YL, Loo MR, Li CL, Chu TW, et al. (2010) Valproic acid enhances Oct4 promoter activity in myogenic cells. J Cell Biochem 110: 995–1004. [DOI] [PubMed] [Google Scholar]
- 58. Simonsson S, Gurdon J (2004) DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat Cell Biol 6: 984–990. [DOI] [PubMed] [Google Scholar]
- 59. Ludwig T, Eggenschwiler J, Fisher P, D'Ercole aJ, Davenport ML, et al. (1996) Mouse mutants lacking the type 2 IGF receptor (IGF2R) are rescued from perinatal lethality in Igf2 and Igf1r null backgrounds. Dev Biol 177: 517–535. [DOI] [PubMed] [Google Scholar]
- 60. Lau MM, Stewart CE, Liu Z, Bhatt H, Rotwein P, et al. (1994) Loss of the imprinted IGF2/cation-independent mannose 6-phosphate receptor results in fetal overgrowth and perinatal lethality. Genes Dev 8: 2953–2963. [DOI] [PubMed] [Google Scholar]
- 61. Perecin F, Méo SC, Yamazaki W, Ferreira CR, Merighe GKF, et al. (2009) Imprinted gene expression in in vivo- and in vitro-produced bovine embryos and chorio-allantoic membranes. Genet Mol Res 8: 76–85. [DOI] [PubMed] [Google Scholar]
- 62. Feil R, Khosla S, Cappai P, Loi P (1998) Genomic imprinting in ruminants: allele-specific gene expression in parthenogenetic sheep. Mamm Genome 9: 831–834. [DOI] [PubMed] [Google Scholar]
- 63. Hagemann LJ, Peterson AJ, Weilert LL, Lee RS, Tervit HR (1998) In vitro and early in vivo development of sheep gynogenones and putative androgenones. Mol Reprod Dev 50: 154–162. [DOI] [PubMed] [Google Scholar]
- 64. Young LE, Schnieke AE, McCreath KJ, Wieckowski S, Konfortova G, et al. (2003) Conservation of IGF2-H19 and IGF2R imprinting in sheep: effects of somatic cell nuclear transfer. Mech Dev 120: 1433–1442. [DOI] [PubMed] [Google Scholar]
- 65. Baqir S, Smith LC (2006) Inhibitors of histone deacetylases and DNA methyltransferases alter imprinted gene regulation in embryonic stem cells. Cloning Stem Cells 8: 200–213. [DOI] [PubMed] [Google Scholar]
- 66. Hu JF, Pham J, Dey I, Li T, Vu TH, et al. (2000) Allele-Specific histone acetylation accompanies genomic imprinting of the insulin-like growth factor II receptor gene. 141: 4428–4435. [DOI] [PubMed] [Google Scholar]
- 67. Ono T, Li C, Mizutani E, Terashita Y, Yamagata K, et al. (2010) Inhibition of class IIb histone deacetylase significantly improves cloning efficiency in mice. Biol Reprod 83: 929–937. [DOI] [PubMed] [Google Scholar]
- 68. Xu W, Wang Y, Li Y, Wang L, Xiong X, et al. (2012) Valproic acid improves the in vitro development competence of bovine somatic cell nuclear transfer embryos. Cell Reprogram 14: 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kim YJ, Ahn KS, Kim M, Shim H (2011) Comparison of potency between histone deacetylase inhibitors trichostatin A and valproic acid on enhancing in vitro development of porcine somatic cell nuclear transfer embryos. In Vitro Cell Dev Biol Anim 47: 283–289. [DOI] [PubMed] [Google Scholar]
- 70. Akagi S, Geshi M, Nagai T (2013) Recent progress in bovine somatic cell nuclear transfer. Anim Sci J 84: 191–199. [DOI] [PubMed] [Google Scholar]
- 71. Chavatte-Palmer P, Camous S, Jammes H, Le Cleac'h N, Guillomot M, et al. (2012) Review: Placental perturbations induce the developmental abnormalities often observed in bovine somatic cell nuclear transfer. Placenta 33 Suppl: S99–S104. [DOI] [PubMed] [Google Scholar]
- 72. Hill JR, Burghardt RC, Jones K, Long CR, Looney CR, et al. (2000) Evidence for placental abnormality as the major cause of mortality in first-trimester somatic cell cloned bovine fetuses. Biol Reprod 63: 1787–1794. [DOI] [PubMed] [Google Scholar]
- 73. Kang JD, Li S, Lu Y, Wang W, Liang S, et al. (2013) Valproic acid improved in vitro development of pig cloning embryos but did not improve survival of cloned pigs to adulthood. Theriogenology 79: 306–11. [DOI] [PubMed] [Google Scholar]
- 74. Johnson AK, Clark-Price SC, Choi YH, Hartaman DL, Hinrichs K (2010) Physical and clinicopathologic findings in foals derived by use of somatic cell nuclear transfer: 14 cases (2004–2008). J Am Med Vet Assoc 236: 983–990. [DOI] [PubMed] [Google Scholar]
- 75. Ogura A, Inoue K, Wakayama T (2013) Recent advancements in cloning by somatic cell nuclear transfer. Philos Trans R Soc Lond B Biol Sci 368: 20110329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Galli C, Duchi R, Colleoni S, Lagutina I, Lazzari G (2014) Ovum pick up, intracytoplasmic sperm injection and somatic cell nuclear transfer in cattle, buffalo and horses: from the research laboratory to clinical practice. Theriogenology 81: 138–151. [DOI] [PubMed] [Google Scholar]